Abstract
Poly (ADP-ribose) polymerase-1 (PARP-1) is an abundant, ubiquitously expressed NAD+-dependent nuclear enzyme that has prognostic value for a multitude of human cancers. PARP-1 activity serves to poly (ADP-ribose)-ylate the vast majority of known client proteins and affects a number of cellular and biological outcomes, by mediating DNA damage response (DDR), base-excision repair (BER), and DNA strand break (DSB) pathways. PARP-1 is also critically important for the maintenance of genomic integrity, as well as chromatin dynamics and transcriptional regulation. Evidence also indicates that PARP-directed therapeutics are “synthetic lethal” in BRCA1/2-dieficient model systems. Strikingly, recent studies have unearthed exciting new transcriptional-regulatory roles for PARP-1, which has profound implications for human malignancies and will be reviewed herein.
Introduction
Poly (ADP-ribose) polymerase-1 (PARP-1) is an enzyme responsible for ~90% of the ADP-ribosyl transferase activity (poly (ADP-ribose) ylation (PARylation)) in both non-transformed and malignant human cells (1), the majority of which is self-directed (1, 2). The PARP family of enzymes contains eighteen family members, PARP-1 being the first to be characterized (3), which PARylate client proteins utilizing NAD+ as a cofactor, and thereby control a diverse set of biological functions (4). The first defined role for PARP-1 was to orchestrate DNA damage resolution, especially in the context of base excision repair (BER) (5). However, subsequent studies implicated PARP-1 as harboring pleiotropic cellular functions, including: DNA repair/maintenance of genomic integrity, DNA methylation, regulation of circadian clocks, chromatin regulation and histone modification (2, 6–8). Parp-1 deficient mouse models are viable and demonstrate increased sensitivity to genotoxic stress, resistance to DNA damage induced cell death, increased tumorigenesis in chemically or genetically induced models (reviewed in (2)) and altered hypoxic response(9). Cell models of Parp-1 deficiency demonstrate altered transcription of p53 targets ((10) heat shock factor 1(11). Most recently, means by which PARP-1 regulates gene transcription have been identified (2, 6, 7, 12); the present review will address the function and consequence of PARP-1-regulated transcription in the context of human malignancies.
Regulation of PARylation
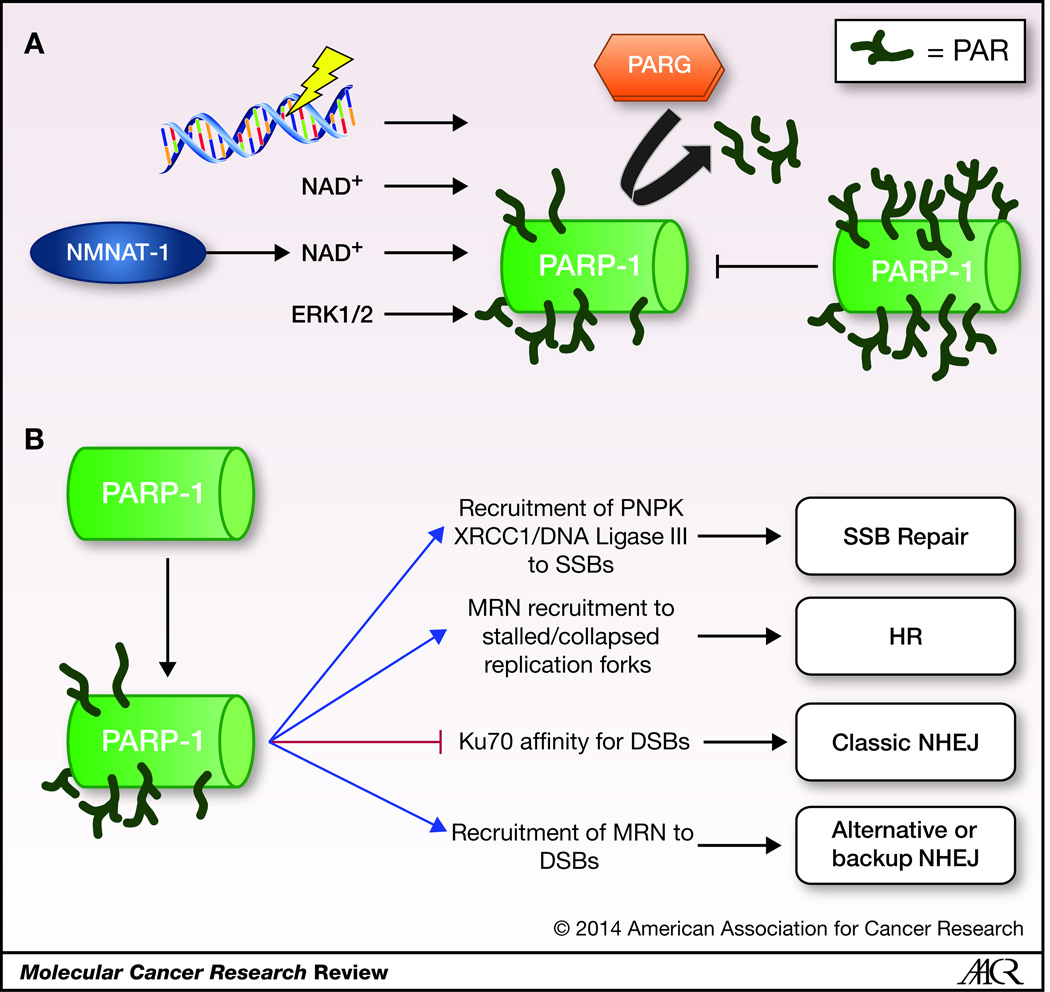
PARP-1 is a DNA-dependent ADP-ribosyl transferase that is localized in the nucleus and is frequently associated with chromatin (1, 2, 12). The capacity of PARP-1 to associate with DNA is manifest via direct binding and/or interacting with nucleosomes and other chromatin-associated proteins, including transcription factors(13), the transcriptional machinery (14, 15), and chromatin modifiers (1, 2, 12). The enzymatic activity of PARP-1 is regulated by what it is bound to, such as damaged DNA or other nuclear proteins (1, 12, 16–21), as well as post-translational modifications, such as autoPARylation (inhibitory) (22–26) and phosphorylation by ERK1/2 (activating, DNA independent) (27, 28). Additionally, an NAD+ synthase (nicotinomide mononucleotide adenylyltransferase-1 (NMNAT-1)) associates with PARP-1, thus allowing for a proximal source of NAD+ cofactor and increasing PARP-1 activity (29) (Figure 1A).
Figure 1.
The first identified roles of PARP-1 were associated with DNA damage and genomic maintenance, with specific roles in BER, single-strand (SSB) and double-strand break (DSB) repair(5). In response to DNA damage, PARP-1 enzymatic function is activated, and persists in a correlative manner with the extent of the damage. When DNA breaks are repairable, PARP-1 regulates repair and cell survival, but in response to catastrophic damage, PARP-1 regulates the induction of cell death(30–32). In addition to playing roles in SSB and DSB repair(33–35), PARP-1 has been implicated in homologous recombination (HR) at stalled or collapsed replication forks (36, 37), as well as regulating non-homologous end-joining (NHEJ) (38–41). While the PARP family of enzymes is responsible for PAR anabolism, the PAR glycohydrolases (PARGs) regulate PAR catabolism. PAR hydrolysis is regulated by a number of isoforms of the single PARG gene that arise from splicing. The long isoforms of PARG shuttle between the cytosol and the nucleus, while the short isoform is exclusively cytoplasmic(42–45). Although the PARP superfamily and PARG play clearly important and distinct roles in PAR metabolism, the role of PARG/PARP-1 interplay in cancer remains poorly described. Additionally, PAR catalysis in the mitochondria has been discovered to be performed by ADP-ribosylhydrolase 3 (ARH3), not PARG, for which very little is known in the context of cancer (46–48).
Current understanding of PARP-1 driven PARylation is that in response to stimuli (such as DNA damage) (Figure 1B), PARP-1 enzymatic function is robustly and rapidly induced, utilizing NAD+ as the donor for ADP-ribose resulting in multiple biological outcomes, and PAR is degraded by PARG to ADP-ribose monomers. These terminal ADP-ribose monomers are then removed by the recently discovered TARG1, whose deficiency results in neurodegenerative disease (49). PAR moieties generated can non-covalently interact with multiple domains that result in controlling of multiple pathways (reviewed in(46, 50)). Additionally, recent proteomics work has begun to define the PARylated proteome upon various stimuli and how these PARylated proteins are often involved in chromatin organization and transcriptional regulation, as well as the DNA damage response (51–53). Here, the major transcriptional regulatory functions of PARP-1, and the downstream biological consequence(s) of these events for human malignancies will be discussed. Specific areas of focus will address: 1) modulation of tumor suppressor and oncogene function, 2) regulation of effectors of the metastatic process, 3) regulation of cell survival and adaptation, and 4) transcriptional regulation in hormone-dependent cancers.
Basic Mechanisms of PARP-1-mediated transcriptional control
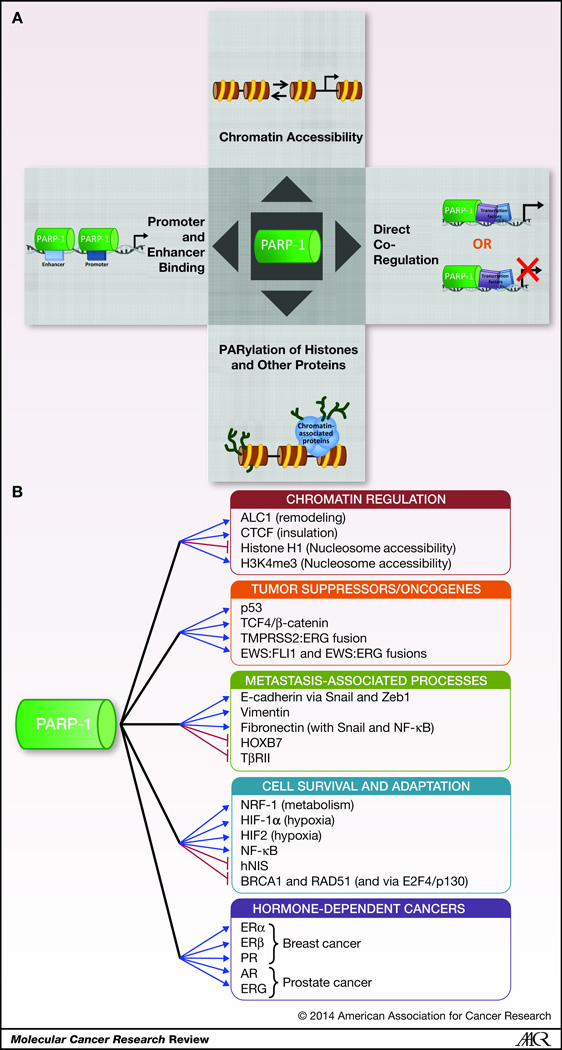
Although initially characterized as a factor intimately involved in regulation of DNA repair/genomic maintenance, PARP-1 was recently demonstrated to exert pleiotropic roles in transcriptional regulation in cancer and non-cancer model systems (2, 6, 7, 12). Under NAD+ depleted conditions, PARP-1 compacts chromatin by binding to and bringing together nucleosomes and can also affect chromatin structure through PARylation of histones, thus disrupting nucleosomes and resultant chromatin architecture (1, 24, 54–56). PARP-1 has also been found to localize to promoters of actively transcribed genes and prevent binding of Histone H1, thus promoting transcriptionally active chromatin (57). The transcriptional regulatory functions of PARP-1 are multi-fold, do not universally require enzymatic activity, and are manifest through divergent functions including: enhancer binding, association with insulators, modulation of chromatin structure, and/or direct transcription factor regulation (as either a context-dependent co-activator or co-repressor). PARP-1 binding at regulatory loci of genes does not always correlate with activation of transcription. In fact, PARP-1 binding can sometimes correlate with transcriptional repression (2, 6, 7, 12). As such, transcriptional regulation by PARP-1 can be either positive or negative, occur through multiple mechanisms, and is complex and cell- and context-specific (Figure 2A). The cancer-context dependent mechanisms by which PARP-1 modulates transcription will be discussed below (Figure 2B), particularly as related to chromatin remodeling, modulation of tumor suppressor and oncogene function, transcriptional regulation of the metastatic process, modifying cell survival and adaptation, and nuclear receptors in hormone-dependent cancers.
PARP-1 regulates select ATPases that control chromatin remodeling, such as amplified in liver cancer 1 (ALC1), an ATPase in the SNF2 superfamily that is frequently deregulated in hepatocellular carcinoma. ALC1 demonstrates sequence similarity to other chromatin remodelers (such as SNF2, ISWI, and CHD1), but lacks any domains with known function in chromatin architecture regulation. However, ALC1 has a macrodomain, which has been demonstrated to serve as a binding domain for PAR(58). Moreover, ALC1 was found to have ATPase activity dependent upon both PARP-1 and NAD+. This ATPase function was correlated with ALC1 binding to and remodeling of chromatin, likely due to activation by PAR moieties involved in PARP-1 automodification (58). While there is no explicit link to transcription, this study demonstrates that PARP-1 activates the ATPase capacity of ALC1, which is frequently deregulated in human hepatocellular carcinoma. Further mechanistic insight was gained upon the discovery of and ALC1-PARP-1-nucleosome intermediate that was stable and required for activation of ALC1 activation and chromatin remodeling(59). Additional research has demonstrated a critical connection between PARP-1 and ALC1 in regulation chromatin remodeling in the context of the DNA damage response(60–62), serving as a model of the dual roles of PARP-1 in regulating DNA damage and chromatin structure, even in the context of the same partnering molecule.
Distinct from these roles, PARP-1 controls the function of selected insulators that modulate chromatin architecture in models of cancer. For example, CTCF (CCCTC-binding factor) is a transcription factor that performs a multitude of transcriptional-regulatory roles dependent on its posttranslational modification status and interactions with other molecules. PARylation is requisite for the ability of CTCF to serve as an insulator (blocking interaction between regulatory loci) and a barrier (inhibiting the spread of heterochromatin). In addition, it was shown that CTCF activates PARP-1, resulting in DNA hypomethylation, which has been linked to cancer initiation and progression. Interestingly, comparison of existing data sets of genome-wide CTCF and PARP-1 residence on chromatin revealed that sites of overlap (deemed “hot spots”) are both intergenic as well as intragenic, and varied between chromosomes. Since these hot spots were clustered within the genome(63), it was suggested that this might be linked to specificity of PARP-1 and CTCF at these loci. These data correlate PARP-1 to the insulating capacity of CTCF.
In addition to chromatin remodeling factors and insulators, PARP-1 can impinge upon nucleosome accessibility. Seminal work in understanding the relationship between PARP-1 and transcription showed that PARP-1 and histone H1 compete for binding to nucleosomes and have an inverse binding pattern at actively transcribed genes(57). Further delineation of the mechanisms by which PARP-1 positively controls gene transcription revealed that PARP-1 promotes the binding of RNA polymerase II (RNApolII) and associated transcriptional machinery to actively transcribed genes in MCF-7 breast cancer (BrCa) cells. This increase in residence correlated with increased chromatin accessibility at the transcriptional start sites of these genes and trimethylation of histone H3 lysine 4 (H3K4me3), a hallmark of active transcription. Further mechanistic insight led to the discovery that PARP-1 blocks the ability of the KDM5B demethylase to demethylate H3K4, thus promoting transcription of these PARP-1 regulated genes. It was determined that demethylation of H3K4 allows for histone H1-driven expulsion of RNA polymerase II from active promoters, and that PARP-1-dependent inhibition of KDM5B-mediated H3K4 demethylation permits a transcriptionally competent chromatin environment. PARP-1 enzymatic activity was determined to be required for limiting KDM5B binding to active promoters, and KDM5B was shown to be a target of PARylation; thus, deregulation of PARP-1 enzymatic activity could lead to inhibition of KDM5B demethylase activity(64). These findings provide an elegant delineation of the means by which PARP-1 regulates transcription, and exemplifies how the transcriptional regulatory capacity of PARP-1 can be assessed.
Although PARP-1 is responsible for the majority of PARylation, the role of PARG must also be considered. Based on the opposing functions of PARP-1 and PARG with respect to PARylation, it might be predicted that transcription is disparately regulated by PARP-1 and PARG. Genome-wide analyses using isogenic models of PARP-1 or PARG depletion in BrCa cells revealed an approximate 50% overlap in genes regulated by PARP-1 (~1200 genes) and PARG (~1100 genes) (both positive and negative regulation), which was typically in the same direction (up or down) and of similar magnitudes. This intersect of ~500 genes was enriched for ontologies associated with metabolism and the stress response, suggesting that PARP-1 and PARG may separately or coordinately regulate these processes. Further examination of the PARP-1- and PARG-regulated transcriptome revealed that PARP-1 and PARG localize to the promoters of both up-regulated and down-regulated genes, and binding of both were proportional in all genes examined. Additional studies demonstrated that both PARP-1 and PARG can regulate the chromatin occupancy of the other, and there was gene context-specific dependency on the enzymatic activity of PARP-1 and PARG in transcriptional regulation. Together, these studies indicate that despite the opposing functions of PARP-1 and PARG (catabolism vs. anabolism of PAR), they bind to target promoters and tend to act in a similar fashion in their capacity to regulate transcription(65).
Taken together, it is clear that there are both context-specific and general mechanisms and consequences of PARP-1 regulated transcription and chromatin remodeling in human malignancy. Although the functions of PARP-1 in regulating a specific set of transcription factors and chromatin modulators (ALC1, CTCF, KDM5B) has been outlined above, whether these observations hold true for other transcriptional regulatory processes remains to be assessed, and would lead to not only greater mechanistic understanding of PARP-1-dependent transcriptional regulation, but could uncover new therapeutic opportunities in the context of cancer management.
Modulation of tumor suppressor and oncogene function by PARP-1
Complementing the transcriptional and chromatin regulatory functions of PARP-1 (described above), PARP-1 also directly modulates sequence-specific transcription factors, including several of high relevance for human malignancies. Notably, PARP-1 is in transcriptional repressive complexes with p53, and PARylation of p53 within in this context results in recruitment of HDAC1 and HDAC2. This transcriptional repressor complex blocks the expression of metastasis-associated protein 1 (MTA1), which is involved in nucleosome remodeling and transcriptional repression, is frequently enriched in a number of cancers, and is associated with disease progression and metastasis. Abrogation of PARP-1-dependent MTA1 repression results in elevated levels of HIF-1α and VEGF, suggesting that PARP-1 mediated p53 transcriptional function negatively regulates MTA1 expression and cancer-associated genes and phenotypes(66). As such, in addition to maintaining genomic integrity, part of the tumor suppressive roles for both p53 and PARP-1 may include regulation of MTA1. Other studies have implicated a functional interaction between PARP-1 and p53 in multiple biological functions (reviewed in(2, 67). Specifically, it has been identified that PARP-1 regulates the p53-mediated DNA damage response via stabilization of p53 in response to radiation(10) and PARP inhibition suppresses the activation of p53 in response to radiation(68), PARP-1 activation results in ATP depletion and subsequent reduced TAF1 kinase activity and p21 activation(69), Parp-1 null mouse embryonic fibroblasts (MEFs) have ~2x lower basal p53 expression and DNA damage dependent reduction than wild type MEFs(70) and functional PARP-1 is required for p53-dependent cytotoxicity in response to proteasome inhibitors(71),
In addition to p53, PARP-1 regulates organ site-specific tumor suppressors. For example, loss of function of the APC (Adenomatous polyposis coli) tumor suppressor gene is frequently associated with familial and sporadic colorectal cancer (CRC), resulting in accumulation of β-catenin and activation of T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factors. PARP-1 interacts with TCF-4, and in complex with TCF-4/β-catenin in CRC; through this function, PARP-1 increases transcriptional activation of TCF/LEF by β-catenin(72). Conversely, Ku70 has also been observed to associate with TCF-4/β-catenin and repress TCF/LEF function, and Ku70 competes with PARP-1 for binding to the complex. Consonantly, PARP-1 mRNA and protein levels are elevated in FAP (familial adenomatous polyposis) and sporadic CRC clinical specimens, suggesting a possible causative role of PARP-1-regulated transcriptional activation of TCF/LEF in CRC. Furthermore, it was observed that Ku70 mRNA was decreased in four of five cases of sporadic CRC compared to matched normal tissue, suggesting that PARP-1-mediated TCF/LEF activation may be increased in human disease. Finally, increased nuclear PAR is observed in colorectal adenoma clinical specimens as compared to matched normal tissue(73). Together, these studies demonstrate that PARP-1 can positively regulate the transcriptional activity of TCF/LEF in the context of CRC, and that PARP-1 is both expressed to a higher degree and is more active in CRC(72, 73), thus suggesting that PARP-1 transcriptional-regulatory function may be worthy of further examination as a therapeutic target in future management of CRC.
As described above, PARP-1 regulates the function of classical tumor suppressor genes (p53 and APC), but there are many roads to tumorigenesis, and consequently many cancers overexpress oncogenic E-twenty six (ETS) transcription factors via gene fusions. In prostate cancer (PCa), gene fusions occur between the TMPRSS2 and ERG (an ETS transcription factor) genes in more than 50% of PCa. As TMPRSS2 is a well-described androgen receptor (AR) target gene, this places ERG expression under control of androgen receptor (AR), which plays a critical role in PCa. Generation of these fusions seems to be an early event in prostate tumorigenesis, and the TMPRSS2-ERG gene fusion has been found to induce cancer cell growth and invasion. Recent studies revealed that the TMPRSS2-ERG fusion gene product interacts with PARP-1, and that PARP-1 expression is required for the transcriptional activation function of ERG. PARP-1-dependent ERG activity was found to drive invasive phenotypes, and PARP inhibition diminished ERG-driven cell invasion, intravasation, and metastasis, as well as xenograft tumor growth in fusion positive tumor cells. Interestingly, ETS function was shown to promote DNA damage, and this could be exacerbated by PARP inhibition, suggesting that in the context of TMPRSS2-ERG positive PCa(74), PARP-1 serves in both DDR as well as the transcriptional regulation of oncogenic fusion protein function. As such, targeting both of these functions with PARP inhibitors may serve therapeutic benefit in PCa.
ETS fusions are not solely found in PCa, as Ewing’s sarcomas are tumors of bone and soft tissue that are characterized by chromosomal translocations that fuse a portion of Ewing’s sarcoma breakpoint gene 1 (EWS) (responsible for transcriptional activation) to the DNA binding domain (DBD) of either FLI1 or ERG. The resulting translocation-induced chimeric proteins regulate cell proliferation, invasion, and tumorigenesis; thus development of means to target these proteins therapeutically could provide clinical benefit. Notably, both the EWS-FLI1 and EWS-ERG fusion gene products interact with and depend on PARP-1 function. Expression of either fusion product enhanced DNA damage and increased their invasive potential (events that were mitigated by PARP inhibition). Additionally, PARP inhibition was found to reduce EWS-FLI1-dependent tumor growth and metastasis in vivo. Finally, knockdown of either PARP-1 or DNA-PK resulted in diminished EWS-FLI1 target gene expression in a similar fashion as EWS knockdown, demonstrating that PARP-1 potentially plays a key role in EWS-FLI1 fusion protein transcriptional function. It was proposed that PARP-1 and EWS-FLI1 function in a feed-forward mechanism in Ewing’s sarcoma, whereby the fusion protein directly induces PARP-1 mRNA and protein expression, culminating in increased EWS-FLI1 transcriptional activity (75). This study provides an example of the capacity of PARP-1 to regulate both DDR as well as transcription, ultimately demonstrating that targeting both facets of PARP-1 function pharmacologically may significantly alter cancer therapy.
Although limited in number, these studies collectively suggest that PARP-1 modulates the transcriptional function of both tumor suppressors and oncogenes, exemplifying the capacity of PARP-1 to elicit context-specific pro- or anti-tumor effects. Further understanding of the underlying mechanisms of PARP-1 function will thereby be of utmost importance for refined utilization of PARP inhibitors in cancer therapy.
Regulation of effectors of the metastatic process by PARP-1
Intriguingly, the transcriptional regulatory function of PARP-1 has also been implicated in EMT (epithelial to mesenchymal transition), a cellular process thought to promote metastatic events. Loss of E-cadherin expression is believed to represent a key event in EMT, resulting in disorganization of cell-to-cell junctions. While multiple factors control E-cadherin expression (including promoter methylation, mutation, and dysregulated transcription), the transcription factors Snail and Zeb1 are key effectors of E-cadherin expression. In brief, Snail and Zeb1 are up-regulated in a subset of malignancies and suppress E-cadherin expression. PARP-1 negatively controls cancer-associated Snail and Zeb1 expression resulting in E-cadherin expression, thus providing some of the first evidence that PARP-1 may serve to inhibit EMT through its role in transcriptional regulation(76). In addition to regulating E-cadherin expression through Snail, PARP-1 also collaborates with Snail and NF-κB to drive expression of another EMT factor, fibronectin(77). Although the functional interaction of Snail, NF-κB, and PARP-1 in the context of fibronectin regulation has been delineated in a small selection of cancer models, the biological consequence(s) and contribution to EMT are yet to be explored.
Along with loss of E-cadherin, vimentin expression is also associated with EMT. Consistent with a role in EMT regulation, PARP-1 binds to and directly regulates transcription from the vimentin promoter. Interestingly, this transcriptional activation was independent of the enzymatic activity of PARP-1, and could be suppressed by H2O2, which is an activator of PARP-1 enzymatic function. H2O2 treatment resulted in diminished PARP-1 protein expression as well as vimentin protein expression, and induced overexpression of PARP-1 resulted in greater H2O2-induced repression of vimentin promoter activity, suggesting that PARP-1 may play an active role in inhibition of vimentin expression(78). Although the study described here clearly implicates PARP-1 in the transcriptional regulation of vimentin, further study as to the biological consequence is needed.
Further evidence that PARP-1 transcriptionally controls EMT is by regulation of HOXB7, which is overexpressed in some instances of breast and ovarian cancer and is capable of inducing EMT. HOXB7 associates with PARP-1 in models of breast cancer (SKBR3), and HOXB7 is also a target of PARylation. PARylation of HOXB7 by PARP-1 reduces HOXB7 affinity for DNA and subsequently the transcriptional activity of HOXB7. Thus, at least in the context of one HOX family member (HOXB7), PARP-1 serves to negatively regulate HOX transcriptional activity(79). Finally, the means by which PARP-1 regulates EMT may be due, in part, to regulation of TGF-β (transforming growth factor beta) signaling, which is capable of inducing EMT. In non-transformed cells, TGF-β inhibits cell proliferation and induces differentiation. In certain contexts, tumor cells bypass TGF-β signaling by deregulation of the receptor for TGF-β, TGF-β receptor type II, whose expression is negatively regulated by PARP-1(80).
Combined, it is clear that PARP-1 plays a role in the transcriptional regulation of events associated with EMT by regulating the expression (E-cadherin, fibronectin, vimentin) or function (HOXB7, TGF-β) of key players in the process of EMT. However, it is not clear whether the net effect of PARP-1-regulated transcription is to drive or block EMT, and may be contingent on the type of malignancy studied. As such, this complicated network merits further examination in the context of human malignancy.
Regulation of cell survival and adaptation
Intriguingly, the transcriptional regulatory function of PARP-1 has also been implicated in cell survival and adaptation processes, largely associated with metabolism, hypoxia, and DNA damage response.
With regard to metabolism, nuclear respiratory factor 1 (NRF-1) is a transcription factor involved in the regulation of mitochondrial biogenesis, translation/protein stability, DNA synthesis, DDR, and proliferation. NRF-1 can interact with DNA and PARP-1 simultaneously, and PARP-1 PARylates NRF-1, thus causing decreased interaction between PARP-1 and NRF-1. However, PARP-1 expression was found to be required for optimal transcriptional activation of NRF-1 target genes(81), suggesting that PARP-1 plays a key role in the transcriptional regulation of cellular metabolism.
A second adaptive process that PARP-1 is thought to influence is the hypoxic response. The hypoxic response is regulated transcriptionally by hypoxia-inducible factor (HIF) subunits α and β, the α subunits being continuously transcribed and translated and regulated by the von Hippel-Lindau (VHL) tumor suppressor in an oxygen availability-specific manner. In an initiation/promotion model of skin carcinogenesis, PARP-1 inhibition delayed tumor promotion, and is associated with decreased inflammatory infiltration, reduced mitosis, diminished apoptosis (in non-cancerous tissue, but not in cancerous tissue), and decreased tumor vasculature. These phenotypes correlated with diminished AP-1 DNA binding, but not NF-κB DNA binding, as well as decreased HIF-1α mRNA and protein expression. Consequently, HIF-1-dependent gene regulation of the hypoxic-responsive transcriptional network was severely compromised(82). In a model of chronic myelogenous leukemia (CML), it was found that PARP-1 and HIF-1α interact and cooperate to activate HIF target gene expression. This was dependent upon PARP-1 enzymatic activity, but did not result in altered HIF-1α stability or DNA binding capacity. PARP-1 knockdown caused diminished HIF-1α target gene expression, which correlated with increased necrotic tumor cell death and diminished tumor vascularization, but paradoxically no change in CML tumor growth(83). Thus, PARP-1 functions to positively regulate HIF-1α in multiple models of oncogenesis. While HIF-2α has ~50% sequence identity to HIF-1α and there is some overlap in the transcriptional programs regulated by HIF-1 and HIF-2, some targets are differentially regulated. In a cell model system of renal cell carcinoma lacking VHL, it was found that HIF-2 and PARP-1 form a complex under hypoxic conditions dependent upon PARP-1 enzymatic activity. Further examination in other model systems indicated that depletion of PARP-1 protein results in diminished HIF-2α mRNA expression, reduced hypoxia-induced HIF-2α protein expression, and subsequent HIF-2α target gene expression(84). In sum, PARP-1 appears to be a positive regulator of both HIF-1 and HIF-2 expression and transcriptional function, and the subsequent hypoxic response.
With regard to DNA damage response, NF-κB represents a group of transcription factors that regulate genes responsible for cell death and proliferation, and is frequently deregulated in cancer. Upon DNA damage, NF-κB targets include antiapoptotic genes, thus blocking cell death. Conversely, loss of NF-κB signaling renders cells more radiosensitive. In model systems of BrCa, PARP-1 inhibition resulted in diminished radiation-induced NF-κB binding to target gene loci. This decrease in NF-κB chromatin occupancy was not due to altered IκB degradation or NF-κB nuclear localization, but rather decreased radiation-induced NF-κB transcriptional activity (determined by reporter assay)(85). While the mechanism of this apparent co-activator function for PARP-1 for NF-κB is not yet defined, the biological consequence of PARP-1 inhibition in BrCa cell lines is an increase in radiation-induced apoptosis and radiosensitization.
In addition to modifying the response to externally applied radiation, PARP-1 transcriptionally regulates systemic radiotherapy in selected contexts. Radioiodine is the only effective therapy for disseminated thyroid cancer, as the thyroid absorbs most of the iodine present in the body, but upon dedifferentiation, tumors no longer respond to this therapeutic modality due to loss of expression of human sodium-iodide symporter (hNIS). hNIS is a transmembrane protein that facilitates the concentration of iodide in both normal and transformed thyroid follicular cells. A number of potential mechanisms for loss of hNIS have been reported, including CpG island methylation and activation of a trans-acting repressor. Upon characterization of the trans-acting repressor, it was found that PARP-1 is a constituent of the complex. PARP-1 occupies the hNIS promoter, and PARP inhibition results in increased hNIS reporter activity as well as endogenous hNIS mRNA expression, suggesting that PARP-1 enzymatic activity may repress hNIS expression(86). Although the mechanism and biological consequence is unclear, PARP inhibition may sensitize disseminated refractory thyroid tumors to radioiodine.
Finally, with regard to the response to radiotherapy, PARP-1 plays a key role in base excision repair (BER), and homologous recombination (HR)-deficient cells rely on BER as regulated by PARP-1. In fact, among BRCA-deficient tumors, use of PARP inhibitors has demonstrated some efficacy due to synthetic lethality. It has been observed that under hypoxic conditions, there is down-regulation of BRCA1 and RAD51 gene transcription via accumulation of a suppressive complex containing E2F4/p130 at regulatory loci. As such, it could be predicted that hypoxia would render cells sensitive to PARP inhibition. Consequently, it was found that colon and lung cancer cells under hypoxic conditions were more sensitive to PARP inhibition than cells under normoxic conditions. Inhibition of PARP activity resulted in diminished BRCA1 and RAD51 protein expression in cell models of lung cancer, breast cancer, and osteosarcoma. This suppression of BRCA1 and RAD51 could be reversed by either HPV E7 expression or p130 knockdown, and was associated with diminished E2F4 and p130 occupancy at the regulatory loci, indicating that PARP inhibitor-mediated regulation of BRCA1 and RAD51 is due, in part, to E2F4/p130-mediated suppression. Further mechanistic studies indicated that PARP inhibition results in an increase in E2F4/p130 complex formation and p130 hypophosphorylation which inactivates its function. Further biological studies demonstrated that PARP inhibition sensitizes cancer cells to radiation by suppressing DNA damage repair in a p130-dependent mechanism(87). Together, PARP-1 not only regulates the DDR response to radiation, but also the transcriptional events associated with this therapeutic modality, and utilization of pharmacological PARP-1 inhibitors may be clinically relevant in the administration of radiation.
Transcriptional regulation in hormone-dependent cancers
Nuclear receptors (NRs) are transcription factors that function in many processes including homeostasis, development, reproduction, metabolism, and cancer. Hormone receptors act as ligand-dependent transcription factors, serving as the means by which steroid signals generate biological responses. Given the fact that many cancers display aberrant NR signaling and have properties that make them amenable to pharmacologic targeting via endocrine therapy, NRs play a significant role in many cancer types. Several studies have examined the role of PARP-1 in mediating tumor-associated NR activity.
In breast cancer (BrCa), PARP-1 elicits disparate functions in estrogen receptor biology, depending on estrogen receptor-α (ERα) or estrogen receptor-β (ERβ) status. ERα is a ligand-dependent nuclear hormone receptor, and when activated in the context of BrCa serves a pro-proliferative role. ERα is expressed in ~70% of BrCa cases, and serves as a therapeutic target for some metastatic BrCa patients (such as through the use of tamoxifen therapy, an ERα antagonist). However, not all patients respond uniformily, and all will eventually relapse, resulting in the generation of hormone-independent BrCa that is resistant to endocrine therapy. Thus, understanding the mechanisms that regulate ER-mediated signaling in BrCa is of importance. The transcriptional response to 17β-estradiol (E2) in MCF-7 BrCa cells results in transient double-strand breaks (DSBs), and the subsequent recruitment of PARP-1 and Topoisomerase IIβ (TopoIIβ) to the promoters of ERα target genes. Abolishing either PARP-1 or TopoIIβ function resulted in diminished ability of E2 to activate the expression of classical ERα target genes, demonstrating that in this context, PARP-1 is required for ERα transcriptional activity(88). By contrast, ERβ exhibits anti-proliferative and pro-differentiative functions in several organ systems, including lung, colon, prostate, and mammary gland. It has been suggested that the ratio of ERα to ERβ determines whether BrCa tissue is proliferative and how the tissue will respond to hormone therapy (tamoxifen), however the role of ERβ in BrCa remains incompletely defined(89). A study that sought to illuminate the mechanism by which ERβ drives transcription in BrCa cells determined that tamoxifen treatment of MCF-7 BrCa cells served to protect cells from E2-induced oxidative DNA damage. This tamoxifen-induced protection was due to recruitment of ERβ to EpREs (electrophile response elements), which in turn induced the expression of antioxidative enzymes, including NQO1 (NAD(P)H quinone oxidoreductase), which required a number of cofactors, including PARP-1. In fact, it was found that upon depletion of PARP-1, the tamoxifen-dependent expression of antioxidative enzymes was compromised, demonstrating that in this context, PARP-1 is potentially a co-activator for ERβ(90). Together, it appears that PARP-1 is a positive regulator of both ERα and ERβ in models of BrCa, and that this positive regulation requires TopoIIβ. As both ERα and ERβ play significant roles in BrCa biology, future analyses of the biological impact of PARP-1 regulation of both ERα and ERβ in BrCa is critical, due to the differential functions of these nuclear receptors.
Progesterone receptor (PR) function is activated by the ovarian steroid progesterone, and serves to regulate differentiation of the endometrium, maintenance of pregnancy, and proliferation of the mammary gland. Nuclear PR acts as a transcription factor, while cytosolic PR acts as a rapid signal transducer. Progesterone can stimulate proliferation independently of estrogen, and is considered a risk factor for BrCa. In models of BrCa, it has been demonstrated that PAR accumulates after progestin stimulation, indicating the activation of PR induces PARP-1 activity. It was found that progestin induced a physical interaction between cyclin-dependent kinase 2 (CDK2) and PARP-1, followed by phosphorylation and increased enzymatic function of PARP-1. Genome-wide analyses indicated that both CDK2 and PARP-1 are enriched at PR binding sites in response to progestin stimulation, and that the majority of PR regulated genes required CDK2 and/or PARP-1 for proper activation or repression(91). These data indicate that in models of BrCa, progestin stimulates CDK2 to interact with and activate PARP-1, and this complex serves as a co-regulator of PR.
In prostate cancer (PCa), the androgen receptor (AR) plays a key role in cell proliferation and maintenance of PCa-associated phenotypes. AR serves as the target of first-line therapy for disseminated disease, but upon relapse AR activity is resurgent despite continued therapeutic targeting. There are limited options for patients with castrate-resistant PCa (CRPC). Therefore, defining novel means of AR regulation is of critical importance.
PARP-1 is recruited to sites of AR transcriptional function, and PARP-1 enzymatic activity is required for AR-driven gene expression and subsequent PCa cell proliferation in both the context of hormone therapy-sensitive and CRPC models of disease. The decrease in AR activity in response to PARP inhibition was associated with diminished AR and PARP-1 residency at sites of AR function, as well as altered capacity of androgen stimulation to elicit pro-transcriptional changes in histone modifications and chromatin architecture. Further analyses indicated that PARP-1 enzymatic activity was increased as a function of transition to CRPC, implying a role for PARP-1 in the evolutionary progression of PCa. Ultimately, pharmacological inhibition of PARP-1 resulted in diminished AR activity and diminution of subsequent tumor growth in vivo and ex vivo, implicating PARP-1 enzymatic activity in the maintenance of the CRPC phenotype in vivo. Together, these data demonstrate that in the context of PCa, PARP-1 appears to serve as an activator of AR function and effector of downstream biological consequences. As described above, PARP-1 also regulates the activity of ETS transcription factors in models of PCa, which is of clinical significance, given the high percentage of prostate tumors that harbor fusions that put ETS expression under the control of AR activity (as through the TMPRSS2:ERG fusion). As such, the studies demonstrating regulation of both AR and ETS transcription factors by PARP-1 are now being translated into the clinic in a trial combining PARP inhibition and an AR-directed therapeutic (abiraterone acetate) for patients with metastatic CRPC (NCT01576172)(92). Combined, the studies outlined above implicate PARP-1 in the regulation of several NRs that have significant roles in human cancer.
With respect to NRs in cancer, PARP-1 has an apparent positive role in regulating transcriptional events. As such, further analyses of mechanisms of PARP-1 responsive transcription by NRs and the biological impact of PARP inhibition should be considered.
Conclusions and future directions
Modifying transcription factor function has long been a goal of cancer research, with some successes, especially in the context of ligand-dependent transcription factors such as nuclear receptors. While it has been long understood that selected transcription factor activities can drive tumor formation and progression, many have proven to be difficult to develop modalities to target them directly, but understanding mechanisms of how “druggable” enzymes regulate transcription factors may yield clinically translatable results. Gains in our understanding of how PARP-1 regulates transcription in human cancer may bring new appreciation of the mechanisms that support aberrant transcriptional events and downstream processes in human disease. The transcriptional roles of PARP-1 should be considered not only in future design of basic and clinical investigation in the utility of PARP inhibitors for cancer therapy, but also the impact that PARP-1 has on transcription in the way that previous studies are interpreted. While there is obviously no unifying theory of transcriptional regulation in cancer by PARP-1, given the divergent and context-specific functions and outcomes of PARP-1-dependent transcription (Figure 2), findings discussed herein underscore the major impact of PARP-1-mediated transcriptional control on human tumor biology. Attaining greater mechanistic insight to these events, discerning the cause and effect of the context-specific PARP-1 functions, and using this knowledge for development of new trials is likely to have significant clinical impact.
Figure 2.
While there have been major advances in understanding of PARP-1 transcriptional regulatory functions, key questions remain that must be addressed to delineate the complex role of PARP-1 in human malignancy. First, what are the context-dependent molecular determinants of transcriptional regulation by PARP-1? While it is apparent that PARP-1 regulates transcription, the molecular mechanisms by which PARP-1 selectively supports or represses transcription is poorly understood and the results to date are disparate in nature. It is not known whether the transcriptional roles for PARP-1 outlined above may be universal, and it is not yet established which divergent roles of PARP-1 influence the tumorigenic program. Second, do other PARP family members compensate for PARP-1 in transcriptional regulation, and how are other PARP family members affected by PARP inhibition? PARP-1 and PARP-2 are the closest family members, and double null mice are embryonic lethal, speaking to the overlap and importance of PARP-1 and PARP-2. However, as of yet it has not been determined whether other PARP family members can contribute to transcription to a similar degree and in similar contexts as PARP-1. Additionally, some of the PARP inhibitors in trial for human malignancies exhibit less specificity than may be desired. As such, determining the impact of these drugs on other PARP members in vivo, and assessment of downstream cellular and biological consequence would be of great value. Third, is it possible to specifically target the transcriptional regulatory function of PARP-1 in cancer therapy? Recent evidence suggests that specific modules of PARP-1 regulate allosteric communication, and abrogation of this communication supports the contention of context-dependent transcriptional regulation by PARP-1, but not the DDR function of PARP-1 (93). Given these compelling findings, pursuit towards more specifically targeting PARP-1 transcriptional regulation is currently underway. Fourth, given the significant body of evidence suggesting that PARP-1 controls critical transcriptional events in models of cancer, do these events alter cancer biology, in the lab and in the clinic? Many of the studies described herein put forth compelling mechanistic observations about how PARP-1 regulates specific transcriptional events in cancer, yet few assess causation to more fully understand the true impact of PARP-1, and by extension, PARP inhibitors in the field of cancer biology, and these deficiencies must be addressed. Fifth, within the field there is a controversy as to whether transcription-associated DNA damage is a true phenomenon. Some literature points to transcription causing transient double strand breaks(88, 94–98). However, whether this is a true biological outcome, what the explicit role that PARP-1 plays in causing/maintaining these breaks remains unclear. Finally, does the concept of transcriptional regulation by PARP-1 alter the interpretation and implications of ongoing and concluded oncology clinical trials, and can the transcriptional regulatory roles of PARP-1 be fully harnessed for clinical benefit? Initial PARP inhibitor clinical trials were rationally developed based on the synthetic lethal interaction of HR deficiency and PARP inhibition. However, given the complex, diverse, and context-specific roles of PARP-1 in regulating key transcriptional events in cancer, the implications thereof for therapeutic response should be considered. In conclusion, PARP-1 functions to regulate many key transcriptional events in cancer biology, and while much is known, further mechanistic insight may lead to better utilization of PARP inhibitors in human malignancies.
Acknowledgements
The authors are grateful the K. Knudsen lab for critical commentary and ongoing discussions. While every effort was made to include all relevant primary research articles, we apologize for any oversight that may have occurred. This work was supported by a Prostate Cancer Foundation Young Investigator Award (to MJS) and a Movember-Prostate Cancer Foundation Challenge Award (to KEK).
Literature cited
- 1.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 3.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 4.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 5.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Flores A, Aguilar-Quesada R, Siles E, Pozo S, Rodriguez-Lara MI, Lopez-Jimenez L, et al. Interaction between PARP-1 and HIF-2alpha in the hypoxic response. Oncogene. 2014;33:891–898. doi: 10.1038/onc.2013.9. [DOI] [PubMed] [Google Scholar]
- 10.Valenzuela MT, Guerrero R, Nunez MI, Ruiz De Almodovar JM, Sarker M, de Murcia G, et al. PARP-1 modifies the effectiveness of p53-mediated DNA damage response. Oncogene. 2002;21:1108–1116. doi: 10.1038/sj.onc.1205169. [DOI] [PubMed] [Google Scholar]
- 11.Fossati S, Formentini L, Wang ZQ, Moroni F, Chiarugi A. Poly(ADP-ribosyl)ation regulates heat shock factor-1 activity and the heat shock response in murine fibroblasts. Biochem Cell Biol. 2006;84:703–712. doi: 10.1139/o06-083. [DOI] [PubMed] [Google Scholar]
- 12.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 13.Droit A, Hunter JM, Rouleau M, Ethier C, Picard-Cloutier A, Bourgais D, et al. PARPs database: a LIMS systems for protein-protein interaction data mining or laboratory information management system. BMC Bioinformatics. 2007;8:483. doi: 10.1186/1471-2105-8-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, et al. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 16.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 18.Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J Mol Biol. 2005;348:609–615. doi: 10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 21.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 22.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 23.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 24.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 26.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 27.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc Natl Acad Sci U S A. 2006;103:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, et al. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem. 2012;287:12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carson DA, Carrera CJ, Wasson DB, Yamanaka H. Programmed cell death and adenine deoxynucleotide metabolism in human lymphocytes. Adv Enzyme Regul. 1988;27:395–404. doi: 10.1016/0065-2571(88)90028-3. [DOI] [PubMed] [Google Scholar]
- 31.Huang Q, Shen HM. To die or to live: the dual role of poly(ADP-ribose) polymerase-1 in autophagy and necrosis under oxidative stress and DNA damage. Autophagy. 2009;5:273–276. doi: 10.4161/auto.5.2.7640. [DOI] [PubMed] [Google Scholar]
- 32.Huang Q, Wu YT, Tan HL, Ong CN, Shen HM. A novel function of poly(ADP-ribose) polymerase-1 in modulation of autophagy and necrosis under oxidative stress. Cell Death Differ. 2009;16:264–277. doi: 10.1038/cdd.2008.151. [DOI] [PubMed] [Google Scholar]
- 33.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 36.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Audebert M, Salles B, Calsou P. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem Biophys Res Commun. 2008;369:982–988. doi: 10.1016/j.bbrc.2007.11.132. [DOI] [PubMed] [Google Scholar]
- 39.Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010;38:6065–6077. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 43.Oka J, Ueda K, Hayaishi O, Komura H, Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J Biol Chem. 1984;259:986–995. [PubMed] [Google Scholar]
- 44.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 45.Okayama H, Honda M, Hayaishi O. Novel enzyme from rat liver that cleaves an ADP-ribosyl histone linkage. Proc Natl Acad Sci U S A. 1978;75:2254–2257. doi: 10.1073/pnas.75.5.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 47.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niere M, Mashimo M, Agledal L, Dolle C, Kasamatsu A, Kato J, et al. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J Biol Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krietsch J, Rouleau M, Pic E, Ethier C, Dawson TM, Dawson VL, et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med. 2013;34:1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012;40:7788–7805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 54.Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, et al. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 55.Mathis G, Althaus FR. Release of core DNA from nucleosomal core particles following (ADP-ribose)n-modification in vitro. Biochem Biophys Res Commun. 1987;143:1049–1054. doi: 10.1016/0006-291x(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 56.Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosylation) J Biol Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- 57.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 58.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gottschalk AJ, Trivedi RD, Conaway JW, Conaway RC. Activation of the SNF2 family ATPase ALC1 by poly(ADP-ribose) in a stable ALC1.PARP1.nucleosome intermediate. J Biol Chem. 2012;287:43527–43532. doi: 10.1074/jbc.M112.401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulkarni A, Oza J, Yao M, Sohail H, Ginjala V, Tomas-Loba A, et al. Tripartite Motif-containing 33 (TRIM33) protein functions in the poly(ADP-ribose) polymerase (PARP)-dependent DNA damage response through interaction with Amplified in Liver Cancer 1 (ALC1) protein. J Biol Chem. 2013;288:32357–32369. doi: 10.1074/jbc.M113.459164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199:235–249. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farrar D, Rai S, Chernukhin I, Jagodic M, Ito Y, Yammine S, et al. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol. 2010;30:1199–1216. doi: 10.1128/MCB.00827-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee MH, Na H, Kim EJ, Lee HW, Lee MO. Poly(ADP-ribosyl)ation of p53 induces gene-specific transcriptional repression of MTA1. Oncogene. 2012;31:5099–5107. doi: 10.1038/onc.2012.2. [DOI] [PubMed] [Google Scholar]
- 67.Alvarez-Gonzalez R. Genomic maintenance: the p53 poly(ADP-ribosyl)ation connection. Sci STKE. 2007;2007:e68. doi: 10.1126/stke.4152007pe68. [DOI] [PubMed] [Google Scholar]
- 68.Wieler S, Gagne JP, Vaziri H, Poirier GG, Benchimol S. Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J Biol Chem. 2003;278:18914–18921. doi: 10.1074/jbc.M211641200. [DOI] [PubMed] [Google Scholar]
- 69.Wu Y, Lin JC, Piluso LG, Dhahbi JM, Bobadilla S, Spindler SR, et al. Phosphorylation of p53 by TAF1 inactivates p53-dependent transcription in the DNA damage response. Mol Cell. 2014;53:63–74. doi: 10.1016/j.molcel.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agarwal ML, Agarwal A, Taylor WR, Wang ZQ, Wagner EF, Stark GR. Defective induction but normal activation and function of p53 in mouse cells lacking poly-ADP-ribose polymerase. Oncogene. 1997;15:1035–1041. doi: 10.1038/sj.onc.1201274. [DOI] [PubMed] [Google Scholar]
- 71.Wesierska-Gadek J, Bohrn E, Herceg Z, Wang ZQ, Wurzer G. Differential susceptibility of normal and PARP knock-out mouse fibroblasts to proteasome inhibitors. J Cell Biochem. 2000;78:681–696. [PubMed] [Google Scholar]
- 72.Idogawa M, Yamada T, Honda K, Sato S, Imai K, Hirohashi S. Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/beta-catenin complex. Gastroenterology. 2005;128:1919–1936. doi: 10.1053/j.gastro.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 73.Idogawa M, Masutani M, Shitashige M, Honda K, Tokino T, Shinomura Y, et al. Ku70 and poly(ADP-ribose) polymerase-1 competitively regulate beta-catenin and T-cell factor-4-mediated gene transactivation: possible linkage of DNA damage recognition and Wnt signaling. Cancer Res. 2007;67:911–918. doi: 10.1158/0008-5472.CAN-06-2360. [DOI] [PubMed] [Google Scholar]
- 74.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res. 2012;72:1608–1613. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McPhee TR, McDonald PC, Oloumi A, Dedhar S. Integrin-linked kinase regulates E-cadherin expression through PARP-1. Dev Dyn. 2008;237:2737–2747. doi: 10.1002/dvdy.21685. [DOI] [PubMed] [Google Scholar]
- 77.Stanisavljevic J, Porta-de-la-Riva M, Batlle R, de Herreros AG, Baulida J. The p65 subunit of NF-kappaB and PARP1 assist Snail1 in activating fibronectin transcription. J Cell Sci. 2011;124:4161–4171. doi: 10.1242/jcs.078824. [DOI] [PubMed] [Google Scholar]
- 78.Chu S, Xu H, Ferro TJ, Rivera PX. Poly(ADP-ribose) polymerase-1 regulates vimentin expression in lung cancer cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1127–1134. doi: 10.1152/ajplung.00197.2007. [DOI] [PubMed] [Google Scholar]
- 79.Wu X, Ellmann S, Rubin E, Gil M, Jin K, Han L, et al. ADP ribosylation by PARP-1 suppresses HOXB7 transcriptional activity. PLoS One. 2012;7:e40644. doi: 10.1371/journal.pone.0040644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sterling JA, Wu L, Banerji SS. PARP regulates TGF-beta receptor type II expression in estrogen receptor-positive breast cancer cell lines. Anticancer Res. 2006;26:1893–1901. [PubMed] [Google Scholar]
- 81.Hossain MB, Ji P, Anish R, Jacobson RH, Takada S. Poly(ADP-ribose) Polymerase 1 Interacts with Nuclear Respiratory Factor 1 (NRF-1) and Plays a Role in NRF-1 Transcriptional Regulation. J Biol Chem. 2009;284:8621–8632. doi: 10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin-Oliva D, Aguilar-Quesada R, O'Valle F, Munoz-Gamez JA, Martinez-Romero R, Garcia Del Moral R, et al. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Res. 2006;66:5744–5756. doi: 10.1158/0008-5472.CAN-05-3050. [DOI] [PubMed] [Google Scholar]
- 83.Elser M, Borsig L, Hassa PO, Erener S, Messner S, Valovka T, et al. Poly(ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol Cancer Res. 2008;6:282–290. doi: 10.1158/1541-7786.MCR-07-0377. [DOI] [PubMed] [Google Scholar]
- 84.Gonzalez-Flores A, Aguilar-Quesada R, Siles E, Pozo S, Rodriguez-Lara MI, Lopez-Jimenez L, et al. Interaction between PARP-1 and HIF-2alpha in the hypoxic response. Oncogene. 2013 doi: 10.1038/onc.2013.9. [DOI] [PubMed] [Google Scholar]
- 85.Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-kappaB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009;28:832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li W, Ain KB. Human sodium-iodide symporter (hNIS) gene expression is inhibited by a trans-active transcriptional repressor, NIS-repressor, containing PARP-1 in thyroid cancer cells. Endocr Relat Cancer. 2010;17:383–398. doi: 10.1677/ERC-09-0156. [DOI] [PubMed] [Google Scholar]
- 87.Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc Natl Acad Sci U S A. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 89.Warner M, Gustafsson JA. The role of estrogen receptor beta (ERbeta) in malignant diseases--a new potential target for antiproliferative drugs in prevention and treatment of cancer. Biochem Biophys Res Commun. 2010;396:63–66. doi: 10.1016/j.bbrc.2010.02.144. [DOI] [PubMed] [Google Scholar]
- 90.Sripathy SP, Chaplin LJ, Gaikwad NW, Rogan EG, Montano MM. hPMC2 is required for recruiting an ERbeta coactivator complex to mediate transcriptional upregulation of NQO1 and protection against oxidative DNA damage by tamoxifen. Oncogene. 2008;27:6376–6384. doi: 10.1038/onc.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wright RH, Castellano G, Bonet J, Le Dily F, Font-Mateu J, Ballare C, et al. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012;26:1972–1983. doi: 10.1101/gad.193193.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steffen JD, Tholey RM, Langelier MF, Planck JL, Schiewer MJ, Lal S, et al. Targeting PARP-1 Allosteric Regulation Offers Therapeutic Potential against Cancer. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- 95.Hendriks G, Jansen JG, Mullenders LH, de Wind N. Transcription and replication: far relatives make uneasy bedfellows. Cell Cycle. 2010;9:2300–2304. doi: 10.4161/cc.9.12.11987. [DOI] [PubMed] [Google Scholar]
- 96.Park C, Qian W, Zhang J. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 2012;13:1123–1129. doi: 10.1038/embor.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khobta A, Epe B. Interactions between DNA damage, repair, and transcription. Mutat Res. 2012;736:5–14. doi: 10.1016/j.mrfmmm.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 98.Tchurikov NA, Kretova OV, Fedoseeva DM, Sosin DV, Grachev SA, Serebraykova MV, et al. DNA double-strand breaks coupled with PARP1 and HNRNPA2B1 binding sites flank coordinately expressed domains in human chromosomes. PLoS Genet. 2013;9:e1003429. doi: 10.1371/journal.pgen.1003429. [DOI] [PMC free article] [PubMed] [Google Scholar]