Abstract
Regulation of gene expression at the level of transcription involves the concerted action of several proteins and protein complexes committed to dynamically alter the surrounding chromatin environment of a gene being activated or repressed. ATP-dependent chromatin remodeling complexes are key actors in chromatin remodeling, and the SWI/SNF complex is the founding member. While many studies have linked the action of these complexes to specific transcriptional regulation of a large number of genes and much is known about their catalytic activity, less is known about the nuclear elements that can enhance or modulate their activity. A number of studies have found that certain High Mobility Group (HMG) proteins are able to stimulate ATP-dependent chromatin remodeling activity, but their influence on the different biochemical outcomes of this activity is still unknown. In this work we studied the influence of the yeast Nhp6A, Nhp6B and Hmo1 proteins (HMGB family members) on different biochemical outcomes of yeast SWI/SNF remodeling activity. We found that all these HMG proteins stimulate the sliding activity of ySWI/SNF, while transient exposure of nucleosomal DNA and octamer transfer catalyzed by this complex are only stimulated by Hmo1. Consistently, only Hmo1 stimulates SWI/SNF binding to the nucleosome. Additionally, the sliding activity of another chromatin remodeling complex, ISW1a, is only stimulated by Hmo1. Further analyses show that these differential stimulatory effects of Hmo1 are dependent on the presence of its C-terminal tail, which contains a stretch of acidic and basic residues.
Keywords: SWI/SNF, HMGB proteins, chromatin remodeling, nucleosome remodeling, Hmo1, Nhp6
1. Introduction
Chromatin dynamics has a profound influence on transcriptional regulation. Several proteins and protein complexes involved in regulation of gene expression exert their action through remodeling of the local chromatin environment accompanying gene activation. Among them are ATP-dependent chromatin remodeling machines, which use the energy of ATP hydrolysis for mobilizing nucleosomes or altering their composition [1]. The founding member of this family of chromatin remodelers is the yeast SWI/SNF complex. The activity of this complex results in different biochemical outcomes, including transient exposure of nucleosomal DNA, nucleosome movement in cis (sliding) and trans-displacement of nucleosomes (related to octamer transfer activity and to nucleosome eviction or ejection) [1]. A number of elements that can influence the activity of this complex have been described, including histone covalent modifications [2,3], transcription factors [4] and High Mobility Group (HMG) proteins.
HMG proteins are small proteins involved in nuclear activities such as transcription, replication and DNA repair. These proteins are divided in three families: HMGA, HMGB and HMGN [5]. The presence of the HMG box domain is characteristic of members of the HMGB family. Mammalian HMGB proteins, whose most prominent member is HMGB1, contain two tandem HMG boxes. Yeast members of this family contain one or two HMG boxes [6,7]. There are seven genes encoding HMGB proteins in Saccharomyces cerevisiae, including NHP6A, NHP6B and HMO1 [8]. Nhp6A, Nhp6B and Hmo1 are abundant proteins in the yeast nucleus [9,10]. Hmo1 contains two HMG boxes, while Nhp6A and Nhp6B contain only one [6,11,12]. It has been observed that these three proteins participate in different events related to transcriptional regulation of gene expression [9,13-18]. Nhp6A/B proteins are required for activation of certain inducible genes [8,9]. In the same context, it has been demonstrated that Nhp6 is required for transcriptional initiation fidelity of RNA polymerase III [17]. A similar role has been recently assigned to Hmo1 [14]. Johnson and colleagues observed that Nhp6A binds to nearly 23% of all RNA polymerase II promoters and also demonstrated that the DNA bending properties of this protein are critical for its role in transcriptional regulation [18].
A small number of studies have analyzed the effect of HMG proteins on ATP-dependent chromatin remodeling. First, it was demonstrated that HMGB1 stimulates sliding activity of ACF and CHRAC complexes [19] and Nhp6 was shown to interact both physically and functionally with the RSC complex [20]. Later, it was observed that the SWI/SNF complex enhances V(D)J cleavage on 5S arrays and that this effect is further stimulated by HMGB1 [21]. Additionally, it has been recently shown that the activity of the yeast SWI/SNF complex is stimulated by rat HMGB1 and HMGB2 [22]. There are no studies addressing the influence of yeast HMG proteins on the catalytic activity of the ySWI/SNF complex. However, several independent studies suggest a functional connection between Nhp6 and the ySWI/SNF complex. Triple mutants nhp6a nhp6b swi2 (catalytic subunit of ySWI/SNF) are lethal [16]. Early high throughput gene expression analyses revealed a subset of yeast genes whose expression is affected by both Nhp6A/B and SWI/SNF [15,23]. Recently, Pugh and colleagues performed ChIP-chip and ChIP-seq analyses for a large number of proteins involved in transcriptional regulation, including the Nhp6A protein and SWI/SNF subunits. Several genes with a relative high co-occupancy of both Nhp6A and SWI/SNF can be observed from their data [24].
The studies demonstrating stimulation of ATP-dependent nucleosome remodeling activity have been mainly focused on determination of sliding activity. It is currently unknown whether these proteins affect other biochemical outcomes of SWI/SNF activity. In this context, a role for Nhp6 on nucleosome eviction has been described, but with respect to its association with the yeast FACT complex [8,25]. In order to gain insight into the aspects influencing the different biochemical outcomes of ATP-dependent chromatin remodeling, in our present work we studied the effect of Nhp6A, Nhp6B and Hmo1 proteins on ySWI/SNF activity, by analyzing the impact of these HMG proteins on nucleosome sliding, octamer transfer and transient exposure of nucleosomal DNA to restriction enzymes. Our studies demonstrate that all the yeast HMG proteins analyzed in our study enhance ySWI/SNF sliding activity and that only Hmo1 stimulates ySWI/SNF binding to the nucleosome, its octamer transfer activity and the transient exposure of nucleosomal DNA generated by this complex. Hmo1 also displays a differential stimulatory effect on sliding activity of the ISW1a complex, another ATP-dependent chromatin remodeling complex. In addition, our results indicate that the C-terminal tail of Hmo1 appears to be required for these stimulatory properties.
2. Materials and methods
2.1. Recombinant proteins, protein complexes and probes
Recombinant proteins Nhp6A, Nhp6B, Hmo1, HMGB1, Hmo1Δ (deletion mutant lacking residues 212 to 246), Nhp6A-Ct, Nhp6B-Ct (corresponding to Nhp6A and Nhp6B proteins fused to Hmo1 residues 212-246 on their C-terminal end) were purified as N-terminal His-tag fusion proteins using Ni-NTA agarose resin (cat. 30210, Qiagen), according to the manufacturer’s instructions. Dilutions of the protein stocks were made using HMG buffer [100 mM KCl, 10 mM Hepes-KOH (pH 7.9) and 15% glycerol]. The chromatin remodeling complexes (CRC) ySWI/SNF, RSC and yISW1a were obtained by tandem affinity purification as described elsewhere [2,26] and quantified as previously described [4]. The pGEM-3Z/601-Gal4 plasmid [4] was used to generate a 216 bp or a 147 bp DNA fragment by PCR (Figure 1A). Before PCR amplification, one of the primers used in each reaction was labeled on its 5′ end using [γ-32P] ATP. Nucleosome reconstitution was carried out by the octamer transfer method, as previously described [4]. All the reconstitution reactions used for this study were carried out using 1 pmol of probe and 3 μg of oligonucleosomes (histone donors in the reconstitution reaction; amount of oligonucleosomes in terms of DNA content of these particles). After reconstitution, the nucleosome probes (and mock reconstituted probes) contain 4 fmol/μL of probe and 12 ng/μL of non-labeled oligonucleosomes (in terms of their DNA content).
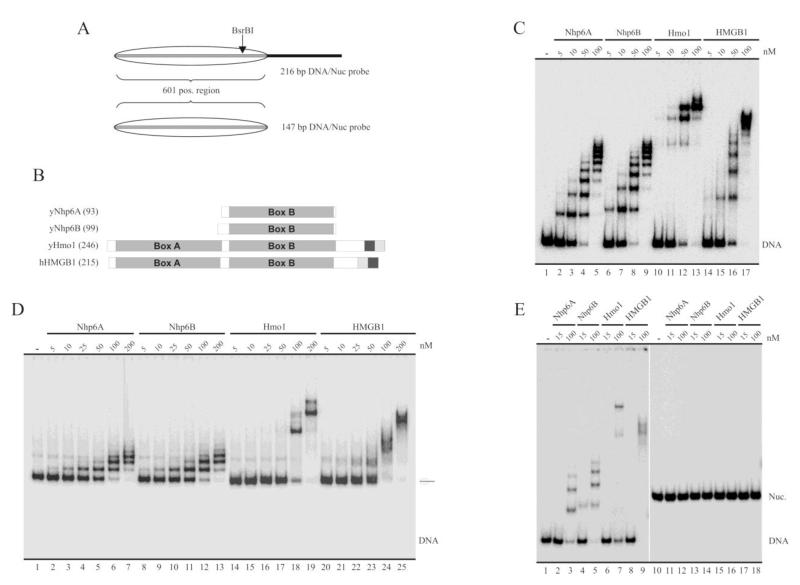
Fig. 1.
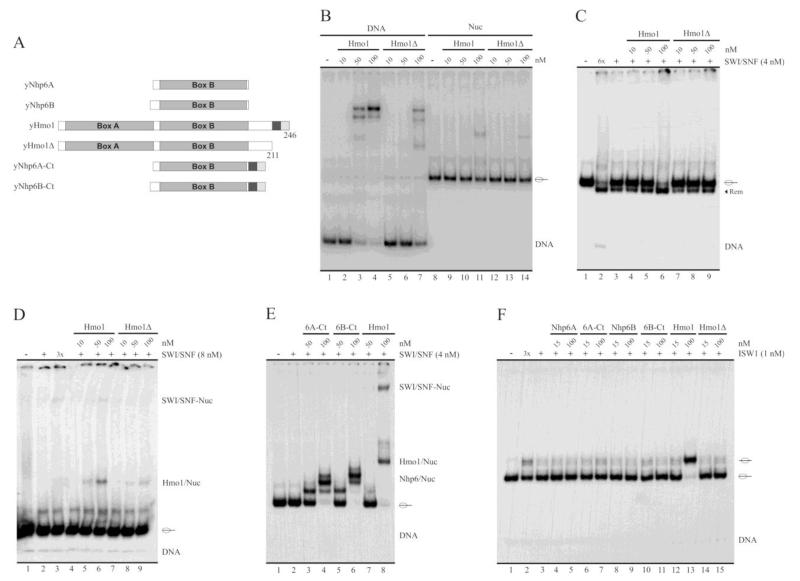
Binding properties of the yeast HMGB proteins under study. A, Schematic representation of the probes used in the analyses, represented here at the form of nucleosomal DNA. The transparent oval represents the position adopted by the histone octamer after reconstitution. 601 pos. region = nucleosome positioning sequence 601 (147 bp). B, Schematic representation of the proteins studied in the present work. The number of amino acids of each protein is indicated in parenthesis. The large gray squares termed Box A and Box B indicate the location of HMG-box domains. The dark gray and soft gray squares represent the acidic and basic stretches (respectively) present in the C-terminal region of these proteins. C-E, Electrophoretic analyses of binding patterns of the HMG proteins using non-denaturing polyacrylamide gels (5%, AA:Bis 40:1). The assays in (C) and (D) were performed using the 216 bp probe as naked DNA (C) or as a reconstituted mononucleosome (D). The assay in (E) used the 147 bp probe at the form of naked DNA (lanes 1-9) or as a reconstituted mononucleosome (lanes 10-18). The HMG protein used in each particular reaction is depicted on top of the figures, as well as the different concentrations tested.
2.2. Binding assays
A mix containing 7.4 μL remodeling buffer [70 mM KCl, 20 mM Hepes-KOH (pH 7.9), 2 mM DTT, 0.5 mM PMSF, 10% glycerol, 0.05% NP-40, 10 mM MgCl2 and 100 μg/mL BSA], 0.6 μL deionized water, 0.5 μL of buffer FCR [100 mM NaCl, 10 mM Tris-Cl (pH 7.4), 1 mM EDTA (pH 8.0), 5 mM DTT, 0.5 mM PMSF, 0.05% NP-40, 10% glycerol and 100 μg/mL BSA], 3 μL of SWI/SNF, CRC buffer [150 mM NaCl, 10 mM Tris-Cl pH 8.0, 1 mM Mg(CH3COO)2, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10% glycerol, 1 mM DTT and 0.5 mM PMSF] or SWI/SNF+CRC buffer, 0.5 μL Gal4 buffer [100 mM KCl, 10 mM Hepes-KOH (pH 7.4), 1 mM DTT, 0.2 mM PMSF, 10 μM ZnCl2, 20% glycerol and 100 ng/μL BSA], 0.5 μL of varying concentrations of HMGB proteins (or HMG buffer) and 2.5 μL probe [nucleosome or DNA (mock reconstituted) probe] was incubated for 30 min at 30°C. The samples were then subjected to electrophoresis in a non-denaturing polyacrylamide gel, in cold room (0.3× TBE buffer; acrylamide concentrations and AA:Bis proportions are indicated in each figure legend). Afterwards, the gel was dried and scanned using phosphor screen and Molecular Imager FX (BioRad). Film autoradiography was also performed.
2.3. Nucleosome sliding assays
These assays were performed by mixing 2.5 μL of the 216 bp probe (as a reconstituted mononucleosome), 0.5 μL oligonucleosomes (or buffer FCR), 0.6 μL 50 mM ATP (Roche, 1140965), 0.5 μL Gal4 buffer, 0.5 μL of varying concentrations of HMGB proteins (or HMG buffer), 3 μL of CRC (or CRC+CRC buffer or CRC buffer) and 7.4 μL remodeling buffer, incubating for 30 min at 30°C. After this incubation, a mix (0.7 μL) containing 750 ng salmon sperm DNA and 500 ng long oligonucleosomes was added, incubating for 20 additional minutes at 30°C. Afterwards, the samples were subjected to gel electrophoresis and subsequent analyses as described above.
2.4. Restriction enzyme accessibility assays
These assays proceeded exactly as described for the sliding assays, with the difference that 0.3 μL of BsrBI (20 U/mL) were added to the mix before starting the remodeling incubation. After this incubation, 1 volume of stop buffer [20 mM Tris-Cl (pH 7.4), 40 mM EDTA (pH 8.0), 1% SDS, 250 ng/μL tRNA, 200 μg/mL proteinase K] was added, incubating for 1 hour at 42°C. Afterwards, the DNA content was precipitated and then resuspended in 10 μL of TE buffer. After adding regular gel loading buffer for DNA, the samples were subjected to gel electrophoresis and subsequent analyses as described above.
2.5. Octamer transfer assays
These assays were performed by mixing 2.5 μL of the 147 bp DNA probe (diluted in buffer FCR), 0.5 μL oligonucleosomes (30 ng/μL), 0.6 μL 50 mM ATP (Roche, 1140965), 0.5 μL Gal4 buffer, 0.5 μL of varying concentrations of HMGB proteins (or HMG buffer), 3 μL of SWI/SNF (or SWI/SNF+CRC buffer or CRC buffer) and 7.4 μL remodeling buffer, incubating for 60 min at 30°C. Subsequently, a mix (0.7 μL) containing 750 ng salmon sperm DNA and 500 ng long oligonucleosomes was added, incubating for 20 additional minutes at 30°C. Next, the samples were subjected to gel electrophoresis and subsequent analyses as described above. The binding assay accompanying the octamer transfer assay in Figure 3 (Figure 3B) was performed as the other binding assays described above, with the exception that here this naked DNA probe (147 bp) does not correspond to a mock reconstituted probe, but just to the DNA probe diluted in buffer FCR.
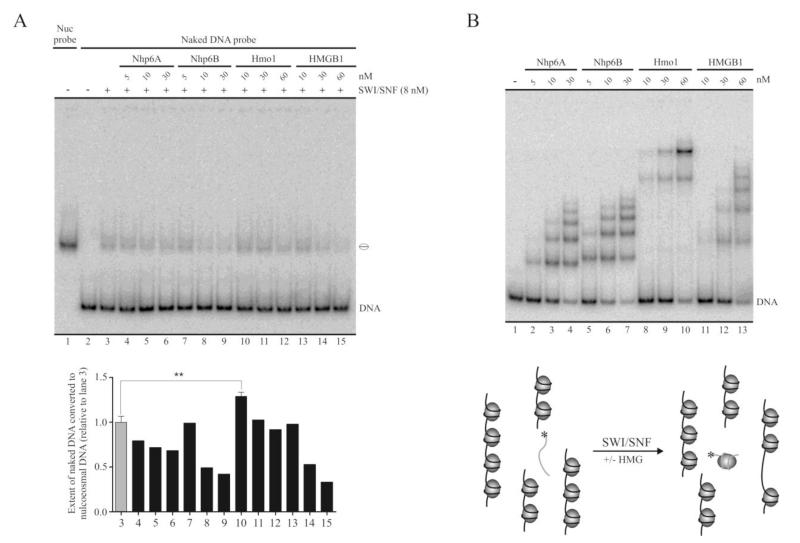
Fig. 3.
Hmo1 enhances the octamer transfer activity of SWI/SNF. A, The 147 bp DNA probe was incubated in the presence of oligonucleosomes. SWI/SNF and each of the HMG proteins were added to the reactions as depicted on the top of the gel picture. The samples were analyzed by electrophoresis in a non-denaturing polyacrylamide gel (5%, AA:Bis 40:1). See the text for a more detailed explanation of the assay. A schematic representation of this assay is shown on the bottom part of (B). The values in the graph correspond to the fraction of naked DNA probe converted to nucleosomal DNA, relative to lane 3 (octamer transfer activity of SWI/SNF alone). The average values for the reactions corresponding to lanes 3 and 10 were obtained from 4 independent assays. Error bars represent one standard deviation. Asterisks denote a statistically significant difference (**p < 0.01), as deducted from the t test. Migration of the mononucleosome is represented schematically at the right of the picture. Lane 1 corresponds to the same probe reconstituted as a mononucleosome before performing the assay. B, Binding assay analyzing the extent of interaction of the HMG proteins with the DNA probe, performed under the same conditions used in the octamer transfer assay shown in (A). The gel picture corresponds to electrophoresis in a non-denaturing polyacrylamide gel (5%, AA:Bis 40:1).
3. Results
In order to study the effect of yeast HMG proteins on ySWI/SNF remodeling activity in vitro, we used 32P-labelled probes of 147 or 216 bp, which were used as reconstituted mononucleosomes or naked DNA probes (mock reconstitution, see Materials and methods for details). The sequence of the 147 bp probe corresponds to the 601 nucleosome positioning sequence [27,28]. The 216 bp probe contains the 601 sequence, starting from one of the ends of this probe. Hence, after reconstitution of this probe, the histone octamer is positioned towards one of its ends, with linker DNA extending to only one side of the reconstituted nucleosome (Figure 1A). HMGB proteins, as well as the SWI/SNF complex, bind nonspecifically to DNA and nucleosomes. Thus, the non-labeled oligonucleosomes present with the probe [30 ng in each remodeling reaction (2 ng/μL)] work in the remodeling reactions as competitors for the interaction of SWI/SNF and the HMG proteins with the probe.
3.1. Binding properties of yeast HMG proteins
Bacterially expressed recombinant yNhp6A, yNhp6B, yHmo1 and human HMGB1 proteins, the latter used in the present study as a positive control for stimulation of nucleosome remodeling activity [19,22], were affinity purified (Supplementary Figure S2) and used throughout the studies presented in this work. Before performing nucleosome remodeling assays, we wanted to determine their strength of binding under our assay conditions. Incubation of these HMG proteins with the 216 bp DNA probe (mock reconstituted probe) and electrophoretic analysis on non-denaturing polyacrylamide gels results in several retardation bands (Figure 1C). A 50 nM concentration of any of the HMG proteins tested is almost saturating (considering saturation as the absence of free DNA). Johnson and co-workers, using a 98 bp DNA probe, have demonstrated that Nhp6A binds to DNA as a monomer in a stepwise manner [29]. The number of retardation bands would be dependent on probe length. To this respect, it has been determined that binding of Nhp6A to DNA extends over 11 bp [30]. These properties are consistent with a reduction in the number of retardation bands when using the 147 bp DNA probe (see below) instead of the 216 bp DNA probe. We observed a very similar binding pattern for both Nhp6A and B. Bindings patterns of Hmo1 and HMGB1 are similar to other obtained in previous studies, with fewer retardation bands than those observed for Nhp6A/B [12,19]. For all the HMG proteins tested, binding to the 216 bp nucleosome probe (Figure 1D) presented fewer retardation bands than those observed when using this probe as naked DNA. The interaction of the HMG proteins with this nucleosome probe appears to be dependent on the extranucleosomal DNA stretch (linker DNA) present in this nucleosome probe (69 bp) because, under our assay conditions and in the range of HMG protein concentrations used in our work, none of the HMG proteins were able to bind to the 147 bp nucleosome probe, which contains no linker DNA (Figure 1E, right panel). Although binding of the HMG proteins to the 147 bp DNA probe (Figure 1E, left panel) presented fewer retardation bands than those observed using the 216 bp DNA probe (Figure 1C), the binding strength was very similar between both DNA probes.
3.2. The yeast HMG proteins Nhp6A, Nhp6B and Hmo1 stimulate ySWI/SNF sliding activity
We next wanted to analyze whether the activity of ATP-dependent chromatin remodeling complexes is stimulated by Nhp6A/B and Hmo1, testing first their influence on SWI/SNF sliding activity with the use of the 216 bp nucleosome probe. As detailed in the Materials and methods section, the assays testing sliding activity incorporate the addition of a mix of cold DNA and nucleosomes after the incubation where remodeling proceeds and before the electrophoretic analysis. This mix of cold DNA and nucleosomes compete SWI/SNF and the HMG proteins from the probe, allowing visualization of the remodeling pattern of the nucleosome probe instead of binding patterns of SWI/SNF and HMG proteins. The non-denaturing gel electrophoresis presented in Figure 2A shows that all these proteins are able to stimulate SWI/SNF sliding activity, reflected by the appearance of a faster migrating band of nucleosomal DNA, which corresponds to a slid nucleosome. This remodeled state denotes a nucleosome where all the histone-DNA interactions have been disrupted and then reestablished in a different translational position [4,31]. Under our assay conditions, a minimal fraction of the nucleosome was remodeled (slid) in the presence of SWI/SNF alone (Figure 2A, lane 3). In contrast, a stronger sliding activity was observed in the presence of SWI/SNF plus any of the HMG proteins tested, at concentrations of 50 nM and higher (Figure 2A, compare lane 3 to lanes 5, 6, 9, 10, 13, 14, 17 and 18). In this assay there was no effect of the HMG proteins on the nucleosome probe in the absence of SWI/SNF, at concentrations where these proteins strongly stimulated the sliding activity of the complex (Figure 2A, lanes 7, 11, 15 and 19).
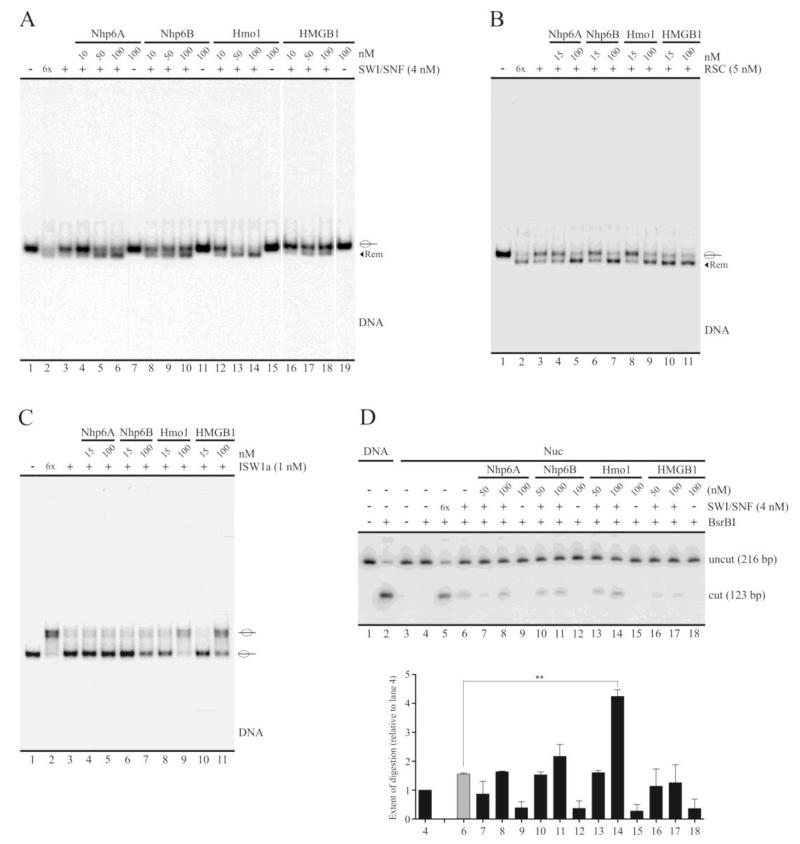
Fig. 2.
The yeast HMGB proteins Nhp6A/B and Hmo1 stimulate SWI/SNF sliding activity. A-C, Electrophoretic analyses of nucleosome remodeling using non-denaturing polyacrylamide gels (5%, AA:Bis 40:1). The assays tested the influence of the HMG proteins on sliding activity of SWI/SNF (A), RSC (B) and ISW1a (C) complexes. All the reactions of these assays used the 216 bp nucleosome probe. The results shown in each figure are representative of the corresponding assay performed at least three times. For all figures 6x stands for a remodeling complex concentration 6 times higher than in the rest of the reactions. Migrations of the different histone octamer translational positions in the nucleosome probe are indicated schematically at the right of the pictures. The term “Rem” stands for remodeled (slid) nucleosome. D, The influence of these HMG proteins on SWI/SNF remodeling activity was also studied by restriction enzyme accessibility assays, using the BsrBI site (see scheme in Figure 1A). After the remodeling reaction in the presence of BsrBI, the DNA content of the samples was purified and analyzed by electrophoresis in a non-denaturing polyacrylamide gel (5%, AA:Bis 40:1). The graph at the bottom of this figure corresponds to a quantification of the percentage of digested probe by BsrBI under each condition (relative to lane 4, which corresponds to digestion of nucleosomal DNA in the absence of SWI/SNF and HMG proteins). Digestion percentages were averaged from two independent assays. The digestion extent corresponding to lane 5 was not included in the graph as it represents an excess (6x) of SWI/SNF complex. Error bars represent one standard deviation. Asterisks denote statistically significant differences (**p < 0.01), as deducted from the t test. The assay used the 216 bp probe, at the form of naked DNA (lanes 1-2) or reconstituted mononucleosome (lanes 3-18).
We also tested whether sliding activity of the yeast RSC and ISW1a chromatin remodeling complexes was affected by these HMG proteins. The yeast HMG proteins stimulated RSC sliding activity, with HMGB1 reproducibly stimulating sliding activity from slightly lower concentrations than its yeast counterparts (Figure 2B). Interestingly, we observed stimulation of ISW1a sliding activity by only Hmo1 and HMGB1 proteins, but not by Nhp6A and Nhp6B (Figure 2C). Several additional concentrations of Nhp6A and Nhp6B were also assayed, ranging from 5 nM to 100 nM, with the same result of no stimulation of ISW1a sliding activity (data not shown).
3.3. Differential stimulation of ySWI/SNF activity by the yeast HMG proteins
We next tested the influence of the HMG proteins under study on SWI/SNF-mediated exposure of nucleosomal DNA during the remodeling reaction through a restriction enzyme accessibility assay, using the enzyme BsrBI. The recognition site for this enzyme is located in the nucleosomal DNA portion of the 216 bp nucleosome probe, at 25 bp from one of the nucleosome edges (Figure 1A). With the exception of Hmo1, we detected no enhancement of SWI/SNF-mediated exposure of nucleosomal DNA during the remodeling reaction (Figure 2D). A similar result was obtained by testing a different position in the nucleosomal DNA portion of this probe, using the restriction enzyme HhaI (Supplementary Figure S3). In some cases we even observed a reduction in the accessibility to the restriction site tested, as compared to the effect of SWI/SNF alone (e.g. compare lane 6 to lane 7 in Figure 2D). This effect could be derived from the nature of the assay itself rather than to an inhibition of SWI/SNF remodeling activity, since there is a strong binding of the HMG proteins to the nucleosome probe at the concentrations tested for these proteins (Figure 1D) and this binding could be hindering the access to the recognition site of the restriction enzyme. In fact, all the HMG proteins reduced BsrBI digestion in the absence of SWI/SNF (Figure 2D, compare lane 4 to lanes 9, 12, 15 and 18). Inhibition of restriction enzyme digestion by Nhp6A has been previously observed in other studies [20,32]. Nevertheless, Hmo1 enhanced the SWI/SNF-mediated exposure of nucleosomal DNA at a concentration where its binding strength is very similar to that of Nhp6A, Nhp6B and HMGB1 (100 nM), denoting a differential effect of Hmo1 on SWI/SNF activity which is not derived from the extent of binding to the nucleosome probe.
Previous studies have shown that the SWI/SNF complex, and a number of other ATP-dependent chromatin remodeling complexes, possess octamer transfer activity [33,34]. We analyzed whether the HMG proteins used in our study have an impact on the octamer transfer activity of SWI/SNF. This assay measures transfer of octamers from unlabeled oligonucleosomes onto the labeled probe. The procedure of this analysis consisted of incubation of the 147 bp probe in the form of naked DNA with 15 ng of oligonucleosomes, in the presence or absence of SWI/SNF and each of the HMG proteins. The extent of octamer transfer activity was measured by quantifying the fraction of naked DNA probe converted to nucleosomal DNA, which is generated by the transfer of histone octamers from the oligonucleosomes to the probe (see a schematic representation of the assay in Figure 3). As in the case of the sliding assays, the octamer transfer assays also includes a step of incubation with a large amount of cold DNA and nucleosomes after the incubation where the remodeling reaction proceeds, in order to observe remodeling patterns instead of the binding patterns of the HMG proteins and SWI/SNF (see Materials and methods for details). In this analysis we found a small but statistically significant stimulation of SWI/SNF octamer transfer activity only in the presence of Hmo1, at the lowest point of the concentration range tested in the assay shown in Figure 3A (10 nM, lane 10). Further assays analyzing Hmo1 concentrations ranging from 5 to 30 nM confirmed this stimulation at 10-15 nM, under our assay conditions (Supplementary Figure S4). Hmo1 concentrations of 30 nM or higher did not stimulate octamer transfer and this activity was even reduced at the higher concentration points of the other HMG proteins (Figure 3A). This reduction could be related to an effect of reduced access of the DNA probe (in this case to the histone octamers being transferred from the oligonucleosomes) at the higher concentration points of the HMG proteins, since these concentrations correlate with a strong binding to the DNA probe, as determined in binding assays carried-out under the same conditions of the octamer transfer assay (Figure 3B). This interpretation is consistent with the effect observed in the restriction enzyme accessibility assays, of HMG binding to the nucleosome probe (see above). Nevertheless, it appears that this differential effect of Hmo1 on octamer transfer activity does not rely on its extent of binding to the DNA probe relative to the other HMG proteins tested, since this extent of binding that correlates with octamer transfer stimulation was also covered in the analysis performed for the other HMG proteins (Figure 3B, lanes 2, 5, 8 and 11).
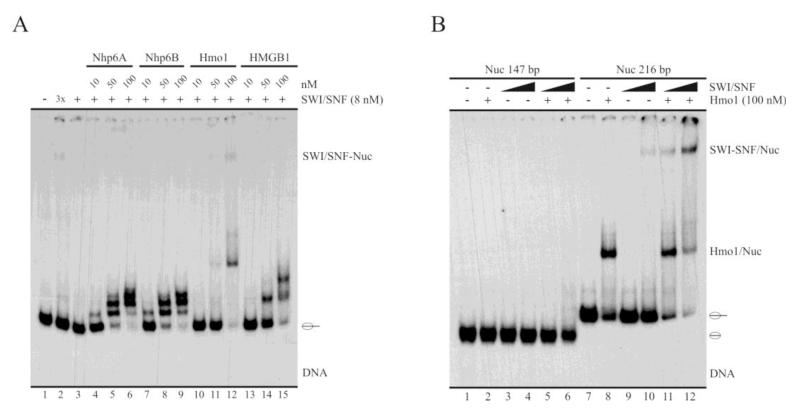
Nhp6 has been shown to facilitate binding of general and specific transcription factors to their cognate sequences ([8] and references therein). Similar properties have been observed for HMGB1. Additionally, this protein has been shown to enhance binding of the ACF chromatin remodeling complex to nucleosomes [19]. Considering these facts, we wanted to test whether the HMG proteins under study enhance binding of ySWI/SNF to nucleosomes. To ascertain this point we performed binding assays (EMSA) in the presence of SWI/SNF and increasing concentrations of each of the HMG proteins, using the 216 bp nucleosome probe. Remarkably, Hmo1 was the only protein able to enhance SWI/SNF binding to the nucleosome probe (Figure 4A, compare lanes 3 and 12). The concentrations of Hmo1 that enhance SWI/SNF binding to the nucleosome are consistent with those stimulating the sliding activity of this complex (compare Figures 2A and 4A). As Hmo1, and the other HMG proteins, shown no binding to a nucleosome probe containing no linker DNA (Figure 1E), we wanted to analyze whether Hmo1 stimulates SWI/SNF binding to this type of nucleosome. To do this, we assayed in parallel the binding of SWI/SNF to the 147 bp nucleosome probe (which contains no linker DNA) and to the 216 bp nucleosome probe (which contains linker DNA), in the absence or presence of Hmo1. As expected from the results of the previous binding assay, Hmo1 strongly stimulated binding of SWI/SNF to the 216 bp nucleosome probe (Figure 4B, compare lane 9 to 11 and lane 10 to 12). Conversely, we did not detect stimulation of SWI/SNF binding to the 147 bp nucleosome (Figure 4B, compare lane 3 to 5 and lane 4 to 6). To this respect, in the absence of Hmo1 the highest concentration of SWI/SNF gave a binding strength to the 147 bp nucleosome probe which was similar to that obtained for the 216 bp nucleosome probe using the lowest concentration of this complex, result that was more clearly observed by film overexposure (data not shown). This result suggests that Hmo1 requires its interaction with the nucleosome in order to enhance SWI/SNF binding.
Fig. 4.
Hmo1 stimulates SWI/SNF binding to the nucleosome. A-B, EMSA analyses comparing the effect of multiple concentrations of each HMG protein on SWI/SNF binding to the nucleosome. The 216 bp nucleosome probe was used in (A). The assay shown in (B) used the 147 bp (lanes 1-6) and 216 bp (lanes 7-12) nucleosome probes. The samples were analyzed by electrophoresis in non-denaturing polyacrylamide gels (3.5%, AA:Bis 60:1). Migration of the mononucleosome bound to SWI/SNF is indicated at right of the pictures, where migration of the nucleosome probe is indicated schematically. The results shown in figures (A) and (B) are representative of three independent assays in each case. 3× in figure (A) stands for a SWI/SNF concentration 3 times higher than in rest of the reactions where the complex is present. In figure (B), SWI/SNF concentrations used were 4 and 12 nM.
Taken together, the binding (EMSA), restriction enzyme accessibility and octamer transfer assays denote a differential effect of Hmo1 on SWI/SNF activity, as compared to the other HMG proteins tested in our study.
3.4. The Hmo1 effect on SWI/SNF is dependent on its C-terminal tail
To further study the influence of Hmo1 on SWI/SNF activity, we generated a deletion mutant of this protein which lacks the last 35 residues (Hmo1Δ, Figure 5A). Mammalian HMGB proteins (HMGB1-3) display a stretch of acidic residues in their C-terminal region, termed acidic tails [35]. In contrast to these HMGB proteins, the Hmo1 C-terminal tail consists of a basic, lysine-rich, region preceded by a short acidic stretch (Figure 1B and Supplementary Figure S1). It has been shown that the acidic tail of HMGB1 plays a role in its interaction with other proteins and chromatin [36]. It also modulates the interaction of HMGB1 with DNA and is required for stimulation of ATP-dependent nucleosome remodeling activity of the ACF complex [19]. On the other hand, it has been observed that the C-terminal tail of Hmo1 is required for the DNA bending activity of this protein [37,38]. Considering this evidence, we wanted to explore whether the C-terminal tail of Hmo1 plays a role in its binding properties (to DNA and nucleosomes) and in stimulation of SWI/SNF remodeling activity. As observed in Figure 5B, Hmo1Δ presents a weaker binding to DNA and nucleosomes than its WT counterpart. Moreover, Hmo1Δ displays a minimal or even non-detectable stimulation of SWI/SNF sliding activity at concentrations where WT Hmo1 strongly stimulates this activity (Figure 5C, compare lane 3 to 6 and 9). In addition, Hmo1Δ stimulates neither transient exposure of nucleosomal DNA, as assessed by a restriction enzyme accessibility assay performed with BsrBI (Supplementary Figure S5), nor octamer transfer activity (Supplementary Figure S4). Consistent with these results, Hmo1Δ does not stimulate SWI/SNF binding to the nucleosome (Figure 5D, compare lane 2 to 6 and 9). These results suggest that the stimulation of SWI/SNF activity by Hmo1 depends on the ability of this protein to interact with the nucleosome, a concept that is reinforced by the fact that wild type Hmo1 does not bind to a nucleosome with no linker DNA (Figure 1E) and concomitantly does not stimulate SWI/SNF binding to this type of nucleosomes (Figure 4B).
Fig. 5.
Hmo1’s stimulation of SWI/SNF remodeling activity and binding to the nucleosome is dependent on the C-terminal tail of this HMG protein. A, Schematic representation of WT Hmo1, its deletion mutant (Hmo1Δ), WT Nhp6A, WT Nhp6B and Nhp6 chimeric versions. Hmo1Δ corresponds to a C-terminal deletion mutant of Hmo1, lacking residues 212 to 246. Nhp6A-Ct and Nhp6B-Ct correspond to this 35 residues stretch fused to the C-terminal end of Nhp6A and B, respectively. The dark gray and soft gray squares represent the acidic and basic stretches (respectively) characteristic of Hmo1. B, Analysis of binding strength of Hmo1 and Hmo1Δ to the naked DNA and nucleosome probes, determined by electrophoresis in a non-denaturing polyacrylamide gel (5%, AA:Bis 40:1). C, Stimulation of SWI/SNF sliding activity by Hmo1 and Hmo1Δ. See legend of figure 2 for a general description of the assay. D, Analysis of stimulation of SWI/SNF binding to the nucleosome by Hmo1 and Hmo1Δ. E, Analysis of stimulation of SWI/SNF binding to the nucleosome by the chimeric forms of Nhp6A and Nhp6B, compared to Hmo1. See legend of figure 4 for a general description of the assays shown in (D) and (E). F, Stimulation of ISW1a sliding activity by WT and chimeric forms of Hmo1, Nhp6A and Nhp6B. See legend of figure 2 for a general description of the assay. The assays corresponding to figures B-D are representative of at least three independent assays. Migration of naked DNA is indicated at right of the pictures, where migration of the reconstituted mononucleosome is indicated schematically. The term “Rem” stands for remodeled (slid) nucleosome.
The studies performed with Hmo1Δ also indicate that the C-terminal tail of Hmo1 is required for the stimulatory effects exerted by this protein on SWI/SNF activity, raising the question of whether this region of Hmo1 would confer Nhp6A and Nhp6B with the stimulatory properties observed only for Hmo1 in our studies. To answer this question we generated chimeric versions of Nhp6A and Nhp6B proteins, fusing the last 35 residues of Hmo1 (C-terminal tail) to their C-terminal ends (see schematic representations in Figure 5A), and tested their ability to stimulate SWI/SNF binding to the 216 bp nucleosome probe. As observed in figure 5E, the Hmo1 C-terminal tail does not endow either Nhp6A or Nhp6B with the ability of stimulating SWI/SNF binding to the nucleosome. Consistently, the Hmo1 C-terminal tail is not sufficient to endow either Nhp6A or Nhp6B with the stimulatory effect exerted by Hmo1 on ISW1a sliding activity (Figure 5F). Still, no stimulation of ISW1a sliding activity was observed in the presence of Hmo1Δ (Figure 5F, compare lane 13 to 15), indicating that the C-terminal tail of this protein is also required for its stimulatory effect on ISW1a activity. To date, the ability to stimulate SWI/SNF sliding activity is not disrupted in these chimeric forms of Nhp6A and Nhp6B (Supplementary Figure S6).
4. Discussion
In this work we have demonstrated that the yeast HMGB-type proteins Nhp6A, Nhp6B and Hmo1 enhance the nucleosome remodeling activity of the yeast SWI/SNF complex. Our results indicate that all these proteins stimulate the sliding activity of this complex but only Hmo1 stimulates SWI/SNF octamer transfer activity and transient exposure of nucleosomal DNA, as well as binding of this complex to the nucleosome. The C-terminal tail of Hmo1 appears to be required for the effect of this HMG protein on SWI/SNF activity.
The enhancement of SWI/SNF remodeling activity by Nhp6 proteins observed in our in vitro studies is in agreement with evidence pointing to a connection between these HMG proteins and ySWI/SNF in vivo (see Introduction). Consistently, in vitro and in vivo studies performed by Szerlong et al. [20] revealed a functional connection between Nhp6 and the chromatin remodeling complex RSC. In the same context, it is known that SWI/SNF complexes of higher eukaryotes contain subunits harboring HMG domains, while there are no HMG domain-containing subunits in the yeast SWI/SNF complex [39,40]. Moreover, it has been shown that the presence of these HMG domains in subunits of SWI/SNF complexes of higher eukaryotes is required for different in vivo actions of these complexes [41,42]. These evidences, together with the stimulatory effects of Hmo1 on ySWI/SNF activity observed in our in vitro studies, suggest a relevant relationship between this HMG protein and ySWI/SNF in vivo.
It has been recently shown that HMGB1 by itself can alter the structure of a mononucleosome in vitro [43]. In our studies we did not observe any effect on nucleosome structure generated by the HMG proteins analyzed in the absence of a chromatin remodeling complex. In the case of similar experimental approaches used in the cited work, this discrepancy could be due to differences in our assay conditions with respect to those assays. For instance, our assays included the presence of non-labeled nucleosomes in addition to the nucleosome probe and the concentrations of the HMG proteins tested are significantly lower in our studies. Alteration of nucleosome structure has also been described for Nhp6 [32]. This work also used higher HMG protein concentrations than those used in our work. However, no effects on nucleosome sliding were observed in the presence of Nhp6 [32]. Taken together, our findings and the previous studies cited here suggest that, although HMG proteins might alter nucleosome structure by themselves, they are able to play their relevant role of facilitating ATP-dependent nucleosome remodeling at significantly lower concentrations than those required for a stand-alone effect.
Our study indicates that linker DNA is required for binding of the HMG proteins studied to the nucleosome. In this context, Formosa and colleagues have shown that affinity of Nhp6 for mononucleosomes is not affected by the presence of linker DNA [44]. Again, this discrepancy could rely on the use of different assay conditions and on the concentration range analyzed in our work. Consistent with our results, it has been shown that binding of HMGB1 to nucleosomes depends on the presence of linker DNA [19]. Additionally, we found that Hmo1 does not stimulate SWI/SNF binding to the nucleosome in the absence of linker DNA.
Remarkably, different assays determined a differential effect of Hmo1 on SWI/SNF activity. We also found a differential effect of Hmo1 on the sliding activity of ISW1a, compared to Nhp6A and Nhp6B. These differential effects should not rely on differences in nucleosome binding strength of the HMG proteins studied, as our assays included concentrations yielding similar binding levels. Our results indicate that the C-terminal tail of Hmo1 is required for its stimulation of SWI/SNF remodeling activity and suggest that the absence of SWI/SNF stimulation by Hmo1 lacking its C-terminal tail (Hmo1Δ) is due to a lower affinity for DNA and nucleosomes of this deletion mutant. Interestingly, it has been shown that stimulation of the ACF complex by HMGB1 is dependent on the C-terminal tail of this protein. However, HMGB1 lacking this region displays a significantly stronger binding to DNA and nucleosomes [19]. This differential feature might rely on the essentially opposite characteristics of HMGB1 and Hmo1 C-terminal tails, which mainly correspond to an acidic tail in the case of HMGB1 and to a basic tail in the case of Hmo1. In this respect, Grove and colleagues demonstrated that Hmo1 lacking its C-terminal tail has reduced DNA bending activity and, to some extent, a reduction of DNA binding can also be observed from their results [37,38].
Taking in account the current knowledge of the mechanisms of ATP-dependent chromatin remodeling [1,45], the mechanism proposed for stimulation of nucleosome remodeling by HMG proteins (facilitation of remodeler action through DNA bending at its entry/exit sites on the nucleosome) [6,19] and the results of our study, we speculate that the DNA bending properties of the HMG proteins analyzed in this work are the common denominator where stimulation of sliding activity relies on. On the other hand, the differential effects exerted by Hmo1 on SWI/SNF activity would rely, at least in part, on its ability to facilitate the binding of this complex to the nucleosome. The C-terminal tail of Hmo1 is required for its ability to stimulate ATP-dependent chromatin remodeling, but this region of Hmo1 is not sufficient to endow Nhp6A nor Nhp6B with the differential stimulatory properties observed for Hmo1. Therefore, the Hmo1 C-terminal tail would not be acting as a moiety for recruiting SWI/SNF to the nucleosomes [46-48] and the presence of two HMG boxes in Hmo1 appears to be also required for its differential stimulatory effects. Thus, we favor a role of the C-terminal tail of Hmo1 relying on an influence on the conformation of the HMG boxes of this protein. In this regard, it has been proposed that the C-terminal tail of Hmo1 has an influence on the conformation of the protein and concurrently on its DNA bending properties [37], suggesting that Hmo1 lacking its C-terminal tail might have a conformation with a reduced ability to facilitate the binding of a remodeling complex to the nucleosome and, eventually, a reduced ability to facilitate remodeling activity.
Our results and the aspects discussed above support the concept that, although stimulation of sliding activity appears to be a general feature of HMG proteins, a number of these proteins would be specialized in stimulation of particular remodeling complexes and/or exert a differential stimulation of the biochemical outcomes of ATP-dependent nucleosome remodeling activity.
Supplementary Material
Highlights.
-
-
Nhp6A, Nhp6B and Hmo1 stimulate the sliding activity of the yeast SWI/SNF complex.
-
-
Hmo1 differentially stimulates SWI/SNF and ISW1a activity, as compared to Nhp6A/B.
-
-
The C-terminal tail of Hmo1 is required for its stimulatory effect on SWI/SNF action.
-
-
Hmo1 C-terminal tail does not endow Nhp6 with the stimulatory properties of Hmo1.
Acknowledgements
This work was supported by grant CONICYT, FONDECYT/Regular 1130818 to JLG, DIUC 211.037.014-1.0 to JLG, NIH Grant R37 GM047867 and support from the Stowers Institute for Medical Research to JLW. MIH was supported by a CONICYT scholarship for Ph.D. students and CONICYT scholarships AT-24100076, P-75110059.
Abbreviations
- SWI/SNF
switching defective/sucrose non-fermenting
- HMG
high mobility group
- Nhp6A/B
non-histone protein 6A/B
- Hmo1
high mobility group family 1
- HMGB1
high mobility group box 1 protein
- EMSA
electrophoretic mobility shift assay
- RSC
remodels the structure of chromatin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- [2].Chandy M, Gutiérrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryotic Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chatterjee N, Sinha D, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 2011;39:8378–8391. doi: 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gutiérrez JL, Chandy M, Carrozza MJ, Workman JL. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 2007;26:730–740. doi: 10.1038/sj.emboj.7601524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reeves R. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Travers AA. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 2003;4:131–136. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- [8].Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010;1799:175–180. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Paull TT, Carey M, Johnson RC. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- [10].Ghaemmaghami S, Huh W, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- [11].Lu J, Kobayashi R, Brill SJ. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 1996;271:33678–33685. doi: 10.1074/jbc.271.52.33678. [DOI] [PubMed] [Google Scholar]
- [12].Kamau E, Bauerle KT, Grove A. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J. Biol. Chem. 2004;279:55234–55240. doi: 10.1074/jbc.M409459200. [DOI] [PubMed] [Google Scholar]
- [13].Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kasahara K, Ohyama Y, Kokubo T. Hmo1 directs pre-initiation complex assembly to an appropriate site on its target gene promoters by masking a nucleosome-free region. Nucleic Acids Res. 2011;39:4136–4150. doi: 10.1093/nar/gkq1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moreira JM, Holmberg S. Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J. 2000;19:6804–6813. doi: 10.1093/emboj/19.24.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol. Cell. Biol. 2004;24:8312–8321. doi: 10.1128/MCB.24.18.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kassavetis GA, Steiner DF. Nhp6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo. J. Biol. Chem. 2006;281:7445–7451. doi: 10.1074/jbc.M512810200. [DOI] [PubMed] [Google Scholar]
- [18].Dowell NL, Sperling AS, Mason MJ, Johnson RC. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev. 2010;24:2031–2042. doi: 10.1101/gad.1948910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bonaldi T, Längst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Patenge N, Elkin SK, Oettinger MA. ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J. Biol. Chem. 2004;279:35360–35367. doi: 10.1074/jbc.M405790200. [DOI] [PubMed] [Google Scholar]
- [22].Ugrinova I, Pashev IG, Pasheva EA. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry. 2009;48:6502–6507. doi: 10.1021/bi9004304. [DOI] [PubMed] [Google Scholar]
- [23].Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- [27].Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- [28].Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- [29].Yen YM, Wong B, Johnson RC. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem. 1998;273:4424–4435. doi: 10.1074/jbc.273.8.4424. [DOI] [PubMed] [Google Scholar]
- [30].Allain FH, Yen YM, Masse JE, Schultze P, Dieckmann T, Johnson RC, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- [32].Rhoades AR, Ruone S, Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- [34].Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- [36].Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochem. Soc. Trans. 2012;40:341–346. doi: 10.1042/BST20120014. [DOI] [PubMed] [Google Scholar]
- [37].Bauerle KT, Kamau E, Grove A. Interactions between N- and C-terminal domains of the Saccharomyces cerevisiae high-mobility group protein HMO1 are required for DNA bending. Biochemistry. 2006;45:3635–3645. doi: 10.1021/bi0522798. [DOI] [PubMed] [Google Scholar]
- [38].Xiao L, Williams AM, Grove A. The C-terminal domain of yeast high mobility group protein HMO1 mediates lateral protein accretion and in-phase DNA bending. Biochemistry. 2010;49:4051–4059. doi: 10.1021/bi1003603. [DOI] [PubMed] [Google Scholar]
- [39].Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- [41].Papoulas O, Daubresse G, Armstrong JA, Jin J, Scott MP, Tamkun JW. The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5728–5733. doi: 10.1073/pnas.091533398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- [43].Joshi SR, Sarpong YC, Peterson RC, Scovell WM. Nucleosome dynamics: HMGB1 relaxes canonical nucleosome structure to facilitate estrogen receptor binding. Nucleic Acids Res. 2012;40:10161–10171. doi: 10.1093/nar/gks815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ruone S, Rhoades AR, Formosa T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 2003;278:45288–45295. doi: 10.1074/jbc.M307291200. [DOI] [PubMed] [Google Scholar]
- [45].Längst G, Becker PB. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta. 2004;1677:58–63. doi: 10.1016/j.bbaexp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- [46].Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- [48].Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.