Abstract
AMPA-type glutamate receptors mediate fast, excitatory neurotransmission in the brain, and their concentrations at synapses are important determinants of synaptic strength. We investigated the post-transcriptional regulation of GluA2, the calcium-impermeable AMPA receptor subunit, by examining the subcellular distribution of its mRNA and evaluating its translational regulation by microRNA in cultured mouse hippocampal neurons. Using computational approaches, we identified a conserved microRNA-124 (miR-124) binding site in the 3'UTR of GluA2 and demonstrated that miR-124 regulated the translation of GluA2 mRNA reporters in a sequence-specific manner in luciferase assays. While we hypothesized that this regulation might occur in dendrites, our biochemical and fluorescent in situ hybridization (FISH) data indicate that GluA2 mRNA does not localize to dendrites or synapses of mouse hippocampal neurons. In contrast, we detected significant concentrations of miR-124 in dendrites. Overexpression of miR-124 in dissociated neurons results in a 30% knockdown of GluA2 protein, as measured by immunoblot and quantitative immunocytochemistry, without producing any changes in GluA2 mRNA concentrations. While total GluA2 concentrations are reduced, we did not detect any changes in the concentration of synaptic GluA2. We conclude from these results that miR-124 interacts with GluA2 mRNA in the cell body to downregulate translation. Our data support a model in which GluA2 is translated in the cell body and subsequently transported to neuronal dendrites and synapses, and suggest that synaptic GluA2 concentrations are modified primarily by regulated protein trafficking rather than by regulated local translation.
Keywords: microRNA, miR-124, local translation, post-transcriptional regulation, dendritic mRNA localization, AMPAR
INTRODUCTION
Precise control of gene expression at synapses is important for proper communication between neurons. Among the proteins that are tightly controlled are members of the 2-amino-3-(3-hydroxy-5-methylisoxazol-4-yl) propanoic acid (AMPA)-type glutamate receptor subunit family, GluA1–4 (Shepherd and Huganir, 2007). AMPA receptors (AMPARs) are mediators of fast, excitatory transmission between neurons, and their concentration at the synapse plays a central role in determining synaptic strength (Malinow and Malenka, 2002). Increased synaptic AMPAR levels are correlated with increased synaptic strength and vice versa. Given their importance, AMPARs have been heavily studied and have been shown to undergo nuanced regulation at many levels of gene expression (Derkach et al., 2007; Jackson and Nicoll, 2011; Lu and Roche, 2012; Nicoll et al., 2006; Santos et al., 2009; Shepherd and Huganir, 2007).
How AMPARs arrive at their synaptic locations is an active area of investigation with several, non-mutually exclusive theories (Shepherd and Huganir, 2007). One theory is that that the receptors are synthesized on the rough endoplasmic reticulum and assembled in the cell body. They are then trafficked to synaptic sites along the cytoskeleton (Hirokawa and Takemura, 2005; Setou et al., 2002; Shin et al., 2003; Wyszynski et al., 2002). Alternatively, the assembled AMPARs may be inserted into the plasma membrane at the cell body, and then transported to synapses via lateral diffusion (Adesnik et al., 2005). Another theory is that AMPARs are locally translated in dendrites and processed in Golgi outposts before being inserted at synapses (Horton and Ehlers, 2004). In support of local translation, GluA1 and GluA2 messenger RNA (mRNA) transcripts have been reported to localize to dendrites of cultured rat neurons (Cajigas et al., 2012; Grooms et al., 2006). Furthermore, stimulation of cultured hippocampal neurons has been shown to alter the dendritic localization of both GluA1 and GluA2 mRNAs (Grooms et al., 2006), and overexpression studies have revealed local translation of GluA1 and GluA2 in dendrites that have been severed from the cell body (Ju et al., 2004; Kacharmina et al., 2000).
The occurrence of stimulus-responsive local translation indicates that regulatory mechanisms exist to ensure that translation occurs when and where the encoded proteins are needed. One potential regulatory mechanism is through the microRNA pathway. MicroRNAs (miRNAs) are non-coding, endogenous RNAs of about ~22 nucleotides in length that downregulate gene expression via the RNA-induced silencing complex (RISC). Within RISC, the 5' end of the miRNA has a “seed” site that recognizes targets by partial complementarity to sequences in the 3' untranslated region (3'UTR) of target mRNAs (Carthew and Sontheimer, 2009; Yates et al., 2013). Upon recognition of a target mRNA, miRNAs repress translation either by reducing translational efficiency or by destabilizing the transcript (Djuranovic et al., 2011; Fabian et al., 2010; Huntzinger and Izaurralde, 2011). The post-transcriptional and potentially reversible mode of action of miRNAs makes them well suited to regulate local translation.
In this study, we focus on the GluA2 subunit and investigate its post-transcriptional regulation. Among the AMPAR subunits, GluA2 is unique because its inclusion in an AMPAR makes the receptor calcium impermeable (Burnashev et al., 1992). Hence, GluA2 levels at the synapse influence calcium influx through AMPARs after glutamate binding to the receptor (Geiger et al., 1995). We used a computational algorithm to predict potential miRNA target sites in the GluA2 3'UTR, and identified miR-124 as a favorable candidate. The prediction was first validated in 293T cells using luciferase assays, and then further tested in dissociated hippocampal cultures using lentivirus-mediated miR-124 overexpression in dissociated hippocampal cultures. Fluorescence in situ hybridization (FISH) and reverse transcription quantitative polymerase chain reaction (RT-qPCR) were used to determine the subcellular localization patterns of miR-124 and GluA2-mRNA. Our results support miR-124 regulation of GluA2 in neurons, but indicate that this interaction regulates GluA2 translation primarily in the somatic cytoplasm rather than in dendrites.
MATERIALS AND METHODS
Target Prediction Process
We used several miRNA target prediction programs to predict miRNAs that target GluA2 mRNA: PicTar (Krek et al., 2005), TargetScan (Grimson et al., 2007), PITA (Kertesz et al., 2007), and Miranda (Sethupathy et al., 2006). We applied additional filters to narrow down the large number of miRNA/mRNA interactions that were identified. First, we selected miRNAs that had previously been shown to be expressed in rodent brain (Thomson et al., 2004), (Landgraf et al., 2007), (Deo et al., 2006), (Hohjoh and Fukushima, 2007; Thomson et al., 2004). Second, we considered only miRNAs predicted by at least two programs (Sethupathy et al., 2006). Third, since each target prediction tool defines conservation differently, we used phastCons scores to determine the conservation of the target site and surrounding bases (Kertesz et al., 2007; Siepel et al., 2005). Fourth, we examined genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) long-term potentiation pathway. The miRNAs with frequently occurring target sites in this KEGG pathway were ranked more favorably (Stark et al., 2005). Finally, we ranked predicted interactions less favorably if they involved G-U wobble base pairing (Brennecke et al., 2005). These filters led us to identify miR-124/GluA2 as a highly favorable predicted interaction.
Luciferase assays
The 3' UTR of GluA2 was cloned downstream of the renilla luciferase coding region in plasmid pRL-TK (Promega). The sequences cloned correspond to nucleotides 3,203 – 3,298 of the flip and flop isoforms (NM_001083806.1 and NM_013540.2), which have identical 3' UTRs. Reporter constructs with the predicted miR-124 target site deleted or point-mutated were generated by site-directed mutagenesis. A mixture of renilla luciferase reporter plasmid (0.35 μg), firefly luciferase control plasmid (0.05 μg) (pGL3, Promega), carrier plasmid (0.4 μg) (pBSK), and miRNA mimic (25 nM final concentration; Thermo Scientific Dharmacon; mimic-124: UAAGGCACGCGGUGAAUGCCA, mimic-124*: GCAUUCACCGCGUGCCUUAUU, mimic-124PM: UAACGGACGCGGUGAAUGCCA, mimic-124 PM*: GCAUUCACCGCGUCCGUUAUU) was transfected using Lipofectamine 2000 (Invitrogen) into one well of HEK293T cells that were plated at a density of 50,000 cells/well in a 24-well plate the day before. At 24 hours post-transfection, luciferase expression was assayed with the Dual-Luciferase® Reporter Assay System (Promega) according to manufacturer's instructions and measured on a Molecular Devices Analyst AD microplate reader (Analyst AD 96–384). Renilla luciferase signals were first normalized to firefly luciferase signals, and then normalized to the control samples (each construct, without miRNA added). The assays were performed four times in triplicate.
Synaptosome fractionation
Synaptosome fractions were prepared from 2–3 month old C57BL/6J mice. Ten mice were used for each fractionation. The mice were anesthetized with isofluorane and sacrificed by cervical dislocation. All centrifugation steps were performed at 4°C and solutions were kept on ice. Forebrains were dissected and homogenized in solution A (0.32 M sucrose, 1 mM NaHCO3, 1 mM MgCl2, 0.5 mM CaCl2, 10 mM Na4P2O7). An aliquot of the total homogenate was removed at this point for RNA and protein extraction. The remaining homogenate was spun at 1,400 × g and the supernatant was saved. The pellet was resuspended in solution A and spun at 700 × g. The supernatants from both spins were pooled and an aliquot representing the cytosolic fraction was removed. The supernatants were then spun at 13,800 × g to pellet synaptosomes and other organelles. The pellet was resuspended in solution B (0.32 M sucrose, 1 mM NaHCO3), and layered onto a discontinuous gradient with 1.2 M, 1.0 M, and 0.8 M sucrose layers prepared in 1 mM NaHCO3. The gradient was spun at 82,500 × g for 2 hours. Synaptosomes separate into the band between the 1.0 M and 1.2 M layers. This band was collected and spun at 100,000 × g for 20 min to pellet the synaptosomes.
Protein extraction and immunoblotting
RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1 % SDS, 0.5 % Na deoxycholate, 0.1 % SDS, 1 % NP-40) with cOmplete protease inhibitor (Roche) and benzonase (Sigma #E1014) was used for protein extraction from synaptosomes and neuronal cultures. For synaptosome preparations, total protein levels were determined with a BCA protein assay (Pierce) and 5 mg of protein was loaded per lane. For neuronal cultures, proteins were extracted and the same volumes were loaded for each condition. All protein samples were run on 10 % polyacrylamide gels and transferred to a PVDF membrane for immunoblotting. Blots were scanned using the Odyssey imaging system (Li-Cor) and quantified with ImageJ software.
Antibodies
We used the following antibodies. Vendors, catalog numbers, and dilutions are shown within parenthesis. PSD-95 (Abcam #18258; 1:1000), GluA2 (Invitrogen #32-0300; 1:200), MAP2 (PhosphoSolutions #1100-MAP2; 1:20,000), copGFP (Evrogen #AB513; 1:3,000), synapsin I (Abcam #AB8; 1:1000), GFAP (Millipore #MAB360; 1:1000), Tuj1 (Millipore #AB15708; 1:1000), POD-conjugated DIG (Roche #1207733; 1:1000), Alexa Fluor 488 goat anti-rabbit (Invitrogen #11008; 1:20,000), Alexa Fluor 488 goat anti-mouse (Invitrogen #11001; 1:20,000), Alexa Fluor 555 goat anti-mouse (Invitrogen #A21422; 1:20,000), Alexa Fluor 633 goat anti-chicken (Invitrogen #A21103; 1:20,000), IRDye 680LT goat anti-mouse (Li-Cor #926-68020; 1:20,000), IRDye 800CW goat anti-rabbit (Li-Cor #926-32211; 1:20,000)
RNA extraction and quantification
The miRNeasy Mini kit (Qiagen) was used according to manufacturer's protocol for extraction of total RNA and DNaseI treatment. The amount of RNA extracted was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). For miRNA RT-qPCR assays, RNA was diluted to below 10 ng/uL and measured with the Qubit RNA assay kit on a Qubit fluorometer (Invitrogen).
Reverse transcription and quantitative PCR (RT-qPCR)
For mRNA RT-qPCR, the amount of starting material in each reaction was normalized to total RNA. Transcripts were primed with random hexamers and reverse transcribed with SuperScript III (Invitrogen). The resulting cDNA was then used for comparative qPCR with SYBR Green (Applied Biosystems) and gene specific primers (GluA2 – fwd: CCATCGAAAGTGCTGAGGAT, rev: AGGGCTCTGCACTCCTCATA; Camk2α – fwd: TCTGAGAGCACCAACACCAC, rev: CCATTGCTTATGGCTTCGAT; Fads3 – fwd: ATGACCTACCAGGCGACAAG, rev: CAATCAACAGGGGTTTCAGG; pre-miR-124 – fwd: GTGTTCACAGCGGACCTTG, rev: ATTCACCGCGTGCCTTAAT). Looped primers specific for mature miRNAs were used for RT, and TaqMan® probes were used for qPCR (Applied Biosystems). Standard curves were determined for all genes quantified, all of which had amplification efficiencies within 100 ± 10%.
Dissociated hippocampal cultures
Hippocampi were dissected from postnatal day 0 C57BL/6J mice (Jackson Laboratory) and dissociated by trypsin treatment and trituration. The dissociated neurons were plated on culture plates or on HCl-etched coverslips (Deckgläser #1001/12). Culture plates and coverslips were coated with 0.1 mg/mL poly-DL-lysine (Sigma #P-9011) overnight at 37 °C. Plating medium consisted of B-27 supplement (1 mL/50 mL media), 0.5 mM glutamine (Gibco #21103-015), 25 μM glutamate (Sigma #G-5889), and β-mercaptoethanol (25 μM; Sigma #G-57522) diluted in Neurobasal-A media (Gibco #21103). Neurons for fluorescence in situ hybridization and immunocytochemistry were plated at low density (approximately 210 cells/mm2 or 40,000 cells/well in a 24-well plate). Neurons for RNA or protein extraction were plated at high density (approximately 630 cells/mm2 or 240,000 cells/well in a 12 well plate).
Immunocytochemistry (ICC)
Neurons grown on coverslips were fixed in 4% paraformaldehyde for 10 minutes, permeabilized in 0.1% Triton-X for 5 minutes, blocked in 10% goat serum for 30 minutes, and incubated in primary antibody for 16–24 hours at 4 °C. Secondary antibodies and Hoechst (1:1000, Invitrogen #H3570) were incubated for 1 hour in the dark. Coverslips were mounted using Aqua PolyMount (Polysciences #18606) and allowed to dry overnight before imaging. All steps were performed at room temperature unless otherwise noted. For surface staining of GluA2, neurons were first incubated live at 37 °C for 30 minutes in GluA2 antibody diluted with artificial cerebrospinal fluid (ACSF; 119 mM NaCl, 26.2 mM NaHCO3, 2.5 mM KCl, 1 mM NaH2PO4, 1.3 mM MgCl2, 10 mM glucose, 2.5 mM CaCl2). The ACSF used was first bubbled with carboxygen (5 % CO2/95 % O2) to pH 7.35, adjusted to 293 mmOsm with glucose, and filtered with a 0.22 μm filter. Then, neurons were fixed, permeabilized, and stained for intracellular proteins (i.e. MAP2, synapsin) as described above.
Fluorescence in situ hybridization (FISH)
Neurons (14–21 DIV) were processed for FISH using the QuantiGene ViewRNA (QGV) mRNA and miRNA ISH Cell Assay Kits according to manufacturer's protocol (Affymetrix). Each QGV mRNA probe set consists of 20 oligonucleotide pairs that are complementary to different regions of the transcript. Each pair hybridizes to the target mRNA at adjacent sites and supports a branched DNA signal amplifier that is assembled via a series of sequential hybridization steps. The resulting signal comes from fluorophore-conjugated label probes and provides up to 8,000-fold signal amplification. The QGV miRNA probes undergo similar amplification steps with branched DNA molecules, however differs in that only one pair of oligonucleotides is used. Also, the final amplification step for the miRNA FISH utilizes alkaline phosphatase label probes in conjunction with Fast Red chromogenic substrate for enzymatic amplification. The QGV miRNA probe sets are designed using proprietary nucleic acid chemistry to increase the melting temperature, allowing for better probe affinity and specificity for short targets like miRNAs. After the FISH amplification and wash steps, neurons were blocked in 10% goat serum and processed as described for ICC.
For FISH with riboprobes, digoxigenin (DIG)-labeled riboprobes against GluA2 mRNA (511–658 nt) were in vitro transcribed from plasmids adapted with a T7 or SP6 RNA polymerase site using the DIG RNA labeling kit (Roche). Riboprobes were then purified on G-50 micro columns (GE #28903408). FISH was performed on 14 DIV dissociated hippocampal neurons as described in Poon et al. (2006). Probe hybridization was performed overnight at 35 °C. The Cy3-TSA kit (Perkin Elmer #NEL744) was used for signal amplification.
Image acquisition
Images were acquired on a Zeiss LSM700 microscope by an experimenter blind to the treatment. Confocal images of FISH and ICC experiments were acquired with a 40 x/1.3 oil objective. For FISH images, Z-stacks were acquired of fields containing the cell bodies and dendritic processes of the same neurons. Neurons were randomly selected by their MAP2 staining. Images of surface GluA2 ICC experiments were acquired in single sections with a pinhole of 1 airy unit. For total GluA2 ICC images, single sections were acquired with different pinhole settings. The pinhole was first set to 5.62 airy units to capture dendritic signal from 5 μm sections. Then, for the same field, the pinhole was maximized to 14.07 airy units and the plane of focus was adjusted to capture somatic signal from 12.4 μm sections. The copGFP transduction marker was partially quenched by fixation, but could still be seen in the cell body and this signal was used to confirm transduction. The gains for signals to be quantified were set at subsaturation levels. Live images to assess transduction efficiency were acquired with a 10 x/0.3 air objective.
Image analysis
Images were analyzed by an experimenter blind to the treatment and quantified with ImageJ software. To measure FISH puncta distribution, stack images of FISH staining were converted to maximum projection intensities. Somatodendritic compartments were linearized using the “Straighten” function in ImageJ with a width setting of 25 pixels (corresponding to ~4 μm). Somatodendritic compartments were selected on the basis of MAP2 staining. Measurements of dendritic distance began at the center of the cell body and extended out along the dendrite until the boundary of the image. Only dendrites that did not overlap with cell bodies were selected. Then, the ImageJ “Find Maxima” function with a noise tolerance of 10 was used to identify individual puncta and report the distance of the puncta from the center of the cell body. Since dendrites of different lengths were imaged, only dendrites longer than 150 μm and only puncta within 150 μm were included in the analyses. To measure total somatic and proximal GluA2 protein expression by ICC, regions of interest were defined manually based on MAP2 staining. For dendrites >20 μm from the soma, regions of interest were defined based on dilated MAP2 masks. The mean pixel intensity and integrated densities were determined using the “Measure” function and corrected for background signal. To measure synaptic GluA2 protein expression by ICC, synapsin puncta were used as synaptic markers in images of surface GluA2 staining. Dendrites greater than 20 μm from the soma were linearized with the “Straighten” function and a width setting of 40 pixels (corresponding to ~6 μm). The “Analyze Particles” function was used to select synapsin puncta that were between 4–500 pixels and with a circularity between 0.5–1.0. The number of puncta identified was used as an approximation of the number of synapses. Then, the “Find Peaks” function in the GDSC plugin was used to identify GluA2 puncta and report their total intensity. Finally, the “Match Calculator” function in the GDSC plugin was used to identify GluA2 puncta that were within 8 pixels (corresponding to ~1 μm) of synapsin puncta. Matched GluA2 puncta were considered synaptic and unmatched puncta were considered non-synaptic.
Statistical methods
To compare the distributions of FISH puncta across cell groups, we performed two types of analysis. First, we developed a mixed effects analysis of variance (ANOVA) model of mean puncta distance, with variation between cell groups represented as a fixed effect, and variation between dendrites represented as a nested random effect. The analysis therefore controls for between-dendrite variation in estimating cell group effects on mean distance from cell body. Second, we classified puncta as either somatic, proximal, or distal according to distance from cell body, and developed one-way ANOVA models of the mean proportion of puncta in each of the three bins. Variation between cell groups was represented as a fixed effect in these models. This second analysis enables us to compare puncta density between cell groups for particular dendrite segments. SAS version 9.3 was used to fit the models, and SPSS version 21 was used to produce Figures 3B and 3C. One-tailed t-tests were used to compare GluA2 ICC intensity between control and miR-124 overexpression conditions.
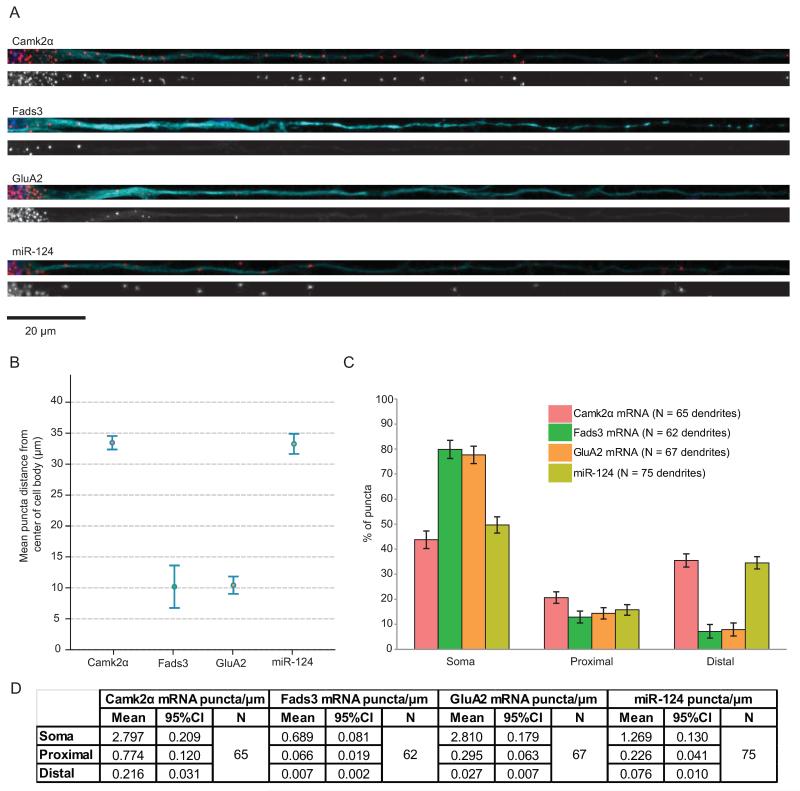
Figure 3.
GluA2 mRNA and miR-124 have different patterns of distribution.
(A) Representative fluorescence in situ hybridization (FISH) images of straightened dendrites. Top: red, FISH puncta; cyan, MAP2; blue, Hoechst. Bottom: FISH puncta in gray scale. Scale bar = 20 μm. (B) Group data for mean puncta distance from the center of the cell body. Error bars show 95% confidence intervals. (C) Puncta were classified into three subcompartments and the percentage of puncta in each subcompartment is shown. “Soma” represents the cell body; “proximal” represents from 0 to 20 μm from the cell body; and “distal” represents greater than 20 μm from the cell body. Error bars show 95% confidence intervals. (D) Table showing the absolute number of puncta per micron in each subcompartment, along with number of dendrites measured for each gene. Dendrites were imaged from 4 to 7 independent experiments.
Lentiviral transduction
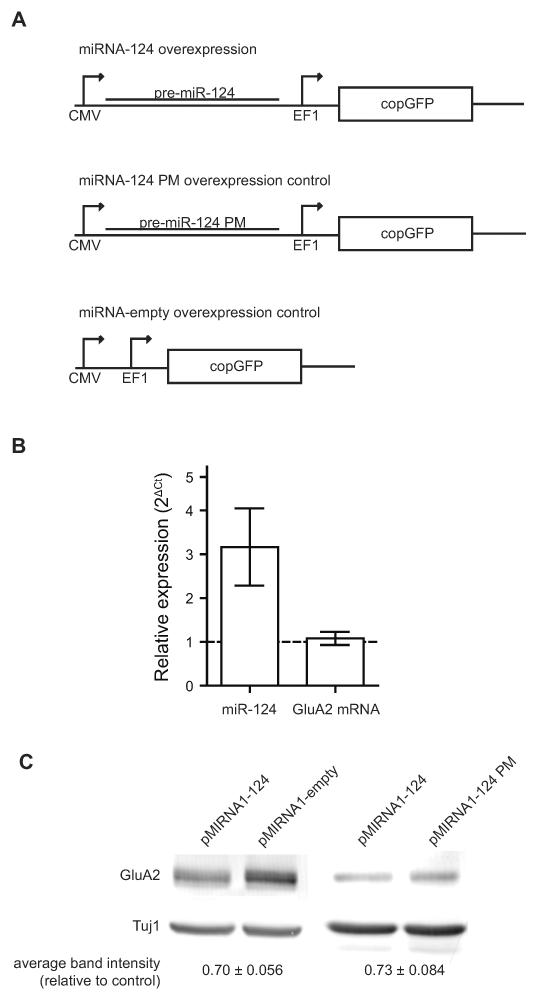
The pMIRNA1-124 overexpression plasmid encoding the precursor sequence of miR-124 was purchased from Systems Biosciences. This plasmid was modified to introduce two point mutations in the seed site for the overexpression control plasmid (pMIRNA1-124PM). Another control plasmid with the precursor sequence completely deleted was also cloned (pMIRNA1-empty). For the sponge plasmids, the pre-miR-124 sequence was deleted and sponge sites were cloned into the 3'UTR of copGFP, as described by Ebert et al. The plasmids were packaged in HEK293T cells with Δ8.9 and VSVG, and concentrated by ultracentrifugation. Neurons were transduced at 13 DIV. During transduction, half of the media was removed and stored at 4°C. Virus was mixed with the remaining media and applied to the neurons. Transduction media was replaced with the saved media the next day (16–24 hour incubation). At 21 DIV (8 DIV post transduction), transduced neurons were harvested for RNA or protein extraction, or fixed for ICC.
RESULTS
Prediction and initial validation of the GluA2/miR-124 interaction
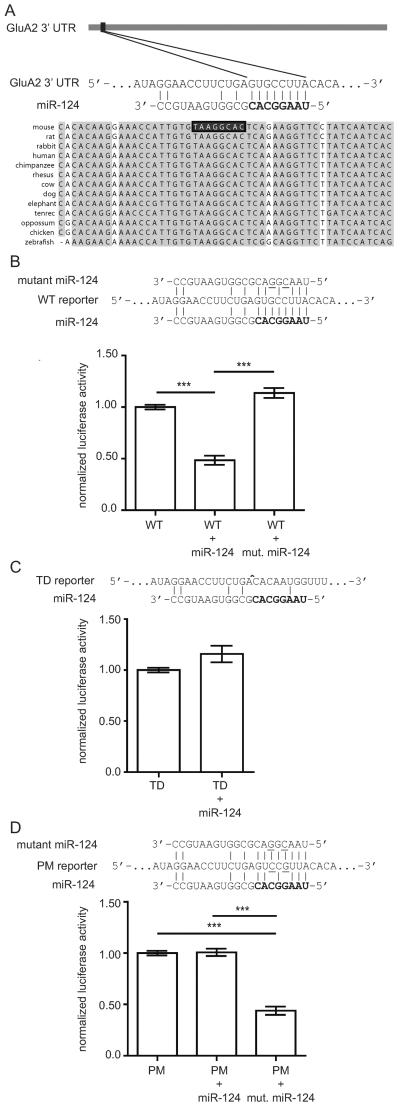
We identified a conserved target site for miR-124 in the 3'UTR of GluA2 mRNA using several prediction algorithms and a set of filtering rules as described in the materials and methods section (Figure 1A). To validate the prediction, we used a dual luciferase reporter assay system in 293T cells (Figure 1B). In one set of assays, the GluA2 3'UTR containing the miR-124 target site was fused to the 3' end of a luciferase reporter construct. Co-transfection of this reporter with a synthetic miR-124 duplex resulted in a 50% knockdown of luciferase signal (Figure 1B). This interaction depended on the miR-124 seed sequence, as transfection with a mutant miR-124 duplex with two point mutations did not produce significant knockdown in luciferase expression (Figure 1C). In another set of assays, the miR-124 target site was deleted from the reporter. Co-transfection of this construct with miR-124 did not reduce luciferase expression compared to transfection of the target-deleted construct alone (Figure 1C). To confirm the importance of sequence complementarity between the miR-124 seed and GluA2 target site, a luciferase construct with point mutations complementary to those in the mutant miR-124 was made. With seed-target complementarity restored, the transfected miRNA once again repressed luciferase expression (Figure 1D). These studies indicate that miR-124 inhibits translation of target GluA2 mRNA reporters in a sequence-specific manner.
Figure 1.
The GluA2 3' untranslated region (UTR) has a functional miR-124 target site.
(A) Schema showing the position and conservation of the fully complementary miR-124 target site at the 5' end of the mouse GluA2 3' UTR. The seed region of miR-124 is shown in bold. Vertical bars depict Watson-Crick base pairs.
(B–D) Wild type and mutant reporter constructs were transfected into HEK293T cells with miR-124 mimics or mutant miR-124 mimics with two point mutations in the seed region (underlined). Luciferase activities are reported relative to reporter-only controls. (B) Transfection of the wild type reporter (“WT”) with miR-124 resulted in robust knockdown of luciferase activity while transfection with mutant miR-124 did not. Significance determined by one-way analysis of variance (ANOVA) with Bonferroni correction. (C) Transfection of miR-124 with a reporter lacking the miR-124 target site (“TD”) did not reduce luciferase activity. ^ site of deletion. Significance determined by two-tailed t-test. (D) Transfection of a mutant reporter with two point mutations in the miR-124 target site (“PM”) did not show reduced translation by miR-124. Translational reduction was restored when the mutant reporter was transfected with the complementary mutant miR-124. Significance determined by one-way ANOVA with Bonferroni correction. ***p<0.001; error bars show standard error of the mean (S.E.M.); N = 4 independent experiments per group.
Determining the subcellular distribution of GluA2 mRNA and miR-124
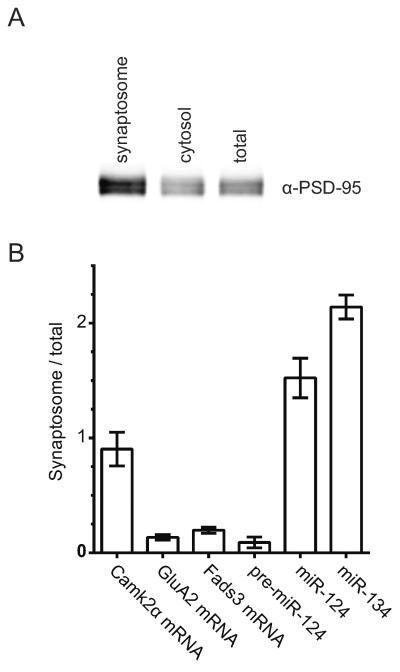
GluA2 mRNA and miR-124 must be present in the same subcellular compartment to be able to interact. To determine whether they are both present at synapses, we prepared synaptosome fractions from mouse forebrain. The enrichment of synaptic terminals was confirmed by western blotting for the post-synaptic scaffold protein, PSD-95 (Figure 2A). RNA was extracted and relative concentrations of GluA2 mRNA and mature miR-124 were measured by comparative RT-qPCR (Figure 2B, S1). The synaptosome:total ratios for GluA2 mRNA and miR-124 were 0.14 and 1.52 respectively. To help put these ratios into context, we measured Camk2α mRNA (ratio = 0.90), which has been widely reported to be present in distal compartments (Benson et al., 1992; Blichenberg et al., 2001; Burgin et al., 1990; Miller et al., 2002; Mori et al., 2000). We also measured miR-134 (ratio = 2.14) as a positive miRNA control for distal localization (Schratt et al., 2006). For a negative control, we chose Fads3 mRNA (ratio = 0.20), which has been reported to be somatically-restricted (Cajigas et al., 2012). These results suggest that GluA2 mRNA is as de-enriched from synapses as Fads3 mRNA, and that miR-124 is more synaptically-enriched than Camk2α mRNA.
Figure 2.
miR-124 is enriched at synapses while GluA2 mRNA is de-enriched.
(A) Synaptosome fractions were prepared from adult forebrain. Immunoblotting shows enrichment for PSD-95 in the synaptosome fraction (5 μg of total protein was loaded in each lane). (B) Comparative RT-qPCR on RNA extracted from total and synaptosome fractions of mouse forebrain. Relative ratios are plotted on the y-axis. Significance was determined by one-way ANOVA with Bonferroni correction. Pairwise comparisons between the ratios for Camk2α vs. GluA2 and GluA2 vs. miR-124 were significantly different (p<0.01 and p<0.001 respectively). GluA2 and miR-124 were not significantly different (p>0.05). Error bars show S.E.M. N = 3 independent experiments.
Since synaptosome preparations are subject to contamination by closely-associated glial components, we also performed FISH on dissociated hippocampal cultures. FISH allowed us to visualize the subcellular distribution patterns of GluA2 mRNA and miR-124 in individual cells. Again, Camk2α and Fads3 mRNA were used for comparison. Using the QuantiGene® (Affymetrix) detection system, all four RNAs appear as discrete puncta that are present at high concentrations in the cell body (Figure 3A).
The extent of dendritic localization varied greatly between genes and, to a lesser extent, between individual neurons and dendrites. Because of the variability between different neurons and dendrites, we imaged many neurons to determine the overall distribution pattern for each gene. In Figure 3B, we estimated the distance of a typical puncta from the center of the cell body. The mean distances for miR-124 and Camk2α mRNA were 33.27 μm (95% CI [31.64, 34.89]) and 33.46 μm (95% CI [32.36, 34.55]) respectively, placing them in the dendritic subcompartment. The mean distances for GluA2 and Fads3 mRNA were 10.42 μm (95% CI [9.02, 11.82]) and 10.18 μm (95% CI [6.75, 13.62]) respectively, which lie in the cell body or proximal dendrite. Pairwise comparisons of the average distance between genes showed that there was sufficient evidence to distinguish between the location of miR-124 and GluA2 puncta, while there was insufficient evidence to distinguish between miR-124 and Camk2α puncta or GluA2 and Fads3 puncta locations (Figure S2A).
For further analysis, we divided the puncta into three subcompartments (cell body; proximal dendrite, up to 20 μm from cell body; and non-proximal dendrite, 20–140 μm from cell body), which were chosen based on the distribution patterns of the different RNAs (Figure S2B,C). We quantified the proportion of puncta in each subcompartment to account for differences in expression levels between the transcripts. The proportion of FISH puncta in the soma and non-proximal dendrites are shown in Figure 3C (see also Figure S2D and mean number of puncta in each compartment in Figure 3D). This analysis reveals a significant difference in the somatic/dendritic distribution of miR-124 and GluA2 puncta, with no significant difference between the somatic/dendritic distribution of miR-124 and Camk2α (known to be dendritically localized) or between GluA2 and Fads3 (known to be somatically localized). We conclude that GluA2 is a predominantly somatically restricted mRNA, while miR-124 is present in somata and in dendrites.
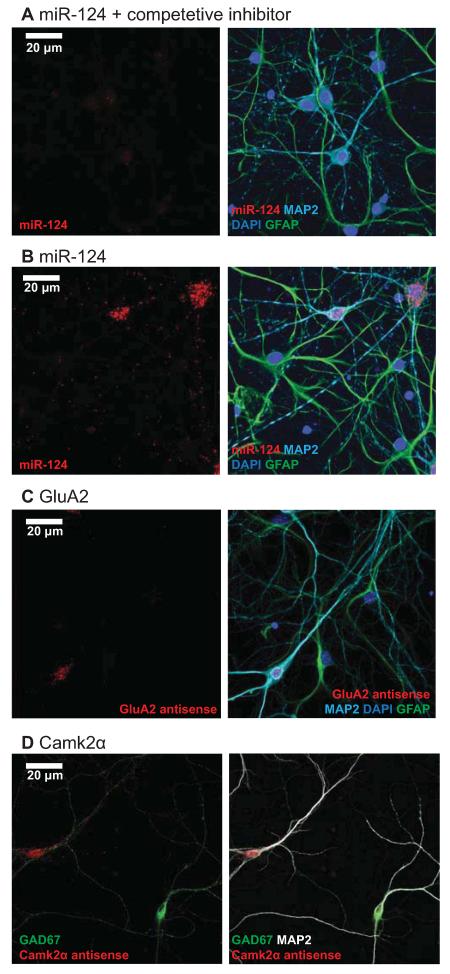
We performed several controls to confirm that the FISH signals we observed were specific. The specificity of GluA2 and Fads3 probes were verified by hybridization with sense probes, which did not produce any signal (Figure S3A,B). The specificity of the miR-124 probe was verified by hybridization in the presence of a competitive inhibitor that has full complementarity to miR-124, which produced a marked reduction in signal (Figure 4A). In addition, a bactillus subtillis dihydropicolinate reductase (DapB) negative control probe for miRNA FISH did not produce any signal (Figure S3C). As yet another indication of probe specificity, both miR-124 and GluA2 antisense probes hybridize to MAP2 positive neurons and do not produce signal in GFAP positive astrocytes (Figures 4B,C). Finally, signal from the Camk2α antisense probe was absent from GAD67 positive inhibitory cells (which do not express Camk2α, Figure 4D). Taken together, these controls indicate that the FISH probes are highly specific.
Figure 4.
Control experiments demonstrate the specificity of the FISH signals.
(A) miR-124 signal is drastically reduced in the presence of a fully complementary competitive inhibitor at 10X concentration. Red, miR-124; green, GFAP; cyan, MAP2; blue, Hoechst. Scale bar = 20 μm. (B) miR-124 signal is absent from astrocytes. Red, miR-124; green, GFAP; cyan, MAP2; blue, Hoechst. (C) GluA2 mRNA signal is absent from astrocytes. Red, GluA2 mRNA; green, GFAP; cyan, MAP2; blue, Hoechst. (D) Camk2α mRNA signal is absent from inhibitory neurons. Red, Camk2α mRNA; green, GAD67; white, MAP2.
Since miRNA FISH probes recognize both mature and precursor miR-124 (pre-miR-124), we measured pre-miR-124 in synaptosome fractions. The RT-qPCR results show that pre-miR-124 is depleted from synaptosomes (ratio = 0.09), indicating that the miR-124 FISH signal in non-proximal dendrites is likely from mature miR-124 (Figure 2B).
Since our findings conflict with previous reports showing that GluA2 mRNA localized to dendrites, we asked whether activity altered the distribution pattern of miR-124 or GluA2 mRNA by silencing neuronal cultures with the sodium channel antagonist tetrodotoxin (TTX; 1 μM), or by stimulation with the GABAA receptor antagonist bicuculline (BIC; 40 μM), which drives glutamatergic transmission. Neither silencing nor stimulation altered the dendritic localization of GluA2 puncta at any time point examined (15 minutes, 1 hour, or 3 hours, Figure S4A–D). As a positive control, we probed for cFos mRNA, an immediate early gene that is strongly induced by activity (Herrera and Robertson, 1996), which was absent in the TTX- treated cultures, but was highly expressed in the cell body of bicuculline-treated cultures (Figure S4E).
The effects of overexpressing miR-124 levels on endogenous GluA2 levels
To test the effects of manipulating miR-124 concentrations on GluA2 expression in neurons, we transduced dissociated hippocampal cultures with lentivirus to overexpress either pre-miR-124 (pMIRNA1-124), which is processed by the Dicer pathway into mature miR-124 (Tomari and Zamore, 2005), or a control with point mutations in the seed region similar to the mutant used in the luciferase assays (pMIRNA1-124P.M.) (Figure 5A). Co-expression of copGFP from the same vector indicated that transduction efficiency was high, with over 90% of neurons being transduced (Figure S5). Transduction of the overexpression construct increased mature miR-124 expression by 3.2-fold while GluA2 mRNA levels were not significantly changed at 1.1-fold relative to control (Figure 5B). However, total GluA2 protein levels were reduced by 27% (Figure 5C), as determined by western blotting on whole cells lysates. This result indicates that miR-124 represses translation of endogenous GluA2 in a manner that is dependent on the seed region.
Figure 5.
Overexpression of miR-124 down-regulates total GluA2 protein in neurons.
(A) Overexpression and control lentiviral constructs. (B) Comparative RT-qPCR measurement of miR-124 and GluA2 mRNA fold changes in neurons transduced with pMIRNA1-124 relative to control. Error bars show S.E.M. N = 3 independent experiments. (C) Immunoblot analysis of protein lysates from transduced cultures. Band intensities were quantified and normalized to Tuj1. The differences relative to controls are shown below with standard error. N = 4 independent experiments.
To complement this approach with experiments in which we reduced endogenous miR-124, we designed sponge constructs with bulged miR-124 binding sites in the 3' UTR of copGFP (sponge-124) as well as a previously published control sponge that does not recognize miRNAs (sponge-CXCR) (Figure S6) (Ebert et al., 2007). By having multiple miRNA binding sites, sponges are believed to divert miRNAs from binding their endogenous targets. However, when we transduced cultures with sponge-124, we noticed that the copGFP marker was almost exclusively expressed in non-neuronal cells and no change in GluA2 protein was observed (Figure S6B). Visual inspection of the cultures and Tuj1 immunoblotting did not indicate reduced neuronal viability (Figure S6C). Instead, we suspect that the endogenous concentration of miR-124 was so high that it repressed copGFP expression and overwhelmed the transduced sponge-124. This possibility is supported by tests in HEK293T cells that show that increasing concentrations of miR-124 do repress copGFP expression from sponge-124 (data not shown). Of note, miR-124 is the most abundant miRNA in the adult mouse brain (Lagos-Quintana et al., 2002). As a result, we were not able to knockdown miR-124 in hippocampal neurons with our sponge-124 construct.
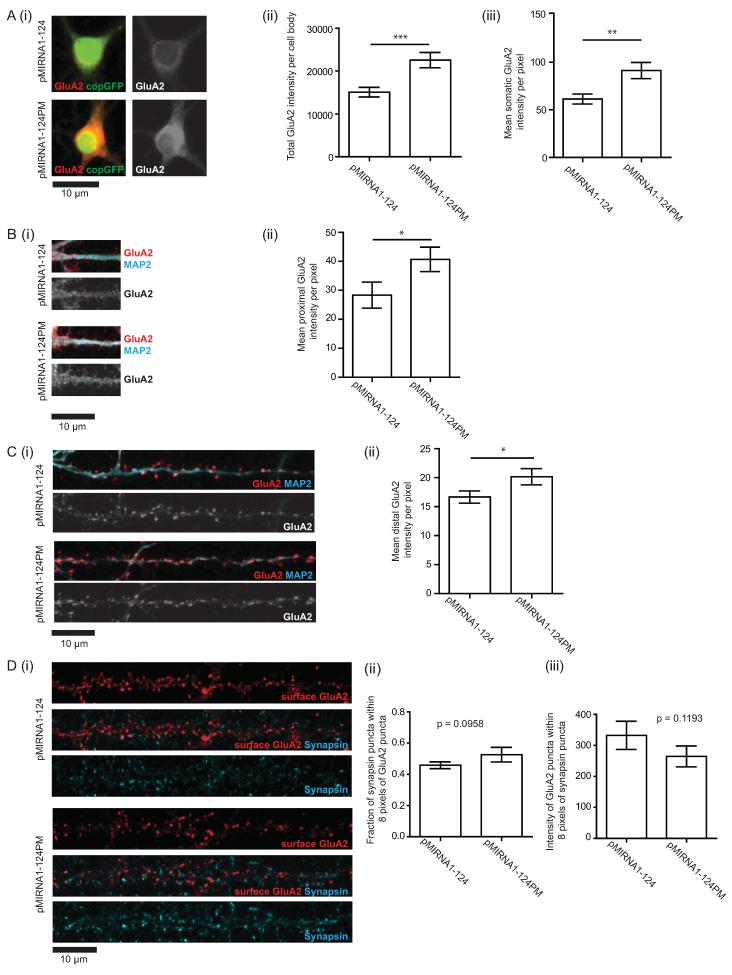
In addition to immunoblotting whole cell lysates, subcompartment-specific GluA2 expression was also measured using quantitative ICC. As with the FISH analysis, the cell body, proximal, and non-proximal dendritic regions were analyzed. GluA2 immunostaining was performed on permeabilized neurons to measure total GluA2 expression. Quantification of mean GluA2 signal intensity shows that miR-124 overexpression significantly reduced GluA2 expression in all three regions of the neuron: the cell body, proximal, and non-proximal regions showed 33%, 30%, and 17% reductions in mean pixel intensity respectively (Figure 6A–C). This observation is consistent with the immunoblotting results.
Figure 6.
Overexpression of miR-124 down-regulates cytoplasmic but not synaptic GluA2 protein levels.
(A–C) Measurement of total GluA2 protein expression by immunocytochemistry (ICC). N = 5 independent experiments, 3 to 5 fields per experiment. Quantification of the cell body and dendritic signals were performed on the same neurons. (A) GluA2 expression in the cell body. (i) Representative images of cell bodies of transduced neurons. Red/grayscale, GluA2 protein; green, copGFP. (ii) Mean GluA2 intensity per pixel. (iii) GluA2 intensity per cell body. (B) GluA2 expression in proximal dendrites (i) Representative images. Red/grayscale, GluA2 protein; cyan, MAP2 (ii) Mean GluA2 intensity per pixel. (C) GluA2 expression distal dendrites (i) Representative images. Red/grayscale, GluA2 protein; cyan, MAP2 (ii) Mean GluA2 intensity per pixel. (D) Measurement of synaptic GluA2 protein expression by surface labeling of GluA2 in live neurons followed by fixation and staining for synapsin. (i) Representative images of surface expressed GluA2. Red, GluA2; cyan, synapsin. (ii) Fraction of synapsin puncta that appose GluA2 puncta. (iii) Integrated intensity of GluA2 puncta that appose synapsin puncta. N = 3 independent experiments, each with dendrites from 5 different neurons quantified. P values were determined by one-tailed t-tests. *P<0.05, **P<0.01, ***P<0.001. Error bars show S.E.M.
To measure synaptic GluA2 expression, GluA2 immunostaining was performed on non-permeabilized neurons using an antibody that recognizes an extracellular N-terminal epitope. In contrast to labeling total GluA2 protein in which a fixed epitope is used, surface GluA2 labeling is performed on live epitopes as fixing the neurons would lead to a degree of permeabilization. In the absence of permeabilization and fixation, only surface-expressed GluA2 proteins are labeled. Since functional synapses should have both pre- and post-synaptic compartments (Micheva et al., 2010), we then fixed the GluA2-antibody labeled neurons and used antibodies against the pre-synaptic protein, synapsin I, to mark presynaptic compartments, and focused on GluA2 signals that were apposed to synapsin-immunoreactive puncta. Quantification of synaptic GluA2 puncta intensities did not reveal a significant difference between miR-124 overexpression and control (Figure 6D). On the other hand, consistent with the finding that miR-124 decreased total concentrations of GluA2, we found that miR-124 overexpression did reduce non-synaptic GluA2 expression (Figure S7). Together, these data indicate that overexpression of miR-124 decreases the total expression of GluA2 throughout the neuron but does not alter the concentration of GluA2 at synapses.
DISCUSSION
This study was aimed at determining whether and how miR-124 regulates translation of GluA2 in neurons. Computational identification of a conserved miR-124 site in the 3'UTR of GluA2 gave rise to the hypothesis that miR-124-mediated regulation of GluA2 could produce rapid changes in GluA2 expression. Since the GluA2 subunit is calcium-impermeable, such local changes would have important functional consequences on synaptic strength and connectivity. We were particularly intrigued by the possibility that local regulation could occur in dendrites given previous reports that GluA2 mRNA localized to dendrites (Grooms et al., 2006), and that GluA2 underwent local, activity-dependent translation in dendrites (Ju et al., 2004).
While we show that translation of GluA2 is regulated by miR-124, both in luciferase assays (Figure1) and following overexpression of GluA2 in neurons (Figures 5 and 6), our data indicate that this regulation occurs in cell bodies and not in dendrites or at synapses. Our experiments using RT-qPCR of synaptosome fractions and FISH analyses of cultured neurons indicate that although miR-124 is detected in synapses and dendrites, GluA2 mRNA is largely restricted to cell bodies (Figures 2B and 3). Furthermore, overexpression of miR-124 decreases GluA2 protein concentrations significantly more in somata than in dendrites (Figures 6A–C), which is consistent with translational regulation in the soma followed by transport into the dendrite. We also find that while overexpression of miR-124 decreases the expression of total GluA2 concentrations in neurons, it does not affect the concentration of the surface GluA2 expression at synapses. Taken together, our findings are most consistent with a model in which GluA2 protein is synthesized in the soma and subsequently transported into dendrites and synapses (Hirokawa and Takemura, 2005; Setou et al., 2002; Shin et al., 2003; Wyszynski et al., 2002). They further indicate that translational regulation primarily controls the overall concentrations of GluA2 in neurons, with post-translational mechanisms functioning to maintain normal levels of synaptic GluA2 expression. These mechanisms may include enhanced trafficking of GluA2-containing AMPARs, which is a well-studied mechanism of AMPAR regulation involving post-translational modifications and several interacting proteins (Anggono and Huganir, 2012; Hanley, 2010; Hirling, 2009; Malenka, 2003). Our findings that miR-124 overexpression decreases the concentration of surface GluA2 at non-synaptic but not synaptic sites favors a model in which the lateral movement of plasma membrane AMPARs, along with membrane trafficking, is a highly regulated step in determining synaptic AMPAR concentrations (Makino and Malinow, 2009). We achieved only a modest reduction of total GluA2 protein in our neuronal cultures (27% by immunoblot; Figure 5C). It is possible that post-translational mechanisms may be inadequate to ensure appropriate synaptic GluA2 levels in the event of drastic reductions of total protein.
Several previous studies have suggested that GluA2 is a likely candidate for post-transcriptional regulation. GluA2 translation following pilocarpine-induced status epilepticus has been shown to be regulated in a 3' UTR-dependent manner (Irier et al., 2009). Furthermore, GluA2 mRNA immunoprecipitates with FMRP, a RISC component, and GluA2 translation following DHPG treatment is dysregulated in FMRP knock-out mice (Muddashetty et al., 2007). miR-124 has also been found to associate with FMRP in the mouse brain (Edbauer et al., 2010). The work presented here further suggests a role for post-transcriptional regulation of GluA2 mRNA by miR-124, and indicates that majority of this interaction likely takes place in the cell body. While our luciferase assays support a direct interaction between miR-124 and GluA2 mRNA, our assays in neuronal cultures do not preclude the possibility of other interactions indirectly decreasing total GluA2 protein levels. This question can be firmly addressed only by mutating or deleting the miR-124 target site in the GluA2 3'UTR, which is beyond the scope of this study. Our studies also show that miR-124 overexpression decreases GluA2 protein concentration without altering GluA2 mRNA concentration, indicating that the regulation is at the level of translation. Although we find that miR-124 does not affect synaptic GluA2 expression by itself, there are several other predicted miRNA target sites in its 3' UTR, including a validated interaction with miR-181. Saba et al. have found that transfection of a miR-181 duplex into hippocampal cultures reduces surface GluA2 puncta size (Saba et al., 2012). Collectively, these findings indicate that the concentration of GluA2 in neurons and at synapses is fine-tuned by multiple mechanisms, some of which occur in the cell body and others of which occur locally at synapses. Our findings suggest that the local mechanisms do not involve local translation, but rather consist predominantly of post-translational processes such as regulated receptor trafficking. We were unable to complement the experiments in which we overexpressed miR-124 in neurons (Figures 5 and 6) with experiments in which we knocked down miR-124 expression. While previous reports have used sponge constructs to decrease miR-124 expression in neurons (Akerblom et al., 2012; Cheng et al., 2009), these experiments were done in neural stem cells and not in mature neurons, where the concentrations of miR-124 are significantly higher (Cheng et al., 2009; Deo et al., 2006; Makeyev et al., 2007). In addition to using sponge constructs to decrease miR-124, we used lentivirally expressed short hairpins (pmiRZip lentivirus, System Biosciences, Mountain View, CA) to inhibit miR-124. Again, we were unable to detect any significant decrease in the concentration of miR-124 in mature neurons using this technology. We conclude from these results, and from published reports that highly abundant miRNAs can require unachievable amounts of inhibitors to knockdown (Arvey et al., 2010; Ebert et al., 2007; Ebert and Sharp, 2010), that we were not able to knock down miR-124 in mature neurons because it is expressed at such high concentrations.
The results of our biochemical fractionation (Figure 2) and FISH (Figure 3) experiments conflict with previous reports that GluA2 mRNA is dendritically localized in neurons (Grooms et al., 2006; Kye et al., 2007). We explored a number of explanations for this discrepancy. We initially performed traditional FISH using digoxigenin-labeled riboprobes for GluA2, but did not detect any signal in dendrites (Figure S8). This encouraged us to perform FISH using branched cDNA probes (Player et al. 2001), which have been shown to be sensitive enough for single molecule FISH (Player et al., 2001; Itzkovitz and van Oudenaarden 2011). This methodology was used by Cajigas et al (2013) to show that GluA2 mRNA was detected at low levels in proximal dendrites of hippocampal neurons, but was not detected in distal dendrites. Despite the increased sensitivity of branched cDNA probes, we were not able to detect significant concentrations of GluA2 mRNA in either proximal or distal dendrites (Figure 3). To ensure the specificity of the branched cDNA probes, we included several controls, the most compelling of which confirmed that signals from the neuron-specific GluA2 mRNA was present in neurons but not in glia, and that CamK2α mRNA was detected in excitatory but not in inhibitory neurons (Figure 4). We considered the possibility that the extent of dendritic GluA2 mRNA might be regulated by neuronal activity, but were unable to detect any increases in GluA2 signal in dendrites following silencing with TTX or activation with BIC (Figure S4). In considering the lack of dendritic localization of GluA2 mRNA, it is possible that RNA binding proteins on some transcripts may block probe recognition resulting in an underestimation of dendritic localization. However, we do not think this is likely as the branched cDNA probes used recognize 20 different regions on each transcript. Additionally, pre-treating the neurons with proteinase K to improve probe accessibility did not increase the number of transcripts detected (data not shown). We also considered the possibility that the discrepancy between our results and those of Grooms et al. derived from the fact their FISH experiments were performed on rat hippocampal neurons while we used mouse hippocampal neurons; however, we were unable to detect dendritic GluA2 mRNA in rat hippocampal neurons (data not shown). Together with the fact that we could not detect significant concentrations of GluA2 mRNA in synaptosome fractions (Figure 2), we are confident in our conclusion that GluA2 mRNA is somatically restricted, with minimal concentrations in dendrites.
Our results also conflict with published reports that miR-124 mRNA is somatically restricted (Kye et al. 2007). We first assayed miR-124 localization with the locked nucleic acid (LNA) approach used in Kye et al, but the results were inconclusive, as the signal for the antisense LNA was not significantly different from the signal from the sense LNA (data not shown). A possible explanation for the lack of detection of miR-124 in dendrites in Kye et al. is that the locked nucleic acid probe used did not hybridize well with miR-124. This possibility is supported by the low level of miR-124 signal detected in the cell body, which was only slightly higher than their negative control; one would expect a much stronger signal, considering that miR-124 is the most abundant miRNA in the adult mouse brain (Lagos-Quintana et al., 2002). We thus also used branched cDNA probes to increase the sensitivity of miRNA FISH. Using these probes, we detected clear dendritic signals for miR-124. Demonstrating the specificity of the signal, we did not detect any signal in astrocytes, which do not express miR-124, and showed that the signal was greatly reduced in the presence of a competitive inhibitor (Figures 4B–C). Performing FISH and RT-qPCR in silenced and stimulated neurons revealed that miR-124 localization and levels were not affected by activity (Figures S4, S9). Together with the fact that RT-qPCR experiments revealed significant concentrations of miR-124 in synaptosome fractions (Figure 2), our results provide strong evidence that miR-124 is dendritically localized in mouse hippocampal neurons.
The discrepancy between our results and previously published studies highlight a set of technical concerns inherent to studies of mRNA localization and regulated translation in neurons. One concern has to do with the potential differences in results obtained when a transcript is overexpressed in neurons. For example, three studies that have reported activity-dependent dendritic translation of GluA2 (Kacharmina et al 2000; Ju et al 2006; Kim et al 2013) were all based on overexpression of GluA2 reporter constructs rather than on translation of endogenously localized mRNA. While it is possible that the small amount of endogenous GluA2 mRNA that we detect in dendrites undergoes regulated translation in response to stimulation, the sparse distribution of transcripts suggest that the contribution to dendritic concentrations of GluA2 protein is minimal (although we cannot rule out the possibility that synapses in proximity to GluA2 mRNA are more susceptible to regulated changed in GluA2 concentrations). Taken together, however, these studies underscore the need to develop methods to monitor translation of endogenous mRNAs, or of exogenous mRNAs expressed at endogenous levels, to complement those obtained in overexpression studies.
A second concern has to do with the extent to which setting a threshold for detection of localized mRNAs affects experimental results. All mRNAs examined to date are present at significantly lower concentrations in dendrites than in somata, and as a result, the detection of localized mRNAs has often required the saturation of somatic signals in FISH analyses. We sought to evaluate dendritic localization of RNA in a manner that was independent of concentration using imaging parameters in which somatic and dendritic signals were in the linear range for quantification. This allowed us to compare somatic and dendritic concentrations within a single neuron, rather than saturating the somatic signal in order to detect signal in the dendrites. This allowed us to quantify and compare the proportion of FISH signal in three compartments: the soma, the proximal dendrite (0 to 20 μm from the soma) and the distal dendrite (greater than 20 μm from the soma). These analyses revealed that Camk2α mRNA and miR-124 were significantly more dendritically-localized than either GluA2 or Fads3 mRNAs (Figure 3). Since we compared the proportion of each transcript in the three compartments, these differences are likely due to an RNA-specific mechanism, rather than reflecting transcript or miRNA abundance. We also note that even when we saturated the signal for somatic GluA2 mRNA, we were unable to detect increased signal in the dendrite; rather than detecting an increased number of puncta, the few puncta simply became brighter (data not shown). We hope that these methods and methods of analyses will provide a useful reference for future studies on transcript and miRNA localization in neurons.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S Hasan for preparation of mouse hippocampal cultures, TH Ch'ng, J Lee, and K Olofsdotter-Otis for comments on the manuscript, and members of the Martin lab for helpful discussions. We thank Dr. Alex Herbert (University of Sussex) for developing the GDSC ImageJ plugin. Support comes from NIH grant R01 NS045324 (to K.C.M.), the Medical Scientist Training Program (NIH T32 GM008042), the Neurobehavioral Genetics Training Program (NIH grant T32 MH073526) (to V.M.H.), NIH/NCATS/UCLA CTSI grant UL1TR000124 (to T.G. and S.V.), the Marie Curie Fellowship of the EU (PIOF-GA-2008-219622 to P.P) and the Research Funding Program: Heracleitus II. Investing in knowledge society through the ESF (to N.K.).
ABBREVIATIONS
- miRNA
microRNA
- miR-124
microRNA-124
- 3' UTR
3' untranslated region
- AMPA
2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl) propanoic acid
- AMPAR
AMPA receptor
- RISC
RNA-induced silencing complex
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- FISH
Fluorescence in situ hybridization
- ICC
Immunocytochemistry
REFERENCES
- Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. doi: 10.1016/j.neuron.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Current opinion in neurobiology. 2012;22:461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Molecular systems biology. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DL, Gall CM, Isackson PJ. Dendritic localization of type II calcium calmodulin-dependent protein kinase mRNA in normal and reinnervated rat hippocampus. Neuroscience. 1992;46:851–857. doi: 10.1016/0306-4522(92)90189-9. [DOI] [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. The European journal of neuroscience. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS biology. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L-C, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature neuroscience. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nature reviews. Neuroscience. 2007;8:101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science (New York, N.Y.) 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Current biology : CB. 2010;20:R858–861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JG. Endosomal sorting of AMPA receptors in hippocampal neurons. Biochemical Society transactions. 2010;38:460–465. doi: 10.1042/BST0380460. [DOI] [PubMed] [Google Scholar]
- Herrera DG, Robertson HA. Activation of c-fos in the brain. Progress in neurobiology. 1996;50:83–107. doi: 10.1016/s0301-0082(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Hirling H. Endosomal trafficking of AMPA-type glutamate receptors. Neuroscience. 2009;158:36–44. doi: 10.1016/j.neuroscience.2008.02.057. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nature reviews. Neuroscience. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, Fukushima T. Expression profile analysis of microRNA (miRNA) in mouse central nervous system using a new miRNA detection system that examines hybridization signals at every step of washing. Gene. 2007;391:39–44. doi: 10.1016/j.gene.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Secretory trafficking in neuronal dendrites. Nature cell biology. 2004;6:585–591. doi: 10.1038/ncb0704-585. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Irier HA, Shaw R, Lau A, Feng Y, Dingledine R. Translational regulation of GluR2 mRNAs in rat hippocampus by alternative 3' untranslated regions. Journal of neurochemistry. 2009;109:584–594. doi: 10.1111/j.1471-4159.2009.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nature neuroscience. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kacharmina JE, Job C, Crino P, Eberwine J. Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11545–11550. doi: 10.1073/pnas.97.21.11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nature genetics. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nature genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA (New York, N.Y.) 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology : CB. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Roche KW. Posttranslational regulation of AMPA receptor trafficking and function. Current opinion in neurobiology. 2012;22:470–479. doi: 10.1016/j.conb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco M.a., Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Molecular cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Malinow R. AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron. 2009;64:381–390. doi: 10.1016/j.neuron.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC. Synaptic plasticity and AMPA receptor trafficking. Annals of the New York Academy of Sciences. 2003;1003:1–11. doi: 10.1196/annals.1300.001. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annual review of neuroscience. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3' untranslated region of alpha-CaMKII regulate its dendritic targeting. Nature neuroscience. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science (New York, N.Y.) 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba R, Storchel PH, Aksoy-Aksel A, Kepura F, Lippi G, Plant TD, Schratt GM. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Molecular and cellular biology. 2012;32:619–632. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Carvalho AL, Caldeira MV, Duarte CB. Regulation of AMPA receptors and synaptic plasticity. Neuroscience. 2009;158:105–125. doi: 10.1016/j.neuroscience.2008.02.037. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nature methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annual review of cell and developmental biology. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Shin H, Wyszynski M, Huh KH, Valtschanoff JG, Lee JR, Ko J, Streuli M, Weinberg RJ, Sheng M, Kim E. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. The Journal of biological chemistry. 2003;278:11393–11401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nature methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes & development. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Kim E, Dunah AW, Passafaro M, Valtschanoff JG, Serra-Pages C, Streuli M, Weinberg RJ, Sheng M. Interaction between GRIP and liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron. 2002;34:39–52. doi: 10.1016/s0896-6273(02)00640-2. [DOI] [PubMed] [Google Scholar]
- Yates LA, Norbury CJ, Gilbert RJ. The Long and Short of MicroRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.