Abstract
It has been well established that an aligned matrix provides structural and signaling cues to guide cell polarization and cell fate decision. However, the modulation role of cells in matrix remodeling and the feedforward effect on stem cell differentiation have not been studied extensively. In this study, we report on the concerted changes of human decidua parietalis placental stem cells (hdpPSCs) and the highly ordered collagen fibril matrix in response to cell-matrix interaction. With high-resolution imaging, we found the hdpPSCs interacted with the matrix by deforming the cell shape, harvesting the nearby collagen fibrils, and reorganizing the fibrils around the cell body to transform a 2D matrix to a localized 3D matrix. Such a unique 3D matrix prompted high expression of β-1 integrin around the cell body that mediates and facilitates the stem cell differentiation toward neural cells. The study offers insights into the coordinated, dynamic changes at the cell-matrix interface and elucidates cell modulation of its matrix to establish structural and biochemical cues for effective cell growth and differentiation.
Keywords: Collagen, Fibril alignment, Cell polarization, Neural differentiation, 3D matrix, Atomic force microscopy (AFM)
1. Introduction
Cell-matrix interaction plays a critical role in various physiological and pathological processes. Collagen type I represents one of the most abundant structural proteins that form the extracellular matrix (ECM) of vertebrates. It provides the mechanical stability for tissues and serves as a functional environment for cells [1,2]. Fibrillar collagen type I is typically aligned in parallel arrays in connective tissues, either locally or extensively [3-5]. Thus, it is attractive to construct aligned collagen fibril arrays to mimic the native tissue environment for in vitro studies. It has been reported that collagen molecules assemble into arrays of ordered fibrils when guided by the crystalline orientation of mica substrates [6,7]. The aligned matrices have been used to study β-1 integrin mediated cell adhesion, polarization, and migration. Regardless of cell type, it was observed that cells expressing α2β1-integrin are capable of dislocating the highly ordered collagen fibrils and depositing the loose fibrils around cell [4,8-10]. While it has been suggested that the 3D-like collagen matrix might induce specific signaling for cell development [4], it has not been studied explicitly.
In this study, we report on the two-way regulation between human decidua parietalis placental stem cells (hdpPSCs) and highly ordered collagen fibril array by directly monitoring the cell-matrix interaction via high-resolution AFM imaging. Since hdpPSCs are robust and easily derived, they are preferable for in vitro studies and clinical therapies [11,12]. In our previous study, we found that these cells are capable of neural differentiation on a collagen-coated substrate in a non-selective medium [13]. In this study, we probed the coordinated, dynamic cell-matrix interaction to reveal the matrix prompted cell polarization and cell prompted local 3D matrix formation. The concerted changes were found to accelerate neural differentiation of hdpPSCs.
2. Materials and Methods
2.1. Collagen Matrix Preparation
Collagen type I solution (9.82 mg/ml) derived from rat-tail tendon was purchased from BD Biosciences. The solution in 0.1% acetic acid was diluted to 35 μg/ml in 10× PBS buffer containing 1N NaOH to adjust the pH to 9 for effective collagen fibril assembly [13,14]. 400 mM KCl was added to promote collagen alignment on mica [7]. A drop of 30 μl collagen solution was cast on a freshly cleaved surface of a Muscovite mica disk (Grade V1, Ted Pella, Inc., Redding CA), and incubated at 37 °C overnight to achieve collagen gelation. After rinsed with PBS, the samples were subjected to AFM imaging at high resolution or serving as a matrix for cell culture. Blank plastic substrate, cut from a cell culture dish, was used as a control. Electro-spun (E-spun) collagen fibers were also prepared (see Supplementary Data), and were used in comparative studies to examine the cell response to 2D vs. 3D matrices.
2.2. Cell Culture
Undifferentiated hdpPSCs (passage 2-3) were obtained from Dr. Strakova's lab and propagated in a self-renewal media according to their pre-defined protocol [12]. For differentiation experiments, the undifferentiated cells at passage 3-6 were plated at a density of 6000 cells/ cm2 on various matrices in non-selective, spontaneous differentiation medium (DMEM+ 10% FBS+ 1% non-essential amino acids).
2.3. Atomic Force Microscopic (AFM) Imaging
AFM imaging was performed using a multimode Nanoscope IIIa AFM (Veeco Metrology, Santa Barbara CA), equipped with a J-scanner. Amplitude images of the aligned collagen matrices and the hdpPSCs were recorded in 1× PBS buffer in fluid tapping mode using Si3N4 tips at a resonance frequency of 8-10 kHz. hdpPSCs were gently fixed with 4% paraformaldehyde or ice-cold methanol for 3 min.
2.4. Immunofluorescence Staining
A Nikon U-2000 microscope was used to collect the immunofluorescent images. The expression of F-actin, Collagen-I and β1-integrin were tracked at 6-32 h post-plating. The expression of β3-tublin and Neu-N were examined in cells at Day 1 and Day 5 of differentiation. The primary antibodies used in this study include mouse anti-F-actin (Millipore, Temecula CA, 1:100 dilution), rabbit anti-collagen-I (Abcam, Cambridge, MA 1:100 dilution), rabbit anti-β3-Tublin (Tuj1, Abcam, 1:200 dilution), mouse anti-NeuN (Millipore, 1:100 dilution) and rabbit anti-β1-integrin (Santa Cruz Biotechnology, 1:100 dilution). Secondary antibodies were purchased from Invitrogen (Carlsbad, CA) and used at a dilution of 1:200.
The images were quantitatively analyzed by ImageJ (NIH). In determining the percentage of positively stained cells, we set the threshold at 50% of the highest staining intensity across the samples. Cell length-to-width ratio was analyzed to reveal cell polarization. Since cell width varies along an elongated cell, average cell width was evaluated from the ratio of cell area and cell length.
3. Results
3.1. Collagen fibril alignment and matrix induced cell polarization/differentiation
As shown in the AFM image in Figure 1a, collagen molecules are assembled into collagen fibrils of 6±1 nm in diameter and 3.4±0.4 μm in length. They exhibit unidirectional alignment and uniform coverage across the mica substrate. The characteristic collagen D-period of 68±1 nm is highlighted in the inset of Figure 1A, and is consistent with our previous observations [13,14]. A blank plastic substrate is rather smooth with a maximum height difference of 4 nm, despite the observed line scratches (Figure 1B). We monitored hdpPSC elongation in 2-32 h post-plating on collagen/mica and plastic, and the changes in length-to-width ratio, derived from optical images (Figs. 1C, D & S1), were summarized in Fig. 1G. Within 2 h post-plating of hdpPSCs on a collage/mica matrix, many cells have started to elongate. All cells are bipolar after 18 h post-plating (Fig. 1C). However, cells on plastic appear non-polar in shape (Fig. 1D). With extended cell culture to Day 5, while cells on plastic show random shapes (Fig. 1F), most cells on collagen-mica appear with long filaments extending from a small cell body (Fig. 1E), resembling neural progenitors [15,16]. Thus, we examined the neural differentiation profile of hdpPSCs. The staining in Figs. 1E & F was against βIII-Tublin (green), a neuron-specific isoform expressed by immature neurons and outgrowth neurites, and NeuN (red), expressed by neural progenitors and mature neurons [17,18]. Results from quantitative analysis of the staining data (Figs. 1H and 1I) indicate that βIII-Tublin positive cells are predominant on collagen/mica even on Day 1 of differentiation. These cells are mostly NeuN negative and express βIII-Tublin at a high level. By Day 5 of differentiation, NeuN positive cells become dominant (69%). Importantly, these cells express NeuN at a substantially higher level and express βIII-Tublin at a relatively lower level, suggesting successful differentiation and maturation of the polarized hdpPSCs toward neurons [18]. On the contrary, nominal cells on plastic are NeuN positive by Day 5 of differentiation even though the level of βIII-Tublin expression has increased. For comparison, we also prepared aligned collagen fibers by electrospinning method. These fibers are much thicker (∼ 1 μm in diameter), and similarly promote cell polarization as collagen fibrils do on mica. However by Day 5 of differentiation, the cells on E-spun fibers express NeuN at a significantly lower level despite a higher level expression of βIII-Tublin. The results from the current study corroborate a dramatically faster neuronal differentiation and maturation process on collagen/mica substrates.
Figure 1.
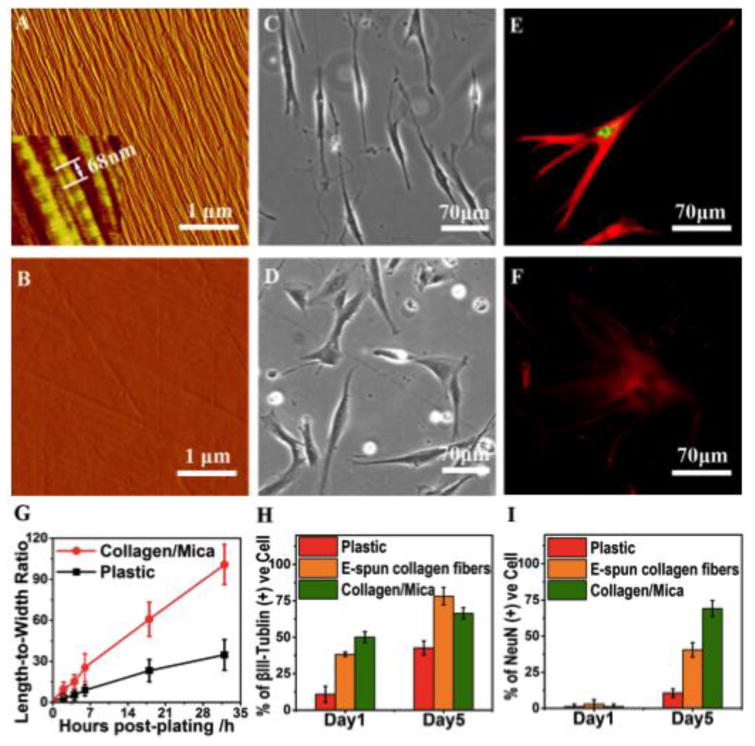
hdpPSCs cultured on unidirectionally aligned collagen fibrils on muscovite mica. (A) AFM image showing highly aligned collagen fibrils on mica. The inset (450 × 450 nm2) highlights the characteristic collagen D-period of 68 ± 1nm. (B) AFM image of a blank plastic substrate. (C,D) Bright-field images of hdpPSCs cultured on collagen/mica (C) and plastic (D) at 18 h post-plating showing distinct cell shapes and alignments. (E,F) Immunofluorescent images of typical cells on Day 5 of differentiation on collagen/mica (E) and plastic (F), staining against βIII-Tublin (red) and NeuN (green). (G) Change of length-to-width ratio with time for cells cultured on collagen/mica and plastic. (H,I) ImageJ quantification of the percentage of positively staining cells for βIII-Tublin (H) and NeuN (I) on Day 1 and Day 5 of differentiation on plastic, E-spun collagen fibers and collagen/mica.
3.2. Examination of cell-collagen fibril interaction at high-resolution
AFM images in Fig. 2 and 3 provide insights into the cell-collagen fibril interaction through the extended lamellipodia and filopodia. Cells use sheet-like lamellipodia and finger-like filopodia to probe the environment for cues, function as pathfinders, and play critical roles in cell adhesion and migration [19-21]. On collagen/mica substrates and before a cell is fully differentiated, long high-density filopodia extend from both ends of a polarized cell preferentially along the direction of collagen fibril alignment (Fig. 2A). In contrast, cells on a plastic substrate show a number of filopodia at the periphery of a cell in random directions (Figs. 2B & S2). It is known that filopodia and lamellipodia bind to collagen through collagen-β1-integrin specific interaction to form focal adhesion complexes, which assist in targeting the location of actin filaments and signaling components [19,21]. The aligned collagen fibrils are expected to coax the deposition of focal adhesion complexes that cause the cytoskeleton structure to stretch [3,5,22], leading to cell polarization and migration along the collagen fibrils (see Fig. 4).
Figure 2.
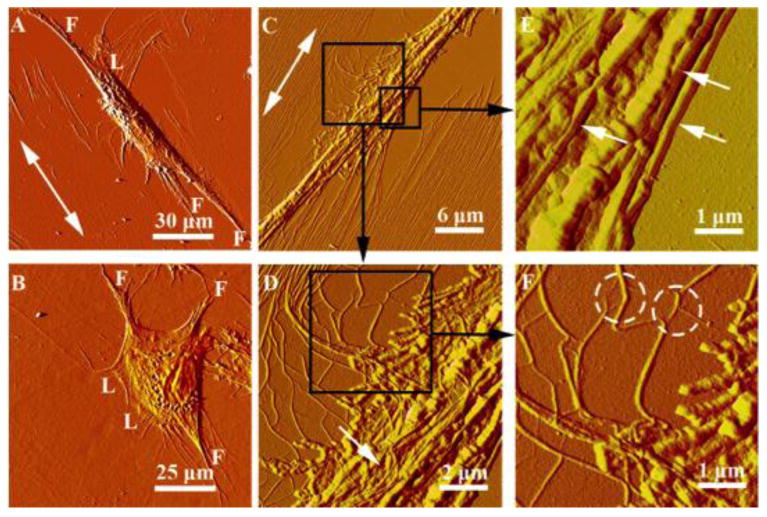
AFM images illustrating the cell-matrix interaction. (A,B) Individual hdpPSCs cultured on collagen/mica (A) and plastic (B) at 18 h post-plating. “F” and “L” represent filopodia and lamellipodia, respectively. (C-F) Sequential images captured at 30 × 30 μm2 (C), 10 ×10 μm2 (D) and 5 × 5 μm2 (E,F), illustrating the interaction between an elongated hdpPSC and the nearby highly aligned collagen fibrils alongside of the cell. The double arrows indicate the collagen fibril orientation. The black frames in (C) and (D) identify the corresponding areas shown in (D), (E) and (F). The white arrows in (E) highlight the collagen fibers (with characteristic D-period) on top and alongside of the cell. Splitting and separation of fibrils from the collagen network are indicated by the dashed circles in (F).
Figure 3.
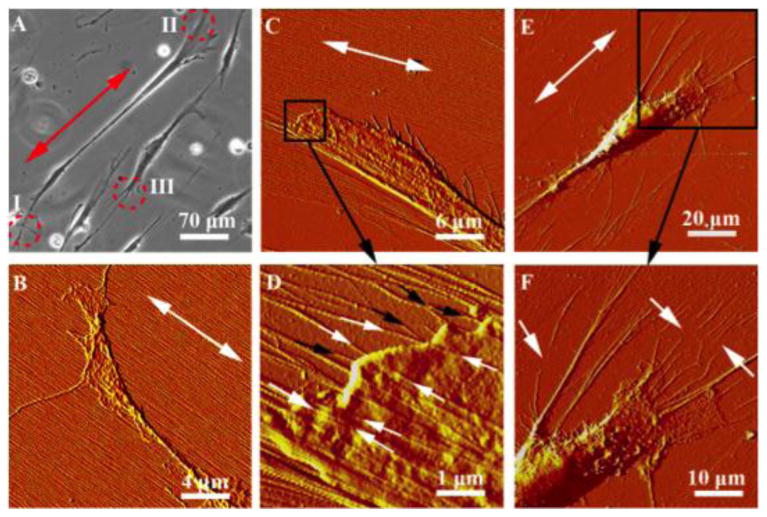
(A) Optical image of hdpPSCs aligned on collagen/mica at 18 h post-plating. “I”, “II”, “III” together with the red circles label the typical cell regions shown in the AFM images in (B), (C) and (E), repsectively. The double arrows highlight the collagen fibril orientation. (B) The axon terminal of a cell with filopodia and lamellipodia stretching on the intact collagen fibril matrix. (C) The dendrite terminal of a cell that evidently pulls the collagen fibrils in the vicinity. A zoom-in image of the framed region is shown in (D). The white and black arrows indicate fibrils stretching from the substrate to the top and the bottom of the cell, respectively. (E) The dendrite terminal of a different cell that extends long filaments to collect, bundle and haul the collagen fibers. Ruptured fibers are shown in the zoom-in image in (F), as pointed by the white arrows.
Figure 4.
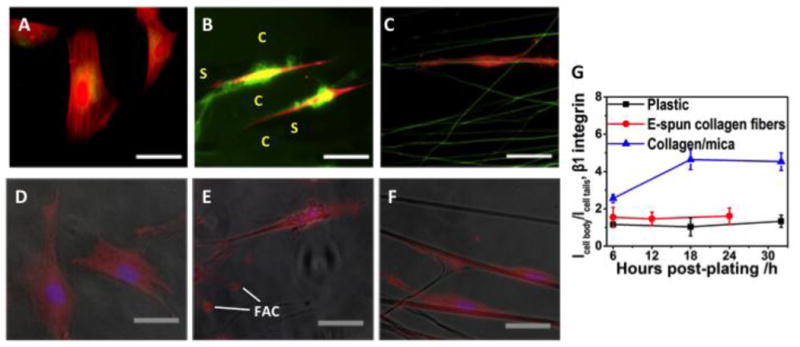
(A-C) Immunofluorescent images of typical hdpPSCs at 18 h post-plating on plastic (A), collagen/mica (B) and E-spun collagen fibers (C), staining against F-actin (red) and collagen (green). Bar size: 40 μm. (D-F) Overlay of optical and immunofluorescent images of hdpPSCs at 18 h post-plating on plastic (D), collagen/mica (E) and E-spun collagen fibers (F) staining against β-1 integrin (red) and DAPI (blue). Bar size: 30 μm. (G) ImageJ quantification of the ratio of β-1 integrin staining intensity at the cell body and at the cell tails, Icell body / Icell tails, for cells on various matrices.
As shown in Fig. 2C, when a cell elongates along the collagen fibrils, fibril alignment near the cell is disrupted. The originally orderly assembled and densely packed fibrils are pulled toward the cell (Fig. 2D), causing the fibrils to split and separate from the well-organized fibril network (white circles in Figure 2F). This is consistent with others' reports that cells are able to dislocate and peel collagen fibrils from mica substrates [4,8,9], suggesting a weaker interaction between collagen and mica than between collagen and a cell. Presumably, when a cell deforms during polarization, the cytoskeleton exerts a pulling force to the collagen matrix in the direction perpendicular to cell polarization while the cell membrane withholds the collagen fibrils through the collagen-integrin interaction. The pulling force is sufficiently high to drag the collagen fibrils toward the cell, disrupt the fibril network, and even rupture the fibrils (Fig. 2C). The rupture of collagen fibrils is assisted by cell-secreted enzymes to degrade the matrix [5,22,23]. These fibrils are accumulated in the vicinity of the cell at high local density and re-assembled into thicker fibers (22±3 nm in diameter), which are readily distinguishable from cell filaments by the characteristic D-period (Figure 2E). These thick fibers are relatively rigid and lay barriers alongside of the cell to resist the cytoplasm mobility perpendicular to the fiber orientation. Such a restriction promotes the unidirectional cellular traction and further enhances cell polarization. Some collagen fibers appear on top of the cell, likely due to cell translocation and mobility of a cell membrane that carries the attached fibers to various destinations. Another interesting observation is that, hdpPSCs were reproducibly observed to take different route in harvesting the collagen fibrils from left and right (Figure 2C and 3C). On the left, flexible and curly collagen fibrils were peeled off and separated from the fibril network; on the right, straight and stretched fibrils were dragged toward the cell at an angle. While further study is needed, we attribute the phenomenon to the right-handed helical structure of collagen fibrils that form a continuous network and appear asymmetric on the left and the right of a cell.
After 32 h of differentiation, many hdpPSCs showed typical neural progenitor cell shape, revealing the long axon shaft, a small cell body, and the dendrites (Fig. 3A). At the two tails, cells show distinct interaction pattern with collagen fibrils. The axon terminal (“I” in Fig. 3A) appeared thin and flat, showing extensive span of filopodia and lamellipodia on finely aligned undisturbed collagen fibrils (Fig. 3B). This is consistent with the pathfinder function of filopodia and lamellipodia in this region. On the other hand, the dendrites (“II” and “III” in Fig. 3A) showed profuse branches and appeared with distorted collagen fibrils (Figure 3C-F). When a cell contracts, cellular cytoskeleton generates shearing and traction forces against the collagen fibrils [4,5,10]. In this region, the dendrites hold the fibrils tightly, allowing the cell to haul the fibrils when it retracts. Analogous to previous observation, these fibrils are seen deposited either on top of the cell or beneath the cell, as indicated by white and black arrows in Fig. 3D. The collagen fibrils are identified by the characteristic D-period and the unremitting stretch from the substrate to the cell. Ruptured fibrils are frequently seen (white arrows in Fig. 3F) due to the high pulling force and the enzyme assisted collagen degradation. This leaves behind the less adhesive, bare mica substrate, which favors further formation of dendrites in the region [23].
3.3. Formation of local 3D matrix and its role in hdpPSCs differentiation
Our results imply that while the aligned collagen matrix prompted cell polarization, the cell prompted collagen fibril reorganization surrounding the cell, leading to the formation of 3D-like matrix. To provide further evidence, we collected immunofluorescent images of hdpPSCs on plastic, collagen/mica, and E-spun collagen fiber matrices, respectively, stained against F-actin (red) and collagen (green) (Fig. 4 A-C). With the staining on plastic to define the background level of collagen expression, staining of E-spun collagen fibers became apparent. Since the aligned collagen fibrils on mica were 6±1 nm in diameter, and they covered the entire surface, they showed uniform green fluorescence against the dark substrate where the fibrils were absent (“C” and “S” regions, respectively). Evidently, collagen expression was remarkably high around the cell body, exhibiting local 3D fibril network. Such a unique 3D-like matrix was absent around cells on E-spun fiber matrix, although cells similarly polarized and aligned along the collagen fibers. This is likely because the E-spun fibers are too bulky and rigid for the cells to manage.
In our previous study, we attributed the neural differentiation of stem cells on collagen-coated substrates to a β-1 integrin mediated β-catenin signal pathway [13]. The 3D matrix is expected to up-regulate β-1 integrin expression level of cells on collagen/mica, leading to accelerated hdpPSC differentiation, as shown in Fig. 1H&I. The comparative immunofluorescent analysis shown in Fig. 4 D-G confirmed the augmentation of β-1 integrin expression on collagen/mica. The expression level was particularly high around the cell body where the 3D collagen fibril network was apparently present (Fig. 4E). The high integrin expression at the focal adhesion complexes (labeled with “FAC”) is also notable. We further quantified the ratio of β-1 integrin staining intensity at the cell body and at the cell tails for cells on various matrices. As shown in Fig. 4G, on collagen/mica, the ratio jumped from 2.5±0.2 at 6 h post-plating to 4.6±0.5 at 18 h and remained essentially the same until 32 h. On the other hand, the ratio for cells on E-spun collagen fibers and plastic was constant over time (1.5 ±0.6 and 1.6 ±0.4, respectively). The lack of change between 18 h and 32 h on collagen/mica is consistent with the AFM observations, which indicate that cells start to peel and reorganize collagen fibrils as early as 2 h post-plating, causing dramatic changes in early hours and essentially unchanged after 18 h.
4. Discussion
A cell responds to matrix compositions and physical properties by modulating the amount and distribution of matrix binding membrane proteins and reshaping its cytoskeleton. They play crucial roles in cell signal transduction and cell development [3,11,20]. Aligned matrices provide unique structural and signaling cues to induce cell polarization and modulate cell fate decision. Previous studies have investigated the interaction between cells and collagen fibrils on mica with a focus on understanding the β-1 integrin mediated cell adhesion, polarization, and migration [4,5,10]. To our knowledge, the current study is the first to reveal the unique two-way regulation between hdpPSCs and collagen fibrils, catalyzing the neural differentiation. Considering all results, we propose the following mechanism. The arrayed collagen fibrils guide the deposition of focal adhesion complexes via collagen-β-1 integrin specific interaction, inducing cell polarization, at as early as 2 h post-plating. When a cell elongates, the traction force perpendicular to fibril alignment causes the cell to drag the fibrils alongside of the cell. These fibrils are bundled to thicker fibers, deposited either along the cell to further assist cell polarization or on top of the cell, through membrane lateral movement. While the filopodia and lammelipoida at the axon terminal protrude on collagen matrix and direct the cell growth, dendrite extensions haul the collagen fibrils upon cellular contraction. When the cell retracts, collagen fibrils are ruptured and broken up with the fibril matrix [22]. Fibrils adhere to the cell, move with the membrane, and accumulate around the cell body, transforming a 2D matrix into a 3D-like matrix. The cell keeps exploring the new path of intact fibril matrix through the axon terminal. Subsequently, it develops in the fibril direction and repeats the dynamic cell-matrix interaction. The last observation is different from those in other studies in which arrayed fibrils were distorted at both tails of polarized cells [3-5]. We ascribe this difference to the cell type. Unlike other cell types, the thin axon terminal and the bulky dendritic terminal of differentiating hdpPSCs are strikingly different, imparting the distinct functions.
The dynamic interaction between the cell and the matrix during the cell development constantly demands and commands the cell and the matrix to respond. It resembles the in vivo environment, in which 3D ECM is constantly remodeled and dynamically interacts with cells, creating the favorable environment for effective cell development. An intriguing observation regarding hdpPSCs on collagen/mica is that the 3D matrix preferentially formed around the cell body, causing the upregulation and local accumulation of β-1 integrin to promote the collagen-β-1 integrin binding near the nucleus for effective signal transduction through the β-catenin pathway [13], accelerating hdpPSCs differentiation. The new insight from this study is that a cell is capable of manipulating the matrix to suit its need. Thus, instead of steady and unvarying matrices that would provide cell anchoring and adhesion, a dynamic, manageable matrix is in demand.
The effect of the 3D-like matrix was validated by the parallel experiments using E-spun collagen fibers, which were found to provide similar structural and biochemical cues for hdpPSC cell polarization and neural differentiation. However the bulky and rigid collagen fibers hinder the cell regulation of the matrix, preventing the formation of the local 3D matrix to up-regulate β-1 integrin expression as in collagen/mica. This attributes to the much slower rate of neural differentiation (Fig. 1H&I). This is consistent with other reports, which suggested that a 3D matrix favors stem cell differentiation [24,25]. 3D collagen gel was applied as a physiologically relevant matrix to studies of cancer cell invasion and stem cell differentiation [24,26]. These matrices consist mostly of randomly oriented fibrils. The derived information is less relevant to cellular responses in vivo where collagen fibrils are aligned in local or extended 3D arrays. The design and construction of aligned 3D matrix is highly desirable.
It should be noted that by Day 3 of differentiation, well-aligned collagen fibrils disappeared from the substrate. They were likely consumed by the cells to create the 3D matrix and subsequently degraded by enzymes. Thus, the 3D matrix environment coaxes and accelerates the hdpPSCs' commitment to neural progenitors in the early stage of differentiation, even in the absence of pro-neural soluble factors.
In conclusion, aligned collagen fibrils can induce unidirectional cell polarization and development. In response, dynamic cell growth prompts the construction of local 3D matrix surrounding the cell body, forming a feedforward loop to support fast and efficient neuronal differentiation of stem cells mediated by β-1 integrin. It is expected to efficiently generate functional neurons applicable in areas such as neural cell injury repair and neural cell based bio-devices.
Supplementary Material
Highlights.
We examined the two-way regulation between cells and aligned collagen fibrils.
Aligned collagen fibrils prompted cell polarization.
Dynamic cell growth prompted the construction of a local 3D matrix.
The local 3D matrix induced upregulation of β-1 integrin around the cell body.
The concerted changes supported fast and efficient neural differentiation.
Acknowledgments
This research was supported by NIH (R01 NS047719).
Footnotes
Supplementary Data: Supplementary data associated with this article can be found in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bosman FT, Stamenkovic I. Functional structure and composition of the extracellular matrix. J Pathol. 2003;200:423–428. doi: 10.1002/path.1437. [DOI] [PubMed] [Google Scholar]
- 2.Grover CN, Cameron RE, Best SM. Investigating the morphological, mechanical and degradation properties of scaffolds comprising collagen, gelatin and elastin for use in soft tissue engineering. J Mech Behav Biomed Mater. 2012;10:62–74. doi: 10.1016/j.jmbbm.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 3.R-C I, Schluter Daniela K, Chaplain Mark AJ. Computational Modeling of single-Cell migration: The leading Role of Extracellular Matrix Fibers. Biophys J. 2012;103:1141–1151. doi: 10.1016/j.bpj.2012.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedrichs J, Taubenberger A, Franz CM, Muller DJ. Cellular remodelling of individual collagen fibrils visualized by time-lapse AFM. J Mol Biol. 2007;372:594–607. doi: 10.1016/j.jmb.2007.06.078. [DOI] [PubMed] [Google Scholar]
- 5.Kirmse R, Otto H, Ludwig T. Interdependency of cell adhesion, force generation and extracellular proteolysis in matrix remodeling. J Cell Sci. 2011;124:1857–1866. doi: 10.1242/jcs.079343. [DOI] [PubMed] [Google Scholar]
- 6.Leow WW, Hwang W. Epitaxially Guided Assembly of Collagen Layers on Mica Surfaces. Langmuir. 2011;27:10907–10913. doi: 10.1021/la2018055. [DOI] [PubMed] [Google Scholar]
- 7.Loo RW, Goh MC. Potassium Ion Mediated Collagen Microfibril Assembly on Mica. Langmuir. 2008;24:13276–13278. doi: 10.1021/la803041v. [DOI] [PubMed] [Google Scholar]
- 8.Gruschwitz R, Friedrichs J, Valtink M, Franz CM, Mueller DJ, Funk RHW, Engelmann K. Alignment and Cell-Matrix Interactions of Human Corneal Endothelial Cells on Nanostructured Collagen Type I Matrices. Invest Ophthalmol Visual Sci. 2010;51:6303–6310. doi: 10.1167/iovs.10-5368. [DOI] [PubMed] [Google Scholar]
- 9.Ulbrich S, Friedrichs J, Valtink M, Murovski S, Franz CM, Mueller DJ, Funk RHW, Engelmann K. Retinal Pigment Epithelium Cell Alignment on Nanostructured Collagen Matrices. Cells Tissues Organs. 2011;194:443–456. doi: 10.1159/000323653. [DOI] [PubMed] [Google Scholar]
- 10.Taubenberger AV, Woodruff MA, Bai H, Muller DJ, Hutmacher DW. The effect of unlocking RGD-motifs in collagen I on pre-osteoblast adhesion and differentiation. Biomaterials. 2010;31:2827–2835. doi: 10.1016/j.biomaterials.2009.12.051. [DOI] [PubMed] [Google Scholar]
- 11.Ihnatovych I, Hu W, Martin JL, Fazleabas AT, de Lanerolle P, Strakova Z. Increased phosphorylation of myosin light chain prevents in vitro decidualization. Endocrinology. 2007;148:3176–3184. doi: 10.1210/en.2006-1673. [DOI] [PubMed] [Google Scholar]
- 12.Strakova Z, Livak M, Krezalek M, Ihnatovych I. Multipotent properties of myofibroblast cells derived from human placenta. Cell Tissue Res. 2008;332:479–488. doi: 10.1007/s00441-008-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sridharan I, Kim T, Strakova Z, Wang R. Matrix-specified differentiation of human decidua parietalis placental stem cells. Biochem Biophys Res Commun. 2013;437:489–495. doi: 10.1016/j.bbrc.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sridharan I, Kim T, Wang R. Adapting collagen/CNT matrix in directing hESC differentiation. Biochem Biophys Res Commun. 2009;381:508–512. doi: 10.1016/j.bbrc.2009.02.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzeng SF. Neural progenitors isolated from newborn rat spinal cords differentiate into neurons and astroglia. J Biomed Sci. 2002;9:10–16. doi: 10.1007/BF02256573. [DOI] [PubMed] [Google Scholar]
- 16.Balenci L, Saoudi Y, Grunwald D, Deloulme JC, Bouron A, Bernards A, Baudier J. IQGAP1 regulates adult neural progenitors in vivo and vascular endothelial growth factor-triggered neural progenitor migration in vitro. J Neurosci. 2007;27:4716–4724. doi: 10.1523/JNEUROSCI.0830-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tublin isotype during chick embryogenesis, Cell Motil. Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 18.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear-protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 19.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 20.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 21.Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adhesion & Migration. 2011;5:421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol. 2011;3:267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 23.Kirfel G, Rigort A, Borm B, Herzog V. Cell migration: mechanisms of rear detachment and the formation of migration tracks. Eur J Cell Biol. 2004;83:717–724. doi: 10.1078/0171-9335-00421. [DOI] [PubMed] [Google Scholar]
- 24.Kothapalli CR, Kamm RD. 3D matrix microenvironment for targeted differentiation of embryonic stem cells into neural and glial lineages. Biomaterials. 2013;34:5995–6007. doi: 10.1016/j.biomaterials.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 25.Yla-Outinen L, Joki T, Varjola M, Skottman H, Narkilahti S. Three-dimensional growth matrix for human embryonic stem cell-derived neuronal cells. J Tissue Eng Regen Med. 2014;8:186–194. doi: 10.1002/term.1512. [DOI] [PubMed] [Google Scholar]
- 26.Beck JN, Singh A, Rothenberg AR, Elisseeff JH, Ewald AJ. The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination. Biomaterials. 2013;34:9486–9495. doi: 10.1016/j.biomaterials.2013.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


