Abstract
Previously we described the identification of a Plasmodium falciparum signal peptide peptidase (PfSPP) functioning at the blood stage of malaria infection. Our studies also demonstrated that mammalian SPP inhibitors prevent malaria parasite growth at the late-ring/early trophozoite stage of intra-erythrocytic development. Consistent with its role in development, we tested the hypothesis that PfSPP functions at the endoplasmic reticulum of Plasmodium falciparum where it cleaves membrane-bound signal peptides generated following the enzyme activity of signal peptidase. The localization of PfSPP to the endoplasmic reticulum was confirmed by immunofluorescence microscopy and immunogold electron microscopy. Biochemical analysis indicated the existence of monomer and dimer forms of PfSPP in the parasite lysate. A comprehensive bioinformatics screen identified several candidate PfSPP substrates in the parasite genome. Using an established transfection based in vivo luminescence assay, malaria heat shock protein 101 (HSP101) was identified as a substrate of PfSPP, and partial inhibition of PfSPP correlated with the emergence of gametocytes. This finding unveils the first known substrate of PfSPP, and provides new perspectives for the function of intra-membrane proteolysis at the erythrocyte stage of malaria parasite life cycle.
Keywords: Malaria, Signal peptide peptidase, Endoplasmic Reticulum, Intramembrane cleaving protease, ER stress
Introduction
Plasmodium falciparum represents the most lethal parasite causing human malaria. Millions of infections occur each year, and show a particularly high mortality in young children [1]. Due to increasing drug resistance, and the lack of an effective vaccine, there is an urgent need to identify new drug targets. Proteases have long been considered viable drug targets. Recent studies have identified a single gene encoded aspartic protease essential for parasite development and survival [2,3,4,5]. This enzyme termed Plasmodium falciparum signal peptide peptidase (PfSPP) may serve as a promising drug target; however, its precise cellular function and natural substrates are not yet known. Eukaryotic signal peptide peptidases (SPPs) are known to process signal peptide “stubs” left behind in the endoplasmic reticulum (ER) during protein processing [6,7]. The human homologue of malaria SPP is known to function as a dimer in vivo, and localizes to the endoplasmic reticulum where it plays a key role in processing the histocompatibility proteins as well as the hepatitis C virus polyprotein [7,8]. In several other species including Drosophila, Caenorhabditis elegans, and zebrafish, SPPs play a key role in development [9,10,11]. Additional functions of SPPs in protein processing include a key role in the stabilization of transmembrane (TM) proteins at the endoplasmic reticulum [12,13,14]. Since human and malaria signal peptide peptidases share identical catalytic site motifs, it is speculated that they may perform similar functions by targeting conserved substrates [7].
Genetic knockout experiments of PfSPP in human erythrocytes did not yield any viable parasites suggesting an essential role PfSPP at the blood stage of parasite life cycle [3]. Moreover, using pharmacological inhibitors of SPP, our studies demonstrated an essential role of PfSPP during the intracellular development of malaria parasite in human erythrocytes [3]. Therefore, analogous to the function of mammalian SPPs, we hypothesized that PfSPP may cleave membrane-embedded signal peptides in the parasite endoplasmic reticulum (ER) during protein processing. Our results demonstrate that PfSPP is an ER protease that cleaves malaria heat shock protein 101 (HSP101), the first known substrate of PfSPP. Implications for the functional role of malaria signal peptide peptidase and HSP101 at the blood stage of parasite life cycle will be discussed.
Materials and Methods
Plasmodium falciparum (Pf) strain 3D7 was cultured as previously described [2,15].
Monospecific PfSPP-CT Antibody
A short peptide sequence corresponding to the C-terminal amino acids 393–412 (EIPKIQETPVSNAKKRITNK) of PfSPP was synthesized, and conjugated to Keyhole Limpet Hemocyanin (KLH) via the N-terminus. The anti-peptide antibody against the PfSPP-CT region was generated by Alpha Diagnostic International (ADI, San Antonio).
Co-Immunoprecipitation
Parasite lysate was prepared in lysis buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, 5 mM EDTA, 5mM EGTA. The lysate was pre-cleared with Protein A beads, and then 300 μL of supernatant was incubated with PfSPP-CT antibody, and pre-immune rabbit IgG, overnight at 4°C.
Immunofluorescence Microscopy
Methanol fixed parasite slides were incubated with the affinity purified anti-PfSPP-CT antibody and anti-PMV antibody. Slides were washed and incubated with respective secondary antibodies, anti-rabbit Alexa 488 (green) and anti-mouse Alexa 594 (red). Immunofluorescence images were captured using the Zeiss LSM510 confocal microscope (Germany).
Immunogold Electron Microscopy
Plasmodium falciparum infected erythrocytes were washed in RPMI 1640, fixed in 4% paraformaldehyde and 0.1% glutaraldehyde for 1 h at 4°C in 0.1M sodium phosphate buffer (pH 7.2), and embedded in white London resin. Ultra-thin sections were blocked and incubated with the affinity purified anti-PfSPP-CT antibody. Secondary antibody conjugated to gold particles was used (15 nm). Labeled sections were stained with uranyl acetate, lead citrate, and visualized using a Philips FEI Tecnai F30 transmission electron microscope at the University of Chicago EM facility.
Bioinformatics Screen
We performed a bioinformatics search for PfSPP substrate candidates by selecting genes in the PlasmoDB that match a selected criteria. This criterion includes seven steps. We selected for protein coding, single copy genes in Plasmodium falciparum, which contain a signal peptide, one type II transmembrane domain protein motif pattern 6666996666 (where 6 represents a hydrophobic residue, and 9 represents a small residue), and the signal peptide and the transmembrane domain overlap.
PfSPP Luminescence Assay
The cleavage assay was performed and analyzed according to standard methods [16]; however, here the pCGN-HA-ATF6 (1–363)-HSP101 substrate plasmid was used. Inhibitor concentrations used were consistent with previous studies [16,17]. Mutant constructs of PfSPP were generated using the QuikChange mutagenesis kit (Agilent Technologies) to replace residues D228A and D269A. A pCDNA6-PfSPP-GFP construct was generated by cloning GFP out of the pEGFP-C1 plasmid and inserting it at the C-terminal end of PfSPP.
Transfection of malaria parasite by PfSPP-GFP construct
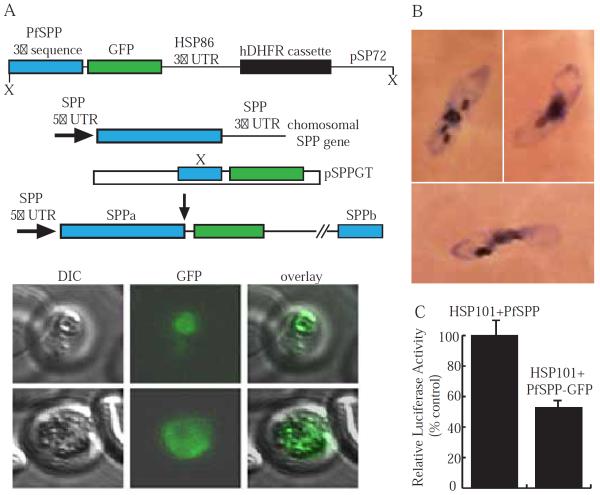
An 883 bp fragment from the C-terminal region of the PfSPP genomic DNA sequence was amplified. This region was inserted into the pPM2GT vector with the C-terminal GFP tag as described previously [18]. We termed this vector, pSPPGT (Fig. 4A). Transfections were performed according to standard protocols [19,20].
Fig 4.
Generation of chromosomal PfSPP-GFP chimera and its effect on parasite life cycle. (A) Schematic illustration of PfSPP vector with GFP inserted at the C-terminus. The integration event produced a full-length PfSPP coding frame fused to GFP. The bottom panel shows parasites displaying GFP fluorescence along with the DIC images of live parasites. (B) A montage of gametocyte-like parasites recovered after transfection and selection in the presence of WR99210. (C) Quantification of HSP101 cleavage activity by PfSPP and PfSPP-GFP constructs.
Results
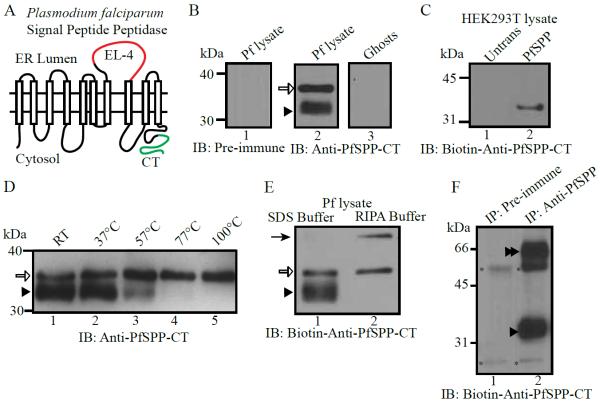
In previous studies, we raised an antibody against the exofacial loop 4 (EL4) of PfSPP (Fig.1A, red line) [3]. Due to weak signal in immunolocalization studies, we generated a new polyclonal antibody against the last 20 amino acids of PfSPP (Fig. 1A, green line). The anti-PfSPP-CT antibody detected specific bands of PfSPP corresponding to 35 kDa and 32 kDa as indicated by the white arrow, and black arrowhead, respectively (Fig 1B, D, E). A similar pattern of PfSPP bands was observed using a biotin-conjugated anti-PfSPP-CT antibody (Fig. 1C, E). The predicted MW of PfSPP is 47 kDa, however a previous study expressed a codon-optimized version of PfSPP in mammalian cells and detected a 35 kDa band [4]. The gel mobility of PfSPP bands was markedly affected by the parasite lysis conditions. For example, detection of the 32 kDa band was dependent upon temperature, possibly due to degradation or aggregation (Fig. 1D). Furthermore, parasites lysed directly in SDS sample buffer showed the two aforementioned 35 and 32 kDa bands, whereas lysate prepared in RIPA buffer showed the presence of a 47 kDa band, and elimination of the 32 kDa band (Fig. 1E). We expressed recombinant PfSPP in HEK293T cells and detected a single 35 kDa band (Fig. 1C). The 35 kDa and 32 kDa bands are likely either the same PfSPP species or a consequence of some unknown post-translational modification [21].
Fig 1.
Immunoblotting of PfSPP using the anti-C-terminal PfSPP antibody. (A) Predicted membrane topology of PfSPP according to Li et al. [3]. Highlighted area in red is the exofacial loop 4 of PfSPP. Highlighted area in green corresponds to the C-terminal domain of PfSPP, where the antibody used in this study was raised. (B) Immunoblotting detected two bands of PfSPP as indicated by arrows (white arrow for 35 kDa, and black arrowhead for 32 kDa, lane 2). (C) Transient transfection of a synthetic PfSPP construct (pCDNA6-PfSPP) resulted in PfSPP expression in HEK293T cells. (D) Sample incubation at different temperature alters the mobility of PfSPP with the disappearance of the 32 kDa band at higher temperature (arrowhead). (E) Lysis conditions significantly alter the mobility of PfSPP. (F) Immunoprecipitation of PfSPP from parasite-infected erythrocyte lysate. Asterisks correspond to heavy and light chain bands in the pre-immune IgG negative control (Lane 1).
Immunoprecipitated material was detected by immunoblotting using the biotinylated anti-PfSPP-CT antibody (Fig. 1F). Bands of 35 and 65 kDa were detected (Fig. 1F). This 65 kDa band corresponds to the approximate position of a putative PfSPP dimer. While we did not detect a dimer in the direct immunoblotting experiments (Fig. 1B–E), immunoprecipitation of PfSPP from parasite lysate solubilized under mild detergent conditions likely preserved the dimer species. This observation is consistent with prior evidence showing that homologous signal peptide peptidases form dimers in vivo [16,22]. Furthermore, our finding is consistent with published evidence demonstrating recombinant PfSPP at 35 and 70 kDa bands when expressed in HEK293T cells [4].
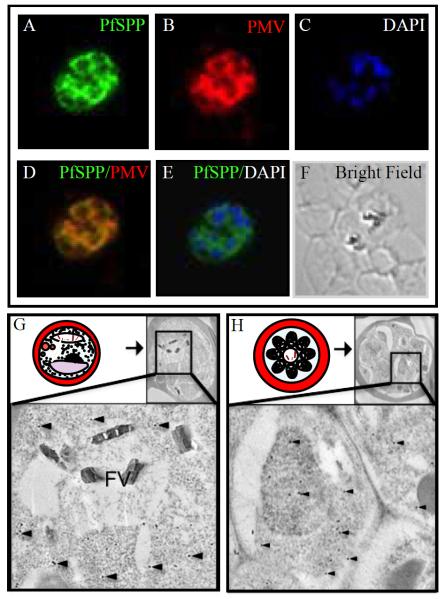
We examined the localization of PfSPP in infected erythrocytes using our affinity-purified anti-PfSPP-CT antibody. Subcellular localization of PfSPP was examined in trophozoite-infected human erythrocytes by indirect immunofluorescence microscopy indicating that PfSPP is co-localized with Plasmepsin V (PMV), a known ER-marker (Fig. 2 A–F). The mesh-like network of PfSPP is consistent with the perinuclear endomembrane system observed at the late stages of intra-erythrocytic parasite development [23,24]. Further analysis using an antibody against the parasite-derived BiP found a similar co-localization pattern with PfSPP (data not shown). To further confirm the ER localization of PfSPP, immunogold electron microscopy was performed (Fig. 2 G, H). Gold particles show a diffuse pattern across the parasite at several stages of parasite development, including the trophozoite and schizont stages (Fig. 2 G, H), as well as the late ring stage (data not shown). PfSPP was not detected in the food vacuole and endosomal compartments of infected erythrocytes (Fig. 2). The diffuse pattern of PfSPP throughout the parasite cytoplasm is consistent with the mesh-like network/or perinuclear endomembrane system shown in previous studies [23,24].
Fig 2.
Co-localization of PfSPP and Plasmepsin V in the endoplasmic reticulum. Indirect immunofluorescence microscopy using specific antibodies demonstrates PfSPP (green) co-localizes with a known parasite ER marker Plasmepsin V (red) in the Plasmodium falciparum (3D7) trophozoite. (A) Staining of PfSPP. (B) Staining of known ER marker, Plasmepsin V. (C) DAPI stain. (D) PfSPP and PMV overlay. (E) Overlay of PfSPP and DAPI stain. (F) Bright field microscopy. (G, H) Immunogold electron microscopy of PfSPP in the parasite infected erythrocytes. PfSPP is indicated by black dots and several spots are identified by black arrowheads. (G) Schematic illustration of a mature trophozoite stage parasite in the infected erythrocyte adapted from Bannister et al. [37]. PfSPP staining in the trophozoite shows a diffuse staining pattern throughout the perinuclear endomembrane system/ER. (H) Illustration of a late schizont-stage parasite in an infected erythrocyte adapted from Bannister et al. [37]. PfSPP staining in the late-schizont stage of parasite development shows staining throughout the perinuclear endomembrane system/ER.
The ER localization of PfSPP suggests potential substrates accessible to PfSPP might include proteins containing a signal peptide processed within the ER. To identify potential substrates of PfSPP, we performed a hypothetical substrate search screen and narrowed down a list of 6 potential substrates in the PlasmoDB (Table 1) [16]. The most intriguing was Plasmodium falciparum heat shock protein 101 (HSP101). Previous evidence indicates that HSP101 encodes an N-terminal ER signal [25], consistent with the ER localization of PfSPP. While HSP101 is essential for parasite protein export and translocation, a functional role of its signal peptide is not yet known [26].
Table 1.
Parasite Substrates Predicted from the Bioinformatics Screen
| Gene Identified | Gene ID | Predicted Function |
|---|---|---|
| Heat Shock Protein 101 (HSP101) | PF11_0175 | Component of protein export machinery |
| Erythrocyte Membrane Protein 3 (PfEMP3) | PFB0095C | Destabilization of erythrocyte membrane skeleton |
| Aspartate Carbamoyltransferase | MAL13P1.221 | Role in pyrimidine biosynthesis |
| Liver Stage Antigen 1 (LSA1) | PF10_0356 | Putative role in liver schizogony and merozoite release |
| Malonyl CoA-acyl Carrier Protein Transacylase Precursor | PF13_0066 | Involved in fatty acid biosynthesis |
| Apicoplast Ribosomal Protein L21 Precursor, putative | PF08_0014 | Member of large ribosomal subunit; role in protein synthesis |
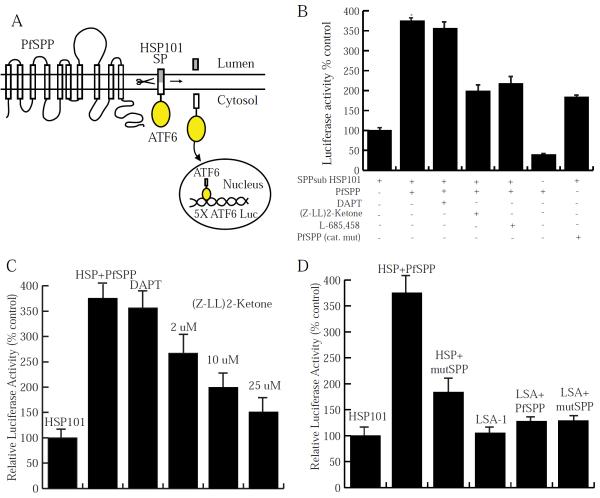
We utilized a transient transfection assay previously used to demonstrate the enzyme activity of PfSPP against a model substrate (Fig. 3A) [4]. The cleavage assay demonstrated cleavage of HSP101 signal peptide by PfSPP (Fig. 3B). PfSPP activity against HSP101 was inhibited by (Z-LL)2 ketone and L-685,458, known SPP inhibitors (Fig. 3B). This inhibition was observed in a dose-dependent manner (Fig. 3C). In contrast, DAPT, a Presenilin-specific inhibitor, showed no effect on cleavage activity (Fig. 3B). These findings indicate that malaria parasite HSP101 serves as a substrate of PfSPP under these conditions. Moreover, we mutated the two active site aspartic acid residues of PfSPP, and the cleavage of HSP101 by PfSPP mutant was markedly reduced to a level similar observed in the presence of SPP inhibitors (Fig. 3B). We also measured the cleavage susceptibility of liver stage antigen-1 (LSA-1) identified as a potential substrate of PfSPP in our bioinformatics screen (Table 1). PfSPP did not cleave LSA-1 (Fig. 3D). Both wild type and catalytically inactive mutants of PfSPP showed the same basal level of protease activity against LSA-1 (Fig. 3D). While a cell-free cleavage assay system has been used previously to measure the protease activity of newly identified substrates of SPP by utilizing their synthetic signal peptide sequences [27], this approach was not feasible because the putative signal peptide sequence of parasite HSP101 (YLKYYIFVTLLFFVQVINNVLCA) is extremely hydrophobic. Genetic tractability of the malaria parasite posed unique technical barriers for biochemical analysis of PfSPP substrates under endogenous conditions. The putative endogenous cleavage of signal peptides is likely to occur rapidly, thus precluding the possibility of detecting PfSPP-associated substrates by conventional pull-down assays. Therefore, a cell-based assay offers the most practical approach to measure the protease activity of PfSPP for potential substrates.
Fig 3.
PfSPP cleaves the signal peptide of HSP101 but not LSA-1. HEK293T cells were transiently transfected with a combination of plasmids expressing PfSPP, HSP101/LSA-1 SP, Renilla luciferase, and Firefly Luciferase. (A) Schematic diagram of PfSPP assay, adapted from Nyborg et al. [16]. (B) Overexpression of PfSPP significantly increased peptidase activity relative to the endogenous activity (*P<0.01). This cleavage activity was inhibited by the SPP inhibitors (Z-LL)2 ketone (10 μM) and L-685,458 (25 μM). DAPT (15 μM) did not show inhibition of PfSPP activity. Mutant PfSPP (D228A and D269A) significantly reduced HSP101 cleavage activity. (C) A dose-dependent analysis was performed using (Z-LL)2 ketone. (D) LSA-1 was tested as a substrate of PfSPP. Luminescence activity of LSA-1 was measured alone, in the presence of the wild type PfSPP, and a catalytically inactive mutant of PfSPP.
Subcellular localization of PfSPP in the micronemes [2] and ER (Fig. 2) suggests delivery and localization of PfSPP to specific organelles is regulated by a defined trafficking pathway. Thus, we generated a PfSPP-GFP (pSPPGT) chimeric construct to monitor the trafficking of PfSPP (Fig. 4A). However, upon transfection, the parasitemia fell dramatically and only a small number of resistant clones could be recovered after 15–20 days of parasite culture. Resistant parasites carrying PfSPP-GFP showed aberrant morphology with essentially no healthy parasites detectable by microscopy. Interestingly, nearly all parasites carrying the chimeric PfSPP gene showed gametocyte-like morphology after 15–20 days of selection in culture (Fig. 4B). We measured the activity of wild type PfSPP and PfSPP-GFP and found the activity of PfSPP-GFP construct was reduced to 50% activity compared to wild type (Fig. 4C). These results indicate partial inhibition of PfSPP may be detrimental to the normal differentiation of malaria parasites.
Discussion
Our previous studies detected PfSPP signal on the merozoite surface and in the interior compartments of unknown origin [2], consistent with the expression of PfSPP in the merozoites as detected by mass spectrometry [28]. The short lifespan of free merozoites often precludes precise distinction between their apical organelles, which are in close proximity to the ER in merozoites. Therefore, a distinction between micronemes and ER compartments is sometimes difficult to discern. Nonetheless, subcellular localization of PfSPP in the ER of trophozoites and schizonts (Fig. 2) is consistent with the localization of signal peptide peptidases in malaria and other species [27,29,30]. Furthermore, we identified HSP101 as a substrate of PfSPP. Upon release of the soluble domain of HSP101 by parasite signal peptidase (SP), PfSPP is expected to cleave the resident signal peptide within the ER. Our findings suggest that PfSPP plays an essential ER clearing function, analogous to other counterparts of SPP [31]. Inhibition of PfSPP may therefore lead to toxic accumulation of signal peptides within the ER. Alternatively, the release of cleaved signal peptides of HSP101 by PfSPP may perform critical signaling functions. It is likely that PfSPP cleaves multiple type II TM signal peptides to ensure optimal ER membrane clearance. For example, a previous study has shown that pharmacological inhibition of malaria SPP inhibits growth of sporozoites in hepatocytes [5].
A recent study reported that PfSPP is an ER-resident protease required for growth and not invasion [21]. Our findings are consistent with the localization of PfSPP to endoplasmic reticulum. We also demonstrate the presence of a major 35 kDa species of PfSPP in parasite lysate (Fig. 1). Furthermore, we provide evidence for dimerization of PfSPP under mild conditions of immunoprecipitation (Fig. 1E). The previous study utilized the E64-treated merozoites to rule out a functional role of PfSPP in erythrocyte invasion [21]. However, global inhibition of cysteine proteases by E64 would render the functionally-impaired merozoites unsuitable for other proteases such as PfSPP to function properly. Since PfSPP is highly expressed in the merozoites, one cannot rule out a functional role of PfSPP in merozoite invasion particularly under conditions where invasion and re-invasion events cannot be distinguished experimentally.
A potentially intriguing finding in our study is the expression of a chimeric form of PfSPP carrying a GFP-tag at the C-terminus resulting in significant conversion of parasites to gametocyte lineage (Fig. 4). Coincidentally, a very recent study demonstrated that ER stress triggers gametocytogenesis [32]. Consistent with this finding and the fact that PfSPP is an ER-resident protease, it is likely that PfSPP plays a key role in ER homeostasis. We hypothesize that impaired enzyme activity of PfSPP causes a significant level of ER stress, triggering a response resulting in substantial elevation of gametocytogenesis. The endoplasmic reticulum is particularly sensitive to stress, and one of the most common hypotheses for inducing gametocytogenesis is stress [33,34,35,36]. However, at this stage we cannot rule out the possibility that PfSPP clears the signal peptides of a critical substrate that is essential for gametocytogenesis. Future studies will test multiple facets of this intriguing finding in clarifying the mechanism of gametocytogenesis.
Highlights.
PfSPP is an ER resident protease
PfSPP is expressed both as a monomer and dimer
The signal peptide of HSP101 is the first known substrate of PfSPP
Reduced PfSPP activity may significantly affect ER homeostasis
Acknowledgments
We are grateful to Drs. Todd Golde for providing the pcDNA-mSPP plasmid, Alan Cowman for pCC1 vector, Daniel Goldberg for Plasmepsin V antibody, and John Adams for BiP antibody. We also thank Donna-Marie Mironchuk for her invaluable assistance with the graphic arts, and Dr. Toshihiko Hanada for technical advice. It is noteworthy that the ER localization of PfSPP was first demonstrated by John Dame's group (Bonilla A, Bonilla T, Yowell, C, and Dame, J. PfSPP as a novel target for malarial chemotherapy. Molecular Parasitology Abstracts 2008).
Funding This study was supported by the NIH Grants HL60961 and HL95050 (A.C.)
Abbreviations
- PfSPP
Plasmodium falciparum signal peptide peptidase
- PMV
Plasmepsin V
- A HSP101
heat shock protein 101
- LSA1
liver stage antigen-1
- ATF6
activating transcription factor 6
- DMSO
dimethyl sulfoxide
- GFP
green fluorescent protein
- PTEX
Plasmodium Translocon of Exported proteins
- SPPL2A
signal peptide peptidase like 2A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Winzeler EA. Malaria research in the post-genomic era. Nature. 2008;455:751–756. doi: 10.1038/nature07361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li X, Chen H, Oh SS, Chishti AH. A Presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol Biochem Parasitol. 2008;158:22–31. doi: 10.1016/j.molbiopara.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li X, Chen H, Bahamontes-Rosa N, Kun JF, Traore B, Crompton PD, Chishti AH. Plasmodium falciparum signal peptide peptidase is a promising drug target against blood stage malaria. Biochem Biophys Res Commun. 2009;380:454–459. doi: 10.1016/j.bbrc.2009.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nyborg AC, Ladd TB, Jansen K, Kukar T, Golde TE. Intramembrane proteolytic cleavage by human signal peptide peptidase like 3 and malaria signal peptide peptidase. FASEB J. 2006;20:1671–1679. doi: 10.1096/fj.06-5762com. [DOI] [PubMed] [Google Scholar]
- [5].Parvanova I, Epiphanio S, Fauq A, Golde TE, Prudencio M, Mota MM. A small molecule inhibitor of signal Peptide peptidase inhibits Plasmodium development in the liver and decreases malaria severity. PLoS ONE. 2009;4:e5078. doi: 10.1371/journal.pone.0005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weihofen A, Lemberg MK, Ploegh HL, Bogyo M, Martoglio B. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J Biol Chem. 2000;275:30951–30956. doi: 10.1074/jbc.M005980200. [DOI] [PubMed] [Google Scholar]
- [7].Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- [8].Nyborg AC, Kornilova AY, Jansen K, Ladd TB, Wolfe MS, Golde TE. Signal peptide peptidase forms a homodimer that is labeled by an active site-directed gamma-secretase inhibitor. J Biol Chem. 2004;279:15153–15160. doi: 10.1074/jbc.M309305200. [DOI] [PubMed] [Google Scholar]
- [9].Casso DJ, Tanda S, Biehs B, Martoglio B, Kornberg TB. Drosophila signal peptide peptidase is an essential protease for larval development. Genetics. 2005;170:139–148. doi: 10.1534/genetics.104.039933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Grigorenko AP, Moliaka YK, Korovaitseva GI, Rogaev EI. Novel class of polytopic proteins with domains associated with putative protease activity. Biochemistry (Mosc) 2002;67:826–835. doi: 10.1023/a:1016365227942. [DOI] [PubMed] [Google Scholar]
- [11].Krawitz P, Haffner C, Fluhrer R, Steiner H, Schmid B, Haass C. Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J Biol Chem. 2005;280:39515–39523. doi: 10.1074/jbc.M501645200. [DOI] [PubMed] [Google Scholar]
- [12].Zucconi A, Dente L, Santonico E, Castagnoli L, Cesareni G. Selection of ligands by panning of domain libraries displayed on phage lambda reveals new potential partners of synaptojanin 1. J Mol Biol. 2001;307:1329–1339. doi: 10.1006/jmbi.2001.4572. [DOI] [PubMed] [Google Scholar]
- [13].Crawshaw SG, Martoglio B, Meacock SL, High S. A misassembled transmembrane domain of a polytopic protein associates with signal peptide peptidase. Biochem J. 2004;384:9–17. doi: 10.1042/BJ20041216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schrul B, Kapp K, Sinning I, Dobberstein B. Signal peptide peptidase (SPP) assembles with substrates and misfolded membrane proteins into distinct oligomeric complexes. Biochem J. 2010;427:523–534. doi: 10.1042/BJ20091005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Trager W, Jenson JB. Cultivation of malarial parasites. Nature. 1978;273:621–622. doi: 10.1038/273621a0. [DOI] [PubMed] [Google Scholar]
- [16].Nyborg AC, Jansen K, Ladd TB, Fauq A, Golde TE. A signal peptide peptidase (SPP) reporter activity assay based on the cleavage of type II membrane protein substrates provides further evidence for an inverted orientation of the SPP active site relative to presenilin. J Biol Chem. 2004;279:43148–43156. doi: 10.1074/jbc.M405879200. [DOI] [PubMed] [Google Scholar]
- [17].Okamoto K, Mori Y, Komoda Y, Okamoto T, Okochi M, Takeda M, Suzuki T, Moriishi K, Matsuura Y. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J Virol. 2008;82:8349–8361. doi: 10.1128/JVI.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Klemba M, Beatty W, Gluzman I, Goldberg DE. Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J Cell Biol. 2004;164:47–56. doi: 10.1083/jcb200307147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Molecular and Biochemical Parasitology. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- [21].Marapana DS, Wilson DW, Zuccala ES, Dekiwadia CD, Beeson JG, Ralph SA, Baum J. Malaria parasite signal peptide peptidase is an ER-resident protease required for growth but not invasion. Traffic. 2012 doi: 10.1111/j.1600-0854.2012.01402.x. [DOI] [PubMed] [Google Scholar]
- [22].Nyborg AC, Herl L, Berezovska O, Thomas AV, Ladd TB, Jansen K, Hyman BT, Golde TE. Signal peptide peptidase (SPP) dimer formation as assessed by fluorescence lifetime imaging microscopy (FLIM) in intact cells. Mol Neurodegener. 2006;1:16. doi: 10.1186/1750-1326-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee MC, Moura PA, Miller EA, Fidock DA. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol Microbiol. 2008;68:1535–1546. doi: 10.1111/j.1365-2958.2008.06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].van Dooren GG, Marti M, Tonkin CJ, Stimmler LM, Cowman AF, McFadden GI. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol Microbiol. 2005;57:405–419. doi: 10.1111/j.1365-2958.2005.04699.x. [DOI] [PubMed] [Google Scholar]
- [25].de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, Sanders PR, Lundie RJ, Maier AG, Cowman AF, Crabb BS. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Matthews K, Kalanon M, Chisholm SA, Sturm A, Goodman CD, Dixon MW, Sanders PR, Nebl T, Fraser F, Haase S, McFadden GI, Gilson PR, Crabb BS, de Koning-Ward TF. The Plasmodium translocon of exported proteins (PTEX) component thioredoxin-2 is important for maintaining normal blood-stage growth. Molecular Microbiology. 2013;89:1167–1186. doi: 10.1111/mmi.12334. [DOI] [PubMed] [Google Scholar]
- [27].Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- [28].Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- [29].Sato T, Nyborg AC, Iwata N, Diehl TS, Saido TC, Golde TE, Wolfe MS. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- [30].Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–20179. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- [31].Martoglio B, Golde TE. Intramembrane-cleaving aspartic proteases and disease: presenilins, signal peptide peptidase and their homologs. Hum Mol Genet. 2003;12(Spec No 2):R201–206. doi: 10.1093/hmg/ddg303. [DOI] [PubMed] [Google Scholar]
- [32].Chaubey S, Grover M, Tatu U. Endoplasmic Reticulum Stress Triggers Gametocytogenesis in the Malaria Parasite. Journal of Biological Chemistry. 2014;289:16662–16674. doi: 10.1074/jbc.M114.551549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dyer M, Day KP. Commitment to gametocytogenesis in Plasmodium falciparum. Parasitol Today. 2000;16:102–107. doi: 10.1016/s0169-4758(99)01608-7. [DOI] [PubMed] [Google Scholar]
- [34].Talman AM, Domarle O, McKenzie FE, Ariey F, Robert V. Gametocytogenesis: the puberty of Plasmodium falciparum. Malar J. 2004;3:24. doi: 10.1186/1475-2875-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Baker DA. Malaria gametocytogenesis. Molecular and Biochemical Parasitology. 2010;172:57–65. doi: 10.1016/j.molbiopara.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harbut MB, Patel BA, Yeung BK, McNamara CW, Bright AT, Ballard J, Supek F, Golde TE, Winzeler EA, Diagana TT, Greenbaum DC. Targeting the ERAD pathway via inhibition of signal peptide peptidase for antiparasitic therapeutic design. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21486–21491. doi: 10.1073/pnas.1216016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bannister LH, Hopkins JM, Fowler RE, Krishna S, Mitchell GH. A brief illustrated guide to the ultrastructure of Plasmodium falciparum asexual blood stages. Parasitol Today. 2000;16:427–433. doi: 10.1016/s0169-4758(00)01755-5. [DOI] [PubMed] [Google Scholar]