Abstract
Genome analysis in several eukaryotes shows a surprising number of transcripts which do not encode conventional messenger RNAs. Once considered noise, these non-coding RNAs (ncRNAs) appear capable of controlling gene expression by various means. We find Drosophila sex determination, specifically the master-switch gene Sex-lethal (Sxl), is regulated by long ncRNAs (>200 nt). The lncRNAs influence the dose sensitive establishment promoter of Sxl, SxlPe, which must be activated to specify female sex. They are primarily from two regions, R1 and R2, upstream of SxlPeand show a dynamic developmental profile. Of the four lncRNA strands only one, R2 antisense, has its peak coincident with SxlPe transcription, suggesting it may promote activation. Indeed, its expression is regulated by the X chromosome counting genes, whose dose determines whether SxlPe is transcribed. Transgenic lines which ectopically express each of the lncRNAs show they can act in trans, impacting the process of sex determination but also altering the levels of the other lncRNAs. Generally, expression of R1 is negative whereas R2 is positive to females. This ectopic expression also results in a change in the local chromatin marks, affecting the timing and strength of SxlPe transcription. The chromatin marks are those deposited by the Polycomb and Trithorax groups of chromatin modifying proteins, which we find bind to the lncRNAs. We suggest the increasing numbers of non-coding transcripts being identified are a harbinger of interacting networks similar to the one we describe.
Keywords: lncRNAs, sex determination, Drosophila, Sex-lethal, transcription
1. Introduction
Transcriptome analysis shows that a large portion of the genome in higher eukaryotes is transcribed, but the majority of these transcripts do not code for protein (reviewed in [1]. Original views were that this transcription is dark matter or trash from the genome, however, more recent analyses suggest the expression is functional and can cover ~85% of the human genome [2]. Non-coding RNAs have limited sequence conservation and are not uniform in length; when longer than 200 nucleotides they have been referred to as long ncRNAs (lncRNAs; [1, 3]). The expanding catalog suggests that most are expressed at levels much lower than protein coding genes, but some can approach the levels of mRNAs. They may or may not be spliced, or polyadenylated. Many are single exon transcripts [4]and even those which are spliced can also be found in their unprocessed form [5, 6].
A few lncRNAs have been analyzed, and the emerging picture is they can have activating or repressing roles, frequently mediated through the modification of chromatin [7–10]. Some are capable of affecting their targets in trans, while others act only in cis [11]. trans-acting lncRNAs can function as signals, guides or scaffolds to alter expression of target genes. cis regulation appears to involve coordinated expression with the neighboring protein coding genes, influencing the related transcripts during their biogenesis. The process of transcription, independent of the lncRNA product itself, can also influence gene activity in cis (reviewed in [12–14]).
We find that Drosophila uses lncRNAs in making the decision of gender; a process which uses an X chromosome counting mechanism to determine the activity state of the master-switch gene, Sex-lethal (Sxl). The initial activation of Sxl in early development is transcriptional [15, 16], at its establishment promoter SxlPe. SxlPe is a dose sensitive promoter that is capable of discriminating between one versus two X chromosomes, and is only transcribed in females (XX animals). It responds to the levels of X-linked activating genes (five known: sisterless-a, sisterless-b, runt, myc and unpaired) which work in conjunction with positive maternal factors such as Daughterless [17], reviewed in [18]. Working against these activators are negative maternal factors, such as Groucho, and autosomal zygotic genes (deadpan, the only identified member) which ensure SxlPe remains off in males (XY animals). The protein from SxlPe transcripts alters the splicing of transcripts from the maintenance promoter, SxlPm, which is transcribed in both sexes as the expression of SxlPe (in females) decays. Without SXL protein, SxlPm transcripts include a translation terminating exon by default so all male SxlPm transcripts do not encode full-length protein. Thus by activating SxlPe, female embryos set in motion a splicing autoregulatory feedback loop which maintains SXL expression and sexual identity, through the rest of the life cycle [19, 20]. In short, the expression state of SxlPe is critical to deciding gender.
Several sense and antisense expressed sequence tags (ESTs) are identified at Sxl by the Fly database (Flybase). We focused on the region upstream of SxlPe as there are several of these lncRNAs, suggesting they may influence the activity of this gender determining promoter. We describe here the effects of ectopically expressing some of these lncRNAs as well as additional lncRNAs we found in the region. The activity of their ectopic expression implicates the lncRNAs with roles beyond that of aberrant transcripts or the byproducts of splicing. Additionally, the different SxlPe lncRNAs show a changing expression profile during development suggesting different roles, which are generally consistent with the effects of their ectopic expression. Surprisingly, they show complex cross interactions between them suggesting they may act combinatorially. They are bound by various Polycomb and Trithorax group protein members, key regulators of histone marks in euchromatin, and can alter the chromatin environment of the promoter to influence its expression.
2. Materials and Methods
2.1. Fly crosses and transgenic lines
Flies were reared under uncrowded conditions on standard cornmeal medium. All crosses were done at 25°C; w1118 was the wild-type control. Progeny were counted 8–9 days out from the first day of eclosion. In all cases the reference class had a minimum of 165 males. Description of genes can be found in Flybase (http://www.flybase.org/).
lncRNA constructs were made by cloning PCR fragments of the SxlPe region into the NotI site of CaSpeR HSP83 ([21]; primers in Table S1). The long R1+R2 combined constructs were created using the R2 F NotI and R1 R NotI primers. Injection of plasmids was done by Genetic Services, Inc. and Rainbow Transgenics. All transgenic lines described were sequenced to confirm orientation and insert identity.
2.2. in situs
These were performed as described in [22]. Two biological replicates for each genotype were performed.
2.3. qRT–PCRs
Embryos were collected on apple juice agar plates for 2h and processed immediately, or for 1h and aged for the appropriate time before processing. They were washed off the plate, dechorionated with 50% bleach, washed extensively in PBS (150 mM NaCl, 10 mM sodium phosphate, pH 7.6) with 0.1% Triton X-100 (PBST), and frozen at −80°C. RNA was extracted from the frozen embryos using tri-reagent™ as per manufacturer's protocol. An additional phenol extraction was performed on the purified RNA, followed by DNAse treatment. A PCR test was performed on the RNA to confirm the lack of DNA, after which 4 ug of the RNA was reverse transcribed (RT) with AMV RT at 55°C for 15 min followed by 1.5 h at 50°C. For each RT 1 ng of R1 or R2, sense or antisense primer (Table S1), was added to 100 ng of oligo-dT. Quantitative PCRs were performed in triplicate on a Bio-Rad iQ5 thermocycler. For each sample a minimum of 2 biological RNA replicates was analyzed, except where noted in Supplemental data. In all cases, each biological sample was analyzed by 3 technical replicates, for both the experimental and reference RNA. Ct values that showed a difference of greater than 0.5 from the other two replicates were discarded. PCR products were between 200 and 300 bp; primers in Table S1.
For the lncRNA developmental profile, the data was corrected for tubulin as well as DNA content across each time point. Tubulin corrections were from quantification data available on Flybase. DNA content correction was done by extracting genomic DNA from embryos at each time point, followed by qPCR and normalizing to the grams of sample. Ore R was the control. SxlPe mRNA quantitation was done as in [22]. Statistical data analysis was completed using Microsoft Excel and GraphPad Prism.
2.4. Chromatin Preparation and Immunoprecipitations
ChIPs and chromatin preparation were performed as described in [17], a modification of the small scale ChIP protocol of Alekseyenko et al. [23] that uses 0.1–0.13g of embryos. Ore R was the control and chromatin was clarified from approximately 80ul of embryos. At least two or more biological replicates were analyzed; only samples which gave a signal with the positive controls were included.
ChIPs were performed using 0.24 ml clarified chromatin, 3.5 µg of anti-H3K4me3 [Active Motif 39915], 2.5 µg of anti-H3K27me3 [Active Motif 39155] or non-immune [Sigma] and 30 µl of Protein A beads [Santa Cruz] in 0.9 ml of RIPA buffer. For the anti-H3K4me3 ChIPs, washes were performed as described in [24]. For the anti-H3K27me3 antibody, beads were incubated with the extract at 4°C for 3 hours. After the incubation, the beads were washed once for 7 minutes with each buffer at 4°C, Wash Buffer 1 (500mM NaCl, 10mM Tris-HCl pH 8.0, 1mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% Sodium-Deoxycholate), Wash Buffer 2 (50mM Tris-HCl pH 8.0, 2mM EDTA, 500mM NaCl, 1% NP-40, 0.1% SDS), Wash Buffer 3 (250mM LiCl, 10mM Tris-HCl pH 8.0, 1mM EDTA, 0.5% NP-40, 0.5% Sodium-Deoxycholate) and TE (10mM Tris-HCl 8.0, 1mM EDTA). The immunoprecipitated material was then eluted as described in [24]. Negative controls used non-immune serum (Sigma). The 1–3h H3K4me3 ChIPs had 3 biological replicates and 1 biological replicate was performed for each of the 1–3 and 2–4h H3K4me3/H3K27me3 ChIPs.
2.5. RNA Immunoprecipitation (RIPs)
Embryos were collected on apple juice plates for 1h and aged for 2.5h (1–3.5h collections). They were washed off, dechorionated in 50% bleach for 3’, washed extensively in PBS (150 mM NaCl, 10 mM sodium phosphate, pH 7.6) with 0.1% Triton X-100 (PBST), and frozen at −80°C. Approx. 55 µl of embryos were used for each experiment. For each genotype, a minimum of 6 RIPs for each protein was performed to identify the lncRNAs consistently bound.
Embryos were homogenized in 150 µl of Buffer C Complete (Buffer C: 150mM NaCl, 50mM Tris-HCl pH 8.0, 0.5% NP-40, 1mM EGTA, 1% Triton X-100, 5mM DTT + 1× Protease Arrest (100× Stock G Biosciences-786–108) + RNase Inhibitor, Murine (New England Biolabs-M0314L)). After homogenizing, another 150 µl of BCC was added and the extract incubated on ice for 10 minutes. The extract was centrifuged for 5 min at 4°C at 8500 g, supernatant transferred into a new tube and centrifuged again for 5 min at 4°C at 8500×g. The extract was then pre-cleared with 5 µl of Protein A or G beads (pre-washed with Buffer C) for 1 hour at RT. After incubation, the extract + beads were centrifuged at RT for 1 minute at 1500×g. The volume was brought up to 0.7 ml. About 20 µg of either anti-ASH1 (Sauer), anti-E(Z) (Jones), or anti-SU(Z)12 (Cavalli) was added. The antibody/extract mixture was then incubated 2 hours at 4°C and 2 hours at RT. The antibody/extract mixture was then added to 5 µl of pre-washed Protein A (rabbit antibodies) or Protein G (rat antibody) beads and rotated for 2 hours at RT, then 4°C overnight.
The extract/bead mixture was then centrifuged at RT for 1 minute at 1500×g. The extract was removed from the beads which were then rinsed with 0.8 ml of Buffer C (150mM NaCl) and centrifuged at RT for 1 minute at 1500×g. Buffer C was discarded and the beads washed by rocking with 0.8 ml of Buffer C (500mM NaCl) for 6 minutes. The beads were centrifuged as before, buffer discarded and washed with rocking 3 times with Buffer C (300mM NaCl) for 6, 8 and 10 minutes at RT, respectively. After the last wash was discarded, 0.4 ml of Buffer C (300mM NaCl) was added to the beads.
2.6. RNA Extraction
Acid Phenol (300 µl) and Chloroform (150 µl) was added to the bead/buffer mixture, mixed thoroughly and incubated on ice for 10 minutes after which it was centrifuged at RT for 5 minutes at 16100×g. The aqueous phase was transferred to a new tube, 450 µl Chloroform added, vortexed and centrifuged at RT for 5 minutes at 16100×g. The aqueous phase was transferred to a new tube and 1 µl of 20mg/ml Glycogen, 80 µl 1M Sodium Acetate and ~900 µl 100% ethanol added, incubated on ice for 10 minutes, centrifuged at 4°C for 10 minutes at 16100×g. The ethanol was removed, pellet washed with 500 µl 75% ethanol and centrifuged at 4°C for 5 minutes at 16100×g. After removing the ethanol the pellet was vacuum dried for 5–10 minutes, resuspended in 12 µl of DEPC-treated water. The RNA was DNase treated (Promega RNase-Free DNase-M6101) and PCR performed on an aliquot with primers specific for Sxl to ensure it was DNA-free.
2.7. Reverse Transcription
About ¼ of the RNA was reversed transcribed (New England Biolabs AMVRT) with 1ng of primers specific to Sxl R1 and R2 sense and antisense strands (see Table S1) at 55°C for 15 min followed by 1.5h at 50°C. Qualitative PCR with 40 amplification cycles for the Sxl R1/R2 sense and antisense strands was performed. Products were analyzed on a 1.2% agarose gel.
3. Results
3.1. Long noncoding RNAs with dynamic developmental profile near SxlPe
A few hundred bases upstream of SxlPe several ESTs are described, suggesting they may have a role in regulating the promoter (Flybase; EP 09229, 484531, 480897, 307363, EK103837, EP05922, EK187542, 151849). These ESTs are unspliced and appear to have no polyA tail, and all but one have no open reading frame (ORF). The exception, 307363, has the potential to code for a peptide of 50 amino acids, well below the 100 amino acid cut off for lncRNAs [25]. The ESTs helped define a region of ~480bp which we termed R1 (Fig. 1A), which excludes the peptide-coding region of EST 307363. At 463bp upstream of SxlPe, R1 is immediately upstream of the previously defined ‘switch region’ which contains most of the elements required for sex-specific expression of the promoter [26]. The ‘switch region’ alone faithfully recapitulates sex-specific expression of reporter constructs, however, it is unable to give the full expression that reporters with additional sequences upto 1.4 kb or more are capable of.
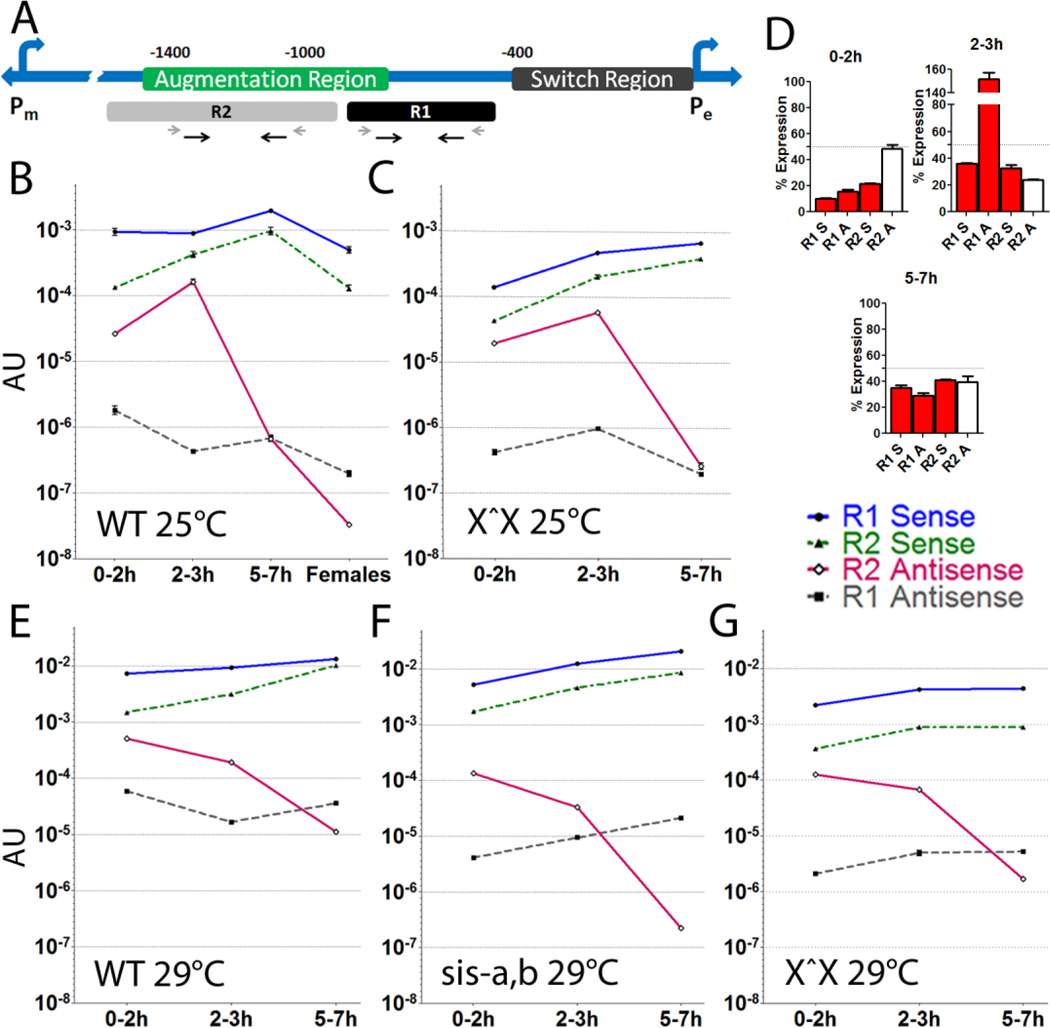
Figure 1. Drosophila sex determination utilizes lncRNAs near SxlPe.
(A) Positions of R1 and R2 lncRNA regions relative to known regulatory elements. The two Sxl promoters are indicated, SxlPm is ~5 kb upstream of SxlPe. Prior deletion studies of SxlPe [26] define the switch region as necessary for sex specific activation (0–0.4 kb upstream), while the augmentation region (0.8–1.4 kb upstream) is required for robust expression. Primers for PCR of R1 and R2 shown as black arrows, RT primers as shorter grey arrows. (B–G) Developmental profiles of the four R1 and R2 lncRNAs strands in different genotypes (key to strands on right). All measured by qRT-PCR and normalized to tubulin to give arbitrary units (AU) of expression. They are also corrected for approximate DNA content at each stage, to give a gauge of the lncRNA to DNA level (removing this correction does not significantly change the plot shapes, it mostly depresses the apparent 0–2h levels). Note the graphs are in log scale, which makes the small error bars difficult to see. (B) wild-type (OreR) 0–2, 2–3, 5–7h embryos and adult females. Expression of the 4 lncRNA strands relative to SxlPe mRNA (2–3 hr) is R1A: 0.000017, R1S: 0.03, R2A: 0.0057, R2S: 0.015. (C) attached-X stock where the free X chromosome has a deletion of Sxl (X^X; SxlfP7B0/Y). This allows scoring the female only signal. (D) Percent female AU for each lncRNA in 0–2, 2–3 and 5–7h embryos from an attached-X stock (data in C) relative to its corresponding lncRNA in wild-type (data in A), to give an indication of the proportion from males. The dotted line at 50% represents equal contribution from males and females. (E–G) Temperature and reduction of X chromosome counting genes depresses relative R2 antisense levels. (E) wild-type, (F) X^X SxlfP7B0, and (G) sis-a1, sis- bsc3-1/FM7 stock at the non-permissive temperature of 29°C. Only 0–2, 2–3 and 5–7h embryos were analyzed in (C, E–G). SxlPe is most active in the 2–3h window. R2 antisense RNA shows a strong positive correlation with SxlPe expression (B and C), and is severely depressed in the background of reduced X chromosome counting genes of sis-a,b/FM7 (G). In all backgrounds, R1 as well as R2 sense RNA show an increase in the 5–7h window, which correlates with the shutdown of SxlPe. The AU level for the genotypes is slightly different; overall females have slightly reduced lncRNA levels and 29°C elevates transcription (* P-value <0.05, ** P-value <0.005, *** P-value <0.0005). Error bars represent mean +/− SEM.
Through a reverse transcription-PCR (RT-PCR) walk in both R1 and the region upstream, we defined a second region of ~650bp which produces RNA in 1–3.5h embryos, when the sex determination decision is being made (R2; Fig. 1A). R2 maps 0.95–1.6 kb upstream of SxlPe, mostly within the ‘augmentation region’. The augmentation region was previously defined by deletion analysis as necessary for enhanced expression [26, 27]. This switch/ augmentation organization is conserved in the distantly related D. virilis which also activates SxlPe in only females, and this specificity is maintained when D. virilis sequences are transposed into melanogaster suggesting the organization is functional [27]. Unlike the R1 region which has no significant ORFs, a few relatively short ORFs are present in the R2 region (one of 101 amino acids in the sense, and two of 66 and 79 amino acids in the antisense orientation). These ORFs, except for the shortest, do not have a good Kozak sequence to suggest they are translated. Moreover, the lncRNAs are of relatively low abundance (see below) and only a small fraction appear to be polyadenylated (oligo-dT versus sequence specific RT ratios of ~5% for R2 antisense and ~2% for R2 sense in both the 0–2h and 2–3h windows; Table S2). Given the requirement for a polyA tail for protein translation, we take this combined evidence to suggest that it is the RNA and not the potential to code for peptides that is relevant, as seen for some mammalian lncRNAs [2].
Quantitation of the 4 RNA strands from the R1 and R2 regions shows that, as seen in mammals [28], the lncRNAs are at much lower levels than their mRNA counterpart. The two sense RNAs are the more abundant, at 3% (R1S) and 1.5% (R2S) of SxlPe mRNA in 2–3h embryos, the peak window of SxlPe expression. The sense RNAs are always present and peak shortly after the expression of SxlPe (5–7h), suggesting they turn off the promoter (Fig. 1B). The antisense strand of R2 is expressed at 0.57% of SxlPe mRNA, and peaks in the same window as the promoter (2–3h, Fig. 1B), suggesting it may serve an activation role. R2 antisense levels are low after that. R1 antisense is the least abundant at 0.0017% of SxlPe mRNA. It is at its highest in 0–2h embryos prior to SxlPe activation, and is transcribed mostly as a long transcript through both regions (Fig. S1A). There is also a long sense transcript spanning R1 and R2, transcribed mostly in the SxlPe preactivation window and at low levels after that (see Suppl. discussion).
3.2. SxlPe lncRNAs levels are affected by the X chromosome counting genes
The wild-type embryos in Fig. 1B are of mixed sex. For the lncRNA expression profile in females, an attached X-chromosome (X^X) stock, where the free X chromosome has a deletion of Sxl (SxlfP7B0), was analyzed. In this stock only females produce the lncRNAs. Extrapolating from wild-type levels suggests that ~50% of the R2 antisense lncRNA in 0–2h embryos is from females (Fig. 1C). The other 3, including the long (R1 + R2) sense lncRNA, are overwhelmingly from males (Fig. 1D). Surprisingly, during the 2–3h window when SxlPe expression peaks, the R2 antisense lncRNA, which continues to increase in females, is at even higher levels in males (Fig. 1D). The process of transcribing SxlPe may decrease the lncRNA levels, as seen for lncRNAs at active mammalian promoters [29]. Alternatively, females may decrease or destabilize the transcripts in the process of activating SxlPe, perhaps through enhanced binding of chromatin proteins or transcription factors to the region.
During SxlPe expression (2–3h), R1 antisense appears to be primarily from females while the two sense RNAs appear to be expressed equally in both sexes. In the 5–7h window when SxlPe is off, the proportions of the lncRNAs are comparable between the sexes, however, the overall levels in females appear slightly lower.
To get the profile of the lncRNAs under the reverse condition where SxlPe expression is inhibited, we took advantage of a stock with mutations in the X chromosome counting genes, sisterless-a and sisterless-b (sis-a1, sis-bsc3-1). This sis-a, b chromosome reduces female viability, however, it can be maintained in the heterozygous condition (over an FM7 balancer chromosome) at temperatures of 25°C or lower, due to the temperature sensitive nature of the sis-bsc3-1 allele [30]. At 29°C however, the reduction of both sis-a and sis-b, even in the heterozygous state, is too severe and almost all the females (>99%) are killed from a failure to properly activate SxlPe. This sis-a,b/ FM7 stock allows collecting embryos the majority of which are compromised for the normal dose of X chromosome counting genes, a more male-like condition where SxlPe is not expressed. Adult females are all heterozygous (sis-a, b/FM7) and adult males are mostly sis-a,b/Y with fewer FM7/Y males, so most of the female embryos are either heterozygous or homozygous for sis-a,b, with some wild-type females (from the FM7 males), while male embryos are equally sis-a,b/Y or FM7/Y.
Correcting for the effect of temperature (note scale bars in Fig. 1B versus E) which appears to elevate the levels of all the lncRNAs in wild-type (except for R2 antisense in the 2–3h window), the most significantly impacted lncRNAs in the sis-a1, sis-bsc3-1 background are the two antisense RNAs with R2 antisense showing the most severe decrease in the 2–3h window and beyond (Fig. 1F, see Fig. S1B for levels relative to wild-type). Prior genetic studies found the X chromosome counting mechanism to be inherently temperature sensitive [16, 31]. This suggests that, in the critical 2–3hr time window, the apparent drop in R2 antisense relative to the sense RNAs in wild-type embryos at 29°C (Fig. 1E), is not conducive to the female sex determination process. Its further decrease when the sis-a and sis-bsc3-1 dose is reduced, is consistent with this view. Indeed, quantifying the levels of SxlPe mRNA at 29°C shows a decrease relative to 25°C in both the wild-type and the sis-a,b/FM7 background with the latter having the more dramatic drop, consistent with the role of X chromosome counting genes (Fig. S1C). When the attached X stock representing females is shifted to 29°C it also shows the effect of temperature, including the drop in SxlPe expression levels (Fig. S1C), and elevates the overall lncRNA levels (compare scale bars in Fig. 1C and G). However, the relationship of the lncRNA levels to wild-type seen at 25°C appears to be generally preserved; in the critical 2–3hr window, the attached X stock continues to show proportionately higher levels of the positively correlated R2 antisense relative to the R1 sense, R1 antisense, and R2 sense lncRNAs. This relationship of R2 antisense is inverted in the sis-a1, sis-bsc3-1 background where its levels are further decreased, while the R1 sense and R2 sense lncRNAs remain comparable to wild-type (compare Figs. 1E–G). We note that the same time windows were used at 29°C as at 25°C, even though development occurs slightly more rapidly at the higher temp. (generation times of approximately 9 and 10 days, respectively). As the time windows are short, we did not correct for the change as it is proportionally small and all the genotypes at 29°C have the same windows.
Combined, these results demonstrate that SxlPe expression correlates positively with the expression of R2 antisense, supporting the hypothesis that R2 antisense is associated with an activation role. R2 antisense (and to a lesser extent R1 antisense) expression is promoted by the X chromosome counting genes while the sense RNAs appear to be ‘constitutive’ (i. e. they are expressed continuously) although their levels may vary and they correlate negatively with SxlPe expression. While the overall levels of all the lncRNAs are a little higher in males than females, the negatively correlated lncRNAs from the R1 region as well as R2 sense are also much lower in females, most particularly in the 0–2h window when the activity state of the promoter is presumably being set up, as it precedes strong expression of SxlPe. Further support for the idea that the two sense lncRNAs may be associated with repression and shut down of SxlPe, is that their levels are elevated in the 5–7 h window when SxlPe is silent. These negatively correlated sense strands are also relatively more abundant in males, which contribute more than 50% of these lncRNAs at all time points (see Fig. 1D). The sense RNAs also show a disproportionate increase on elevation of temperature as well as when the X chromosome counting gene dose is decreased (Fig. 1D, E and Fig. S1B), conditions which do not favor SxlPe expression.
3.3. Ectopic expression of lncRNAs alters SxlPe expression
To determine how the lncRNAs function, transgenic lines which ectopically express each lncRNA were made. The R1 or R2 region in either sense or antisense orientation was placed downstream of the hsp83 heat shock promoter. The hsp83 is strongly expressed in the female germline, so it maternally provides the RNAs in embryos. As this could potentially be lethal to one of the sexes, depending on the role of the lncRNA, we chose to randomly integrate the transgenes as this would increase the chance of recovering insertion lines, in case the unfettered expression from the hsp83 promoter should prove lethal.
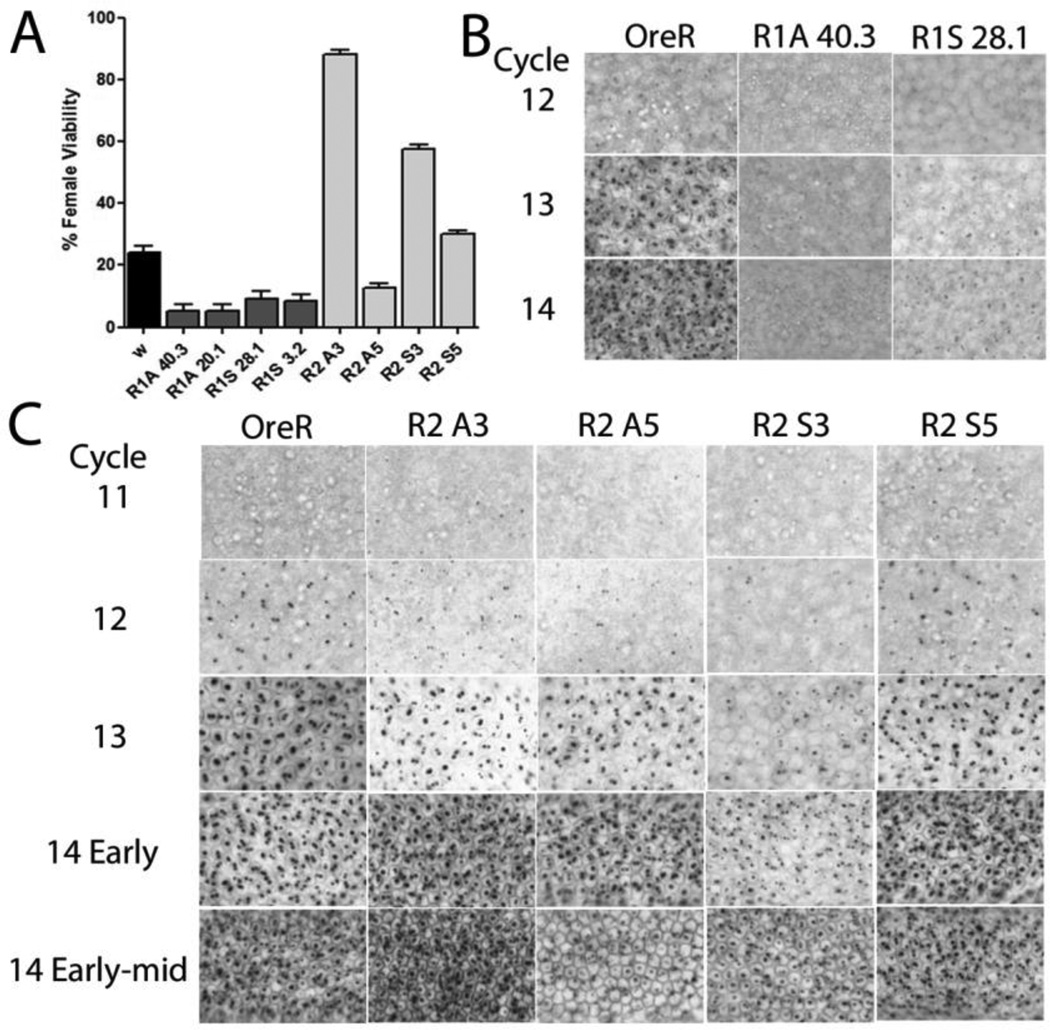
When homozygous most of the R1 lines did not alter the sex ratio, however, some of the R2 lines reduced male viability by 17–26%. The role of the lines in sex determination was further explored using the sis-a1, bsc3-1 chromosome described above which at 25°C compromises, but does not completely eliminate, the ability of females to activate SxlPe. Under this condition, each of the R1 lines was found to further reduce female viability demonstrating a negative trans effect by the R1 sense and antisense lncRNAs (Fig. 2A). The R2 lines gave mixed results. Most improved, but some showed a worsening in female viability. This suggests that while the R2 region can enhance female viability, its effects are complex, likely the outcome of different expression levels from the random integration sites. Indeed, lines which improved female viability as homozygotes showed a decrease in their positive effect when their copy number was reduced, while lines which worsened or had slightly below wild-type female viability as homozygotes either improved or stayed unchanged as heterozygotes (Fig. S2).
Figure 2. Constructs ectopically expressing R1 and R2 lncRNAs affect SxlPe.
(A)Altered female viability induced by ectopic expression of lncRNAs. Viability of eclosed females relative to males expressed as a percent. Mothers homozygous for the indicated lncRNA line were crossed to males with decreased sis-a, b numerator dose. w is w1118 and is the wild-type control (black bar), female viability ~24%. Error bars show percent +/− %SE. (B, C) Ectopic expression of lncRNAs affects SxlPe expression as shown by in situ hybridization of SxlPe transcripts, normally seen as two dots on each of the two X chromosomes as Sxl is on the X. Nuclear cycle number is shown on the left (Cycles 11–14 correspond with ~90–200min after egg laying), genotype at top. OreR is wild-type where the promoter is only on in females from cycle 12 to 14. in situs of R1 lines (B) show weaker staining dots (additional R1 lines in Fig. S2A), and R2 lines (C) which have more varied expression patterns and match the genetics in (A). Quantitation of the SxlPe mRNAs by qRT-PCR supports the in situs – shown in Fig. S2B.
To corroborate the genetic analyses, SxlPe mRNA expression was examined by in situ hybridization in representative lines. For the R1 lines many of the embryos showed reduced SxlPe expression, consistent with their negative effects (Fig. 2B, Fig. S3A). For R2, two lines for each strand, a representative positive and negative, are described. All had a change in both timing and levels (Fig. 2C) with most showing premature expression of SxlPe. This early elevated expression did not persist, in fact the normal SxlPe expression peak was delayed. The negative antisense line (A5) had a short late peak, while the activating line (A3) had a peak which persisted longer than wild-type; the latter extends the time window for activating the autoregulatory splicing feedback loop and would account for the ability of A3 to rescue females with a weakened sex determination signal. For the sense lines, one (S3) had persistent and longer than wild-type expression but few embryos with robust levels. This persistence potentially explains its positive outcome in the genetic analysis, as it also extends the window for activating the autoregulatory feedback loop. The second line (S5) showed relatively normal SxlPe expression levels which persisted a little longer than wild-type. qRT-PCR of SxlPe mRNA over windows of half hour increments corroborated the in situ data (Fig. S3B).
Quantitation of transcripts from the other Sxl promoter, SxlPm, during its activation window showed the lines do not consistently alter its expression (Fig. S3C). We infer that the lncRNAs primarily affect SxlPe with the effects on SxlPm by some of the lines being indirect. Removing SxlPe and the region 1.4kb upstream has been shown to alter the expression of SxlPm [32].
Overall, the inhibitory effect of the R1 ectopic expression lines supports the initial suggestion that this region is repressive. The R2 region gave complex outcomes, but was generally activating. While the enhancement in female viability by the R2 sense lines runs counter to its expression profile, as shown by the results below this activating ability is likely from the complex interactions of the lncRNAs - ectopic expression of the R2 sense lncRNA actually decreases its own total levels.
3.4 lncRNAs affect each other’s levels
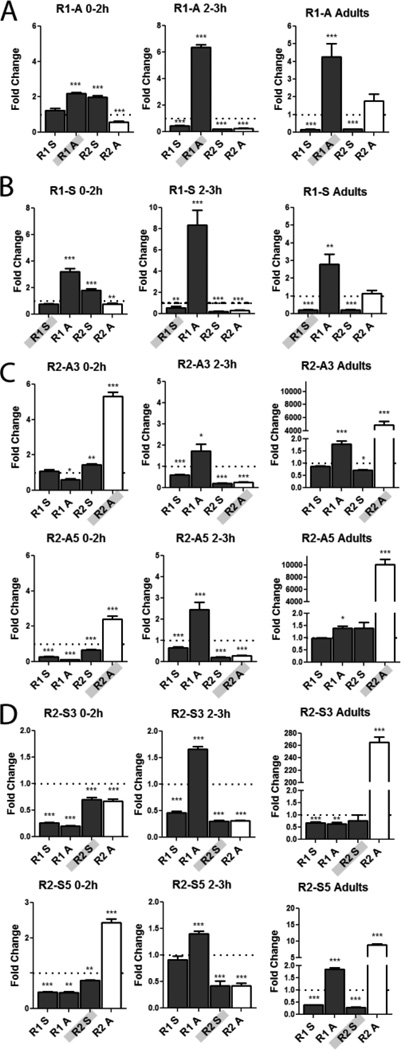
To get a better insight into how the lncRNAs exert their effects, all 4 strands were quantified in the representative transgenic lines. Zero-2h, 2–3h embryos and female adults were scored and the levels compared to wild-type (Fig. 3). The first observation of note is that for all the lines, the levels of the other 3 lncRNAs change regardless of which lncRNA is being expressed. This was true for all time points. Second, despite the fact that the hsp83 promoter is highly expressed in the female germline, the level of the lncRNA being ectopically expressed is not proportionally increased in embryos (hsp83 mRNA is almost as high as tubulin [33]). Including endogenous transcripts the lncRNAs are at best elevated only a few-fold, suggesting that the majority of the RNA from the transgene is being turned over. For the sense transcripts the total levels mostly decreased. The only exception was R2 antisense which in adults was several thousand-fold higher than wild-type (Fig. 3C). However, even these high levels were turned over in 0–2h embryos. To confirm expression, each transgenic line was quantified in ovaries for its contribution to the corresponding lncRNA total (Table S3). All were expressed, although the proportions showed a broad range, from essentially 100% for R1 antisense, down to 18 to 3% for the remaining lncRNAs (Table S3).
Figure 3. R1 and R2 lncRNAs interact and affect the levels of other lncRNAs.
All the lncRNAs change in levels when only one lncRNA is over-expressed. qRT-PCR of R1 and R2 transcripts was done in homozygous transgenic lines expressing each lncRNA. All data was normalized to tubulin and are reported as fold change relative to wild-type. Genotype and time window noted at top, lncRNA being expressed is shaded in grey on the X axis. Bars for the positively correlated lncRNA R2 antisense are shown in white, to facilitate tracking its relative levels. Quantitation of lncRNA levels in lines expressing transgene of R1A (A), R1S (B), two R2A lines (C) and two R2S lines (D). Note the y-axes have different scales. Error bars represent mean +/− SEM. * P-value <0.05, ** P-value <0.005, *** P-value <0.0005.
Examining the effects of the R1 lines shows that both significantly lowered the level of the positively correlated R2 antisense lncRNA in the 0–2 as well as 2–3h windows, while the constitutively expressed R2 sense lncRNA increased in the 0–2h window (Fig. 3A, B). Additionally, both lines resulted in similar lncRNA profiles particularly in 2–3h embryos and adults (Fig. 3A, B), despite the fact they are complementary RNAs. These data highlight the instability of the R1 sense lncRNA which failed to accumulate. By contrast, the synthesis or stability of the complementary R1 antisense RNA was enhanced.
For the R2 lines (Fig. 3C, D), overexpressing the antisense lncRNA did increase its levels in embryos although, as noted above, not in proportion to the strength of the hsp83 promoter. As seen for the R1 sense RNA, the R2 sense lncRNA levels did not increase. In fact they generally showed a decrease, in addition to reducing the levels of R1 sense. However, they increased the complementary R2 antisense RNA, so that the relative levels of the positively correlated R2 antisense RNA appeared higher in adults and 0–2h embryos. These changes potentially explain the premature activation of SxlPe observed in the in situs, if one assumes that the positively correlated R2 antisense transcript facilitates activation. The premature expression of SxlPe in the R2 lines is generally followed by reduced/ delayed transcription, ending ultimately with fairly robust expression (Fig. 2C and Fig. S3B). The decrease in R2 antisense in the 2–3h window potentially accounts for this delay. The only exception was the R2 S3 line which did not increase R2 antisense, however, it decreased the other lncRNAs (Fig. 3D).
The general trends from these experiments suggest that altering the levels of any one of the SxlPe lncRNAs will in turn affect the others at the promoter, alluding to an interactive nature. Interestingly, for all the lines the 0–2h window appeared to be the most predictive of whether SxlPe will be expressed strongly or weakly, correlating with an increase or decrease, respectively, in the levels of R2 antisense relative to the other lncRNAs (Fig. 3). As this window precedes the window of highest SxlPe activity (2–3h), it suggests the lncRNAs may prime the promoter for its strength of expression.
3.5. Interplay and net balance of lncRNA strands determines outcome
The complex changes the lncRNAs induced on each other and their effects on females suggested that not only the strand, but also the relative levels of the lncRNAs affect how strongly SxlPe is transcribed. To test this idea we combined representative R1 and R2 transgenes with each other, as well as transgenes which express both regions (R1+R2) as one lncRNA, in the various possible combinations (Fig. 4, which displays a transgene and all its permutations with the other transgenes, before the next series of permutations with another transgene etc.). Each lncRNA combination was tested against a reduced dose of X chromosome counting genes (sis-a1, bsc3-1) to determine if it was negative or positive to females. Early in development, the primary role of the X chromosome counting genes in sex determination is to turn on SxlPe, after which they are no longer required [30]. This genetic test is, therefore, a good reporter of the efficacy of SxlPe activation.
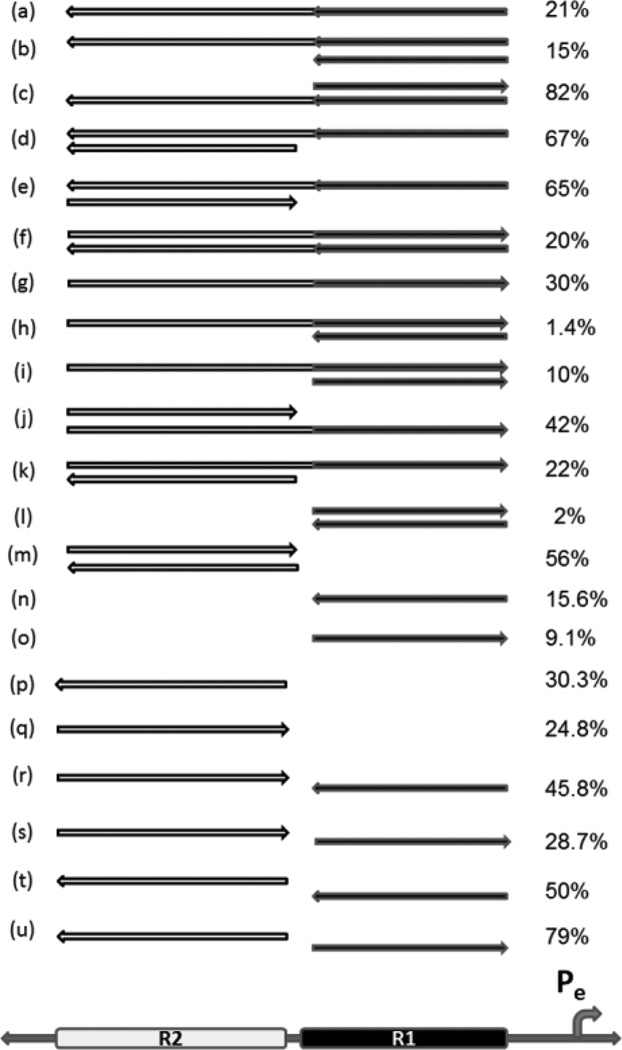
Figure 4. Different combinations of lncRNA transgenes can increase or decrease female viability.
Female viability relative to males as percent, when females with different lncRNA combinations are crossed to males with reduced X chromosome counting genes (sis-a1, sis- bsc3-1). For all crosses, only one copy of the lncRNA transgene is present. Viability of w1118 females without a transgene was 24%. Arrow direction indicates the strand expressed; 5’ is to the left. Graphic of promoter and R1 and R2 regions at bottom shows location of region expressed in each line. Light gray is R2 and dark line represents R1 region. Transgenes of the combined regions (R1 + R2) shown with both tones. They encompass all of the R1 and R2 regions and the short ~35bp region between them. The effect on females with only the (R1+ R2) sense or antisense lncRNA is in lines (a) and (g), and is relatively mild. R1 lines represented: 40.3 and 28.1; R2 lines represented: A3 and S5.
The R1 region, which is negative to females when either strand alone is ectopically expressed, is worse if both strands are simultaneously introduced (Fig. 4, line (l) versus (n) or (o)). Attaching R2 to the R1 region makes it less negative (Fig. 4 line (o) versus (g)). When a transgene of the R1 region is combined with a long (R1 + R2) transgene, the effect is frequently worse than the latter alone (Fig. 4 line (g) versus (h); line (g) versus (i)).
Increasing the R2 region is generally favorable in several combinations (Fig. 4 line (a) versus (d); line (a) versus (e); line (g) versus (j); line (q) versus (m); line (n) versus (r); line (n) versus (t); line (o) versus (s); and line (o) versus (u)). Combining R2 antisense with R1 sense also results in high female viability (line (u)).
Remarkably, the combination of (R1 + R2) antisense with R1 sense results in a dramatic improvement in female viability, even though the individual transgenes are neutral or negative (Fig. 4 line (e) versus lines (a) and (o)). In stark contrast, the combination of their complementary strands is the most negative (Fig. 4 line (h)).
We note that as each transgene has a different insertion site, the female viability of each of the lines tested only approximates the effect of that particular lncRNA. However, it is this same lncRNA that is placed in combination with one of the other representative lncRNAs, so a given pair of lncRNAs gives an indication of how the combination shifts female viability when compared to each lncRNA alone. In some cases, the combinations show large changes from the effect of each individual lncRNA, emphasizing an outcome of an interactive nature.
To confirm that these genetic results reflect a change in SxlPe expression, representative combinations were assessed for their levels of SxlPe mRNA (Fig. S3D). As expected, given the role of the X chromosome counting genes in activating SxlPe, the wild-type control shows that reducing the dose of sis-a,b significantly reduces the levels of SxlPe mRNA. Transgene combinations which improved the drop in female viability from reducing the sis-a,b dose also showed an increase in SxlPe mRNA levels, while a combination which worsened it showed a further decrease. By contrast, a lncRNA combination that did not alter female viability much relative to the wild-type, also did not show a significant change in SxlPe mRNA levels (Fig. S3D). These results support the idea that the changes in female viability which the lncRNA combinations produce are a function of changing SxlPe expression.
Together, these data indicate that in addition to the strand being significant, the relative level of expression of the two regions is also critical. An increase of the R2 antisense region appears positive to females (note that R1 antisense is mostly R1 + R2; Fig. S1A). It bears remarking that the R2 sense transgene, which is also positive, tends to decrease its own levels while elevating R2 antisense, particularly in the 0–2h window (Fig. 3D), supporting the view that a relative excess of R2 antisense is what is favorable. These data explain why deletion analyses of the SxlPe promoter defined R2 as an augmentation region [26, 27].
3.6. Ectopic expression of lncRNAs can rescue effects of PcG/trxG mutations
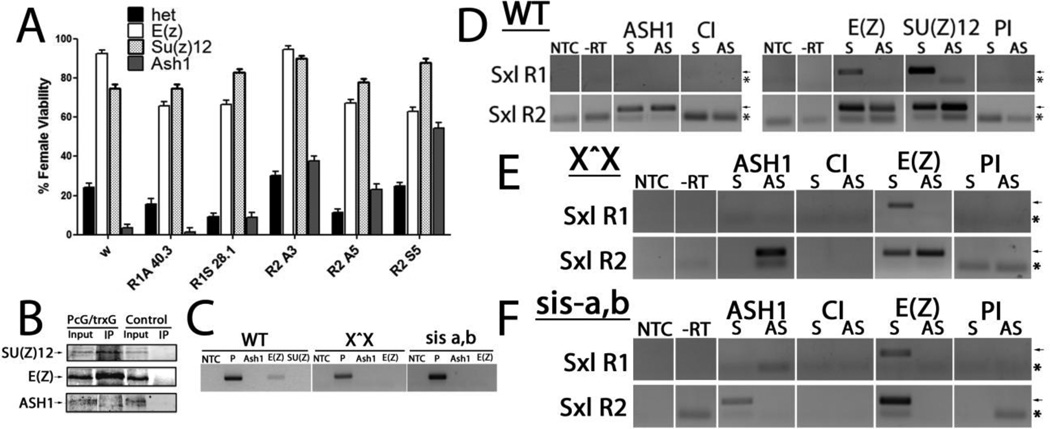
The Polycomb/ trithorax groups (PcG/trxG) of proteins are involved in the epigenetic silencing and activation of several developmental genes. These protein complexes have been shown to interact with lncRNAs and affect target genes in trans [34]. in situ analysis shows mutations in PcG/ trxG members impact SxlPe, functioning as expected with the trxG member, ash1 (absent, small or homoetic discs 1) acting positively, and two PcG members, Suppressor of zeste 12 (Su(z)12) and Enhancer of Zeste (E(z)) acting negatively on the promoter (Fig. S5). To functionally test the lncRNAs with these chromatin modifiers, these PcG/trxG mutants were individually combined with each lncRNA transgene and analyzed in the female-sensitized background of reduced X chromosome counting genes. As demonstrated above (Figs. S1C and S3D), reducing the dose of these X chromosome counting genes compromises SxlPe expression, and females cannot survive without appropriate activation of SxlPe. Female viability thus serves as a readout of proper SxlPe expression. Strong loss of function alleles, each of which is homozygous lethal, were tested (Fig. 5A). ash1, a trxG member which influences methylation of histone H3 at lysine 4, H3K4me3, and two PcG members E(z), which catalyzes methylation of lysine 27 in H3 to generate H3K27me3, and Su(z)12, which modulates the activity of E(Z).
Figure 5. lncRNAs genetically interact with and bind PcG/trxG proteins.
(A) Genetic interaction between transgenes and PcG/trxG mutants. Female viability of trans-heterozygotes with single copy of lncRNA alone (het), one copy of the ash1MB03235, E(z)731 or Su(z)124 mutation over w (to remove the balancer chromosome) or one copy of both the lncRNA shown and either ash1MB03235, E(z)731 or Su(z)124 mutation, crossed to numerator deficient, sis-a,b/Y, males. Viability shown as a percentage, which is relative to the male reference class. white1118 (w) serves as the wild-type control. Error bars show +/− %SE. (B–F) PcG/trG proteins bind lncRNAs. RIPs of lncRNAs at SxlPe. (B) IP of the proteins from RIPS. Input lane has 1% of the extract for E(z) and Su(z)12, 3% for Ash1. Note ASH1 protein runs as a doublet ([53]) (C) RNA control: GS2 mRNA which is expressed a little more abundantly than the lncRNAs, gave little to no signal in RIPs for the PcG and trxG proteins. Genotypes in which RIPs were performed at top. NTC: no template control, P: positive control for RT-PCR shows band for GS2. (D–F) ASH1, E(Z) and SU(Z)12 RIPs in Ore R (D); ASH1 and E(Z) for X^X SxlfP7B0/Y (E) and sis-a1, sisb sc3-1/FM7 (F). Antibody of RIP shown at top of each panel. In sis a-b/FM7 background E(Z) is less consistently found on R2 antisense, probably because its levels are reduced (see Fig. 1F). The arrows indicate protein and PCR product bands. Asterisks mark the position of primer dimers generated, particularly when there is no signal. NTC: no template control; -RT: no RT control; S: sense, AS: antisense. Rabbit serum (PI) served as the control for anti-SU(Z)12 and anti-E(Z) antibodies which are from rabbits, CI monoclonal was used as the control for anti-ASH1 as both are rat monoclonals.
Alone, ash1 heterozygous females have only 3.3% viability when the dose of sis-a,b is reduced (Fig. 5A), indicating it makes a strong contribution to SxlPe activation. Indeed, providing ash1 heterozygotes with one copy of the constitutive Sxl allele, SxlM4, completely rescues this female lethality (105% relative to sibling males) as the SxlM4 allele can activate the autoregulatory splicing feedback loop independently of sis-a,b. This result is also consistent with an early role of ash1 in Sxl activation. The R2 lines were also able to ameliorate the severely negative effect on females from reducing ash1. By contrast, the R1 lncRNAs showed very modest or no improvement (Fig. 5A). The ability of the R2 lines to rescue the detrimental effect of reducing the activator ASH1 positively correlates this region with SxlPe activation.
Reducing the capacity to introduce H3K27me3 through the reduction of either Su(Z)12 or E(Z) improved female viabiity in all the lines (Fig. 5A). The strong loss of function Su(z)124 allele improved female viablity to essentially the level of the Su(z)12 mutant alone, including the negative R1 lines. As SU(Z)12 is in Pc repressive complex 2 (PRC2; [35]) and is required for the stability of E(Z) [36], this effect strongly suggests that PRC2 is critical to the final outcome of the lncRNAs. Decreasing the dose of these PRC2 members enhances SxlPe expression at the critical stage at cycle 14 (Fig. S5). The improvement in female viability with all the lncRNAs when the dose of either Su(z)12 or E(z) is reduced, would suggest that the negative effects of the lncRNAs are dependent on PRC2.
3.7. PcG/trxG proteins bind the SxlPe lncRNAs
These genetic interactions raised the question of whether PcG/trxG proteins directly bind to the SxlPe lncRNAs. To answer this question we performed RNA immunoprecipitations (RIPs) on extracts from 1–3.5h embryo using antibodies to ASH1, SU(Z)12 and E(Z). As shown in Fig. 5B, ASH1 consistently binds to the lncRNAs from R2. Previously, ASH1 was shown to act in trans, activating Ultrabithorax through its trithorax response element RNAs [37]. The ectopic R2 lncRNAs may similarly be recruiting ASH1 to Sxl allowing the lncRNAs to compensate for the reduction in ASH1, so rescuing female viability (Fig. 5A). We find E(Z) also binds to the R2 lncRNAs in addition to the R1 sense transcript, and SU(Z)12 generally binds to the same lncRNAs as E(Z). As E(Z)and SU(Z)12 interact directly in PRC2, this is not unexpected. The negative control, GS2 mRNA which is expressed at comparable levels to the lncRNAs gave little to no signal (Fig. 5C).
As Ore R represents the male and female states combined, the transcriptionally active and inactive states represented by the X^X; SxlfP7B0/Y and sis-a1, sis- bsc3-1/FM7 backgrounds, respectively, were also examined. ASH1 and E(Z), as a representative of PRC2, were tested. In the X^X stock, ASH1 is primarily on R2 antisense (Fig. 5E), consistent with R2 antisense and ASH1 acting as activators, and suggesting that the ASH1 on R2 sense detected in Ore R embryos (Fig. 5D) may be contributed by the male embryos. E(Z) binds R1 and R2 sense, and with more variability, to R2 antisense (some RIPs were negative for R2 antisense). This variability of E(Z) on R2 antisense may reflect ASH1 and E(Z) being in competition, as the promoter transitions from being silent into activation during the window examined. The idea that ASH1 and E(Z) are in competition is consistent with recent studies on mammalian ASH1 which suggest its activity directly counteracts Polycomb silencing [38]. Further, conditions which are negative for females (sis-a1, sis- bsc3-1/FM7 at the non-permissive temperature of 29°C) show ASH1 on only R2 sense, the opposite strand from females (Fig. 5F). E(Z) is on the constitutive R1 and R2 sense lncRNAs. As SxlPe is inactive, it suggests the repressed state has ASH1 absent from R2 antisense, and E(Z)is bound to the R1 and R2 sense lncRNAs.
Taken together, ASH1 on R2 antisense with little to no E(Z) appears to favor the female state. PRC2 has been shown to bind RNA promiscuously [39], so robust SxlPe expression may require ASH1 to compete or displace PRC2 from this lncRNA. The SxlPe lncRNAs may act as a PcG/TrxG response element, serving to recruit the proteins to regulate SxlPe. Indeed they can compensate for decreases in PcG/trxG proteins and a reduction in key X-chromosome counting proteins (Fig. 5A).
3.8. lncRNAs alter chromatin at the SxlPe promoter
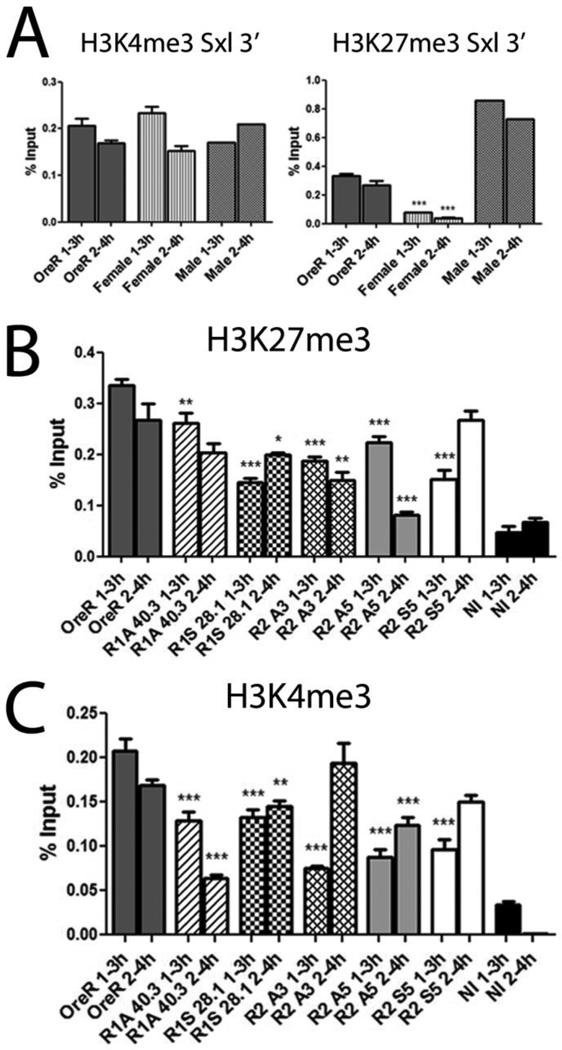
The lncRNAs are bound by PcG/trG proteins raising the question of whether they influence the deposition of their chromatin marks. Our analyses (Cabrera, Olcese and Horabin, unpublished) indicate high levels of H3K4me3 with low H3K27me3 correlate with strong SxlPe expression in females; males maintain high levels of both marks but do not activate the promoter (Fig. 6A).
Figure 6. lncRNAs alter H3K4me3 and H3K27me3 levels.
lncRNA expression alters epigenetic marks associated with PcG/trxG proteins. ChIPs of H3K4me3 and H3K27me3 (A) reported as % input, scored immediately 3’ of SxlPe (+138 bp avg. position) in Ore R and X^X 7BO (females), as well as the projected male signal calculated by subtracting the female contribution from Ore R (obtained by multiplying Ore R signal by 3, giving the total from the avg. of 3 contributing X chromosomes, and subtracting 2 times the female signal which represents the avg. of 2 contributing X chromosomes). ChIP signal of H3K27me3 (B) and H3K4me3 (C) in OreR and lncRNA lines. NI is non-immune serum. Astersisks show significant changes relative to wild-type, *** P-value <0.0005. ChIP experiments were repeated at least twice. Highest P-value compared to non-immune <0.0001. Error bars represent mean +/− SEM.
To determine whether the lncRNAs were influencing these regulatory marks, chromatin immunoprecipitations (ChIPS) were performed on 1–3 and 2–4h embryos for the activating H3K4me3 and repressive H3K27me3 levels (Fig. 6B,C). Ectopic expression of both R1 lines reduced the levels of the activating H3K4me3 mark in both time windows, consistent with their poor SxlPe expression (Fig. 2B). This poor expression is in spite of the slightly reduced levels of H3K27me3. Consistent with it worsening the effect of ash1 (Fig. 5A), R1 antisense reestablished normal H3K27me3 levels in the later 2–4h window while H3K4me3 levels showed a further decrease.
The three R2 lines also changed the chromatin. In the early 1–3h window, all had lowered H3K27me3 and significantly lowered H3K4me3, consistent with a delay in strong SxlPe expression (Fig. 2C). In the later 2–4h window, the antisense lines (A3 and A5) further decreased H3K27me3, while the sense line (S5) reestablished wild-type levels. H3K4me3 levels, however, were elevated to wild-type in the 2 positive lines (S5 and A3), but in the negative A5 line they remained below wild-type. These data show the lncRNAs influence the chromatin marks at the promoter, and the changes correlate with their effects on females.
As the lncRNAs also affect their own and each other’s levels, we examined the chromatin at R1 and R2 (Fig. S4). For all lines, H3K7me3 was decreased in both regions in the two time windows (except for R2 sense which reestablished wild-type levels in 2–4h). Although each line changed the levels slightly differently, for every change the trend was similar for both the R1 and R2 regions. For H3K4me3, a few of the samples showed close to wild-type levels but the rest, like H3K27me3, were generally reduced (R2 antisense line A3 was the only exception increasing it above wild-type at R2). As for H3K7me3, the changes for each line generally showed the same trend in both R1 and R2. These chromatin shifts suggest why the transgenes alter the levels of the other lncRNAs (Fig. 3), and their coordinated effects across the 2 regions reinforce the idea that they are interactive.
4. Discussion
lncRNAs are increasingly being accepted as a major new class of regulatory element. They act in the nucleus and cytoplasm, with activities that can impact all aspects of cellular function including signal transduction, translation and transcription [12, 40]. Their best characterized function is arguably that of epigenetic modulators, which recruit or alter the activity of chromatin modifying proteins at specific sites [11, 40].
This is the first description of an interactive lncRNA network which modulates the chromatin of its associated promoter. Two regions, R1 and R2, upstream of the SxlPe promoter were identified as expressing lncRNAs. Transgenic lines encoding each strand from the two regions indicate they can act on the promoter in trans, and the lncRNAs alter not only their own levels but also those of the others in the network. Ectopic expression was favored over deletion as the latter would also remove transcription factor binding sites (see [29], [30]) and so affect the promoter. On discovery of their interactive nature, individual shRNA knockdown was no longer considered; besides the risk of the opposite strand lncRNA also being knocked down (both strands are potentially available as siRNA), the interactive nature of the lncRNAs suggested that not only the targeted lncRNA would change but so would the others, as seen with the ectopic expression lines. This would make it hard to isolate the effect of reducing any single lncRNA.
4.1. Low levels of stability of the lncRNAs
The endogenous lncRNAs are expressed at low levels, <1/3000 to ~1/30 relative to Sxl mRNA at 2–3h, and appear to have low stability. Quantifying the RNA from only the transgenes gave signals that were much lower than expected for the hsp83 promoter driving their expression. For the sense RNAs, the greater the distance of the RT primers from the lncRNA body in the transgene, the more the signal diminished, supporting the idea they are highly labile (see Table S3). Despite their relatively low steady state levels, which in most part were lower than the endogenous, all the transgene RNAs were able to produce effects on females and on the levels of the endogenous lncRNAs.
The total levels of the R1 and R2 antisense lncRNAs could be elevated to some extent by the ectopic expression, and these lncRNAs showed dynamic changes over development. However, the two sense lncRNAs, which are always present and are the most abundant (Fig. 1B), could not have their levels elevated. On the contrary, their levels dropped. A drop frequently also occurred in the other (non-transgene) sense lncRNA. This reduction suggests a turnover mechanism is being induced. As it occurs in trans we speculate it involves small RNAs; the RNA silencing machinery may also be involved in the other lncRNA cross interactions, an avenue that should be explored in the future.
4.2. Overlapping activating and repressive lncRNAs
Of the four lncRNAs, R2 antisense is the only one which peaks in the 2–3h window when SxlPe is being transcribed. It is also dependent on the X chromosome counting genes, but unlike SxlPe, not in a dose-sensitive manner as it also expressed in males. Coordinated expression of lncRNAs with their mRNAs has also been observed at mammalian promoters in embryonic stem cells suggesting a regulatory link [41].
R2 antisense is normally expressed in the presence of the other lncRNAs, particularly the two sense strands which appear to be constitutive, so it must typically function under the condition of combinatorial lncRNAs. The different transgene combinations in Fig. 4 emphasize this same point, highlighting the impact of the two regions on each other and indicating that the amounts of each strand are significant. Our data suggest that the levels of the lncRNAs relative to each other, determines the outcome. For example, even though the endogenous R2 sense strand does not show a correlation with SxlPe expression, its transgene behaves positively. This is likely from its decrease which results from ectopic expression, while it increases the levels of R2 antisense (discussed above). Similarly, although R2 antisense is present in males and peaks in the same window as females, the fact that males do not transcribe SxlPe indicates that the lncRNA alone is not sufficient for activation. Males express much higher levels of the sense lncRNAs than females, particularly in the 0–2h window (Fig. 1D), which we suggest compete with the activity of R2 antisense. The final outcome must be determined by the lncRNAs working in conjunction with the transcription factors, so that two X chromsomes can activate the ‘switch region’ [26] to drive female specific expression.
4.3. Roles of the lncRNAs in the network
Activation of SxlPe is biphasic; the promoter is first set up in repressive chromatin which is the default or male state, after which it transitions to activation in females [24, 42]. The R1 region and R2 sense lncRNAs appear to favor this repressed state. In the second phase, activation occurs and likely utilizes the lncRNA from R2 antisense to augment the effect of the accumulating X-linked numerator proteins which serve to activate the promoter. This is similar to the ncRNA HOTTIP whose expression alone cannot activate its target promoter. Only when the appropriate transcription factors are present does HOTTIP potentiate expression [7].
Setting up the chromatin phases of SxlPe appears to call for interactions between the lncRNAs, in a complex interplay that includes recruitment of the PcG and trxG histone modifiers (Fig. 7; [43]). Indeed, ectopic expression of the lncRNAs can compensate for reductions in these proteins, as well as alter the chromatin marks they induce (Fig. 5 and 6). We find that depending on the transcription state, the lncRNAs bound by the PcG and trxG chromatin modifiers changes (Fig. 5B, E, and F). Wild-type embryos show PRC2 (E(Z) and SU(Z)12) is on the 3 more abundant lncRNAs (R1 sense, R2 sense and antisense), presumably to establish the silenced or off state through H3K27 methylation (Fig. 7A). As PRC2 appears to bind promiscuously with higher affinity for longer RNAs [39], this is not entirely surprising. Despite this promiscuity, it binds more readily to the SxlPe lncRNAs than the mRNA tested as a control.
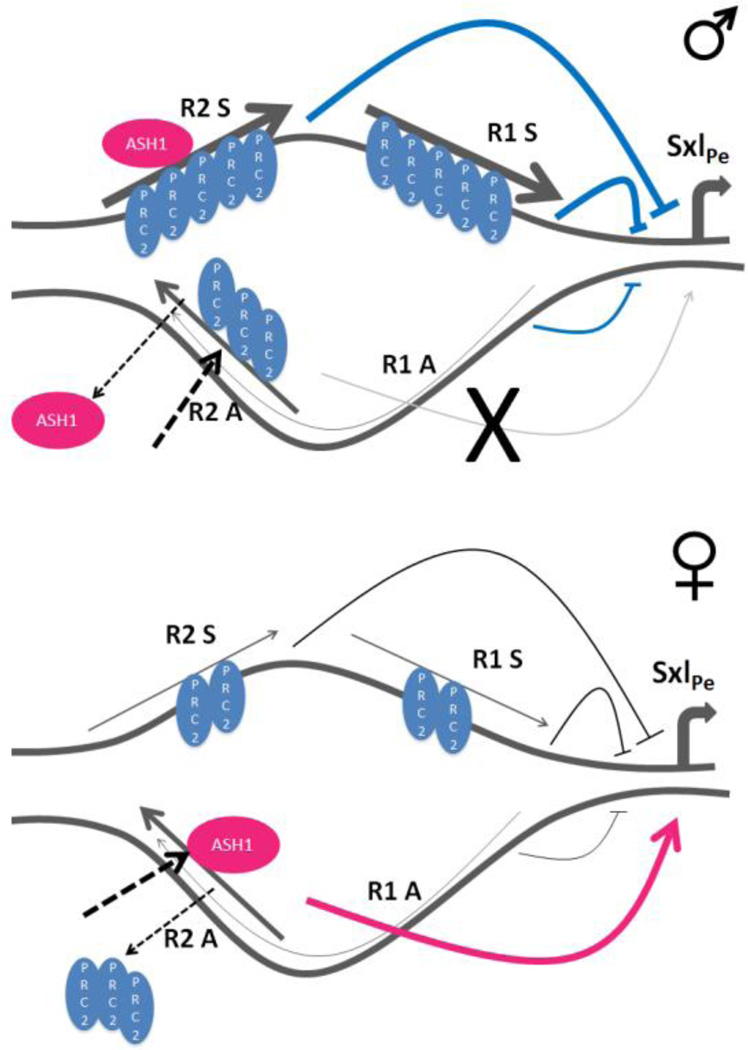
Figure 7. Model of SxlPe repression or activation by lncRNAs and PcG/trxG proteins.
SxlPe in males, indicative of the ‘default’ or transcriptionally repressed state. This is marked by R1 and R2 sense, as well as R2 antisense lncRNAs being bound by PCR2 to ensure SxlPe is not expressed. ASH1 is on R2 sense and unable to displace PRC2 from R2 antisense lncRNA. The ‘X’ represents a failure to activate SxlPe. Activation of SxlPe in females which have reduced levels of R1 and R2 sense, that are also bound by less PRC2. ASH1 is able to evict PRC2 and bind the R2 antisense lncRNA in the process which activates SxlPe (see discussion for more details). Thickness of lncRNA lines is meant to depict their levels.
The transition to activation in females shows ASH1 on R2 antisense, while the repressive E(Z) protein is primarily on the sense lncRNAs (Fig. 7B). In wild-type embryos ASH1 is on both R2 strands, so we infer its presence on R2 sense is contributed by the male embryos. This idea is supported by the condition of reduced X chromosome counting gene dose (sis-a1, sis- bsc3-1/FM7 at non-permissive temperature), where ASH1 is not detected on R2 antisense but on R2 sense, together with PRC2. Recruitment of ASH1 to its targets is thought to occur in a transcription dependent manner, but its role is not dependent on transcription elongation of the mRNA [38, 44]. We suggest part of this recruitment includes lncRNAs, besides the transcription factors. The higher dose of activating transcription factors in females must facilitate the recruitment of ASH1 to the activating lncRNA, R2 antisense (Fig. 7B). ASH1 may then displace E(Z) from R2 antisense to promote SxlPe expression, in keeping with its role of antagonizing PcG repression [45]; the H3K36 mono- and di-methylation which it introduces negatively affects the H3K27 methylation activity of PRC2 [46, 47]. ASH1 and H3K36me2 have been described as overlapping on chromosomes, and both localize to the 5’ end of active genes [48–50], consistent with such a role. Presumably, this effect of ASH1 only occurs when it is on R2 antisense, but not R2 sense, to explain the low levels of H3K27me3 observed in females (Fig. 6A). In the silenced or off configuration, the presence of PRC2 on both R2 sense and antisense must preclude ASH1 function, allowing PRC2 to maintain its activity and generate the high levels of H3K27me3 in males (Fig. 6A). A comparable scenario of ASH1 together with the PRCs on the chromatin of ES cells has been suggested for the repressed state, with ASH1 deposited but only poised for activation [38].
Transcription occurs in both the sense and antisense direction in many parts of the human genome [5]. PRC2 presumably surveys these transcripts, binding promiscuously to them to silence expression until the appropriate developmental cues which recruit transcription factors and trxG protein activators, such as ASH1, to activate expression at the appropriate times. The complex interplay we describe here may well be commonplace and likely reflects the norm rather than exception in gene regulation, a mechanism perhaps for PcG/trxG targets. A good case in point is also sex related; mammalian X chromosome silencing uses multiple lncRNAs as well as PcG members to select which X chromosome is kept active or silenced [51]. This system also employs trans-acting ncRNAs to facilitate transcription; through binding and titration of the repressive zinc finger protein CTCF, the Jpx ncRNA permits transcription of the Xist promoter to trigger X chromosome inactivation [52].
Supplementary Material
Highlights.
Drosophila sex determining promoter, SxlPe, expresses several lncRNAs
Ectopic expression of lncRNAs can alter SxlPe expression
lncRNAs act as a network with complex interplay, affecting each other’s levels
lncRNAs are bound by Polycomb and trithorax group proteins
Ectopic expression of lncRNAs alters the chromatin at SxlPe
Acknowledgements
We thank Drs. F. Sauer, J. Goodliffe, R. Jones, G. Cavalli, R. Holmgren and for anti-ASH1 monoclonal, ASH1 polyclonal, E(Z), SU(Z)12 and CI antibodies, Batory Foods for donating the cornmeal for fly food, our colleagues in the Horabin lab for helpful discussions, and Oscar Cabrera for comments on the manuscript. We thank the Bloomington Stock Center and Dr. J. Muller for Drosophila strains. This work was supported by funding from National Institutes of Health (R01 GM 085165 to JIH), and funds from the Biomedical Sciences dept. at the FSU College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: JIH, BBM and JRC conceived the experiments, BBM, UO, JRC, JIH did the work, JIH and BBM wrote the manuscript.
References
- 1.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 4.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome research. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 10.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 13.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long noncoding RNA transcription. BMC biology. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horabin JI. Long noncoding RNAs as metazoan developmental regulators, Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2013;21:673–684. doi: 10.1007/s10577-013-9382-8. [DOI] [PubMed] [Google Scholar]
- 15.Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- 16.Torres M, Sanchez L. The Sisterless-B Function of the Drosophila Gene Scute Is Restricted to the Stage When the X-a Ratio Determines the Activity of Sex-Lethal. Development. 1991;113:715–722. doi: 10.1242/dev.113.2.715. [DOI] [PubMed] [Google Scholar]
- 17.Kappes G, Deshpande G, Mulvey BB, Horabin JI, Schedl P. The Drosophila Myc gene, diminutive, is a positive regulator of the Sex-lethal establishment promoter, Sxl-Pe. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly. 2010;4:60–70. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cline TW. Autoregulatory functioning of a Drosophila gene product that establish es and maintains the sexually determined state. Genetics. 1984;107:231–277. doi: 10.1093/genetics/107.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 21.Horabin JI, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5' splice site. Mol Cell Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladstein N, McKeon MN, Horabin JI. Requirement of male-specific dosage compensation in Drosophila females--implications of early X chromosome gene expression. PLoS Genet. 2010;6:e1001041. doi: 10.1371/journal.pgen.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Rodriguez J, Yoo Y, Shareef MM, Badugu R, Horabin JI, Kellum R. Cooperative and antagonistic contributions of two heterochromatin proteins to transcriptional regulation of the Drosophila sex determination decision. PLoS Genet. 2011;7:e1002122. doi: 10.1371/journal.pgen.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS computational biology. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estes PA, Keyes LN, Schedl P. Multiple Response Elements in the Sex-Lethal Early Promoter Ensure Its Female-Specific Expression Pattern. Molecular and Cellular Biology. 1995;15:904–917. doi: 10.1128/mcb.15.2.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinks TM, Calhoun G, Schedl P. Functional conservation of the sex-lethal sex determining promoter, Sxl-Pe, in Drosophila virilis. Development genes and evolution. 2003;213:155–165. doi: 10.1007/s00427-003-0304-1. [DOI] [PubMed] [Google Scholar]
- 28.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent Transcription from Active Promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, Jensen TH. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erickson JW, Cline TW. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993;7:1688–1702. doi: 10.1101/gad.7.9.1688. [DOI] [PubMed] [Google Scholar]
- 31.Cline TW. Evidence That Sisterless-a and Sisterless-B Are 2 of Several Discrete Numerator Elements of the X/a Sex Determination Signal in Drosophila That Switch Sxl between 2 Alternative Stable Expression States. Genetics. 1988;119:829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez AN, Lu H, Erickson JW. A shared enhancer controls a temporal switch between promoters during Drosophila primary sex determination. Proc Natl Acad Sci U S A. 2008;105:18436–18441. doi: 10.1073/pnas.0805993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, Carlson JW, Carr A, Jungreis I, Marbach D, Sealfon R, Tolstorukov MY, Will S, Alekseyenko AA, Artieri C, Booth BW, Brooks AN, Dai Q, Davis CA, Duff MO, Feng X, Gorchakov AA, Gu T, Henikoff JG, Kapranov P, Li R, MacAlpine HK, Malone J, Minoda A, Nordman J, Okamura K, Perry M, Powell SK, Riddle NC, Sakai A, Samsonova A, Sandler JE, Schwartz YB, Sher N, Spokony R, Sturgill D, van Baren M, Wan KH, Yang L, Yu C, Feingold E, Good P, Guyer M, Lowdon R, Ahmad K, Andrews J, Berger B, Brenner SE, Brent MR, Cherbas L, Elgin SC, Gingeras TR, Grossman R, Hoskins RA, Kaufman TC, Kent W, Kuroda MI, Orr-Weaver T, Perrimon N, Pirrotta V, Posakony JW, Ren B, Russell S, Cherbas P, Graveley BR, Lewis S, Micklem G, Oliver B, Park PJ, Celniker SE, Henikoff S, Karpen GH, Lai EC, MacAlpine DM, Stein LD, White KP, Kellis M. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. The Journal of biological chemistry. 2004;279:401–406. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]
- 36.Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. Embo Journal. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki H, Higashimoto K, Yada Y, Endo TA, Sharif J, Komori T, Matsuda M, Koseki Y, Nakayama M, Soejima H, Handa H, Koseki H, Hirose S, Nishioka K. Ash1l methylates Lys36 of histone H3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet. 2013;9:e1003897. doi: 10.1371/journal.pgen.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 41.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunders A, Core LJ, Sutcliffe C, Lis JT, Ashe HL. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev. 2013;27:1146–1158. doi: 10.1101/gad.215459.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorighi KM, Tamkun JW. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development. 2013;140:4182–4192. doi: 10.1242/dev.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO reports. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Muller J, Thoma NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 47.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schubeler D. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JT, Bartolomei MS. X-Inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 52.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.