Abstract
Neuroimaging has played an important role in non-invasive diagnosis and differentiation of neurodegenerative disorders, such as Alzheimer's disease and Mild Cognitive Impairment. Various features have been extracted from the neuroimaging data to characterize the disorders, and these features can be roughly divided into global and local features. Recent studies show a tendency of using local features in disease characterization, since they are capable of identifying the subtle disease-specific patterns associated with the effects of the disease on human brain. However, problems arise if the neuroimaging database involved multiple disorders or progressive disorders, as disorders of different types or at different progressive stages might exhibit different degenerative patterns. It is difficult for the researchers to reach consensus on what brain regions could effectively distinguish multiple disorders or multiple progression stages. In this study we proposed a Multi-Channel pattern analysis approach to identify the most discriminative local brain metabolism features for neurodegenerative disorder characterization. We compared our method to global methods and other pattern analysis methods based on clinical expertise or statistics tests. The preliminary results suggested that the proposed Multi-Channel pattern analysis method outperformed other approaches in Alzheimer's disease characterization, and meanwhile provided important insights into the underlying pathology of Alzheimer's disease and Mild Cognitive Impairment.
Keywords: Pattern analysis, Alzheimer's disease, Mild cognitive impairment
1. Introduction
Neuroimaging data are a rich source of information on brain anatomy and physiology. Neuroimaging has been a fundamental component of the neurological disorder diagnosis, and also plays an important role in therapy assessment and disease progression monitoring. Due to the large size of current 3D neuroimaging data, it is difficult to quantitatively analyze the volumetric images for computer-aided-diagnosis (CAD) and clinical-decision-support (CDS) [1–3]. Therefore, researchers usually extract features from the neuroimaging data to efficiently represent them without losing important information.
Various studies on identifying the most discriminative disease-related brain features have been reported. A thorough review of all these studies is beyond the scope of this paper. For interested readers, more details of these studies can be found in several reports [4–12]. These features can be roughly divided into two groups, global and local features. Global features that treat all of the brain regions with no distinction were commonly used. For example, Qian et al. [4] designed a neuroimaging retrieval system with four 3D feature descriptors based on 100 brain Magnetic Resonance Imaging (MRI) studies. Unay et al. [5] proposed a retrieval system for MRI data based on local binary patterns incorporating spatial context information. Ramírez et al. [6] employed Support Vector Machine (SVM) combined with a casting votes technique in their study specifically for early diagnosis of Alzheimer's disease (AD) based on Single Photon Emission Computed Tomography (SPECT).
However, the neurodegenerative disorders usually affect certain brain regions associated with memory or motor functions. We could use these disease-specific patterns to enhance our understanding of AD pathology and facilitate the neuroimaging analysis. Many researchers took advantage of such disease-related pathological pattern by analyzing a set of brain region of interest (BROIs) instead of the whole brain. There are two ways to select BROIs. The first is based on the known pathology of the disorder. Wang et al. [7,8] recently investigated the surface and shape features on MRI data, using multivariate tensor-based morphometry (mTBM) and spherical harmonics (SPHARM) shape analysis pipeline, respectively. Only ventricles and hippocampus regions were selected in their studies. Batty et al. [9] used a predefined knowledge-based mask to segment the five BROIs from the brain and further extracted the Gabor wavelet features for retrieval using 2-[18F]fluoro-2-deoxy-d-glucose (FDG) with Positron Emission Tomography (PET). We have previously proposed a set of disease-oriented masks (DOMs) based on the literature [10–16] and adaptively modified them with t-maps [17]. The second approach to BROI selection is to apply certain pattern analysis techniques on the whole brain and then select the BROIs based on a significance metric or test statistics. Zhang et al. [18] selected a set of BROIs in their study based on the cost sensitive variance score (CostVS) derived from SVM. The PET measurements of the regional average cerebral metabolic rate of glucose consumption (CMRGlc) [19] were used to calculate the CostVS. Heckemann et al. [20,21] employed the two-one-sided-test (TOST) to evaluate the discriminative power of the BROIs based on the regional volume loss using MRI.
Both types of BROI selection approaches were reported to improve the analysis performance by using BROIs instead of whole brain. However, the resultant BROI sets of these studies were not optimally defined and there were several issues that needed to be addressed. The first is that most of the findings were based on single-center studies with small populations [4–6]. Potentially biased conclusions could be drawn based on such single-center populations. In our previous studies, we investigated several advanced feature descriptors, i.e., 3D-GLCM [22], 3D-DCvT [23], 3DGabor [24], and vcLPB [25], based on a small dataset of 209 subjects with 11 subtypes of dementia. Voxel-wise t-test was carried out to refine the predefined DOMs. This method achieved better performance than using conventional DOMs, but this better performance might be specific to our dataset and better performance on other datasets could not be guaranteed. The second problem is the drawbacks of individual pattern analysis techniques. Most researchers used the t-test [7,8,15,20,21] or SVM [16,18] to detect the disorder sensitive regions, but neither t-test nor SVM supported multi-group analysis and they also ignored the correlations between different features. Another issue is the disorder diversity. It is crucial to understand what brain functional regions are involved in distinguishing the disorders from each other, as well as distinguishing the patients with disease (‘patients’) from the normal aging control subjects (NC) who also showed slightly reduced metabolism in certain regions highly related to age. If the dataset contains multiple disorders or progressive disorders, it is difficult to reach a consensus on what brain regions could best distinguish different disorders from each other and also from the normal controls. We could understand this issue by looking at two extreme cases: an overlarge pattern comprising the degenerative patterns of all individual disorders might have a negative impact on distinguishing the patients from normal subjects due to the involvement of many non-disorder-specific regions, whereas a small pattern of associated regions with high agreement by all disorders might decrease the discriminative power between disorders since the abnormalities exist in the same regions. In addition, since neurodegenerative disorders, such as AD, are usually progressive, the degenerative patterns at different progressive stages would be different. AD is an irreversible neurodegenerative disease that results in a loss of mental functions due to loss of brain tissue. As the disease progresses, it robs the patients of their memory, and eventually, overall mental and physical function, leading to death. Mild Cognitive Impairment (MCI) differs from AD and normal age-related memory change. People with MCI have ongoing memory problems but not to the point where their impairment interferes significantly with daily activities. MCI can represent a pre-symptomatic status of AD, conferring a high conversion rate of 16% to AD per annum [26]. A pattern of hypo-metabolism in Hippocampus and Posterior Cingulate Cortex was reported in a study on MCI [27], whereas whole brain was affected in more severe AD [12–15]. The pathological pattern analysis for multiple and progressive disorders has been recognized as a central research area to advance our understanding the AD and MCI pathology.
To address the abovementioned issues, in this paper, we proposed a Multi-Channel analysis approach to analyze the hypo-metabolism patterns of AD and MCI. The innovation of our proposed approach is that it could integrate the multiple patterns derived from different patient groups using different analysis tools. We investigated a variety of pattern analysis approaches and applied them in parallel to analyze a multi-center dataset of 369 participants. The integrated results of individual analyses were then used to characterize AD and MCI patients. Several advanced feature descriptors were also investigated in this study to further improve its performance.
The paper is organized as follows. In Section 2, we will elaborate the proposed Multi-Channel pattern analysis approach and also detail data acquisition, pre-processing, feature extraction and performance evaluation methods used in this study. In Section 3, a multi-phase workflow of experiments will be introduced, together with the preliminary results from each phase of experiments. The findings from the results will be discussed in Section 4. Finally, we will conclude in Section 5.
2. Materials and methods
2.1. Study design overview
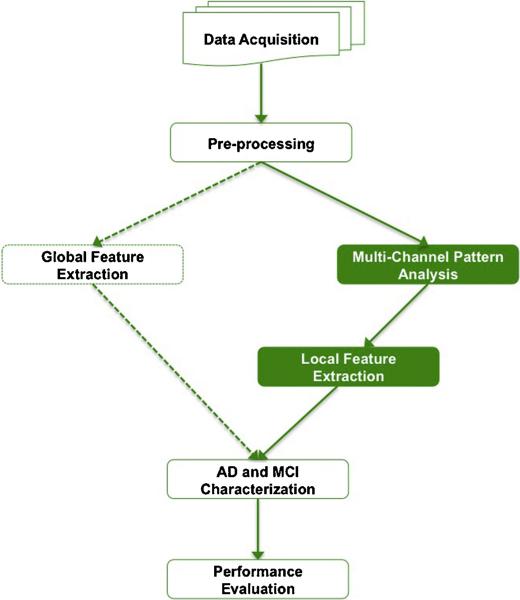
The design of this study is shown in Fig. 1. We first obtained the neuroimaging data of 369 participants from a public multi-center neuroimaging repository, the Alzheimer's Disease Neuroimaging Initiative (ADNI). The data were then pre-processed by spatial normalization, brain functional region segmentation, and gray-tone correction. Both global and local features could be extracted from the pre-processed data, and our focus is local feature extraction. To address the issues discussed in Section 1, we designed the Multi-Channel analysis framework, which could overcome the deficiencies of individual pattern analysis methods and meanwhile highlight the most discriminative brain regions. We used the features derived from the proposed Multi-Channel framework to characterize AD and MCI patients, and compared its performance with global methods and other local feature selection methods.
Fig. 1.
Study design schematic.
2.2. Data acquisition
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. The identification of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55–90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. For up-to-date information, see www.adni-info.org.
To achieve more reliable region-wise pattern analysis and also to extract robust features from the functional regions, accurate registration with MRI data is desired to compensate the low resolution of PET data. Therefore, the recruited subjects should have both FDG-PET and T1-weighted MRI images. 369 subjects from the ADNI baseline cohort met with our requirements, and were consequently selected in this study.
2.3. Pre-processing
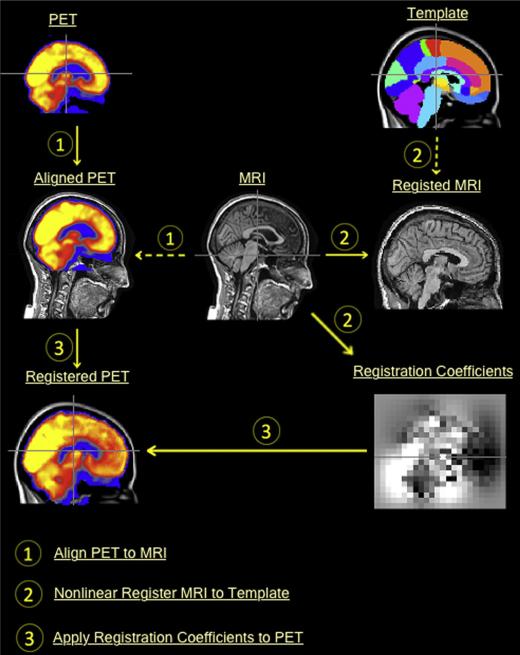
All ‘raw’ PET data were converted to the ADNI format with same voxel size of 1.5 mm3 and a uniform resolution of 8 mm full width at half maximum resolution [26]. The PET images were further aligned to the corresponding MRI image using FSL FLIRT [28], as shown in Fig. 2 – Step 1. We then nonlinearly registered the MRI images to the ICBM 152 template [29], using the NREG algorithm [30,31] provided in Image Registration Toolkit (IRTK) [30]. IRTK works in a coarse-to-fine manner with isotropic control point spaces ranging from 12 mm to 1.5 mm in 4 octaves. Fig. 2 – Step 2 shows an example of nonlinear registration using IRTK. Finally, the resulting registration coefficients were applied to warp the aligned PET image into the template space, as shown in Fig. 2 – Step 3. After the PET images were registered to the ICBM template, we visually checked all registered PET images and found 17 of them had morphometric distortions that could not be corrected. Therefore, we excluded these distorted images in this study and downsized the database to 352 subjects. The participants include 85 patients with AD, 181 subjects with MCI and 86 NCs. Automatic labeling of 83 brain regions was achieved in the template space using the multi-atlas propagation with an enhanced registration (MAPER) approach [21]. The atlas data required for MAPER comprising of 30 T1-weighted MRI images acquired from the National Society for Epilepsy at Chalfont, UK. The segmentation protocols were described by Hammers et al. [32]. The voxel-wise CMRGlc parameters were estimated based on raw uptake values of PET data [17]. To eliminate gray-tone variations between individual scans, we further normalized the CMRGlc parameters using the mean value of the cerebellum, which is spared in AD and MCI.
Fig. 2.
PET registration onto the template space.
2.4. Multi-Channel neurodegenerative pattern analysis
Pattern analysis could be carried out between different group pairs using different analysis tools. We refer to each combination as a channel, and all channels are parallel to each other. Multi-Channel analysis could provide complementary information to individual single channel analyses. Therefore, in this study, we developed a Multi-Channel analysis algorithm to depict the neurodegenerative patterns for AD and MCI. We first divided the ADNI dataset into three groups based on their diagnoses, and then designed 10 parallel channels to analyze the neurodegenerative patterns on all possible group-pairs. Finally, individual channels voted on all brain regions to select the most discriminative regions. Table 1 shows the combination of the group pairs and the analysis techniques, including the Two-One-Sided-Test (TOST), SVM and Elastic Net (EN). In this study, we selected mean CMRGlc value of each brain region as the observation values to carry out the Multi-Channel analysis, as it is simple and effective in capturing brain metabolism patterns and has been widely used in AD and MCI diagnosis [10,13,15,18,33–36].
Table 1.
Confusion table of10 channels; digits in the table represent the channel index.
| Group Pair | TOST | SVM | EN |
|---|---|---|---|
| AD/NC | 1 | 2 | 3 |
| MCI/NC | 4 | 5 | 6 |
| AD/MCI | 7 | 8 | 9 |
| D/MCI/NC | n/a | n/a | 10 |
We first designed three channels (CH_1, CH_4, CH_7) based on the classic TOST on AD/NC group pair, MCI/NC group pair, and AD/MCI group pair, respectively. TOST is a classic statistical test in inference [37] and arguably the most commonly used test in neurodegerative pattern analysis [7,8,15,20,21]. In a TOST, a given null hypothesis of equivalence, H0, will be rejected when the p-value of the test statistic is smaller than a user-defined threshold. In this study, we set the threshold as 0.05. The feature variables with significance test statistics were outputted into one vector as the analysis result.
Another 3 channels (CH_2, CH_5, CH_8) were designed with SVM for same group pairs as TOST. SVM is widely used in classification and regression [38]. When SVM is used with the linear kernal to classify two groups of data, the hyperplane slopes on each dimension could be used as the feature weights. Given a training set of feature-diagnosis pairs (xm, xm), m = 1, . . ., M where xm ∈ RN and ym ∈{−1,1}M, the SVM solves the problem:
| (1) |
subject to ym(wT xm + b)≥1 − ξm; ξm≥0.
In this study, SVM based pattern analysis was performed in two steps. Firstly, we conducted a 10-fold cross validation on the three-category classification using the LIBSVM [39] to obtain the best estimate of C. When C was decided (C = 8 in this study), we then solved the problem in Eq. (1) using CVX, a package for specifying and solving convex programs [40]. Among all the regions, we selected the regions with larger weights than the upper quartile of the weight distribution.
We further designed 4 channels (CH_3, CH_7, CH_9, CH_10) using EN. EN is a well established feature selection algorithm solving the problem:
| (2) |
where y is the response vector of M observations, X is the matrix of M feature vectors, X = {x1, . . ., xm, . . ., xM}T, λ is a positive regularization parameter, and β is the vector with same dimension as xm. EN introduced l1 and squared l2 penalty of β, which could encourage grouping effect on feature variables and remove the limitation on the number of selected features. These characteritics make EN very suitable for studies with a large number of observations and a small number of features, or correlated features [41]. The ADNI data used in this study are suitable for EN since the number of patients is much larger than the number of features/brain regions, and the brain regions were highly correlated. Another characteristic of EN is that it can be used for multi-categorical feature selection, which is not supported by TOST and SVM. Therefore, we performed EN-based analysis on three group pairs and also on the whole database with multiple classes. In this study, the weight of λ1/λ2 was set as 0.5. With no need to predefine the number of results or a statistic threshod for any hypothesis, EN is able to select the features automatically.
Ten channels were set up in parallel. We finally integrated the outputs from individual channels for each brain functional region as:
| (3) |
| (4) |
where vc,n is the output of the nth region in the cth channel, and
| (5) |
This Multi-Channel scheme allowed the individual channels to vote on each brain region, and set higher values on the feature variables recognized by more channels [34]. The votes of each brain region in this study, were highly correlated with the brain structure's discriminative power.
2.5. Feature extraction
As mentioned in Section 2.4, regional CMRGlc parameters are very simple and effective in capturing patterns of brain hypo-metabolism and we used them as the basic features and fused them into a N-element vector:
| (6) |
where xm is the mth image in the database, N is the number of selected regions, and xm,n is the average CMRGlc parameter in the nth region. For a whole brain based method, the feature vector has N = 83 elements. To clarify, region-wise mean CMRGlc parameters were used in Multi-Channel analysis to select the most discriminative brain regions; they were also used as basic features in AD and MCI characterization.
We further investigated three advanced feature descriptors and extracted more sophisicated features incorporated in the Multi-Channel analysis method. The advanced features were used to improve the characterization performance. All of the advanced feature descriptors we used were specific for 3D data, since current neuroimaging data are usually collected in 3D format and 3D feature descriptors are superior to 2D feature descriptors in capturing the spatial variations on volumetric neuroimaging data. We have verified the superiority of 3D feature descriptors vs. their 2D counterparts in previous work [22–24]. The advanced features based on spatial transforms were applied to each selected brain regions with a bounding box where the non-related regions were filtered out using the zero-padding technique.
We extracted 14 Haralick texture features [42] based on the 3D Gray Level Co-occurrence Matrices (GLCMs). GLCM is widely used in texture analysis since it is capable of capturing the spatial dependence of voxel intensities in local neighborhood in an image. In this study, the d parameters for GLCM algorithm were set as three steps (1, 3, and 5 voxels).
We further extracted the frequency features based on 3D Gabor wavelets and 3D discrete curvelets, since they both performed well in general. Gabor wavelets could offer the optimal simultaneous localization of both spatial and frequency information [43].
Different from Gabor wavelets, the Discrete Curvelet Transform (DCvT) is more sensitive to the curvature features along the lines and boundaries [44]. A 3D curvelet can be defined by a scale parameter j, an orientation parameter l, and a translation parameter k. The 3D discrete curvelet bank was defined as having the same scale and bandwidth parameters as the 3D Gabor wavelet bank. More implementation details could be found in our previous studies [22–24].
2.6. Performance evaluation
We used the query by example paradigm in this study, and adopted the leave-one-out cross-validation on the whole database. The similarity between any two feature-vectors was calculated by the normalized mutual information [45].
We evaluated the performance using the mean average precision (MAP), i.e.:
| (7) |
where q is the index of the query, Q is the total number of queries, k is the rank in the sequence of retrieved image, Kq is the number of total retrieved image for the query, pq(k) is the precision at cut-off k in the result list, relq(k) is the relevance score of the kth retrieval result given the query, and Tq is the number of relevant images associated with the query. In this study, MAP is implemented slightly different from the classical way, since the group sizes of AD and MCI do not match. We assume that there are at least 5 relevant cases exist for each query, therefore we set the Tq = 5 for all the queries. We also used elastic relevance criteria in this study, based on fact that MCI usually represents the transition state of a NC to AD. Therefore, we set the relevance criteria as follows:
- if the query is AD and the retrieval result is normal, or vise versa, then the relevance score is 0:
- if the retrieval result is from the same group of query, than the relevance score is 1:
- for other scenarios, we set the relevance score as 0.25:
To verify the effectiveness of the proposed method, we compared it to the global method and other local methods, including the baseline method with whole brain features, the DOM method with 40 preselected regions as listed in Table 2 based on literature [10–16], and three state-of-the-art feature selection methods, i.e., TOST, SVM and EN, which were equivalent to the single channels in our Multi-Channel framework. Same features and evaluation metric were used across all methods.
Table 2.
The voting results for all 83 brain functional regions by Multi-Channel analysis. The first half regions are disease-specific regions and included in DOMs. The second part regions are generally considered spared by AD pathology.
| Region index | Brain functional region label (right, left) | MultiXH scores |
|---|---|---|
| DOM regions: 40 regions based on literatures | ||
| (1.2) | Hippocampus (R, L) | (6,5) |
| (3,4) | Amygdala (R, L) | (3, 6) |
| (5,6) | Anterior temporal lobe (R, L) | (1,0) |
| (7,8) | Anterior temporal lobe, lateral part (R, L) | (0,3) |
| (9,10) | Parahippocampal and ambient gyri (R, L) | (4,1) |
| (11,12) | Superior temporal gyrus, posterior part (R, L) | (2,4) |
| (13.14) | Middle and inferior temporal gyrus (R, L) | (6, 4) |
| (15,16) | Fusiform gyrus (R, L) | (2, 2) |
| (20,21) | Insula (R, L) | (0, 0) |
| (24, 25) | Cingulate gyrus, anterior part (R, L) | (0, 2) |
| (26. 27) | Cingulate gyrus, posterior part (R, L) | (4, 5) |
| (30, 31) | Posterior temporal lobe (R, L) | (2, 1) |
| (32. 33) | Inferiolateral remainder of parietal lobe (R, L) | (6, 5) |
| (54, 55) | Anterior orbital gyrus (R, L) | (2, 1) |
| (60, 61) | Postcentral gyrus (R, L) | (2, 1) |
| (62. 63) | Precuneus included in superior parietal | (4, 4) |
| gyrus (R, L) | ||
| (68, 69) | Medial orbital gyrus (R, L) | (0, 2) |
| (70, 71) | Lateral orbital gyrus (R, L) | (1,2) |
| (72, 73) | Posterior orbital gyrus (R, L) | (2, 2) |
| (82, 83) | Superior temporal gyrus, anterior part (R, L) | (1,2) |
| Non-DOM regions: 43 regions spared by AD pathology | ||
| (17,18) | Cerebellum (R, L) | (1,1) |
| 19 | Brainstem (unpaired) | 4 |
| (22, 23) | Lateral remainder of occipital lobe (R, L) | (0, 3) |
| (28, 29) | Middle frontal gyrus (R, L) | (2, 0) |
| (34, 35) | Inferiolateral remainder of parietal lobe (R, L) | (1,1) |
| (36, 37) | Caudate nucleus (R, L) | (2, 2) |
| (38, 39) | Nucleus accumben (R, L) | (2, 4) |
| (40, 41) | Thalamus (R, L) | (1,0) |
| (42, 43) | Pallidum (R, L) | (1,2) |
| 44 | Corpus callosum (unpaired) | |
| (45, 46) | Lateral ventricle apart temporal horn (R, L) | (0, 4) |
| (47, 48) | Lateral ventricle, temporal horn (R, L) | (3, 3) |
| 49 | Third ventricle (unpaired) | 0 |
| (50, 51) | Precentral gyrus (R, L) | (1,1) |
| (52, 53) | Straight gyrus (R, L) | (0, 0) |
| (56, 57) | Inferior frontal gyrus (R, L) | (1,1) |
| (58, 59) | Superior frontal gyrus (R, L) | (0, 0) |
| (64, 65) | Lingual gyrus (R, L) | (3, 2) |
| (66, 67) | Cuneus (R, L) | (0, 1) |
| (74, 75) | Substantia nigra (R, L) | (2, 5) |
| (76. 77) | Subgenual frontal cortex (R. L) | (6, 6) |
| (78, 79) | Subcallosal area (R, L) | (3, 4) |
| 80, 81) | Pre-subgenual frontal cortex (R, L) | (6, 1) |
3. Experiments and results
This section is divided into three subsections. Section 3.1 describes the pattern analysis experiments (Phase 1) and demonstrates the derived patterns of each single channel approaches and our proposed Multi-Channel approach. Section 3.2 describes the optimization experiments (Phase 2) on identifying the most discriminative regions of the brain based on the results from Phase 1 experiments. To validate the effectiveness of the selected brain regions, we carried out a further series of validation experiments (Phase 3) using the advanced features, as described in Section 2.5. The design of the Phase 3 experiments and the preliminary results were shown in the Section 3.3.
3.1. Multi-Channel pattern analysis
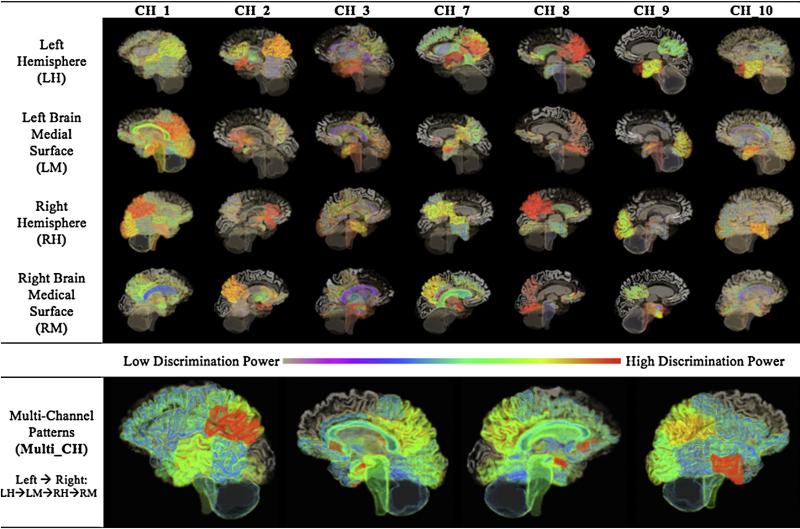
The pattern analysis experiments were carried out in 10 parallel channels in this phase (Phase 1), as described in Section 2.4. Table 2 shows the selected BROIs and their final voting scores of Multi-Channel analysis. The 40 BROIs based on literature studies [10–16] were also listed for comparison. Diverse patterns were derived from different channels providing complementary information to each other. CH_1, CH_2 and CH_3 recoginized the Hippocampus, Posterior Cingulat Gyrus, Parietal Lobe as the sensitive regions to distinguish AD from NC. Few regions were identified to distinguish MCI from NC by CH_4, CH_5 and CH_6. Specificly no regions were selected by Elastic Net (EN) for MCI/NC group pair. CH_7, CH_8, and CH_9 showed strong agreement on Parietal Lobe. The pattern derived from multi-class EN analysis in CH_10 includes 18 regions spreading around the Hippocampus, Amygdala, Cingulate Gyrus, Parietal Lobe. These findings were consistent with known AD pathology. However, we also found the AD tended to affect the Temporal Pole, Brainstem, Subgenual and Pre-subgenual Frontal Cortex. Fig. 3 illustrates the patterns for all channels expected for the MCI/NC pair.
Fig. 3.
The 3D patterns derived from single channel and Multi-Channel analyses. The brain images were generated using 3D Slicer (version 4.1) [46]. The MRI image, acquired from a normal subject at ADNI, was used for illustration purposes only.
Multi_CH captured all regions detected by CH_1 to CH_10, thereby showing a more spreading pattern. Furthermore, Multi_CH could depict the discriminative power of individual regions on a more objetive basis, for more votes on a certain region means stronger agreement among different channels in distinguising the disorders from each other or the NCs. The pattern of Multi_CH involved more functional brain regions when compared to the patterns of individual single-channel analyses, and the pattern's contrast was remarkably increased, too.
3.2. Supervised pattern optimization
Multi-Channel pattern analysis identified the disorder sensitive regions, but resulted in an overlarge pattern containing 67 brain regions with at least 1 vote from these single channels, as shown in Table 2. However, a considerable number of these brain regions were supported by very few channels. For example, 19 out of the 67 brain regions were recognized by single channels. Such regions are undesirable as they might be caused by biased group size or the drawbacks of the pattern analysis algorithm, and hence decrease the characterization performance.
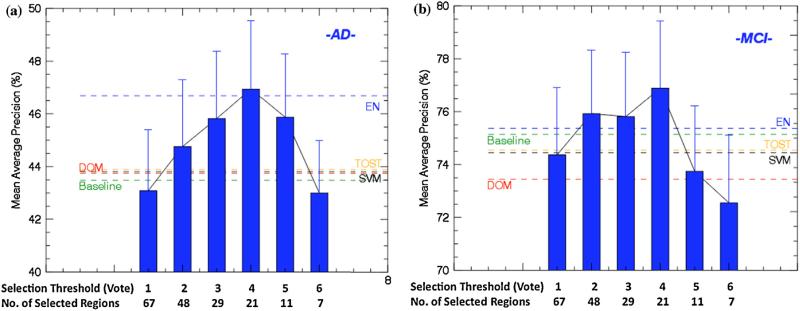
To optimize the pathological pattern derived from Multi-Channel analysis in neuroimaging characterization, we developed a learning-based pattern optimization approach, which tested a set of feature selection criteria and helped to find the optimal subset of the brain regions in the Multi_CH pattern. We first sorted the brain regions according to analysis result, and then carried a series of retrieval experiments to characterize the AD and MCI subjects using subsets of the 67 brain regions. The subset was selected in a ‘many to few’ manner based on the regions’ votes. We designed 6 experiments in this phase (Phase 2). Regions with votes larger than 1 were selected in experiment 1; regions with votes larger than 2 were selected in the experiment 2; and so on.
The results of Phase 2 experiments were shown in Fig. 4, compared with the Baseline, DOM-based, TOST, SVM, and EN methods. The best performance achieved when 21 regions were selected with the selection threshold of 4 for both AD and MCI, as highlighted in Table 2. Overall, DOM did not enhance the AD and MCI characterization. DOM-based method outperformed the baseline method on AD by 0.3%, but did not match baseline performance on MCI with a decrease of 1.9%. The reason for the conflicting performance on AD and MCI is that the DOM contains too many brain regions. On the one hand, the DOM might have a negative impact on distinguishing AD subjects from NCs due to the involvement of many MCI related regions. On the other hand, DOM also helps to distinguish MCI cases from AD cases since many AD related regions might not have an impact on MCI cases. Single pattern analysis methods, i.e., TOST, SVM and EN, were not robust in AD and MCI characteriation. EN, taking the feature correlation into account, could always achive better result than TOST and SVM. However, none of them could achieve better performance than the Multi-Channel method when the vote threshold was set to 4.
Fig. 4.
The MAP for AD and MCI characterization compared to DOM and Baseline methods. Single analysis approaches were also compared. (a) Feature Optimization for AD. (b) Feature Optimization for MCI.
3.3. Validation with advanced features
To validate the effectiveness of the derived pathological pattern, we employed several advanced feature descriptors as discussed in Section 2.5 to extract various features from the selected regions for analysis, and compared the performance to baseline and DOM-based methods.
Tables 3 and 4 show the performances for AD and MCI when using different features, including (a) the region-wise average CMRGlc parameters, (b) 3D-DCvT coefficients, (c) 3D-Gabor coefficients and (d) Haralick features from 3D-GLCM. The optimized Multi-CH based pattern could facilitate the characterization of AD and MCI, compared to baseline and the DOM-based method. The best performance (measured by MAP) was achieved by 3D-Gabor at 56.3%, strikingly improved the baseline retrieval by 13.3%. 3D-GLCM and 3D-DCvT also have comparable performance as 3D-Gabor, and all these three features could better characterize AD subjects than the average CMRGlc parameters. The retrieval of MCI subjects, on the other hand, was less impressive. The advanced features on the selected regions did not lead to better characterization than the baseline or DOM-based method. On the contrary, advanced features combined with Multi-CH patterns resulted in even worse performance than the baseline performance in MCI. In particular, the 3D-Gabor had the poorest performance at 61.47%, which was 11.7% lower than the baseline. However, Table 4 shows an increase of MAP based on the Multi_CH pattern using the average CMRGlc features, which was 1.6% higher than the baseline method and 3.1% higher than DOM-based method. Average CMRGlc features also achieved the best retrieval accuracy of MCI at 76.2%, and outperformed DCvT by 14.7%, Gabor by 6% and GLCM by 6.9%. Therefore, we could conclude that the pattern derived by the proposed Multi-Channel analysis approach overcomes the drawbacks of DOM and could enhance the characterization for AD and MCI, and the improvements were striking when the most suitable features were extracted, i.e.: the average CMRGlc parameters for MCI, and 3D-Gabor coefficients for AD.
Table 3.
The AD patient characterization performance (MAP ± standard deviation, %) of feature descriptors.
| Extraction | Feature |
|||
|---|---|---|---|---|
| Regional CMRGlc | 3D-Gabor | 3D-DCvT | 3D-GLCM | |
| Baseline | 43.48 ± 15.44 | 43.00 ± 12.08 | 43.71 ± 13.20 | 45.26 ± 12.75 |
| DOM | 43.75 ± 17.42 | 46.59 ±11.30 | 46.53 ± 12.55 | 33.48 ± 08.36 |
| Multi-CH | 46.69 ± 17.25 | 56.32 ± 08.72 | 51.63 ± 16.85 | 48.05 ± 17.21 |
Table 4.
The MCI patient characterization performance (MAP ± standard deviation, %) of feature descriptors.
| Extraction | Feature |
|||
|---|---|---|---|---|
| Regional CMRGlc | 3D-Gabor | 3D-DCvT | 3D-GLCM | |
| Baseline | 74.63 ± 15.07 | 73.18 ± 15.17 | 72.11 ± 14.05 | 70.19 ± 13.99 |
| DOM | 73.10 ± 14.86 | 71.69 ± 12.98 | 71.83 ± 14.33 | 71.18 ± 25.01 |
| Multi-CH | 76.20 ± 15.32 | 61.47 ± 06.15 | 70.25 ± 11.87 | 69.27 ± 16.43 |
4. Discussion
ADNI Dataset
The dataset comprised 352 subjects from the ADNI baseline FDG-PET database; more than 50% were diagnosed with MCI; ~25% were diagnosed with AD; and the others were NCs. The disparity of the group sizes means there is a tendency to retrieve more MCI subjects; therefore MCI has a higher retrieval rate than AD. To reduce the negative impact of the large number of MCI subjects, we added a constraint when evaluating the retrieval performance with MAP. We retrieved a fixed number of relevant subjects for all queries, instead of the real number of relevant subjects. We set this number as 5 for this work.
Multi-Channel Analysis
Some brain regions with strong discriminative power might not be detected due to the limits of single channel analyses. For example, SVM failed to detect Lingual Gyrus across all group pairs (CH_2, CH_5 and CH_8), and TOST failed to detect the Left Posterior Cingulate (CH_1, CH_4 and CH_7), although these regions are known as sensitive regions for AD and MCI. Another example is the MCI/NC group pair (CH_4, CH_5 and CH_6) where few brain regions were detected. The proposed Multi-Channel analysis, on the other hand, could compensate for the drawbacks of single channel analyses by integrating the analysis results from individual channels, and meanwhile highlighting the brain regions with strong agreement among many channels thus generating a more reliable and discriminative pattern. There are many ways to integrate single channel analysis results, and we adopted a voting scheme in this study due to its simplicity and effectiveness. However, this voting scheme does not take into consideration the correlation between individual channels, which may result in redundant information. Therefore, our proposed Multi-Channel analysis approach could be further optimized.
Supervised Pattern Optimization
Multi-Channel generated an over extended pattern, which consisted of 67 brain regions in this study. To further optimize the neurodegenerative pattern, we tested a set of feature selection threshold and finally identified 21 brain regions with the most discriminative power. The Multi-Channel analysis results were consistent with other studies on Hippocampus, Cingulate Gyrus, Temporal Lobe, Parietal Lobe, but also suggested some new AD and MCI biomarkers, i.e., Temporal Pole, Brainstem, Subgenual and Pre-subgenual Frontal Cortex. Some sub-cortical regions, such as Nucleus Accumbens, were also detected as sensitive regions to AD and MCI. Further investigation will be required to verify if they were caused by inaccurate registration or acquisition procedures.
Feature descriptors
The proposed Multi-Channel pattern analysis approach greatly benefits the characterization for AD and MCI. However, the characterization required careful selection of the features to describe different groups of subjects. The average CMRGlc features were very simple features requiring no higher-order calculations, and could measure the metabolic rate of brain regions. It was the most suitable feature descriptor for MCI, because the functional changes measured by CMRGlc for MCI subjects were moderate. However, the anatomical changes in MCI were hardly noticeable. We further divided the CMRGlc parameters by the mean value of whole brain intensity to remove the gray tone differences. However, for severe AD, the CMRGlc would decrease on a whole brain basis, and gray tone correction might alleviate the imaging differences between severe AD and NC subjects. Therefore, such CMRGlc parameters with gray tone correction may not be suitable for severe AD cases and can only be applied in non-severe AD or MCI subjects. However, if the patients are so severe, we don not need any analysis to make a diagnosis. The advanced feature descriptors, on the other hand, were more suitable to describe AD patients, since anatomical changes began to manifest after AD onset and other features except for the metabolic rate should be applied. GLCM could extract the texture features, whereas Gabor filters and DCvT could capture the boundary and curvature features. Gabor filters convolved with a Gaussian kernel function to localize the impulse responses in both spatial and frequency domain. In this study, we found the average CMRGlc features achieved highest performance for MCI retrieval at 76.2%, and 3D-Gabor coefficients exceeded all other features in AD retrieval with a MAP of 56.3%.
5. Conclusions and future works
In this work we developed a Multi-Channel neurodegenerative pattern analysis approach for characterization of the AD and MCI patients. The main advantage of our algorithm is its capability to overcome the drawbacks of individual analysis techniques on various group pairs. We further optimized the pattern by a supervised thresholding method, and consequently 21 brain regions were selected as the most discriminative biomarkers for AD and MCI. Our findings were congruent with established knowledge about pathological progression in AD and MCI, and also suggested that Temporal Pole, Brainstem, Subgenual and Pre-subgenual Frontal Cortex and Nucleus Accumbens, might also have a role in AD and MCI diagnosis. We evaluated the proposed algorithm by applying the derived pattern on the 352 subjects from the ADNI baseline FDG-PET dataset with various advanced feature descriptors. Our algorithm achieved the highest performance in AD and MCI retrievals, compared to the whole-brain based method and the DOM-based method.
For future work, the impact of gray tone correction on AD will be investigated. In addition, we will further verify our Multi-Channel analysis approach using advanced feature fusion techniques. We also hope to investigate the role of Diffusion Tensor Imaging (DTI) in the differentiation of AD and MCI within our Multi-Channel pattern analysis framework.
Acknowledgements
1. This work was supported in part by ARC, AADRF, NIH NA-MIC (U54 EB005149) and NAC (P41 EB015902).
2. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is Rev March 26, 2012 coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Conflicts of interest
None.
References
- 1.Muller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications – clinical benefits and future directions. Int J Med Inf. 2004;73(1):1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Cai W, Kim J, Feng D. Chapter 4 – Content-based medical image retrieval. In: Feng D, editor. Biomedical information technology. Elsevier; San Diego, USA: 2008. pp. 83–113. [Google Scholar]
- 3.Long LR, Antani S, Deserno TM, Thoma GR. Content-based image retrieval in medicine: retrospective assessment, state of the art, and future directions. Int J Healthcare Inform Syst Inform. 2009;4(1):1–16. doi: 10.4018/jhisi.2009010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian Y, Gao X, Loomes M, Comley R, Barn B, Hui R, Tian Z. Content-based retrieval of 3D medical images. International Conference on eHealth, Telemedicine, and Social Medicine (eTELEMED) 2011:7–12. [Google Scholar]
- 5.Unay D, Ekin A, Jasinschi RS. Local structure-based region of interest retrieval in brain MR images. IEEE Trans Inf Technol Biomed. 2010;14(4):897–903. doi: 10.1109/TITB.2009.2038152. [DOI] [PubMed] [Google Scholar]
- 6.Ramirez J, Gorriz JM, Lopez M, Salas-Gonzalez D, Alvarez I, Segovia F, Puntonet CG. Early detection of the Alzheimer disease combining feature selection and kernel machines advances in neuro-information processing. Lect Notes Comput Sci. 2009;5507:410–7. [Google Scholar]
- 7.Wang Y, Song Y, Rajagopalan P, An T, Liu K, Chou YY, et al. Surface-based TBM. Boosts power to detect disease effects on the brain: an N = 804 ADNI study. Neuroimage. 2011;56(4):1993–2010. doi: 10.1016/j.neuroimage.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong Z, Lu J, Chen J, Wang Y, Yuan Y, Zhang T, et al. Ventricle shape analysis for centenarians, elderly subjects, MCI and AD patients. Multimodal brain image analysis. Lect Notes Comput Sci. 2011;7012:84–92. [Google Scholar]
- 9.Batty S, Clark J, Fryer T, Gao X. Prototype system for semantic retrieval of neurological PET images. Medical imaging and informatics. Lect Notes Comput Sci. 2008;4987:179–88. [Google Scholar]
- 10.Li Y, Rinne JO, Mosconi L, Pirraglia E, Rusinek H, DeSanti S, et al. Regional analysis of FDG and PiB-PET images in normal aging, MCI and AD. Eur J Nucl Med Mol Imaging. 2008;35(12):2169–81. doi: 10.1007/s00259-008-0833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe VJ, Kemp BJ, Jack CR, Jr, Senjem M, Weigand S, Shiung M, et al. Comparison of FDG and PiB PET in MCI. J Nucl Med. 2009;50(6):878–86. doi: 10.2967/jnumed.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosconi L, Mistur R, Switaliski R, Tsui WH, Glodzik L, Li Y, et al. FDG-PET changes in brain glucose metabolism from normal cognition to pathologically verified AD. Eur J Nucl Med Mol Imaging. 2009;36(5):811–22. doi: 10.1007/s00259-008-1039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisoni GB, Lorenzi M, Caroli A, Kemppainen N, Nagren K, Rinne JO. In vivo mapping of amyloid toxicity in Alzheimer's disease. Neurology. 2009;28(72):1504–11. doi: 10.1212/WNL.0b013e3181a2e896. [DOI] [PubMed] [Google Scholar]
- 14.Brand M, Makowitsch HJ. Brain structures involved in dementia. Competence assessment in dementia. 2008:25–34. [Google Scholar]
- 15.Langbaum J, Chen K, Lee W, Reschke C, Randy D, Fleisher AS, et al. Categorical and correlation analyses of baseline FDG-PET images from the ADNI. Neuroimage. 2009;1(45):1107–16. doi: 10.1016/j.neuroimage.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirsra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in mild cognitive impairment patients, and their use in prediction of short-term conversion of Alzheimer's disease: results from ADNI. Neuroimage. 2009;44(4):1415–22. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai W, Liu S, Wen L, Eberl S, Fulham MJ, Feng D. 3D Neurological image retrieval with localized pathology-centric CMRGLC patterns. IEEE international conference on image processing (ICIP) 2010:3201–4. [Google Scholar]
- 18.Zhang D, Shen D. MultiCost:. Multi-stage cost-sensitive classification of Alzheimer's disease. Machine learning in medical imaging. Lect Notes Comput Sci. 2011;7009:344–51. [Google Scholar]
- 19.Cai W, Feng D, Fulton R. Content-based retrieval of dynamic PET functional images. IEEE Trans Inform Technol Biomed. 2000;4(2):152–8. doi: 10.1109/4233.845208. [DOI] [PubMed] [Google Scholar]
- 20.Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving inter-subject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage. 2010;51(1):221–7. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- 21.Heckemann RA, Keihaninejad S, Aljabar P, Gray KR, Nielsen C, Rueckert D, et al. Automatic morphometry in Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;56(4):2024–37. doi: 10.1016/j.neuroimage.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Cai W, Wen L, Eberl S, Fulham MJ, Feng D. A robust volumetric feature extraction approach for 3D neuroimaging retrieval. Annual international conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 2010:5657–60. doi: 10.1109/IEMBS.2010.5627900. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Cai W, Wen L, Eberl S, Fulham MJ, Feng D. Localized functional neuroimaging retrieval using 3D discrete curvelet transform. IEEE International Symposium on Biomedical Imaging From Nano to Macro (ISBI) 2011:1877–80. [Google Scholar]
- 24.Liu S, Cai W, Wen L, Feng D. Multiscale and multiorientation feature extraction with degenerative patterns for 3D neuroimaging retrieval. IEEE International Conference on Image Processing (ICIP) 2012:3201–4. [Google Scholar]
- 25.Liu S, Cai W, Wen L, Feng D. Volumetric congruent local binary patterns for 3D neurological image retrieval. International conference on image and vision computing New Zealand (ICVNZ) 2011:272–6. [Google Scholar]
- 26.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer's disease neuroimaging initiative positron emission tomography core. Alzheimer's Dementia. 2010;6(3):221–9. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosconi L, Tsui WH, Herholz K, Pupi A, Drzezga A, Lucignani G, et al. Multicenter standardized 18F FDG PET diagnosis of mild cognitive impairment, Alzheimer's disease and other dementias. J Nucl Med. 2008;49(3):390–8. doi: 10.2967/jnumed.107.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkinson M, Bannister P, Brady JM, Smith S. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–41. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 29.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc B Biol Sci. 2001;356(1412):1293–322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Non-rigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18(8):712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 31.Schnabel JA, Rueckert D, Quist M, Blackall JM, Castellano-Smith AD, Hartkens T, et al. A generic framework for non-rigid registration based on non-uniform multi-level free-form deformations. Medical image computing and computer-assisted intervention (MICCAI). Lect Notes Comput Sci. 2011;2208:573–81. [Google Scholar]
- 32.Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19(4):224–47. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of clinical scores in alzheimer's disease. Multimodal brain image analysis (MBIA). Lect Notes Comput Sci. 2011;7012:60–7. [Google Scholar]
- 34.Liu S, Cai W, Wen L, Feng D. Multi-channel brain atrophy pattern analysis in neuroimaging retrieval. IEEE international symposium on biomedical imaging from nano to macro (ISBI) 2013:202–5. [Google Scholar]
- 35.Liu S, Cai W, Wen L, Feng D. Neuroimaging biomarker based prediction of Alzheimer's disease severity with optimized graph construction. IEEE international symposium on Biomedical Imaging from Nano to Macro (ISBI) 2013:1336–9. [Google Scholar]
- 36.Liu S, Cai W, Wen L, Eberl S, Fulham MJ, Feng D. Generalized regional disorder-sensitive-weighting scheme for 3D neuroimaging retrieval. Annual international conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 2011:7009–12. doi: 10.1109/IEMBS.2011.6091772. [DOI] [PubMed] [Google Scholar]
- 37.Schuirmann DJ. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J Pharmacokinet Biopharm. 1987;15(6):657–80. doi: 10.1007/BF01068419. [DOI] [PubMed] [Google Scholar]
- 38.Cortes C, Vapnik VN. Support-vector networks. Machine Learn. 1995;20(3):273–97. [Google Scholar]
- 39.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(27):1–27. [Google Scholar]
- 40.CVX Research, Inc. CVX: Matlab Software for Disciplined Convex Programming, Version 2.0. 2011 http://cvxr.com/cvx.
- 41.Zou H, Hastie T. Regularization and variable selection via the elastic net. Philos Trans R Soc B Biol Sci. 2005;67(2):301–20. [Google Scholar]
- 42.Haralick RM, Shanmugam K, Dinstein I. Texture features for image classification. IEEE Trans Syst Man Cybernet. 1973;3(6):610–21. [Google Scholar]
- 43.Daugman J. Complete discrete 2-D Gabor transforms by neural networks for image analysis and compression. IEEE Trans Acoust Speech Signal Process. 1988;36(7):1169–79. [Google Scholar]
- 44.Ying L, Demanet L, Candes E. 3D Discrete curvelet transform. Proc SPIE. 2005:5914. [Google Scholar]
- 45.Kraskov A, Stogbarer H, Andrzejak RG, Grassberger P. Hierarchical clustering based on mutual information. Europhys Lett. 2005;70(2):278–88. [Google Scholar]
- 46.Frdorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30(9):1323–41. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]