Abstract
Poly (ADP-ribose) polymerase-1 (PARP-1) is important for the recognition of both endogenous and exogenous DNA damage, and binds to DNA strand-breaks including intermediates of base excision repair (BER). Once DNA-bound, PARP-1 becomes catalytically activate synthesizing poly-ADP-ribose (PAR) polymers onto itself and other repair factors (PARylation). As a result, BER repair proteins such as XRCC1 and DNA polymerase β (pol β) are more efficiently and rapidly recruited to sites of DNA damage. In the presence of an inhibitor of PARP activity (PARPi), PARP-1 binds to sites of DNA damage, but PARylation is prevented. BER enzyme recruitment is hindered, but binding of PARP-1 to DNA is stabilized, impeding DNA repair and leading to double-strand DNA breaks (DSB). Deficiencies in pol β−/− and Xrcc1−/− cells resulted in hypersensitivity to the PARP inhibitor 4-AN and re-expression of pol β or XRCC1, in these contexts, reversed the 4-AN hypersensitivity phenotype. BER deficiencies also showed evidence of replication defects that lead to DSB-induced apoptosis upon PARPi treatment. Finally, the clinically relevant PARP inhibitors olaparib and veliparib also exhibited hypersensitivity in both pol β−/− and Xrcc1−/− BER-deficient cells. These results reveal heightened sensitivity to PARPi as a function of BER deficiency.
Keywords: DNA polymerase, XRCC1, PARP-1, methyl methanesulfonate, PARP inhibitor, poly(ADP-ribose)
Introduction
The base excision repair (BER) pathway is a primary mechanism for repair of endogenous and exogenous base lesions in DNA. PARP-1, the first discovered member of the PARP family and an abundant nuclear protein, is particularly important for DNA damage recognition involving binding to strand-breaks in DNA (1), including intermediates of BER (2). Once bound to damaged DNA, PARP-1 becomes catalytically activated synthesizing poly(ADP-ribose) (PAR) polymers attached to itself and other repair factors (PARylation). As a result of PARP-1 auto-PARylation, BER repair proteins such as DNA polymerase β (pol β) and the XRCC1-DNA ligase IIIα(lig-IIIα) complex are recruited more efficiently to sites of DNA damage.
XRCC1 is a multi-domain protein with no known catalytic activity. Yet, it interacts with a number of repair proteins, e.g., PARP-1, pol β and lig-IIIα, and is thought to function as a scaffold able to modulate and coordinate the various steps of BER. In this repair pathway, base lesions removed by a lesion-specific monofunctional glycosylase produce abasic sites that are cleaved by AP endonuclease 1. This results in a repair intermediate with a gap containing a 3′-OH and 5′-deoxyribose phosphate (dRP) flap at the margins. DNA pol β removes the 5′-dRP flap through its lyase activity, and conducts gap-filling DNA synthesis leaving a nicked DNA that is a substrate for DNA ligase. Both XRCC1-deficient and pol β-deficient mouse fibroblasts are hypersensitive to the DNA base methylating agent methyl methanesulfonate (MMS), and this is linked to a repair deficiency as measured by accumulation of strand breaks (3).
In the presence of an inhibitor of PARP catalytic activity (PARPi), PARP-1 binds at sites of DNA damage, but auto-PARylation is prevented. In the absence of PAR polymers, BER enzyme recruitment is hindered, but binding of PARP-1 to DNA is stabilized, impeding DNA repair (4). PARPi-treated wild-type cells are highly sensitive to MMS whereas Parp-1−/− cells demonstrate only moderate MMS hypersensitivity (5). Thus, the cellular cytotoxicity phenotype associated with inhibition of the PARP-1 enzyme is not equivalent to PARP-1 deletion (4, 5). Previous results suggested that the DNA-bound and inhibited PARP-1 protein is cytotoxic as a function of formation of replication-dependent doublestrand breaks (DSBs) (6). This type of DSB is preferentially repaired by the homologous recombination (HR) pathway.
BRCA1 and BRCA2 are components of the HR pathway and BRCA1/2-deficient cells were shown to be hypersensitive to treatment with a PARPi (7, 8). This key combination became known as a form of ‘synthetic lethality’ where there is synergy between two otherwise non-lethal events, here a PARPi resulting in DSB formation and a genetic deficiency resulting in loss of the pathway required for DSB repair. A recent model predicts that inactivation of both PARP-1 and BRCA activities at the same time would result in repair through an error-prone non-homologous end-joining mechanism (9). Since carriers of germ-line heterozygous BRCA mutations are predisposed to cancer after loss of the wild-type allele, PARPi were rapidly considered for targeted mono-therapy that should not affect other repair-competent cells (10).
Screening for determinants of PARPi sensitivity has become critical (11, 12). A high throughput siRNA screen targeting 98% of known DNA repair proteins implicated several HR proteins, but additionally identified XRCC1, involved in BER (11). The most striking PARPi-induced enhancement of cytotoxicity in mouse cell lines is observed with alkylating agents resulting in damage repaired by a specific BER sub-pathway involving pol β and XRCC1 (13). With MMS and PARPi combination treatment in mouse fibroblasts, the effects of PARPi and BER deficiency are clearly not epistatic, but instead are additive (5, 14). Thus, PARPi does not target the BER deficiency mediators studied, pol β and XRCC1, but instead works through a separate mechanism.
Sensitization to MMS is consistent with increased PARP binding sites in DNA in the absence of efficient BER. However, the question of PARPi hypersensitivity as a function of BER deficiency has not been widely studied. Since the BER deficiencies mediated by polβ and XRCC1 knockouts are additive with combination treatment, we proposed that cells deficient in these BER factors would be hypersensitive to treatment with a PARPi. We suspected that endogenous DNA damage would effectively substitute for the alkylating agent used in combination treatment. Here, we determined whether BER deficiency can lead to hypersensitivity to 4-amino-1,8-naphthalimde (4-AN), a commercially available PARPi. Results were compared to those obtained with clinically utilized PARPi olaparib and veliparib. We find that BER deficiency as a result of either pol β-deletion or XRCC1-deletion is associated with PARPi hypersensitivity. These results suggest that BER deficiency could represent a therapeutic opportunity for single agent PARPi therapy.
Materials and Methods
Cell culture - pol β and XRCC1 cell variants
The originally characterized wild-type and pol β null (termed pol β−/−) SV40-transformed mouse embryonic fibroblasts (16.3 and 19.4, respectively) have been described previously (15). Wild-type full-length Flag-tagged pol β protein was over-expressed in the wild-type (16.3) background using a pIRESpuro-derived expression vector as described previously (16). Clones were selected in 6 μg/ml puromycin (Invitrogen) and pol β expression level was ascertained by western blotting. Independently derived SV40- transformed wild-type and pol βnull cells (36.3 and 38Δ4) have also been described (16). Stable transfection of 38Δ4 pol βnull cells with wild-type pol β and its 8-kDa dRP lyase domain and selection in G418 (600 μg/ml) or puromycin (6 μg/ml) respectively, has been outlined (16). Cells were routinely grown at 34°C in a 10% CO2 incubator in high glucose Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), glutaMAX 1 and 80 μg/ml hygromycin (Invitrogen, Carlsbad, CA).
Xrcc1+/+ and Xrcc1−/− p53-deficient mouse embryonic fibroblasts were obtained from Dr. Robert Tebbs (17). These cells were maintained in low glucose DMEM (Invitrogen) supplemented with 10% FBS at 37°C. A clone containing the full-length open reading frame of mouse XRCC1 (Invitrogen) was subcloned into the pDONR221 vector and then the pEF-DEST51 vector utilizing Gateway technology (Invitrogen). Xrcc1−/− cells were stably transfected with the full-length wild-type XRCC1 construct (WT17 cells). Other stable cell lines with mutant XRCC1 proteins have been engineered. V88R XRCC1-transfected cells (XV2 cells) are known to have interrupted binding to pol β (14, 18, 19). Reduced C12A XRCC1 protein transfectants (XRE8 cells), unable to form the C12-C20 disulfide bond and also deficient in interaction with pol β (20), have been characterized previously (14). Another XRCC1 mutant, L360R, was introduced by site-directed mutagenesis of the pDONR221 vector and subcloned into pEF-DEST51. The resulting vector, pXL was sequence verified. Transfection was conducted as described, clones were selected with blasticidin (10 μg/ml; Invitrogen), and XRCC1 expression was confirmed by western blotting. Mycoplasma testing was performed routinely on all cell lines using a MycoAlert® Mycoplasma detection kit (Lonza, Rockland, ME).
Determination of total cellular PAR
Cellular PAR levels were quantified using the PARP in vivo Pharmacodynamic Assay 2nd Generation (PDA II) kit (4520-096-K, Trevigen) as previously (21). Cells were seeded in 60-mm dishes at 106 cells per dish and analyzed the following day. Dishes were placed on ice and lysed according to the kit protocol. Cell counts were verified to ensure no growth differential between cell lines. After cell lysis and DNA digestion, total protein amounts were determined for each sample, and 4 μg of pol β−/− and Xrcc1−/− cell extracts and 10 μg of pol β+/+ and Xrcc1+/+ and cell extracts were added to pre-coated capture antibody plates and incubated overnight at 4°C. The following morning, wells were washed four times in PBS with Tween-20 (PBST), then a 1:250 dilution of PAR detecting polyclonal antibody was added and wells were incubated at room temperature for 2 h. Each well was again washed four times in PBST, then a 1:250 dilution of goat anti-rabbit IgG-HRP was added and incubated for 1 h. Cells were washed again four times, then a 1:1 mixture of PARP PeroxyGlow™ A and B was added, and luminescence was measured with a Tropix TR717 Microplate Luminometer.
Measurement of PARP-1 DNA-protein cross-links (DPCs) in genomic DNA
Mouse fibroblasts were mock-treated or treated with 30 μM 4-AN for 24 h. Cells were washed in phosphate-buffered saline (PBS), then isolated by scraping. For isolation of DPCs the method of Barker et al. (22) was followed with slight modifications (23).
Briefly, nuclei were lysed by the addition of 500 ml DNAzol per 6 × 107 cells and vortexing briefly. After lysis, 2 ml pre-warmed (65°C) 10 mM Tris-HCl, pH 7.0, was added and the DNA was sheared by passing the suspension through a 21-gauge needle and then through a 25-gauge needle three times each. 5 M NaCl (9.6 ml) was added to a final concentration of 4 M, and the mixture was incubated at 37°C in a shaking water bath for 20 min. 8 M urea (12 ml) was added to a final concentration of 4 M, and the incubation was continued for 20 min at 37°C in a shaking water bath. After this incubation, an equal volume (24 ml) of 100% ethanol was added and the sample was mixed by inversion. The QIAEX II silica slurry (0.5 ml per 6 × 107 nuclei) was added to each sample, and the samples were gently rocked for 40 min at room temperature to allow DNA to bind. Silica particles were then collected by centrifugation, and the supernatant fraction was carefully removed and discarded. The silica particles were washed four times with 50% ethanol and collected by centrifugation. The DNA-protein cross-linked complexes were eluted from silica by adding 2 ml of 8 mM NaOH and incubation at 65°C for 5 min. The elution process was repeated and the supernatant fractions were combined. An aliquot (10 μl) from each sample was saved for DNA measurement and to verify the concentration of each DPC sample subjected to PARP-1 immunoblotting analysis (see below).
For DNA digestion, the samples (4 ml) were mixed with 1 ml of 5 × digestion buffer (to a final concentration of 10 mM MgCl2, 10 mM ZnCl2, 0.1 M sodium acetate, pH 5.0), 5 units of DNase I and 5 units of S1 nuclease. The samples were then incubated at 37°C for 1 h, and the digestion of DNA was stopped by transferring samples to 65°C for 10 min. After this heat inactivation, 100% ice-cold trichloroacetic acid (TCA) was added to each sample to a final concentration of 15%, and the samples were incubated for 60 min on ice for precipitation of DPC proteins. Samples were centrifuged at 12,000 rpm for 15 min at 4°C and the pellet fraction was washed twice with 15% ice-cold TCA and twice with 100% ice-cold acetone. The pellet was air-dried and dissolved in 10 μl 1 M Tris-HCl, pH 8.0, and 90 μl SDS-PAGE gel-buffer.
For immunoblotting, approximately equal amounts (adjusted by DNA content) of each DPC sample isolated as described above, along with marker purified PARP-1 were separated by Nu-PAGE 4-12% Bis-Tris mini-gel and the proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBST) and then probed with anti-PARP-1 antibody. Goat anti-mouse IgG conjugated to horseradish peroxidase (1:10,000 dilution) was used as secondary antibody, and immobilized horseradish peroxidase activity was detected by enhanced chemiluminescence. The PARP-1 DPC material co-migrated with purified PARP-1 in this analysis.
Cytotoxicity studies
For growth inhibition assays, cells were seeded at a density of 5-40,000 cells per well in six-well dishes in medium without selection antibiotic. The next day, cells were treated with a range of concentrations of 4-AN (Acros) continuously or for the time period indicated. For comparison with 4-AN, cells were treated continuously with the clinically utilized PARPi olaparib and veliparib (Selleckchem) until untreated control cells reached 80% confluence (24). In other studies, cells were treated for 1 h with MMS (Sigma) in the presence or absence of a PARPi. After a further 23 h PARPi incubation and washing with Hanks’ Balanced Salt Solution (HyClone) as needed, growth medium was added and cells were incubated until untreated cells were 80% confluent (24). At this time, triplicate wells for each drug concentration were counted by a cell lysis procedure, and results were expressed as % control growth.
Western blot analysis
For preparation of whole cell extracts, cells were washed with PBS, scraped, collected into PBS and centrifuged. Cell pellets were flash frozen in dry ice and thawed by resuspension in Buffer I (10 mM Tris-HCl, pH 7.8, 0.2 M KCl, 25 mM NaF and complete protease inhibitor cocktail (Roche)). An equal volume of Buffer II (10 mM Tris-HCl, pH 7.8, 0.2 M KCl, 25 mM NaF, 2 mM EDTA, 40% glycerol, 0.2% NP-40 and 2 mM dithiothreitol (DTT)) was then added as described previously. The suspension was rotated for 1 h at 4°C and extracts were clarified by centrifugation in a microcentrifuge at full speed for 15 min at 4°C. Total protein concentration of extracts was determined by the Bio-Rad assay using bovine serum albumin (BSA) as protein standard.
Extract samples (60 μg) were loaded onto 4-12% Bis-Tris NuPAGE gels (Invitrogen) and electrophoresed in NuPAGE MES running buffer at 4°C. Proteins were transferred to nitrocellulose filters at 25 V overnight, in the cold. Following transfer, filters were blocked overnight at 4°C in 5% nonfat dry milk in TBST. Filters were first incubated for 2 h at room temperature or overnight at 4°C with either pol β 18S monoclonal antibody (25) or mouse monoclonal anti-XRCC1 primary antibody (Thermo Fisher Scientific, 33-2-5). After washing with TBST, filters were further incubated with anti-mouse IgG-horseradish peroxidase (HRP) conjugated secondary antibody (1:2,000-1:20,000 dilution, Bio-Rad) and visualized using Super Signal chemiluminescent detection (Thermo Scientific) according to the manufacturer's instructions. Blots were then stripped for up to 30 min at 37°C or room temperature in Restore Western Blot Stripping Buffer (Thermo Scientific), washed three times in TBST, then blocked in 5% nonfat dry milk/TBST overnight. Other primary antibodies utilized were Ligase III-1F3 (GeneTex GTX70143), PARP-1 (BD Pharmingen 51-669GR) and GAPDH (Alpha Diagnostic G3PDH11-M) used as a loading control.
Co-immunoprecipitation
Cells were washed in PBS and collected by scraping, suspended in two volumes of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 25 mM NaF, 0.1 mM sodium orthovanadate, 0.2% Triton X-100, and 0.3% NP-40) containing protease inhibitors, 0.1 mM PMSF, 1 μg/ml aprotinin, and 5 μg/ml leupeptin and incubated on ice for 30 min. After agitating the tubes briefly, the lysates were centrifuged at 20,800 × g for 30 min at 4°C, and the supernatant fraction was removed. Protein concentrations were determined using the Bio-Rad protein assay with BSA as standard.
For co-immunoprecipitations with XRCC1, an equal amount (1 mg protein) of cell extract was mixed with 1-2 μg of anti-XRCC1 antibody (Santa Cruz H-300). The mixture was incubated with rotation for 4 h at 4°C. The immunocomplexes were adsorbed onto protein A-sepharose and protein G-agarose beads (1:1 mixture) by incubating the mixture for 16 h at 4°C. The beads were then washed four times with lysis buffer containing protease inhibitors. Finally, the beads were resuspended in SDS sample buffer, heated at 95°C for 5 min, and briefly centrifuged. The soluble proteins were separated by electrophoresis on 4-12% SDS-PAGE gels in MOPS buffer and then transferred to a nitrocellulose membrane for 2 h. The membranes were blocked, then probed with anti-pol β antibody (Abcam ab3181) or pol β 18S monoclonal antibody, anti-XRCC1 antibody, or anti-ligase III and anti-PARP-1 monoclonal antibodies as described above. In control experiments, the immunoprecipitating antibody was substituted with agarose-conjugated IgG.
Flow cytometry
For cell cycle analysis, wild-type and pol β null cells (16.3 and 19.4) were seeded in 100-mm dishes at a density of 1 × 106/dish. The next day, cells were mock-treated or treated for 24 h with 4-AN (30 μM). Bromodeoxyuridine (BrdU) (Sigma-Aldrich) was added (10 M) for 30 min to pulse-label the cells. Cells were washed with PBS, trypsinized, and harvested in growth medium. The cell pellet was resuspended in 100 μl cold PBS and the cells were fixed by slowly dropping into 70% ethanol while vortexing gently to prevent clumping then storing at 4°C overnight. Samples were washed in PBS, resuspended in 2 N HCl containing 0.5% Triton X-100 to denature DNA and incubated at room temperature for 30 min. The cells were resuspended in 0.1 M sodium borate (pH 8.5) to neutralize the acid and then washed. Cell samples were incubated overnight at 4°C with 20 μl anti-BrdU FITC-conjugated antibody (BD Biosciences) in PBS containing 0.5% Tween 20, 1% BSA and 5 μl of 10 mg/ml RNase A (Sigma-Aldrich). The cells were washed then resuspended in 1 ml PBS containing 5 μg/ml of propidium iodide (PI) for measurement of DNA content. Samples were read on a FACS flow cytometer (BD Biosciences) and analyzed using Cell Quest or ModFit LT software (Verity Software House, Inc.).
For cell death analysis, wild-type and pol β null cells (16.3 and 19.4) were seeded in 100-mm dishes at a density of 0.5 × 106/dish and treated the next day with 30 μM 4-AN for 24 h. At this time and at 24 and 48 h after the 4-AN exposure, cells were harvested using 0.05% trypsin and collected with the culture medium. After centrifuging and washing with PBS, annexin V-fluorescein isothiocyanate (FITC) and PI were added in binding buffer as suggested by the manufacturer (TACS Annexin V Apoptosis Dectection Kit, Trevigen, Gaithersburg, MD). The samples were incubated at room temperature in the dark for 15 min, read on a FACS flow cytometer, and data analyzed using Cell Quest software.
For γH2A.X staining, wild-type and pol β null cells (16.3 and 19.4) were seeded in 100-mm dishes at a density of 2 × 106/dish. The following day, cells were mock-treated or treated for 24 h with 4-AN (30 μM). At scheduled time points as indicated, cells were trypsinized and the pellet washed with PBS. Analysis utilized the H2A.X Phosphorylation Kit for flow cytometry (Millipore) following the manufacturer's instructions. Cells were resuspended in 500 μl 1 × fixation solution and incubated on ice for 20 min. After washing, cells were resuspended in 500 μl 1 × permeabilization solution and further incubated on ice for 30 min. 50 μl of this suspension was mixed with γH2A.X-FITC antibody in PBS containing 0.5% Tween 20, 1% BSA and 5 μg of RNase A. The mixture was incubated on ice for 30 min with periodic mixing. After centrifugation, cells were resuspended in 500 μl of PI solution (5 μg/ml) and then stored in the dark for 30 min. Samples were read on a FACS flow cytometer and data analyzed using Cell Quest software (Becton-Dickinson Immunocytochemistry Systems).
Results
Characterization of PAR and PARP levels in untreated BER-deficient cell lines
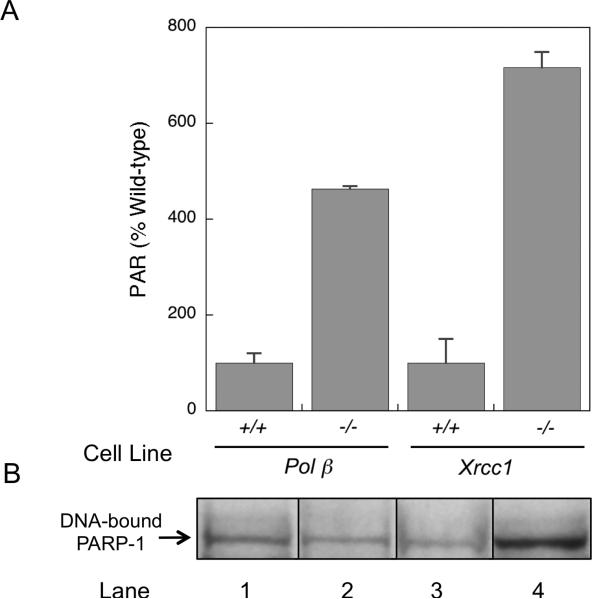
Elevation of cellular PAR levels has been found to correlate with a disruption in the normal process of BER (21). Thus, the higher level of PAR observed here in control non-treated pol β−/− and Xrcc1−/− cells than in repair-proficient pol β+/+ and Xrcc1+/+ cells was anticipated (Fig. 1A). These results are consistent with the hypothesis strand breaks secondary to endogenous DNA damage in pol β−/− and Xrcc1−/− cells accumulate to a higher level than in wild-type cells and result in increased activation of PARP. Both of these gene-deleted cell lines are known to have BER-deficiency and hypersensitivity to monofunctional alkylating agent treatment (3). In the absence of the repair enzyme pol β, cells are deficient in the 5′-dRP lyase activity critical for repair of methylation-induced DNA damage (24). In addition, pol β interacts with the N-terminal domain (NTD) of XRCC1 (18), and in the absence of XRCC1, pol β is less efficiently recruited to sites of DNA damage (14).
Fig. 1.
Analysis of cellular PAR and DNA-bound PARP-1. (A) Cellular PAR levels were quantified using the Trevigen PARP in vivo Pharmacodynamic Assay 2nd Generation (PDA II) kit as described in Materials and Methods. Panel (A) shows PAR levels in untreated pol β and XRCC1 wild-type and -deficient cells. Results are expressed as % of respective wildtype value. Plotted are mean ± SEM of at least 3 independent experiments. (B) PARP-1 bound to endogenous DNA damage was analyzed as DPCs in genomic DNA isolated from untreated pol β and XRCC1 wild-type and -deficient cells as indicated. PARP-1 DPCs were measured by immunoblotting as described in Materials and Methods. Typical results are shown from at least 2 independent experiments, with all samples run on the same gel.
As outlined above, the elevation of PAR levels under conditions of repair deficiency suggested enhanced activation of PARP as a result of increased levels of endogenous DNA damage in the BER-deficient cells. The PAR results are also consistent with an elevation in PARP-1 binding to endogenous repair intermediates in DNA. Figure 1B shows DNA-bound PARP-1 isolated from untreated cell lines as indicated. The levels of DNA links (DPCs) were similar in pol β+/+, pol β−/− and Xrcc1+/+ cell types (lanes 1-3), whereas the level in Xrcc1−/− cells (lane 4) was considerably elevated, consistent with the high amount of PAR detected in these cells (Fig. 1A).
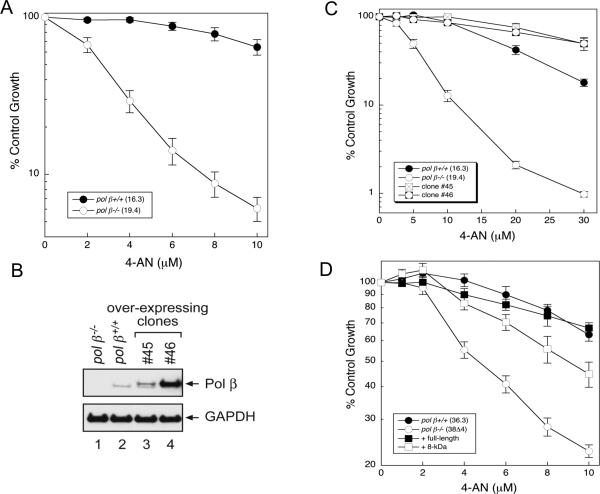
Effect of pol β expression on sensitivity to 4-AN
With exposure to the PARPi 4-AN, pol β−/− cells (19.4) showed hypersensitivity compared with wild-type (16.3) cells (Fig. 2A). This is consistent with increased sensitivity to the PARPi as a function of BER deficiency in the the absence of pol β. Hypersensitivity of pol β−/− cells to 4-AN was similarly observed using a clonogenic survival assay (Supplementary Fig. S1A). Additionally, pol β−/− cells were found to be hypersensitive to two clinically used PARPi, olaparib and veliparib (Supplementary Fig. S7A and B). Next, we derived new stable cell lines from pol β+/+ (16.3) cells that over-express pol β (Fig. 2B). A low level (~ 2- to 3-fold) of pol β over-expression (clone #45) (Fig. 2B) resulted in enhanced resistance to 4-AN (Fig. 2C), but a higher level of pol β over-expression in clone #46 (8-fold) did not further enhance resistance to 4-AN (Fig. 2C). Note that a shorter 4-AN treatment time (24 h vs continuous) was required to observe a substantial difference between pol β+/+ and clone #45. Hypersensitivity to 4-AN in the absence of pol β expression was similarly observed in independent pair of pol β+/+ and −/− cells (36.3 and 38Δ4, respectively) (Fig. 2D). Stable expression of the 8-kDa dRP lyase domain of pol β in these pol β−/− cells resulted in significant, although not complete reversal of the hypersensitivity phenotype (Fig. 2D) and is consistent with PARPi hypersensitivity as a function of the dRP lyase activity of pol β.
Fig. 2.
Sensitivity of pol β-deficient and pol β-over-expressing cells to 4-AN. Survival was measured by growth inhibition assays as outlined in Materials and Methods. (A) Continuous exposure of wild-type (16.3) and pol β null (19.4) cells to 4-AN at doses indicated. (B) Over-expression of pol β in wild-type cells (clone #45 and #46) as determined by western blotting. GAPDH was used as a loading control. (C) 48 h exposure of cells to 4-AN. (D) Continuous exposure of wild-type (36.3) and pol β null (38Δ4) cells to 4-AN at doses indicated, full complementation of hypersensitivity by full-length pol β protein and significant complementation by expression of the 8-kDa domain. Plotted are mean ± SEM of at least 3 independent experiments.
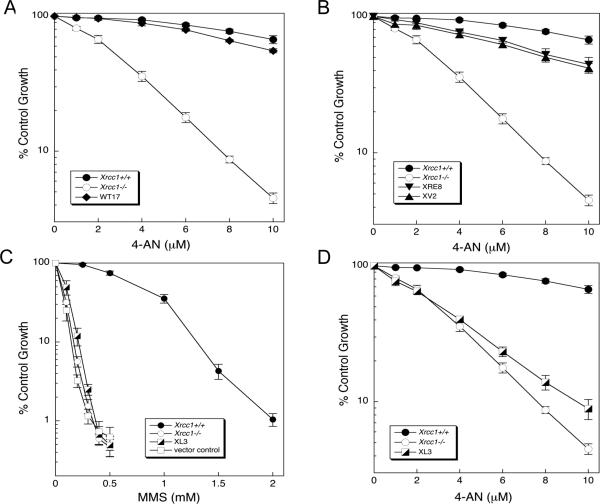
Effect of XRCC1 expression on sensitivity to 4-AN
Following exposure to 4-AN, hypersensitivity was observed in Xrcc1−/− compared with Xrcc1+/+ mouse fibroblasts (Fig. 3A). Similar hypersensitivity of Xrcc1−/− cells to 4-AN was observed using a clonogenic survival assay (Supplementary Fig. S1B). Additionally, Xrcc1−/− cells were found to be hypersensitive to the PARPi olaparib and veliparib (Supplementary Fig. S7C and D). Expression of wild-type XRCC1 in these Xrcc1−/− cells (clone WT17) reversed the hypersensitivity phenotype (Fig. 3A). Two XRCC1 variants that had been described previously (14) also were studied here. One of these, C12A (XRE8 cells), is a disulfide blocking variant that has a lesser binding affinity for pol β than the disulfide bond oxidized form of the wild-type protein, and the other variant, V88R (XV2 cells), is unable to bind pol β. Expression of either variant XRCC1 protein in Xrcc1−/− cells resulted in near-complete reversal of the 4-AN hypersensitivity (Fig. 3B). These results are consistent with previous data indicating that both variant XRCC1 proteins are capable of complementing the BER deficiency of the Xrcc1−/− cells and that the 4-AN hypersensitivity reflects a BER deficiency.
Fig. 3.
Sensitivity of XRCC1 cell variants to 4-AN and MMS. Survival was measured by growth inhibition assays as outlined in Materials and Methods. (A) Hypersensitivity of Xrcc1−/− cells to continuous exposure 4-AN and complementation by expression of wild-type XRCC1 protein (WT17). (B) Near full complementation of 4-AN hypersensitivity by expression of mutant XRCC1 proteins, C12A (XRE8) and V88R (XV2). (C) Absence of complementation of MMS hypersensitivity after expression of the L360R XRCC1 mutant in Xrcc1−/− cells. (D) Limited complementation of hypersensitivity to continuous 4-AN exposure by L360R mutant protein. Plotted are mean ± SEM of at least 3 independent experiments.
A point mutation in the BRCT I domain of XRCC1 (L360R) also was constructed with the aim of interrupting the interaction between XRCC1 and PARP-1 (Supplementary Fig. S2) (26). Characterization of one of the selected clones (XL3) revealed that the L360R XRCC1 protein level was very similar to that in Xrcc1+/+ cells (Supplementary Fig. S3A). Additionally, compared with Xrcc1−/− cells, the XL3 cell line now expressed wild-type levels of DNA lig-IIIα and pol β, suggesting that this mutant XRCC1 is able to bind and stabilize both of these partner repair proteins (Supplementary Fig. S3A). Co-immunoprecipitation with anti-XRCC1 antibody failed to pull down PARP-1 (Supplementary Fig. S3B). This is consistent with the hypothesis that the L360R protein does not bind PARP-1. In a reciprocal experiment, PARP-1 was not observed in an XRCC1 co-immunoprecipitate. Yet, both lig-IIIα and pol β were present in this pull down (Supplementary Fig. S3C), suggesting that, other than the BRCT I domain, the full length L360R protein has interaction sites similar to those in cellular wild-type XRCC1 protein. For confirmation, purified full length L360R protein was further compared with wild-type (WT) XRCC1 protein by controlled proteolysis (Supplementary Fig. S4A) and circular dichroism (CD) spectroscopy (Supplementary Fig. S4B). Neither of these studies of protein conformation indicated a detectable difference between the two proteins, confirming that the L360R protein was properly folded. Yet, the L360R mutation (XL3 cells) did not have any ability to rescue the MMS hypersensitivity of Xrcc1−/− cells (Fig. 3C). A similar observation had been made previously utilizing the L360R mutation in CHO EM9 XRCC1 null cells (26). The results clearly show that an interaction between PARP-1 and XRCC1 is critical for the protective effect of XRCC1 against MMS-induced cytotoxicity.
In the evaluation of 4-AN hypersensitivity, XL3 cells showed only minimal reversal of the hypersensitivity phenotype (Fig. 3D). Thus, the PARP-1/XRCC1 interaction is required for the cellular resistance to the PARP inhibitor 4-AN. It is known that activated PARP-1 and PAR are essential for recruitment of XRCC1 to sites of DNA damage (27, 28), and additionally that XRCC1 is important for recruitment of pol β (14, 29), an enzyme that is known to be critical for efficient BER.
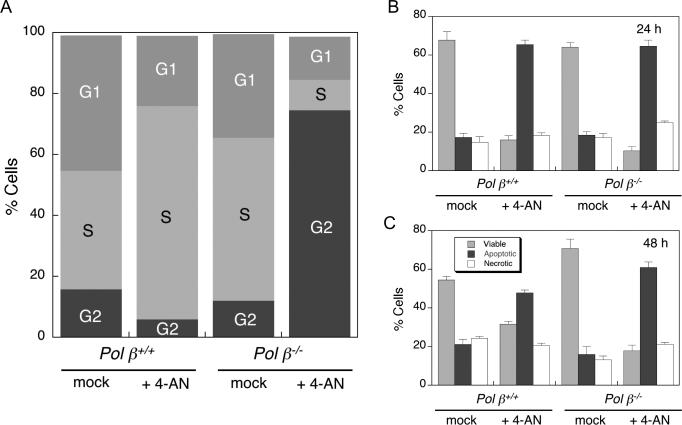
Cell cycle and cell death after exposure to 4-AN
Wild-type and pol β null cells were treated for 24 h with 30 μM 4-AN, then cells were stained with BrdU and harvested for cell cycle analysis by flow cytometry. At this time, a high proportion of pol β+/+ cells (70%) had accumulated in S phase of the cell cycle (Fig. 4A). This result is similar to data obtained following treatment of the same wild-type cells with the combination of MMS plus 4-AN (5). In contrast, 75% of pol β−/− cells were able to progress through S phase and accumulate in G2 after 24 h 4-AN treatment (Fig. 4A).
Fig. 4.
Cell cycle and cell death analysis in 4-AN treated cells. Flow cytometry methods are described in Materials and Methods. Pol β+/+ and pol β−/− cells were untreated, or exposed to 4-AN (30 μM) for 24 h. (A) After 24 h, cell cycle analysis was by BrdU and PI staining. Plotted are proportions of mock- and 4-AN-treated cells in G1, S and G2 phases of the cell cycle. Cell death analysis at (B) 24 h or (C) 48 h was by annexin-FITC and PI staining. Plotted is % of viable, apoptotic or necrotic cells after treatment as indicated (mean ± SEM of at least 3 independent experiments).
Flow cytometric analysis of 4-AN-treated cells following staining with annexin-FITC and PI revealed a primarily apoptotic mechanism of cell death in both wild-type and pol β null cell lines (Fig. 4B and C). At 48 h, apoptotic cells were observed in a higher proportion of pol β−/− than pol β+/+ cells. Necrotic cells were detected, but at a lower level similar to that observed in control mock-treated cells. These results are consistent with data obtained following treatment of the same cell lines with the combination of MMS plus 4-AN (30).
Persistent DNA damage following exposure of cells to 4-AN
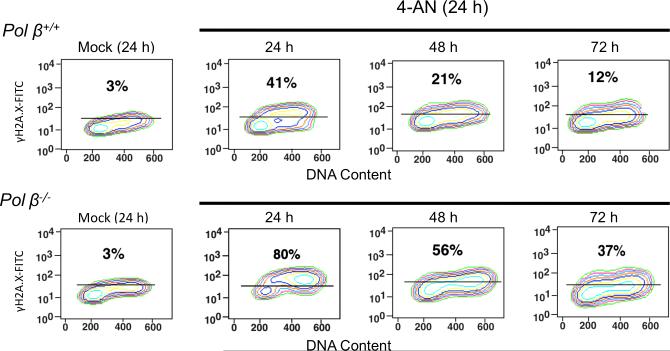
It has been proposed that DNA-bound and catalytically inhibited PARP-1 is cytotoxic as a function of formation of replication-dependent DSBs (6). In this earlier publication, DSBs could be detected by both pulsed-field gel electrophoresis and staining for γH2A.X. Here, wild-type and pol β null cells were treated for 24 h with 30 μM 4-AN and cell populations were analyzed for H2A.X phosphorylation (γH2A.X), a DNA damage signal and marker consistent with formation of DSBs (31-33). The percentage of cells positive for γH2A.X was measured by flow cytometry and, at the same time, staining with PI was employed to measure DNA content (Fig. 5). After 24 h of 4-AN treatment, a larger fraction of γH2A.X positive cells was identified in pol β null cells than in wild-type cells (80 and 41%, respectively). At 24 h, γH2A.X staining was associated with S phase in wild-type cells, consistent with production of replication-dependent damage and the accumulation of cells in S phase (Fig. 4A). In pol β null cells, γH2A.X staining was observed in late S and G2 phases of the cell cycle (Fig. 5), consistent with the observed cell passage through S phase and accumulation in G2 after 24 h 4-AN treatment (Fig. 4A). At later times after removal of 4-AN when cells were allowed to repair, decreasing proportions of γH2A.X positive cells were observed, but the level was always er in pol β-deficient cells. Thus, the hypersensitivity to 4-AN in these pol β null variants correlates with unrepaired DNA damage, most likely DSBs.
Fig. 5.
Flow cytometric analysis of γH2A.X in 4-AN treated cells. Pol β+/+ and pol β−/− cells were mock-treated or exposed to 4-AN (30 μM) for 24 h. Flow cytometry methods are described under Materials and Methods. Shown in each panel is the percentage of cells positive for γH2A.X staining with a FITC conjugated antibody to γH2A.X at the end of 24 h, or at later time points allowing cells to repair. The % γH2A.X staining was set at 3% in mock-treated cells.
Measurement of cellular DNA repair intermediates after 4-AN treatment
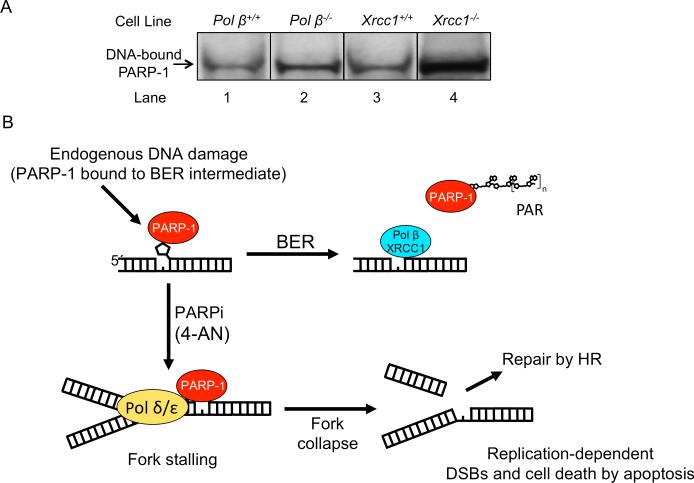
PARPi hypersensitivity has been demonstrated in BER-deficient mouse cell lines (Figs 2, 3 and Supplementary S7). We propose that endogenous DNA damage, such as that arising from spontaneous hydrolysis, is repaired by BER, and in the case of pathway deficiency, more unrepaired intermediates accumulate than in wild-type cells. To test this hypothesis, we measured PARP-1 cross-linking to cellular DNA. PARP-1 DNA-protein cross-links (DPCs) are an indicator of unrepaired AP site BER intermediates (23). Wild-type and BER-deficient cells were treated for 24 h with 30 μM 4-AN. Samples of DPCs were isolated and proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with antibody against PARP-1. In wild-type cells, there was a 2-3-fold increase in DNA-bound PARP-1 compared with untreated cells. A greater increase in PARP-1 binding was observed following 4-AN-treatment of pol β−/− and Xrcc1−/− cells. For both pol β- and XRCC1-deficient cells, the results revealed a higher level of DNA-bound PARP-1 than in the isogenic wild-type cells after 4-AN exposure (compare lanes 1 and 2, 3 and 4; Fig. 6A). These results are consistent with the working model shown in Fig. 6B, where immobilized PARP-1 leads to replication fork disruption and DSB-mediated apoptosis when DSB repair by HR is overwhelmed. The results indicate that the observed increase in PARP inhibitor hypersensitivity correlates with an increase in unrepaired BER intermediates.
Fig. 6.
Measurement of PARP-1 DPCs in genomic DNA and a model for PARPi-induced cytotoxicity. (A) Cells as indicated were exposed to 4-AN (30 μM) for 24 h and then harvested. PARP-1 DPCs in genomic DNA were isolated and immunoblotted with anti-PARP-1 antibody. Experiments were conducted as described under Materials and Methods. The DNA-bound PARP-1 observed in each cell line co-migrated in the gel with purified PARP-1 run in parallel (not shown). Typical results from at least 2 independent experiments are shown. (B) Scheme illustrating a model addressing the mechanism of PARPi-induced hypersensitivity in BER-deficient cells. The scheme is consistent with the results of PARP-1 DPC analysis in panel (A) and with previous results from studies of the mechanism of PARPi-induced cytotoxicity in mouse fibroblasts upon co-exposure with a DNA alkylating agent. DNA-bound immobilized PARP-1 leads to cell cycle arrest (5), replication fork disruption and DSB-mediated apoptosis (6, 30). PARPi hypersensitivity in BER-deficient cells correlates with elevated levels of endogenous unrepaired BER intermediates.
Discussion
Evaluation of pol β- and XRCC1-deficient cells for hypersensitivity to 4-AN revealed that disruption of BER can be a determinant of PARPi hypersensitivity. Thus, the BER deficiency imposed by deletion of either of these BER factors was associated with hypersensitivity to treatment with a PARPi. An XRCC1-deficiency has been implicated previously in a screen for factors mediating PARPi-mediated synthetic lethality (11). Our evaluation of XRCC1 mutants further suggests a requirement for the interactions between PARP-1, XRCC1 and pol β, and for their recruitment to sites of DNA damage, in order for the cell to exhibit resistance to PARPi. One implication of these results may be that determining the BER status of a cancer cell could represent an opportunity for increased PARPi-mediated lethality.
In 2005, BRCA1- and BRCA2-deficient cells were shown to be hypersensitive to PARPi (8). Such inhibitors were rapidly evaluated for targeted single agent therapy that should not affect repair competent non-cancer cells, and the concept of ‘synthetic lethality’ emerged in cancer therapy regimes (34). In an early phase I trial, the PARPi olaparib had antitumor activity in carriers of a BRCA1/2 mutation (35). Phase II studies of targeted treatment with olaparib provided positive proof of concept and a favorable therapeutic index in BRCA-mutated advanced ovarian and breast cancer (36, 37). Following presentations at the 2013 ASCO meeting, a cover story in Chemical and Engineering News (June 17, 2013) outlined new optimism about PARPi treatment of tumors with BRCA mutations. New information provided evidence that olaparib prevented disease progression and improved survival.
On the negative side, it was discovered that selection with a PARPi could result in deletion of the original BRCA mutation, restoration of HR and cellular resistance to PARPi (38). Resistance to another PARPi (AZD2281) was caused by up-regulation of P-glycoprotein efflux pumps (39). Additional determinants of PARPi sensitivity also were discovered (11, 12). In searching for alternate targets, PARPi sensitivity was observed in tumors with other HR defects including RAD51, ATM or the MRN complex (40-42). Mutations in the phosphate and tensin homolog (PTEN) were shown to result in a reduced capacity for HR (43). Phase I treatment in BRCA2 mutation carriers, also negative for PTEN, suggested that the synthetic lethality concept may be useful in prostate cancer patients with germline inactivating mutations in a number of HR repair genes (44). Some cancers exhibit “BRCAness” through an inhibition of the phosphoinositide-3-kinase (PI3K) pathway, down-regulation of BRCA1/2 and promotion of HR deficiency (45).
A high throughput siRNA screen implicated several HR proteins, and also XRCC1 in hypersensitivity to PARPi (11). Additionally, XRCC1-defective CHO cells were shown to be hypersensitive to single agent PARPi (46). Another group concluded that PARPi synthetic lethality was independent of XRCC1 and BER (47), but their studies utilized PARP siRNA-induced PARP depletion rather than a PARPi. XRCC1-deficient CHO and human ovarian cancer cell lines are also hypersensitive to ATR inhibitors that lead to DNA single-strand break formation (48). The authors proposed a mechanism of synthetic lethality. Taken together, these studies suggest that repair factors other than BRCA1/2 can be targets for PARPi.
Deficiency in BER (absence of pol β or XRCC1) is associated with MMS and temozolomide (TMZ) hypersensitivity (3). PARPi-induced enhancement of cytotoxicity is observed following combination with agents like MMS and TMZ resulting in damage repaired by a specific BER sub-pathway involving pol β and XRCC1 (13). Importantly, the effect of a PARPi is not epistatic with BER deficiency since it is observed in both pol β−/− and Xrcc1−/− cell lines (5, 14). We proposed that sensitization occurs because of increased numbers of PARP binding sites in DNA in the absence of efficient BER. An assay of total cellular PAR in untreated cells (Fig. 1A) revealed a higher level of PAR in the pol β and XRCC1 BER-deficient variants. This result is consistent with the idea that these cell types have a greater number of endogenous unrepaired PARP binding and activating DNA damage sites. We proposed that cells deficient in these BER factors would be hypersensitive to treatment with a PARPi alone. We suspected that endogenous DNA damage would be sufficient to effectively substitute for the alkylating agent used in combination treatment.
Pol β−/− and Xrcc1−/− cell lines demonstrated hypersensitivity to 4-AN compared with and cells, respectively (Fig. 2A and 3A). The BER-deficient cell types were also hypersensitive to olaparib and veliparib, PARPi currently under investigation for clinical use (Supplementary Fig. S7). As outlined above, in the absence of pol β, there will be a deficiency in cellular 5′-dRP lyase activity resulting in increased cellular damage sites for binding of PARP. Consistent with this proposal, over-expression of pol β in wild-type cells (clone #45 and #46) resulted in a further increase in 4-AN resistance (Fig. 2C). PARPi hypersensitivity also was observed in an alternate pair of isogenic pol β+/+ and −/− cell lines (Fig. 2D). As anticipated, and strikingly similar to results obtained with MMS (16), expression of full-length pol β reversed the 4-AN hypersensitivity phenotype while expression of the 8-kDa dRP lyase domain allowed for significant reversal (Fig. 2D).
Hypersensitivity to 4-AN observed in Xrcc1−/− cells could be reversed by expression of the wild-type XRCC1 protein (Fig. 3A). This result was expected since XRCC1 is required for recruitment of pol β to DNA damage sites, and as described above, pol β deficiency results in hypersensitivity to 4-AN. The clone (XRE8) expressing C12A mutated XRCC1 with a lesser binding affinity to pol β (14) was able to reverse sensitivity to 4-AN (Fig. 3B). Surprisingly the V88R variant (clone XV2), unable to bind pol β (14), also reversed the hypersensitivity phenotype (Fig. 3B). A newly characterized mouse XRCC1 L360R mutant was expressed in Xrcc1−/− cells and was unable to bind PARP-1 (Supplementary Fig. S3). Additionally, the L360R transfected cells (XL3) lacked the ability to rescue either the MMS or 4-AN hypersensitivity phenotype of Xrcc1−/− cells (Fig. 3C and D). These results are consistent with the idea that XRCC1 is recruited to DNA damage sites by binding to PARP and PAR, and that pol β is recruited by binding to XRCC1.
After 24 h treatment with 30 μM 4-AN, pol β wild-type cells accumulated in S phase of the cell cycle (Fig. 4A). This observation is similar to results obtained previously in this cell type after treatment with the combination of 0.25 mM MMS plus 10 μM 4-AN (5). Consistent with the hypothesis that inhibited PARP-1 bound to damaged DNA is cytotoxic as a function of replication-dependent cellular DSBs (6), we demonstrated γH2A.X staining associated with the S phase wild-type cells 24 h after exposure to 4-AN (Fig. 5). Under the same treatment conditions with 4-AN alone, pol β null cells, exhibiting significantly higher levels of DNA damage than wild-type cells (Fig. 5), accumulated in G2 rather than S phase (Fig. 4A), suggesting that the absence of pol β-dependent repair may prevent the S phase arrest. In this cell line, γH2A.X was associated with late S and G2 phase cells (Fig. 5). Consistent with 4-AN survival data in these pol β+/+ and pol β−/− cell lines (Fig. 2) and the relative increase in apoptotic cell death observed in pol β−/− cell lines (Fig. 5), a higher proportion of γH2A.X positive cells was observed at 24 h in pol β null compared with pol β null compared with pol β wild-type cells (Fig. 5). After removal of 4-AN, the levels of γH2A.X positive cells decreased. However, the proportion of cells positive for γH2A.X was always greater in the pol β−/− cells (Fig. 5).
We propose that endogenous DNA damage is repaired by BER, and in the case of repair deficiency, more unrepaired intermediates accumulate than in wild-type cells. In 4-AN-treated pol β- and XRCC1-deficient cells (Fig. 6A; lanes 2 and 4), the results revealed a higher level of DNA-bound PARP-1 than in the isogenic wild-type cells (Fig. 6A; lanes 1 and 3). DNA-bound immobilized PARP-1 leads to replication fork disruption and DSB-mediated apoptosis (Fig. 6B). The increased PARP inhibitor hypersensitivity observed in BER-deficient cells thus correlates with an increase in unrepaired BER intermediates. In summary, we show that BER deficiency as a result of either pol β- or XRCC1-deletion is associated with PARPi hypersensitivity. Thus, BER deficiency could represent a therapeutic opportunity for PARPi single agent therapy.
Supplementary Material
Acknowledgements
The authors thank Dr. William Beard and Lois Wyrick for help with figure preparation, members of the NIEHS Protein Expression Core Facility for assistance with expression vector construction and CD analysis, and the NIEHS Flow Cytometry Center for help with flow cytometric analyses.
Grant Support
This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (project number Z01 ES050159).
Footnotes
Conflict of Interest Statement: The authors declare that there is no conflict of interest.
References
- 1.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–8. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 3.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase β in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murai J, Huang S-yN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. Poly(ADP-ribose) polymerase activity prevents signaling pathways for cell cycle arrest following DNA methylating agent exposure. J Biol Chem. 2005;280:15773–85. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- 6.Heacock ML, Stefanick DF, Horton JK, Wilson SH. Alkylation DNA damage in combination with PARP inhibition results in formation of S-phase-dependent double-strand breaks. DNA Repair. 2010;9:929–36. doi: 10.1016/j.dnarep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 8.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 9.Metzger MJ, Stoddard BL, Monnat RJ., Jr PARP-mediated repair, homologous recombination, and back-up non-homologous end joining-like repair of single-strand nicks. DNA Repair. 2013;12:529–34. doi: 10.1016/j.dnarep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–9. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair. 2008;7:2010–9. doi: 10.1016/j.dnarep.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J, et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle. 2012;11:3837–50. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton JK, Wilson SH. Predicting enhanced cell killing through PARP inhibition. Mol Cancer Res. 2013;11:13–8. doi: 10.1158/1541-7786.MCR-12-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton JK, Stefanick DF, Gassman NR, Williams JG, Kedar PS, Wilson SH. Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses. DNA Repair. 2013;12:774–85. doi: 10.1016/j.dnarep.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, et al. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–6. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 16.Sobol RW, Prasad R, Evenski A, Baker A, Yang X-P, Horton JK, et al. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–10. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 17.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–29. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 18.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. Domain specific interaction in the XRCC1-DNA polymerase β complex. Nucleic Acids Res. 2000;28:2049–59. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berquist BR, Singh DK, Fan J, Kim D, Gillenwater E, Kulkarni A, et al. Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res. 2010;38:5023–35. doi: 10.1093/nar/gkq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuneo MJ, London RE. Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase β binding affinity. Proc Natl Acad Sci USA. 2010;107:6805–10. doi: 10.1073/pnas.0914077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gassman NR, Stefanick DF, Kedar PS, Horton JK, Wilson SH. Hyperactivation of PARP triggers nonhomologous end-Joining in repair-deficient mouse fibroblasts. PLoS ONE. 2012;7:e49301. doi: 10.1371/journal.pone.0049301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J BiolChem. 2005;280:33826–38. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 23.Prasad R, Horton JK, Chastain PD, II, Gassman NR, Freudenthal BD, Hou EW, et al. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku288. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase β null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair. 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 25.Singhal RK, Prasad R, Wilson SH. DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testes nuclear extract. J Biol Chem. 1995;270:949–57. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 26.Kubota Y, Horiuchi S. Independent roles of XRCC1's two BRCT motifs in recovery from methylation damage. DNA Repair. 2003;2:407–15. doi: 10.1016/s1568-7864(02)00242-2. [DOI] [PubMed] [Google Scholar]
- 27.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–33. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–81. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong H-K, Wilson DM., 3rd XRCC1 and DNA polymerase β interaction contributes to cellular alkylating-agent resistance and single-strand break repair. J Cell Biochem. 2005;95:794–804. doi: 10.1002/jcb.20448. [DOI] [PubMed] [Google Scholar]
- 30.Horton JK, Stefanick DF, Wilson SH. Involvement of poly(ADP-ribose) polymerase activity in regulating Chk1-dependent apoptotic cell death. DNA Repair. 2005;4:1111–20. doi: 10.1016/j.dnarep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Kinner A, Wu W, Staudt C, Iliakis G. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–94. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redon CE, Nakamura AJ, Zhang YW, Ji JJ, Bonner WM, Kinders RJ, et al. Histone γH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res. 2010;16:4532–42. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redon CE, Nakamura AJ, Martin OA, Parekh PR, Weyemi US, Bonner WM. Recent developments in the use of γ-H2AX as a quantitative DNA double-strand break biomarker. Aging. 2011;3:168–74. doi: 10.18632/aging.100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chalmers AJ. The potential role and application of PARP inhibitors in cancer treatment. Br Med Bull. 2009;89:23–40. doi: 10.1093/bmb/ldp005. [DOI] [PubMed] [Google Scholar]
- 35.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-Ribose) polymerase in tumors from BRCA mutation carriers. NEJM. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 36.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. The Lancet. 2010;376:245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 37.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. The Lancet. 2010;376:235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 38.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 39.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AOH, Zander SAL, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA. 2008;105:17079–84. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–9. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Javle M, Curtin NJ. The potential for poly (ADP-ribose) polymerase inhibitors in cancer therapy. Ther Adv Med Oncol. 2011;3:257–67. doi: 10.1177/1758834011417039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ihnen M, Zu Eulenburg C, Kolarova T, Qi JW, Manivong K, Chalukya M, et al. Therapeutic potential of the poly(ADP-ribose) polymerase inhibitor rucaparib for the treatment of sporadic human ovarian cancer. Mol Cancer Ther. 2013;12:1002–15. doi: 10.1158/1535-7163.MCT-12-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim J-S, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Molec Med. 2009;1:315–22. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandhu SK, Omlin A, Hylands L, Miranda S, Barber LJ, Riisnaes R, et al. Poly (ADP-ribose) polymerase (PARP) inhibitors for the treatment of advanced germline BRCA2 mutant prostate cancer. Ann Oncol. 2013;24:1416–8. doi: 10.1093/annonc/mdt074. [DOI] [PubMed] [Google Scholar]
- 45.Ibrahim YH, Garcéa-Garcéa C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–47. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strém CE, Johansson F, Uhlén M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–75. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Nat Acad Sci USA. 2011;108:3406–11. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sultana R, Abdel-Fatah T, Abbotts R, Hawkes C, Albarakati N, Seedhouse C, et al. Targeting XRCC1 deficiency in breast cancer for personalized therapy. Cancer Res. 2013;73:1621–34. doi: 10.1158/0008-5472.CAN-12-2929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.