Abstract
Understanding the pathways by which viral capsid proteins assemble around their genomes could identify key intermediates as potential drug targets. In this work we use computer simulations to characterize assembly over a wide range of capsid protein-protein interaction strengths and solution ionic strengths. We find that assembly pathways can be categorized into two classes, in which intermediates are either predominantly ordered or disordered. Our results suggest that estimating the protein-protein and the protein-genome binding affinities may be sufficient to predict which pathway occurs. Furthermore, the calculated phase diagrams suggest that knowledge of the dominant assembly pathway and its relationship to control parameters could identify optimal strategies to thwart or redirect assembly to block infection. Finally, analysis of simulation trajectories suggests that the two classes of assembly pathways can be distinguished in single molecule fluorescence correlation spectroscopy or bulk time resolved small angle x-ray scattering experiments.
II. INTRODUCTION
In many virus families, the spontaneous assembly of a protein shell (capsid) around the viral nucleic acid (NA) genome is an essential step in the viral life cycle [1]. These families include most viruses with single stranded RNA (ssRNA) genomes, as well as the Hepadnaviridae (e.g. hepatitis B virus, HBV). Understanding the mechanisms which underlie this cooperative assembly process could facilitate efforts to develop anti-viral drugs that block or derail the formation of infectious particles (for reviews see [2, 3]) and promote efforts to reengineer them for biomedical delivery. In this article, we explore how the interactions between the molecular components determine the mechanism of assembly, and how these interactions can be altered by changing solution conditions or mutagenesis to modulate assembly pathways.
The most detailed knowledge of capsid-NA interactions comes from structural analysis of assembled viral particles. Atomic resolution structures of capsids assembled around ssRNA have been obtained by x-ray crystallography and/or cryo-electron microscopy (cryo-EM) (e.g. [4–16]). The packaged nucleic acids are less ordered than their protein containers and thus have been more difficult to characterize. However cryo-EM experiments have identified that the nucleotide densities are nonuniform, with a peak near the inner capsid surface and relatively low densities in the interior[7, 17, 18]. While atomistic detail has not been possible in these experiments, all-atom models have been derived from equilibrium simulations [19–21]. In some cases, striking image reconstructions reveal that the packaged RNA adopts the symmetry of the overlying capsid (e.g. [7, 10, 16, 22, 23]). While it has been proposed that this order arises as a function of the assembly mechanism for several viruses [24–26], computational analysis of polyelectrolyte configurations inside capsids also indicate that capsid-polymer interactions can generically drive spatial organization of the packaged polymer [20, 27–38]. Theoretical works have also characterized the relationship between the NA charge and structure and the length which is optimal for packaging [27, 31, 32, 38–45].
In addition to this structural data on assembled capsids, an extensive combination of mass spectrometry, assembly kinetics experiments, constraints from assembled capsid structures, and mathematical modeling has delineated assembly pathways for several viruses, with a particular focus on the role of interactions between CPs and specific RNA sequences called ‘packaging signals’. Recent single molecule fluorescence correlation spectroscopy (smFCS) experiments indicate that, for these viruses, assembly around the viral genome is more robust and proceeds by a different mechanism as compared to around heterologous RNA [46]. Yet, in other cases capsid proteins show no preference for genomic RNA over heterologous RNA (e.g. HBV [47]), and cowpea chlorotic mottle virus (CCMV) proteins preferentially encapsidate heterologous RNA (from BMV) over the genomic CCMV RNA with equivalent length [48]. Furthermore, experimental model systems in which capsid proteins assemble into icosahedral capsids around synthetic polyelectrolytes or other polyanions [49–61]) demonstrate that specific RNA sequences are not required for capsid formation or cargo packaging. Thus, a complete picture of capsid assembly mechanisms requires understanding how assembly pathways depend on those features which are generic to polyelectrolytes, as well as those which are specific to viral RNAs.
In previous work on assembly around a simple model for a polymer, Elrad and Hagan [37] proposed that mechanisms for assembly around a cargo (i.e. RNA, polymer, or nanoparticle) can be classified on the basis of two extreme limits. In the first (originally proposed by McPherson [62] and then Refs [20, 63]), strong protein-cargo interactions drive proteins to adsorb ‘en masse’ onto the cargo in a disordered manner, meaning there are few protein-protein interactions. Once enough subunits are bound, subunits undergo cooperative rearrangements (potentially including dissociation of excess subunits) to form an ordered capsid. This mechanism has been observed in recent simulations [37, 38, 63–65]. In the second limit, where protein-protein interactions dominate, a small partial capsid nucleates on the cargo, followed by a growth phase in which individual proteins or small oligomers sequentially add to the growing capsid. This class of pathways resembles the nucleation-and-growth mechanism by which empty capsids assemble [66], except that the polymer plays an active role by stabilizing protein-protein interactions and by enhancing the flux of proteins to the assembling capsid [37, 67, 68].
It is difficult to determine assembly mechanisms directly from experiments, due to the small size (≲ 10 of nm) and transience (~ms) of most intermediates. Observations in vitro suggest that both mechanisms may be viable. Kler et al. [69] used time-resolved X-ray scattering (trSAXS) to monitor simian virus 40 (SV40) capsid proteins assembling around ssRNA. The scattering profiles at all time points during assembly could be decomposed into unassembled components (RNA + protein subunits) and complete capsid; the absence of any signal corresponding to a large disordered intermediate suggests this assembly follows the nucleation-and-growth (ordered) assembly mechanism [69]. Other observations suggest that viruses can assemble through the en masse mechanism. Refs. [70, 71] found that in vitro assembly CCMV assembly was most productive when performed in two steps: (1) at low salt (strong protein-RNA interactions) and neutral pH (weak protein-protein interactions) the proteins undergo extensive adsorption onto RNA, then (2) pH is reduced to activate binding of protein-protein binding [70]. Similarly, a recent observation of capsid protein assembly around charge-functionalized nanoparticles found that assembly initially proceeded through nonspecific aggregation of proteins and nanoparticles, followed by the gradual extrusion of nanoparticles within completed capsids [72]. These experiments used viral proteins with relatively weak protein-protein interactions (CCMV and BMV) [73] and moderate salt concentrations (100–150mM). The Kler et al. [69, 74] experiments considered SV40 proteins, which have strong protein-protein interactions [73], and high salt (250mM) Together, these in vitro experiments suggest that productive assembly could proceed by either the en masse or nucleation-and-growth mechanism.
In this work, we use dynamical simulations to investigate the extent to which the assembly mechanism can be controlled by tuning solution ionic strength and protein-protein attractions. We extend a model that was recently used to calculate the thermostability and assembly yields of viral particles as a function of protein charge and nucleic acid (NA) length and structure. Those previous simulations found quantitative agreement between predicted NA lengths that optimize capsid thermostability and viral genome length for seven viruses [38]. Here, we perform extensive new simulations of assembly, in which protein-protein interactions, the sequence of charges in capsid protein-NA binding domains, and the solution ionic strength are varied. We find that, by varying these control parameters, the assembly mechanism can be systematically varied between the two extreme limits described above. Our results suggest that knowledge of protein-protein and protein-NA binding affinities may be sufficient to predict which assembly mechanism will occur, and we estimate relative protein-NA binding interactions for three viruses (based on non-specific electrostatic interactions). These findings suggest that assembly mechanisms can be rationally designed through choice of solution conditions and mutagenesis of capsid protein-protein interfaces and protein-NA binding domains. Finally, by calculating hydrodynamic radii and SAXS profiles associated with assembly intermediates, we show that assembly mechanisms can be distinguished by experimental techniques recently applied to virus assembly, respectively smFCS [46]) and trSAXS [69]. While the NA is represented by a linear polyelectrolyte in most of the simulations, we obtain qualitatively similar results when considering a model for base-paired NAs developed in Ref. [38].
III. RESULTS
To study how capsid assembly around a polyelectrolyte depends on the strength of protein subunit-subunit and subunit-polyelectrolyte interactions, we performed Brownian dynamics simulations with a recently developed model [38] (Figure 1). The capsid is modeled as a dodecahedron composed of 12 pentagonal subunits (each of which represents a rapidly forming and stable homopentamer of the capsid protein, which then more slowly assembles into the complete capsid, as is the case for SV40 in vitro around ssRNA [69, 74, 75]). This model was motivated by the observation [69, 74] that purified SV40 capsid proteins assemble in vitro around ssRNA molecules to form capsids composed of 12 homopentamer subunits. Though SV40 in vivo forms a larger capsid when assembling around its compacted, minichromosomal DNA genome [75], this 12-subunit capsid has provided the most precise experimental characterization of ssRNA-mediated assembly kinetics to date [69, 74], and may provide the opportunity to experimentally test our model predictions (see section 8).
FIG. 1.
(A),(B) Model schematic for (A) a single subunit, and (B) two interacting subunits, showing positions of the attractor (‘A’), Top (‘T’), and Bottom (‘B’) pseudoatoms, which are defined in the Model section and in the Methods. (C) (left) The pentameric SV40 capsid protein subunit, which motivates our model. The globular portions of proteins are shown in blue and the beginning of the NA binding motifs (ARMs) in yellow, though much of the ARMs are not resolved in the crystal structure [117]. Space-filling model of the generic subunit model (middle) and a pentamer from the PC2 model (right). For all simulation snapshots shown in this article, beads are colored as follows: blue=excluders, green=attractors, yellow=positive ARM bead, gray=neutral ARM bead, red=polyelectrolyte.
Model subunits are attracted to each other via attractive pseudoatoms at the vertices (type ‘A’) and driven toward a preferred subunit-subunit angle by repulsive ‘Top’ pseudoatoms (type ‘T’) and ‘Bottom’ pseudoatoms (type ‘B’) (see Fig. 1 and the Methods section). These attractions represent the interactions between capsid protein subunits that arise from hydrophobic, van der Waals, and electrostatic interactions as well as hydrogen bonding [1]). The magnitude of these interactions varies between viral species [73] and can be experimentally tuned by pH and salt concentration [1, 76, 77]. Here, the attraction strength is controlled by the model parameter εss. To relate εss to the free energy of dimerization, we have run a separate series of calculations, where in the absence of cargo, we find that the free energy of subunit of dimerization is gss = 5.0 – 1.5 * εss (see SI section 12). Throughout this article energies are given in units of the thermal energy, kBT.
Capsid assembly around nucleic acids and other polyelectrolytes is primarily driven by electrostatic interactions between negative charges on the encapsulated polyelectrolyte and positive charges on the inner surfaces of capsid proteins [1]. We consider a linear bead-spring polyelectrolyte, with a charge of e per bead and a persistence length comparable to that of ssRNA in the absence of base pairing. Positive charges on capsid proteins are located in flexible polymers affixed to the inner surface of the model subunit, which represent the highly charged, flexible terminal tails known as arginine rich motifs (ARMs) that are typical of positive-sense ssRNA virus capsid proteins (e.g., [16]). We will begin with a simple model capsid in which the ARMs consist of 5 positively charged segments. Below, we consider models that represent specific viruses, with ARMs that can be substantially longer (≲ 50 segments) and include neutral amino acids (and even acidic) amino acids in addition to basic residues.
Electrostatics are modeled using Debye-Hückel (DH) interactions, where the Debye screening length (λD) is determined by the ionic strength I as λD ≈ 0.3/I1/2 with λD in nm and I in molar units. We consider monovalent salt, for which I is given by the salt concentration Csalt. Perlmutter et al. [38] showed that DH interactions compare well to simulations with explicit counterions for the parameter values under consideration; however, we note that the DH approximation is less accurate at lower salt concentrations. For example, the polyelectrolyte lengths that minimize the free energy of the assembled nucleocapsid (the thermodynamic optimal length) computed using the DH approximation or Coulomb interactions with explicit counterions are compared at several ionic strengths in Fig. S1. Furthermore, analysis of the free energy of uniformly charged spheres found strong agreement between DH and Poisson-Boltzmann electrostatics at moderate salt concentrations (≥ 100mM), but weaker agreement at ≤ 10mM [78]. We note that divalent ions can impact NA structure, and sufficiently large concentrations of multivalent ions modify the osmotic pressure of densely packed dsDNA within bacteriophage capsids [79]. However, inclusion of divalent ions (modeled with Coulomb interactions) at physiological relevant concentrations (5mM) had only a small effect on properties such as the optimal genome length (~ 10%) in our model system [38] (Fig. S1). The effect of base-pairing on the optimal length was also considered in that reference [38].
In [38] we demonstrated that the thermodynamic optimum length corresponds to the length which maximizes the finite-time yield. Therefore, we have calculated the thermodynamic optimal length for each salt concentration (supplementary figure S1) and for each dynamical simulation described here we have used the polyelectrolyte with the optimal length for the simulated salt concentration.
A. Kinetic phase diagram
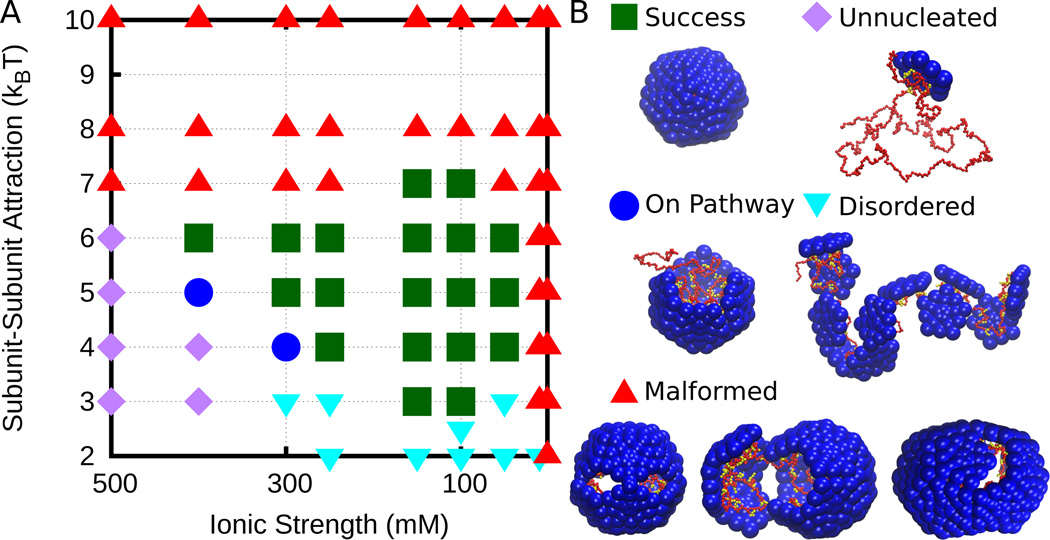
We first consider the predominant assembly products (Fig. 2) and the packaging efficiency (Fig. 3) as a function of Debye length λD and subunit-subunit interaction strength εss. The packaging efficiency is defined as the fraction of trajectories in which the polyelectrolyte is completely encapsulated by a well-formed capsid, which contains 12 subunits each of which interact with 5 neighbors. We refer to this as a kinetic phase diagram [1, 80] because we characterize products at a finite observation time of tf=2 × 108 time steps, which is long enough that assemblages do not vary significantly with time (except for under weak interactions, see below), but is not sufficient to equilibrate kinetic traps if there are large activation barriers [1, 37]. We see that for the range of simulated ionic strengths (1 ≤ Csalt ≤ 500 mM or 10 ≥ λD ≥ 0.4 nm) assembly yields are highest for εss=5kBT and Csalt=100 mM (the parameter values focused on in [38]), and that for moderate subunit-subunit interaction strengths (4 ≤ εss ≤ 6, kBT) yield to remain high as the ionic strength is increased to about 300 mM (λD ≈ 0.6 nm). For higher salt concentrations, yields are depressed by the appearance of long-lived on-pathway intermediates. As will be discussed further below, weakening the electrostatic interaction between the polymer and protein limits the ability of the polymer to promote assembly. Although we expect that these simulations would eventually result in complete capsids, the low yield at our finite measurement time reflects the fact that assembly is less efficient than for lower salt concentrations. At the highest ionic strength considered (Csalt=500 mM), the most prevalent outcome is that no nucleation occurs. At lower salt concentrations (Csalt ≥ 10 mM), rapid adsorption of a super-stroichiometric quantity of subunits results in malformed capsids.
FIG. 2.
(A) Kinetic phase diagram showing the most prevalent final product at the conclusion of assembly simulations (tf=2 × 108 time steps). (B) Snapshots illustrating categories.
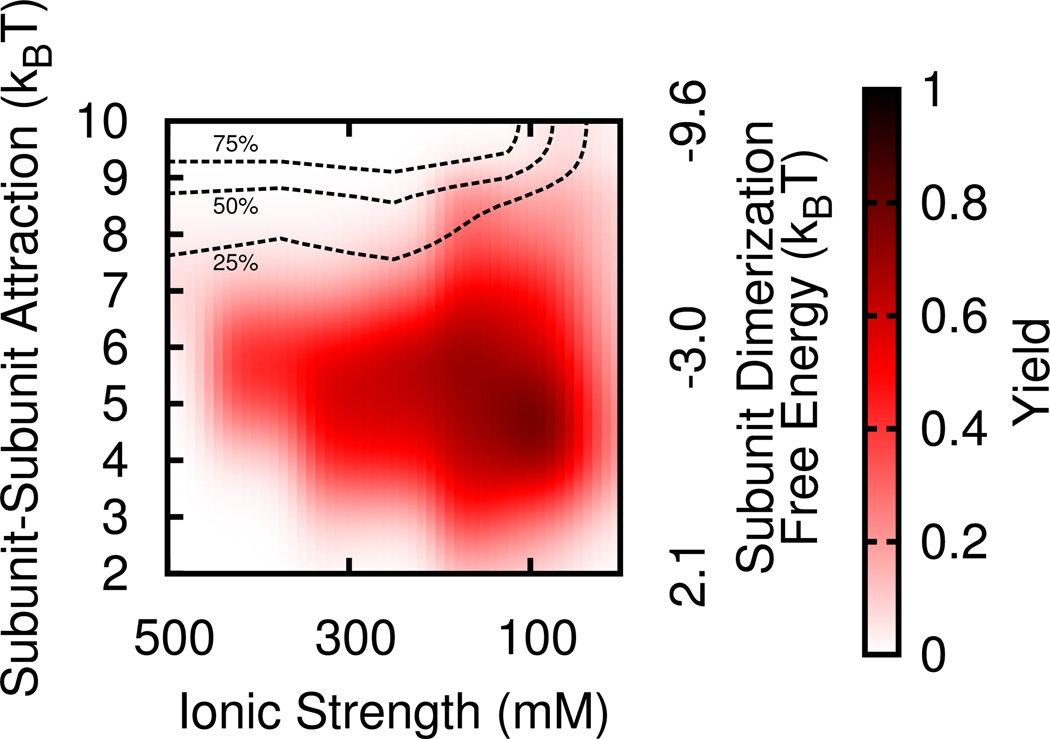
FIG. 3.
Kinetic phase diagram showing fraction of simulations which result in successful assembly of a complete capsid. The black isosurface lines show the fraction of subunits which are not adsorbed to the polymer and show any partial assembly, i.e. above the 75% line, ≥ 75% of the subunits not bound to the polymer are clustered.
At larger-than-optimal protein-protein interaction strengths (εss > 6) assembly yields are lower for two reasons. The first, and more frequent outcome, is long-lived malformed structures with strained interactions. This kinetic trap arises in a wide variety of assembly systems when interactions become strong in comparison to the thermal energy, because strained interactions are unable to anneal before additional subunits lock them in place [1, 81, 82]. In our simulations, we found that these structures frequently result from an incorrect merger between two partial capsid intermediates; even when each individual interaction is too weak to lock-in the non-optimal structure, multiple erroneous interactions formed by two partial capsids are effectively irreversible on our timescale. The tendency for oligomer binding to lead to malformed structures was seen previously in the context of empty capsid assembly [80, 83]. Here, the polymer helps to bring oligomers together, and thus this trap arises when nucleation on the polymer is faster than growth of a nucleus into a complete capsid. This trap resembles Geminivirus particles, which are composed of a union of two nearly complete capsids [84].
The second obstacle to polyelectrolyte encapsulation arises at the highest protein-protein interaction strengths studied (εss ≥ 8kBT), for which subunits not associated with the polyelectrolyte undergo spontaneous assembly. The resulting off-polyelectrolyte assembly depletes the pool of available monomers and small oligomers available for assembly on the polyelectrolyte, leading to a form of the monomer-starvation kinetic trap previously discussed for empty capsid assembly [1, 66]. Triggering formation of empty capsids and thus preventing nucleic acid encapsidation by strengthening subunit-subunit interactions has been suggested as a mode of action for a putative antiviral drug for hepatitis B virus [85, 86].
At smaller-than-optimal protein-protein interaction strengths (εss < 4) assembly is unsuccessful for two reasons, depending upon the ionic strength. At smaller values (Csalt ≤ 300 mM), electrostatic interactions are relatively strong, and many proteins adsorb onto to the polymer. However, because of the weak protein-protein interaction strength, these proteins do not form stable capsids, predominantly because nucleation is slow in comparison to tf (the final observation time, tf=2 × 108 time steps). In a minority of cases, a nucleus will form, but completion is prevented by the excess number of subunits adsorbed to the polyelectrolyte. We refer to the resulting configurations as disordered, due to the lack of ordered binding between protein subunits. At larger ionic strengths (Csalt > 300 mM) electrostatic interactions are relatively weak, and individual subunits rapidly desorb from the polyelectrolyte. In this regime, assembly requires a fluctuation in the number of adsorbed subunits which leads to nucleation of a partial capsid intermediate which has enough subunit-polyelectrolyte interactions to be stable against rapid desorption. The nucleation rate decreases exponentially with subunit-subunit interaction strength [1, 68], and thus most simulations at high salt and weak subunit-subunit interactions never undergo nucleation. (We categorize simulations with fewer than 3 subunits adsorbed to the polyelectrolyte and no subunit-subunit interactions (i.e. no progress towards assembly) as ‘unnucleated’.)
Importantly, we expect that trajectories in this region of parameter space will eventually undergo nucleation. Thus, as the finite assembly time tf is increased the region of successful assembly will expand to lower values of εss and higher ionic strength, until eventually reaching values below which capsid assembly is thermodynamically unstable (see Fig. 7 in Ref. [80]). To confirm this possibility, we used the Markov state model (MSM) approach described in Ref. [65] to characterize assembly with εss=5, 6 and Csalt=500 mM (see Fig. 4C and SI Fig. S5). In contrast, the malformed capsids encountered under large parameter values typically will not resolve on any relevant timescales, since numerous strong interactions would need to be broken [1]. Thus, the boundaries of successful assembly at large εss values are insensitive to tf.
FIG. 4.
(A) Snapshots from trajectories corresponding to the en masse and nucleation-and-growth (ordered) assembly mechanisms, at indicated parameter values. (B) The number of subunits adsorbed (nad) to the polyelectrolyte and the size of the largest cluster (n) are shown as a function of simulation time steps, averaged over all trajectories at the two sets of parameter values shown in (A). The labels ‘a’ through ‘e’ connect each structure pictured in (A) to its corresponding value of nad in (B). (C) Average values of nad and n are shown as a function of time step for parameters that lead to the nucleation-and-growth mechanism with a larger nucleation barrier than in (B).
Several additional conclusions can be drawn from the variation of packaging efficiency (Fig. 3). Firstly, the yield of complete virus-like particles is relatively high for moderate subunit-subunit interaction strengths εss ∈ [4, 6] across a range of ionic strengths (Csalt ∈ [100, 300] mM). Even above (Csalt > 300 mM) and below (Csalt=10 mM) this range we observe moderate yields of complete particles. Secondly, as electrostatic interactions are weakened (moving to the left), the subunit-subunit interaction strength which optimizes the yield increases (i.e. from εss = 5kBT at Csalt = 100 mM to εss = 6kBT at Csalt : 150 – 400 mM to εss = 7kBT at Csalt = 500 mM). This result suggests that one interaction type can compensate for the other within a limited range. However, though all successful capsids contain the same subunit geometry, the mechanism by which they form depends on the relative interaction strengths, as discussed next.
B. Assembly mechanisms
As noted previously [1, 37, 63], pathways for assembly around a central core such as a polyelectrolyte can be roughly separated into two classes. In the first class (Fig. 4a), which we refer to as the ‘en masse’ mechanism, subunits first adsorb onto the polyelectrolyte in a disordered manner, followed by cooperative rearrangements to form an ordered capsid. In the second class (Fig. 4b), referred to as the nucleation-and-growth mechanism, a small partial capsid nucleates on the polyelectrolyte followed by the sequential, reversible addition of subunits or small oligomers until assembly completes. In contrast to the earlier models which considered a qualitative subunit-polyelectrolyte interaction, we study here how assembly pathways depend on the ionic strength.
To quantify the degree of order along assembly pathways, we record the total number of subunits adsorbed to the polyelectrolyte nad and the number of subunits in the largest cluster n. Trajectories that pass through configurations with a large value of nfree = nad – n are disordered, with many adsorbed subunits not participating in ordered assemblies. In Fig. 4, these quantities are shown as a function of time averaged over all simulation trajectories (leading to successful assembly or not), for parameter sets that respectively lead to the en masse mechanism (Fig. 4A) and nucleation-and-growth mechanism (Fig. 4B). In the first case, there are strong subunit-polyelectrolyte interactions (low ionic strength, Csalt=100 mM) and weak subunit-subunit interactions (εss=3kBT). Subunits therefore initially adsorb nonspecifically and form only transient subunit-subunit interactions, leading to a rapid rise in nad with n ≈ 0. Once enough subunits are adsorbed (~ 12 around an optimal-length polyelectrolyte for this model with a 12-subunit capsid), a cooperative fluctuation in subunit configurations eventually leads to a stable nucleus and then rapid completion of the ordered capsid geometry. Since this nucleation process is stochastic, there is a distribution of waiting times and thus a more gradual increase in the average cluster size n (see Fig. S3). We note that more than 12-subunits can adsorb onto the polymer. In some cases, typically under moderate subunit-polyelectrolyte interactions, these excess subunits are shed and well-formed capsids are assembled. Under stronger subunit-polyelectrolyte interactions, adsorption of excess subunits typically leads to ‘disordered’ assemblages (Fig. 2). In the nucleation-and-growth case, on the other hand, the subunit-polyelectrolyte interactions are weak (Csalt=300 mM) and the subunit-subunit interactions are strong (εss=6kBT). There is limited nonspecific subunit adsorption onto the polyelectrolyte, adsorbed subunits form relatively strong associations, and thus nad and n increase nearly in lockstep. Snapshots from typical trajectories for each of these parameter sets are shown in Fig. 4A.
To visualize the degree of order as a function of parameter values, we define a trajectory-averaged order parameter n̄free, which is nfree averaged over all configurations with 4 ≤ n ≤ 6 and over all trajectories at a given parameter set[87]. Large values of this parameter (n̄free & 5) indicate the en masse mechanism, while small values (n̄free ≲ 2) indicate the nucleation-and-growth mechanism. As shown in Fig. 5, the degree of order generally increases with ionic strength and subunit-subunit interaction strength, with the most ordered assembly occurring at Csalt = 500 mM (where on average fewer than one subunit is adsorbed nonspecifically) and εss = 6kBT. However, notice that for εss = 3 assembly is always disordered; for such weak subunit-subunit interactions the critical nucleus size is large and a high density of adsorbed subunits is required to achieve nucleation. On the other hand, for moderate subunit-subunit interactions we do not observe the extreme limit of the en masse mechanism even for low ionic strength. This observation can be explained by the fact that the en masse mechanism requires significant subunit adsorption to occur before cluster nucleation takes place. Though a low ionic strength drives strong nonspecific subunit adsorption, absorbed subunits collide frequently due to cooperative polymer motions and subunit sliding along the polymer [37, 67]. For εss > 3 adsorbed subunits therefore achieve nucleation before nonspecific absorption has time to saturate at all ionic strengths considered.
FIG. 5.
Dependence of the assembly mechanism on the subunit-subunit attraction strength εss and the ionic strength Csalt. The assembly order parameter n̄free (the average number of adsorbed subunits not in the largest partial capsid) is shown as a function of parameter values. As described in the text, n̄free ≲ 2 corresponds to ordered nucleation-and-growth assembly pathways, whereas larger values correspond to disordered pathways. The alternate y-axis shows the subunit-subunit dimerization free energy gss corresponding to εss (see section VII B) and the alternate x-axis shows the linear adsorption density of subunits in the absence of assembly (see Fig. 6 below).
The nucleation-and-growth trajectories can be further classified based on the relative timescales of nucleation and growth. When nucleation is slow compared to growth (Figs. 4C and S5), the reaction is effectively two-state — each critical nucleus rapidly proceeds to completion, leading to low concentrations of intermediates which are essentially undetectable in a bulk experiment. When nucleation and growth timescales are comparable, multiple capsids within a spatial region can be assembling simultaneously, and thus potentially could be detected in bulk experiments. Below, we consider whether it is possible to experimentally distinguish between the latter case, ordered assembly with rapid nucleation, and en masse assembly pathways characterized by disordered intermediates.
Biological ARM sequences
The simulations described to this point demonstrate that the assembly mechanism depends on the subunit-polyelectrolyte binding affinity, which we have controlled by varying the solution ionic strength. However, while we have considered a simplified peptide ARM with five positive charges, the actual number and charges of amino acids in the ARM varies between viruses, which should also affect the binding affinity. Indeed, our previous equilibrium simulations demonstrated that the thermodynamic optimal length for encapsidation depends on the ARM amino acid sequence [38]. To place our results in the context of specific viruses, we calculated the polyelectrolyte binding affinity for subunits containing ARMs representing those of three viruses, SV40, Brome mosaic virus (BMV), and Porcine circovirus 2 (PC2). Specifically, the number and charges of beads in the simulated ARM was determined from the ARM amino acid sequence for each virus, with amino acids classified as neutral, cationic, or anionic. Each ARM sequence is shown in Table I, and Fig. 1C compares the ARMs of the simple model and the SV40 model. The binding affinity was estimated by setting the subunit-subunit attraction strength to zero (but keeping repulsive excluded volume interactions) and recording the equilibrium average number of adsorbed subunits at varying Csalt (or Debye length λD).
TABLE I.
Amino acid sequence of ARMs investigated in Fig. 6. The secondary lines indicate whether model ARM segments are designated neutral (0), positively charged (+), or negatively charged (−).
| Virus | ARM Amino Acid Sequence |
|---|---|
| SV40 | mkmaptkrkgscpgaapkkpke |
| 0+0000+++00000000++0+- | |
| BMV | mstsgtgkmtraqrraaarrnrwtarvqpviveplaagqgkaik |
| 0000000+00+00++000++0+000+000000-0000000+00+ | |
| PC2 | mtyprrryrrrrhrprshlgqilrrrpwlvhprhryrwrrkng |
| 0000+++0++++++0+0+00000+++0000+0+++0+0+++00 |
The measured equilibrium linear densities of adsorbed subunits, ceq, are shown as a function of Csalt in Fig. 6A–C. In all cases the linear density increases monotonically with decreasing Csalt; saturating at a maximum density. The simplified ARM has the largest binding affinity, despite having the lowest net positive charge (+5, 0 neutral segments) of the four species considered. Comparison with SV40 (+6, 14 neutral segments) and BMV (+9, 33 neutral segments) illustrates the expected result that that neutral segments decrease the binding affinity, particularly at high salt. The PC2 subunits, with net charge +22 demonstrate markedly different behavior, with significant subunit absorption at the highest salt concentration simulated (500 mM), but saturating at about 300 mM ionic strength due to subunit-subunit charge repulsions. Variations in adsorption density with subunit concentration are shown in Fig. S2.
FIG. 6.
Average number of subunits adsorbed to polymer (in absence of assembly), depending on Debye Length and ARM sequence. Comparison between simple, +5-ARM and SV40 (A), BMV (B), and PC2 (C). (D) n̄free values obtained during assembly simulations are plotted for varying values of ceq and subunit-subunit dimerization free energy for our simple 5-ARM assembly model (circles) and the SV40 model (squares). ARM net charge and total length: SV40 +6/22, BMV +9/44, PC2 +22/43.
Next, we investigated how the ARM sequence and its corresponding polyelectrolyte binding affinity affects assembly dynamics, by performing dynamical assembly simulations (with finite subunit-subunit attraction strength) for subunits with the SV40 ARM. Based on our analysis of the assembly dynamics for the simplified 5-ARM, we anticipated that the polyelectrolyte binding affinity (as measured by the equilibrium adsorption without subunit-subunit attractions) correlates to the assembly order parameter. To test this hypothesis, we plot the assembly order parameter n̄free as a function of ceq (controlled by varying Csalt), for several values of the subunit dimerization free energy (determined by εss and Csalt) for the 5-ARM and SV40 models. Plotting against ceq rather than Csalt (λD) allows us to overlay data from these two models while accounting for the differences in affinity due to ARM sequence described above. In support of the proposed link between binding affinity and assembly mechanism, we find rough agreement in the measured assembly order parameters between the two models. The results indicate that ARM sequence can significantly influence the assembly mechanism. For example, if we define n̄free ≤ 2 as the nucleation-growth mechanism, Fig. 6D indicates that nucleation-growth occurs for with the threshold value for εss ≥ 2.5kBT. From Fig. 6A–C, we can then identify the threshold values of ionic strength , above which the nucleation-growth mechanism will occur: for the 5-ARM and for the BMV and SV40 models; while PC2 is below the threshold value for all salt concentrations considered. This allows us to predict, for example, that recent experiments on SV40 assembly (at Csalt=250mM and observed strong subunit-subunit attraction) would have a very low n̄free (~ 1), which is consistent with SAXS observations [69].
C. Experimental Observables
We now seek to provide experimental signatures through which the capsid assembly pathways discussed above can be distinguished. We focus on two experimental techniques which have recently been applied to respectively probe the formation of individual capsids and bulk assembly kinetics.
smFCS measurements on individual capsids can distinguish assembly mechanisms
Borodavka et al. [46] used single molecule fluorescence correlation spectroscopy (smFCS) to monitor the hydrodynamic radii RH of nucleocapsid complexes during assembly of MS2 and STNV capsid proteins around cognate RNA or non-cognate RNA. Assembly around cognate RNA was characterized by either constant RH or, in some trajectories, a collapsed complex followed by gradual increase until reaching the size of an assembled capsid. In contrast, assembly around non-cognate RNA led to an increase in RH before eventually decreasing to the size of the capsid. The difference between these two behaviors can be attributed to sequence-specific ‘packaging signals’ on cognate RNA that interact with the capsid proteins. In this article we do not consider the effect of packaging signals (these will be considered in a subsequent article); instead, we consider whether the pathways described in the previous section can be distinguished by this experimental technique.
We estimated the hydrodynamic radii RH for polymer-subunit intermediates using the program HYDROPRO, which has been shown to accurately predict RH for large protein and protein-NA complexes [88]. The resulting RH are shown during representative en masse and ordered assembly trajectories in Fig. 7E. We see that the complex RH first increases as subunits adsorb onto the complex and then decreases as subunits assemble (Fig. 7E). However, the en masse mechanism leads to a much larger and longer duration increase in RH, due to the extensive and long-lived disordered adsorption of unassembled subunits. The difference in RH between en masse and ordered trajectories is conserved across their respective parameter ranges (see SI Fig. S4 for other trajectories), and also occurs for assembly trajectories around the model nucleic acid with intramolecular base-pairing developed in Ref. [38] (see SI Fig. S4C,F). These results suggest that smFCS can distinguish among the classes of assembly pathways observed in our simulations. They are consistent with an interpretation of the Borodavka et al. [46] results in which assembly around the non-cognate RNA proceeds via the disordered mechanism while packaging signals lead to an ordered mechanism.
FIG. 7.
(A),(B) Average polymer radius of gyration during assembly as a function of varying εss with constant Csalt (A) and varying Csalt with constant εss (B). (C),(D) Average polymer radius of gyration during assembly as a function of number of adsorbed subunits nad (C) and size of the largest cluster n (D) for parameters that lead to relatively disordered (squares) and ordered (circles) pathways. (E) Approximate average radius of hydration for representative ordered and disordered trajectories.
To further characterize the differences in polymer conformations between disordered and ordered assembly pathways, we show the polymer radius of gyration, Rg, during assembly trajectories in Fig. 7. In contrast to RH, contributions of the capsid proteins are not included in Rg. While the results in Fig. 7 are averaged over multiple trajectories, example individual trajectories are shown in SI Fig. S3. In all cases of successful assembly, the polymer is gradually compacted from its free size into its encapsidated size. However, at fixed Csalt=100mM, the average rate of compaction increases with εss, with a dramatic increase in rate for εss > 3kBT (Fig. 7A). Similarly, decreasing Csalt increases the rate of compaction (Fig. 7B). Notice that the rate of polymer compaction is not determined by the assembly mechanism | increased order correlates with faster compaction in Fig. 7A but with slower compaction in Figure 7B. When the Rg is plotted as a function of number of adsorbed subunits (nad), the en masse and ordered pathways clearly split into two groups (Fig. 7C). However, this distinction disappears when Rg is plotted as a function of the number of subunits in the largest cluster (n, Fig. 7D). Taken together, these data demonstrate that polymer compaction is driven by adsorbed subunits forming ordered intermediates, with the rate of compaction consequently mirroring the rate of capsid assembly.
trSAXS measurments of bulk assembly kinetics can distinguish assembly mechanisms
While smFCS can detect individual assembly intermediates, Kler et al. [69, 74] recently used time resolved small angle x-ray scattering (trSAXS) to elucidate structures of assembling capsids in bulk. They found that the SAXS profiles at all time points could be decomposed into scattering contributions from the unassembled and complete components, suggesting that assembly proceeded by an effectively two-state reaction with undetectable levels of intermediates (i.e. the nucleation-and-growth pathway with relatively slow nucleation). While it is evident that profiles from profiles from a two-state reaction can be decomposed in this way, we investigated the extent to which SAXS profiles from the other pathway classes (en masse or nucleatation-and-growth with rapid nucleation) can be distinguished from the two-state case.
First, Fig. 8A,B shows SAXS profiles calculated (using CRYSOL [89]) from simulation snapshots along ensembles of en masse and ordered assembly trajectories. For each parameter set, SAXS profiles are averaged over 6 time windows. In both cases, the first profile is dominated by scattering from free subunits (Fig. 8C) and the final time segment shows clear minima and maxima corresponding to the complete capsid (Fig. 8C). For comparison, Fig. 8C presents the SAXS profiles for ordered subunit clusters ranging in size from a single subunit (black) to a complete capsid (yellow). As the capsid grows distinct minima and maxima appear and become more pronounced. We note that the positions of the minima and maxima in the complete model capsid are similar to those observed experimentally for SV40 [69].
FIG. 8.
SAXS profiles for (A) the nucleation-and-growth (ordered) and (B) the en masse (disordered) assembly mechanisms, at indicated parameter values. Simulations were divided into six segments of equal length to describe time evolution from beginning (1) to completion (6). (C) Scattering of subunit clusters from 1 subunit (black) to a 12 subunit capsid (yellow). (D,E,F) Best fit to SAXS profiles at an intermediate time (line 2 in (A,B)) assuming two-state kinetics (a linear combination of complete capsid and unassembled subunits) for ordered (D), disordered (E), and ordered with rate-limiting nucleation (F).
To test the extent to which these trajectories can be distinguished from the two-state case, we attempted to fit SAXS profiles using a linear combination of the profiles for unassembled polymer and subunits and the complete capsid. The resulting fits for the second fifth of the trajectory (where intermediates are most plentiful) are shown in Figs. 8D–F. At this stage, the ordered systems contain mostly incomplete subunit clusters (6–11 subunits), while the disordered simulations contain mostly adsorbed but unassembled subunits. For the ordered simulations, we find that the fit reproduces all of the features of the intermediates, except at low q. In contrast, the intermediates in the disordered trajectory display a shoulder at q ~ 0.3nm−1 that is not observed in any of the unassembled or assembled components. This shoulder is a distinct feature of the disordered intermediate, and thus could be used to identify this class of pathways. Finally, as expected, SAXS profiles from trajectories at parameter sets (Fig. 4C) which lead to two-state kinetics are very well fit by a linear combination of polymer/subunit and complete capsid (Fig. 8F).
A significant distinction between the SAXS profiles is that the ordered pathways lead to an isosbestic point at q ~ 0.3nm−1 (as seen in SV40 experiments[69]), whereas the disordered pathways do not. The presence of an isosbestic point is frequently assumed to indicate two-state behaviour; however, it occurs in our data for ordered trajectories even when the reaction is far from two-state due to rapid nucleation. In these cases the isosbestic point appears due to due to the similarity in scattering from the ordered intermediates and the complete capsid. This suggests that an isosbestic point may distinguish ordered from disordered assemblies, but is less sensitive to the extent to which the reaction kinetics can be approximated as two-state (i.e., how undetectable the intermediate concentrations are).
IV. DISCUSSION
Our simulations demonstrate that capsid assembly around a NA or other cargo can proceed through two mechanisms, and that which mechanism occurs depends on the relative strengths of protein-protein and protein-cargo interactions. The assembly mechanism can be rationally designed by tuning these interactions through solution conditions or mutagenesis of capsid protein-NA binding domains. However, because productive assembly requires weak interactions, the parameters must be tuned within relatively narrow ranges, and alterations which increase (decrease) the strength of one type of interaction must be compensated by a decrease (increase) in the other. Our results suggest that the subunit-cargo dissociation constant is an important parameter, whose value might be used to map specific viruses onto our phase diagrams (Figs. 2, 3, 5), although experimental tests of this capability are required. Finally, we have shown that the nature of assembly pathways can be inferred from the results of recently developed techniques to monitor the assembly of individual capsids or bulk assembly kinetics.
System-dependence of model predictions
An important consideration is to what extent results from this model can be extrapolated to other systems. First, while we have considered the smallest relevant icosahedral capsid geometry (12 pentameric subunits [69, 74]), viruses can assemble from much larger numbers of subunits. However, previous simulations using more simplified models for assembly around polymers or nanoparticles with 20 or 30 subunits observed similar assembly outcomes and also observed the two classes of assembly mechanisms described here [37, 63]. In fact, the en masse and nucleation-and-growth mechanisms can be more easily distinguished for larger capsids, where there can be more adsorbed-but-unassembled subunits and thus a wider range of nfree values. A second factor is the role of interactions between capsid proteins and specific NA sequences [90]. We have performed additional simulations in which model subunits interact with specific sites (‘packaging signals’) on the polyelectrolyte. We find that, while introducing packaging signals can change which assembly mechanism occurs at a specific set of parameter values (i.e. ionic strength and subunit-subunit interaction strength), over the range of relevant parameter values both mechanisms can be observed in the presence of packaging signals. These results will be described in a subsequent article.
The previous arguments suggest that the qualitative trends predicted from our simulations can apply to a wide range of systems. However, the simulations presented here demonstrate that the assembly mechanism and assembly outcome are determined by a balance between different interactions (subunit-polyelectrolyte, subunit-subunit) and different timescales (adsorption, assembly). Thus, predicting the quantitative location of boundaries between different assembly outcomes and different assembly mechanisms in the phase diagrams (Figs. 2, 3, and 5) will require calculating several biophysical parameters in addition to solution conditions. For example, we have shown here that changing the ARM sequence changes the thermodynamic polyelectrolyte binding affinity and thus alters the ionic strength at which the mechanism changes from en masse to nucleation-and-growth (Fig. 6). Factors which change timescales include subunit diffusion constants, subunit-subunit association rates, allosteric or autosteric subunit conformational changes [2, 91], and the number of subunits in a complete capsid (which controls the adsorption timescale and the timescale for a nucleated capsid to grow to completion).
Relationship between model predictions and experiments or potential future experiments
Our simulations predict that a single viral species can be driven to assemble via different mechanisms in vitro by changing solution conditions. In particular, under a constant subunit-subunit binding energy (εss ~ 5–6kBT) robust assembly occurs for a range of solvent conditions, with highly ordered or disordered assemblies occurring depending on salt concentration (Csalt ∈ [50, 400] mM). To our knowledge, this prediction has not yet been realized experimentally, although the signatures of the two classes of assembly pathways have been seen in experiments on different viruses and/or different cargoes [46, 69–72].
One recent experimental study sought to test the role of of Csalt and subunit-subunit attractions (controlled by solution pH) on in vitro assembly of CCMV around RNA [70]. In some regards, these experiments mirror our observations, with malformed capsids observed for strong subunit-subunit attractions, disordered products for weak subunit-subunit attractions, and well-formed capsids at intermediate conditions. However, robust assembly was only observed for a two-step process: first Csalt was reduced (from 1M) to drive RNA-subunit interactions, and then secondly pH was reduced to activate subunit-subunit attractions. The resulting assembly pathways resemble the en masse mechanism described here. On the other hand, one-step assembly led to malformed particles even at moderate pH, suggesting an inability to assemble through a nucleation-and-growth mechanism in these experiments. In our simulations, this outcome would only occur at high salt concentrations (e.g. Csalt ∈ 400 – 500mM, see Figs. 2, 3), where the narrow range of εss leading to successful assembly indicates that parameters must be finely tuned. Reproducing such a lack of successful assembly at moderate salt concentrations would require a reduction in the orientation-dependence of the subunit-subunit interaction potential (see Methods), or introduction of additional factors as discussed below. Experiments in which solution conditions are changed for other viruses which do undergo one-step assembly (e.g. SV40 [69, 74]), may elucidate which of these possibilities to pursue and would directly test our prediction that the assembly mechanism can be controlled by solution conditions.
Understanding capsid assembly mechanisms and their location within the assembly phase diagram has important implications for the design of antiviral agents. As one example, we consider the recently developed class of HBV inhibitors based on phenylpropenamides [85, 86], which act by increasing the strength of subunit-subunit interactions, driving subunits to assemble in the absence of their genome and thus increasing the generation of empty, non-infective capsids [85, 86]. Comparing Figs. 3 and 5 shows that a virus that undergoes ordered assembly (e.g. εss ~ 6 – 7, Csalt ≤ 300 mM), sits close to parameters that support empty capsid assembly, which are demarcated by dashed lines in Fig. 3. Thus only a small increase in subunit-subunit interactions is required to trigger unproductive RNA-free assembly. In contrast, a much larger perturbation would be required to achieve empty capsid assembly for a virus that assembles via the en masse mechanism.
Outlook
We have described two classes of assembly pathways and several distinct failure modes (Fig. 2) that arise when assembly is driven by nonspecific electrostatic subunit-cargo interactions. Our phase diagrams can serve as a starting point to understand how virus-specific features, such as packaging signals [90], allosteric NA-induced [9, 74] or ‘autosteric’ protein-induced conformational changes [91], base pairing-induced NA-structure [38, 46, 92], coordination between assembly and RNA replication [93, 94], or subcellular localization [95] can robustly avoid failure modes amidst the crowded intracellular milieu while enabling selective assembly around the viral genome [23, 94, 96]. For example, allosteric or autosteric conformational changes may allow for strong subunit-subunit interactions on the NA while avoiding the off-cargo assembly we observe at large εss. Systematically studying how these additional factors expand (or contract) regions of parameter space that lead to successful assembly will ultimately reveal how viruses have optimized their structures and interactions for robust assembly in vivo and how their assembly in vivo or in vitro can be manipulated for biomedical or materials science applications.
V. METHODS
Model
We have recently a presented a complete description of our model system, which we summarize here briefly [38] and in the SI. Our model subunits are based upon that previous used to simulate assembly of empty capsids [97–99] which we extended previously to model assembly around cargo [38]. The pseudoatoms in the capsid subunit model are illustrated in Fig. 1. Subunit assembly is mediated through an attractive Morse potential between Attractor (‘A’) pseudoatoms located at each subunit vertex. The Top (‘T’) pseudoatoms interact with other ‘T’ psuedoatoms through a potential consisting of the repulsive term of the Lennard-Jones (LJ) potential, the radius of which is chosen to favor a subunit-subunit angle consistent with a dodecahedron (116 degrees). The Bottom (‘B’) pseudoatom has a repulsive LJ interaction with ‘T’ pseudoatoms, intended to prevent ‘upside-down’ assembly. The ‘T’, ‘B’, and ‘A’ pseudoatoms form a rigid body [97–99]. See Refs. [37, 63, 64, 80, 81, 99–111] for related models.
To model electrostatic interaction with a negatively charged NA or polyelectrolyte we extend the model as follows. Firstly, to better represent the capsid shell we add a layer of ‘Excluder’ pseudoatoms which have a repulsive LJ interaction with the polyelectrolyte and the ARMs. Each ARM is modeled as a bead-spring polymer, with one bead per amino acid. The ‘Excluders’ and first ARM segment are part of the subunit rigid body. ARM beads interact through repulsive LJ interactions and, if charged, electrostatic interactions modelled by a Debye-Hückel potential. We note that repulsion between subunits due to the positive charges does affect the magnitude of the subunit-subunit interaction. Previously, we estimated this repulsion contributes 1kBT to the dimerization free energy [38]. This contribution (and entropic terms) are not included in εss, which is the magnitude of the Morse potential depth. See SI section VII B) for a discussion of binding free energies.
Simulations and units
Simulations were performed with the Brownian Dynamics algorithm of HOOMD, which uses the Langevin equation to evolve positions and rigid body orientations in time [112–114]. Simulations were run using a set of fundamental units. The fundamental energy unit is selected to be Eu ≡ 1kBT. The unit of length Du is set to the circumradius of a pentagonal subunit, which is taken to be 1Du ≡ 5 nm so that the dodecahedron inradius of 1.46Du = 7.3 nm gives an interior volume consistent with that of the smallest T=1 capsids. To calculate the thermodynamic optimal encapsidation length, we placed a very long polymer in or near a preassembled capsid, with one of the capsid subunits made permeable to the polymer and performed unbiased Brownian dynamics. Once the amount of packaged polymer reached equilibrium, the thermodynamic optimum length was measured. We previously [38] found that this strategy closely matched that produced using the Widom insertion method [115] as applied to growing polymer chains [37, 116]. Assembly simulations were run at least 10 times for each set of parameters, each of which were concluded at either completion, persistent malformation, or 2 × 108 time steps. This observation time was chosen based on the time after which assembly yields and outcomes change only logarithmically with time for most parameter values. For all dynamics simulations there were 60 subunits with box size=200 × 200 × 200 nm, resulting in a concentration of 12µM.
SAXS profile and hydrodynamic radius estimations
Small Angle X-ray Scattering (SAXS) analysis was performed using CRYSOL [89]. For this analysis the all-atom structure of an SV40 pentameric subunit [117] was aligned into the position of each coarse-grained subunit and the polymer was replaced with a carbon chain. We note that this entails significant simplification: segments of the protein which were not resolved in the crystal structure were not reconstructed and there is no optimization of structure at the all-atom resolution. We believe this approximation is suitable, given that our analysis is limited to the X-ray scattering profile in the small angle regime, which reflects ~nm scale structural features. Fitting of the scattering profiles was performed using least squares fitting. Hydrodynamic radius analysis was performed using HYDROPRO [88]. This program is capable of calculating the hydrodynamic radius of large protein complexes and protein-NA complexes. Our treatment of the polyelectrolyte chain in this analysis involves two approximations. The first is that we neglect the specific chemistry of their polyelectrolyte, instead treating it as an amino acid chain. Based on our observations, the RH is dominated by the adsorbed capsid subunits and removing the polymer entirely from the RH calculation has a minimal effect on RH (see Fig. S4A). This suggests a more accurate representation of the polyelectrolyte would not qualitatively alter our conclusions. The second approximation is analyzing a flexible polyelectrolyte using an algorithm intended to calculate the hydrodynamics radius of ‘rigid’ molecules (such as globular proteins). However, there is a long precedent for applying the rigid body method to flexible polymers: in these cases an ensemble of conformations is generated, and the hydrodynamic radius is the ensemble average of the ‘instantaneously rigid’ conformations [118]. Note that in Fig. 7E we are presenting the average over many independent assembly simulations. Simulations were visualized using VMD [119].
Supplementary Material
Research Highlights.
The spontaneous assembly of viral capsids is an essential step in the lifecycle of many viruses and provides a paradigm for efficient, high fidelity molecular self-assembly.
Capsid assembly around a negatively charged substrate can proceed through two classes of pathways, depending upon the properties of the capsid proteins and solution conditions.
We perform computer simulations which predict that these pathways can be distinguished using two different experimental techniques
Assembly pathways and their sensitivity to control parameters may be an important factor in determining the effectiveness of anti-viral agents and engineered delivery vectors.
ACKNOWLEDGMENTS
We gratefully acknowledge Adam Zlotnick for insightful discussion and critical reading of the manuscript. This work was supported by Award Number R01GM108021 from the National Institute Of General Medical Sciences. Computational resources were provided by the NSF through XSEDE computing resources (Longhorn, Maverick, Condor, and Keeneland) and the Brandeis HPCC which is partially supported by NSF-MRSEC-0820492.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hagan MF. Modeling Viral Capsid Assembly. Adv Chem Phys. 2014:155. doi: 10.1002/9781118755815.ch01. http://arxiv.org/abs/1301.1657. [DOI] [PMC free article] [PubMed]
- 2.Zlotnick A, Mukhopadhyay S. Virus assembly, allostery and antivirals. Trends Microbiol. 2011;19(1):14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevelige Peter E. New approaches for antiviral targeting of HIV assembly. J. Mol. Biol. 2011 Jul;410(4):634–640. doi: 10.1016/j.jmb.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JM, Johnson JE, Young MJ. RNA/Protein Interactions in Icosahedral Virus Assembly. Seminars in Virology. 1994;5(1):51–60. [Google Scholar]
- 5.Valegard K, Murray JB, Stonehouse NJ, vandenWorm S, Stockley PG, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J Mol Biol. 1997;270(5):724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JM, Willits DA, Young MJ, Zlotnick A. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of Cowpea chlorotic mottle virus. J Mol Biol. 2004;335(2):455–464. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 7.Tihova M, Dryden KA, Le TVL, Harvey SC, Johnson JE, Yeager M, Schneemann A. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J Virol. 2004;78(6):2897–2905. doi: 10.1128/JVI.78.6.2897-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci U S A. 1999 Nov 23;96(24):13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockley Peter G, Rolfsson Ottar, Thompson Gary S, Basnak Gabriella, Francese Simona, Stonehouse Nicola J, Homans Steven W, Ashcroft Alison E. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T=3 viral capsid. J. Mol. BIo. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA. The three-dimensional structure of genomic RNA in bacteriophage MS2: Implications for assembly. J Mol Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 11.Lucas RW, Larson SB, McPherson A. The crystallographic structure of brome mosaic virus. J Mol Biol. 2002;317(1):95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 12.Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an bacteriophage RNA coat protein operator complex. Nature. 1994;371(6498):623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 13.van den Worm SHE, Stonehouse NJ, Valegard K, Murray JB, Walton C, Fridborg K, Stockley PG, Liljas L. Crystal structures of MS2 coat protein mutants in complex with wild-type RNA operator fragments. Nucleic Acids Res. 1998;26(5):1345–1351. doi: 10.1093/nar/26.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grahn E, Moss T, Helgstrand C, Fridborg K, Sundaram M, Tars K, Lago H, Stonehouse NJ, Davis DR, Stockley PG, Liljas L. Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex. RNA-a Publication of the RNA Society. 2001;7(11):1616–1627. [PMC free article] [PubMed] [Google Scholar]
- 15.Helgstrand C, Grahn E, Moss T, Stonehouse NJ, Tars K, Stockley PG, Liljas L. Investigating the structural basis of purine specificity in the structures of MS2 coat protein RNA translational operator hairpins. Nucleic Acids Res. 2002;30(12):2678–2685. doi: 10.1093/nar/gkf371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol. 2006;60:51–67. doi: 10.1146/annurev.micro.60.080805.142304. [DOI] [PubMed] [Google Scholar]
- 17.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: Implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci U S A. 1997;94(18):9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway JF, Steven AC. Methods for reconstructing density maps of"single" particles from cryoelectron micrographs to subnanometer resolution. J of Struct Biol. 1999;128(1):106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 19.Freddolino Peter L, Arkhipov Anton S, Larson Steven B, McPherson Alexander, Schulten Klaus. Molecular Dynamics Simulations of the Complete Satellite Tobacco Mosaic Virus. Structure. 2006;14(3):437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Devkota B, Petrov AS, Lemieux S, Boz MB, Tang L, Schneemann A, Johnson JE, Harvey SC. Structural and Electrostatic Characterization of Pariacoto Virus: Implications for Viral Assembly. Biopolymers. 2009;91(7):530–538. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Y, Larson SB, Heitsch CE, McPherson A, Harvey SC. A Model for the Structure of Satellite Tobacco Mosaic Virus. J Struct Biol. 2012 doi: 10.1016/j.jsb.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakker Saskia E, Ford Robert J, Barker Amy M, Robottom Janice, Saunders Keith, Pearson Arwen R, Ranson Neil A, Stockley Peter G. Isolation of an asymmetric RNA uncoating intermediate for a single-stranded RNA plant virus. Journal of Molecular Biology. 2012;417(1):65–78. doi: 10.1016/j.jmb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford Robert J, Barker Amy M, Bakker Saskia E, Coutts Robert H, Ranson Neil A, Phillips Simon EV, Pearson Arwen R, Stockley Peter G. Sequence-specific, RNA–protein interactions overcome electrostatic barriers preventing assembly of satellite tobacco necrosis virus coat protein. Journal of molecular biology. 2013;425(6):1050–1064. doi: 10.1016/j.jmb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson Steven B, McPherson Alexander. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Current opinion in structural biology. 2001;11(1):59–65. doi: 10.1016/s0959-440x(00)00166-4. [DOI] [PubMed] [Google Scholar]
- 25.Dykeman EC, Grayson NE, Toropova K, Ranson NA, Stockley PG, Twarock R. Simple Rules for Efficient Assembly Predict the Layout of a Packaged Viral RNA. J Mol Biol. 2011;408(3):399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 26.Dykeman Eric C, Stockley Peter G, Twarock Reidun. Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome. Journal of molecular biology. 2013;425(17):3235–3249. doi: 10.1016/j.jmb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Siber A, Podgornik R. Nonspecific interactions in spontaneous assembly of empty versus functional single-stranded RNA viruses. Phys Rev E. 2008;78:051915. doi: 10.1103/PhysRevE.78.051915. [DOI] [PubMed] [Google Scholar]
- 28.Hu Tao, Zhang Rui, Shklovskii BI. Electrostatic theory of viral self-assembly: a toy model. Physica A. 2008;387:3059. [Google Scholar]
- 29.Forrey C, Muthukumar M. Electrostatics of capsid-induced viral RNA organization. J Chem Phys. 2009;131(10) [Google Scholar]
- 30.Harvey SC, Petrov AS, Devkota B, Boz MB. Viral assembly: a molecular modeling perspective. Phys Chem Chem Phys. 2009;11(45):10553–10564. doi: 10.1039/b912884k. [DOI] [PubMed] [Google Scholar]
- 31.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci U S A. 2006;103(46):17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angelescu DG, Bruinsma R, Linse P. Monte Carlo simulations of polyelectrolytes inside viral capsids. Phys Rev E. 2006;73(4):041921. doi: 10.1103/PhysRevE.73.041921. [DOI] [PubMed] [Google Scholar]
- 33.Zhang ZL, Glotzer SC. Self-assembly of patchy particles. Nano Lett. 2004;4(8):1407–1413. doi: 10.1021/nl0493500. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Nguyen TT. Radial distribution of RNA genomes packaged inside spherical viruses. Phys Rev Lett. 2008;100(19) doi: 10.1103/PhysRevLett.100.198102. [DOI] [PubMed] [Google Scholar]
- 35.Angelescu Daniel George, Linse Per. Modelling of icosahedral viruses. Curr Opin Colloid Interface Sci. 2008;13(6):389–394. [Google Scholar]
- 36.Jiang T, Wang ZG, Wu JZ. Electrostatic Regulation of Genome Packaging in Human Hepatitis B Virus. Biophys J. 2009;96(8):3065–3073. doi: 10.1016/j.bpj.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elrad OM, Hagan MF. Encapsulation of a polymer by an icosahedral virus. Phys Biol. 2010;7:045003. doi: 10.1088/1478-3975/7/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perlmutter Jason D, Qiao Cong, Hagan Michael F. Viral genome structures are optimal for capsid assembly. eLife. 2013;2:e00632. doi: 10.7554/eLife.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoot Paul, Zandi Roya. Impact of the topology of viral RNAs on their encapsulation by virus coat proteins. Journal of Biological Physics. 2013;39(2):289–299. doi: 10.1007/s10867-013-9307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu T, Zhang R, Shklovskii BI. Electrostatic theory of viral self-assembly. Physica a-Statistical Mechanics and Its Applications. 2008;387(12):3059–3064. [Google Scholar]
- 41.van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Phys Rev E. 2005;71(6):061928. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- 42.Ting Christina L, Wu Jianzhong, Wang Zhen-Gang. Thermodynamic basis for the genome to capsid charge relationship in viral encapsidation. Proc Natl Acad Sci U S A. 2011;108(41):16986–16991. doi: 10.1073/pnas.1109307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni P, Wang Z, Ma X, Das NC, Sokol P, Chiu W, Dragnea B, Hagan MF, C Kao C. An Examination of the Electrostatic Interactions between the N-Terminal Tail of the Coat Protein and RNA in Brome Mosaic Virus. J Mol Biol. 2012;419:284–300. doi: 10.1016/j.jmb.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siber Antonio, Zandi Roya, Podgornik Rudolf. Thermodynamics of nanospheres encapsulated in virus capsids. Phys Rev E. 2010;81(5):051919. doi: 10.1103/PhysRevE.81.051919. [DOI] [PubMed] [Google Scholar]
- 45.Siber Antonio, Bozic Anze Losdorfer, Podgornik Rudolf. Energies and pressures in viruses: contribution of nonspecific electrostatic interactions. Phys Chem Chem Phys. 2012;14(11):3746–3765. doi: 10.1039/c1cp22756d. [DOI] [PubMed] [Google Scholar]
- 46.Borodavka Alexander, Tuma Roman, Stockley Peter G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc Natl Acad Sci U S A. 2012;109(39):15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porterfield J. Zachary, Dhason Mary Savari, Loeb Daniel D, Nassal Michael, Stray Stephen J, Zlotnick Adam. Full-Length Hepatitis B Virus Core Protein Packages Viral and Heterologous RNA with Similarly High Levels of Cooperativity. J Virol. 2010;84(14):7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comas-Garcia Mauricio, Cadena-Nava Ruben D, Rao ALN, Knobler Charles M, Gelbart William M. In vitro quantification of the relative packaging efficiencies of single-stranded RNA molecules by viral capsid protein. J Virol. 2012 doi: 10.1128/JVI.01695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiebert E, Bancroft JE, Bracker CE. Assembly In Vitro Of Some Small Spherical Viruses, Hybrid Viruses, And Other Nucleoproteins. Virology. 1968;34(3(3)):492–509. doi: 10.1016/0042-6822(68)90069-x. [DOI] [PubMed] [Google Scholar]
- 50.Bancroft Jb, E Hiebert, Ce Bracker. Effects Of Various Polyanions On Shell Formation Of Some Spherical Viruses. Virology. 1969;39(4):924–930. doi: 10.1016/0042-6822(69)90029-4. [DOI] [PubMed] [Google Scholar]
- 51.Dixit SK, Goicochea NL, Daniel MC, Murali A, Bronstein L, De M, Stein B, Rotello VM, Kao CC, Dragnea B. Quantum dot encapsulation in viral capsids. Nano Lett. 2006;6(9):1993–1999. doi: 10.1021/nl061165u. [DOI] [PubMed] [Google Scholar]
- 52.Loo L, Guenther RH, Basnayake VR, Lommel SA, Franzen S. Controlled encapsidation of gold nanoparticles by a viral protein shell. J Am Chem Soc. 2006 Apr 12;128(14):4502–4503. doi: 10.1021/ja057332u. [DOI] [PubMed] [Google Scholar]
- 53.Goicochea Nancy L, De Mrinmoy, Rotello Vincent M, Mukhopadhyay Suchetana, Dragnea Bogdan. Core-like particles of an enveloped animal virus can self-assemble efficiently on artificial templates. Nano Lett. 2007;7(8):2281–2290. doi: 10.1021/nl070860e. [DOI] [PubMed] [Google Scholar]
- 54.Huang Xinlei, Bronstein Lyudmila M, Retrum John, Dufort Chris, Tsvetkova Irina, Aniagyei Stella, Stein Barry, Stucky Galen, McKenna Brandon, Remmes Nicholas, Baxter David, Kao C. Cheng, Dragnea Bogdan. Self-assembled virus-like particles with magnetic cores. Nano Lett. 2007 Aug;7(8):2407–2416. doi: 10.1021/nl071083l. [DOI] [PubMed] [Google Scholar]
- 55.Loo LiNa, Guenther Richard H, Lommel Steven A, Franzen Stefan. Encapsidation of nanoparticles by Red Clover Necrotic Mosaic Virus. J Am Chem Soc. 2007 Sep 12;129(36):11111–11117. doi: 10.1021/ja071896b. [DOI] [PubMed] [Google Scholar]
- 56.Sikkema FD, Comellas-Aragones M, Fokkink RG, Verduin BJM, Cornelissen Jjlm, Nolte RJM. Monodisperse polymer-virus hybrid nanoparticles. Org Biomol Chem. 2007;5(1):54–57. doi: 10.1039/b613890j. [DOI] [PubMed] [Google Scholar]
- 57.Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, Kao CC, Dragnea B. Core-controlled polymorphism in virus-like particles. Proc Natl Acad Sci U S A. 2007;104(4):1354–1359. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu LH, Grosberg AY, Bruinsma R. Are DNA transcription factor proteins Maxwellian Demons? Biophys J. 2008;95(3):1151–1156. doi: 10.1529/biophysj.108.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comellas-Aragones Marta, de la Escosura Andres, Dirks AJ, van der Ham Anne, Fuste-Cune Anna, Cornelissen Jeroen JLM, Nolte Roeland JM. Controlled Integration of Polymers into Viral Capsids. Biomacromolecules. 2009 Nov;10(11):3141–3147. doi: 10.1021/bm9007953. [DOI] [PubMed] [Google Scholar]
- 60.Crist RM, Datta SAK, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. Assembly Properties of Human Immunodeficiency Virus Type 1 Gag-Leucine Zipper Chimeras: Implications for Retrovirus Assembly. J Virol. 2009;83(5):2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang Connie B, Knobler Charles M, Gelbart William M, Mason Thomas G. Curvature dependence of viral protein structures on encapsidated nanoemulsion droplets. ACS Nano. 2008 Feb;2(2):281–286. doi: 10.1021/nn700385z. [DOI] [PubMed] [Google Scholar]
- 62.McPherson A. Micelle formation and crystallization as paradigms for virus assembly. Bioessays. 2005;27(4):447–458. doi: 10.1002/bies.20196. [DOI] [PubMed] [Google Scholar]
- 63.Hagan MF. Controlling viral capsid assembly with templating. Phys Rev E. 2008;77:051904. doi: 10.1103/PhysRevE.77.051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahalik JP, Muthukumar M. Langevin dynamics simulation of polymer-assisted virus-like assembly. J Chem Phys. 2012;136(13):135101. doi: 10.1063/1.3698408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perkett Matthew R, Hagan Michael F. Using Markov State Models to Study Self-Assembly. doi: 10.1063/1.4878494. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Endres D, Zlotnick A. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys J. 2002;83(2):1217–1230. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Tao, Shklovskii BI. Kinetics of viral self-assembly: Role of the single-stranded RNA antenna. Phys Rev E. 2007;75:051901. doi: 10.1103/PhysRevE.75.051901. [DOI] [PubMed] [Google Scholar]
- 68.Kivenson A, Hagan MF. Mechanisms of viral capsid assembly around a polymer. Biophys J. 2010;99(2(2)):619–628. doi: 10.1016/j.bpj.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kler Stanislav, Asor Roi, Li Chenglei, Ginsburg Avi, Harries Daniel, Oppenheim Ariella, Zlotnick Adam, Raviv Uri. RNA Encapsidation by SV40-Derived Nanoparticles Follows a Rapid Two-State Mechanism. J Am Chem Soc. 2012;134(21):8823–8830. doi: 10.1021/ja2110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garmann Rees F, Comas-Garcia Mauricio, Gopal Ajaykumar, Knobler Charles M, Gelbart William M. The Assembly Pathway of an Icosahedral Single-Stranded fRNAg Virus Depends on the Strength of Inter-Subunit Attractions. Journal of Molecular Biology. 2013;(0) doi: 10.1016/j.jmb.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cadena-Nava Ruben D, Comas-Garcia Mauricio, Garmann Rees F, Rao ALN, Knobler Charles M, Gelbart William M. Self-Assembly of Viral Capsid Protein and RNA Molecules of Different Sizes: Requirement for a Specific High Protein/RNA Mass Ratio. J Virol. 2012;86(6):3318–3326. doi: 10.1128/JVI.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malyutin Andrey G, Dragnea Bogdan. Budding Pathway in the Templated Assembly of Viruslike Particles. J Phys Chem B. 2013 Sep; doi: 10.1021/jp405603m. [DOI] [PubMed] [Google Scholar]
- 73.Zlotnick Adam, Porterfield J. Zachary, Wang Joseph Che-Yen. To Build a Virus on a Nucleic Acid Substrate. Biophys J. 2013;104(7):1595–1604. doi: 10.1016/j.bpj.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kler Stanislav, Wang Joseph Che-Yen, Dhason Mary, Oppenheim Ariella, Zlotnick Adam. Scaffold properties are a key determinant of the size and shape of self-assembled virus-derived particles. ACS Chem. Biol. 2013 Dec;8(12):2753–61. doi: 10.1021/cb4005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li PP, Nakanishi A, Clark SW, Kasamatsu H. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc Natl Acad Sci U S A. 2002;99(3):1353–1358. doi: 10.1073/pnas.032668699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ceres P, Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41(39):11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 77.Kegel WK, van der Schoot P. Competing hydrophobic and screened-Coulomb interactions in hepatitis B virus capsid assembly. Biophys J. 2004;86(6):3905–3913. doi: 10.1529/biophysj.104.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siber Antonio, Podgornik Rudolf. Role of electrostatic interactions in the assembly of empty spherical viral capsids. Phys Rev E. 2007;76:061906. doi: 10.1103/PhysRevE.76.061906. [DOI] [PubMed] [Google Scholar]
- 79.Evilevitch Alex, Fang Li Tai, Yoffe Aron M, Castelnovo Martin, Rau Donald C, Parsegian V Adrian, Gelbart William M, Knobler Charles M. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophysical journal. 2008;94(3):1110–1120. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hagan MF, Chandler D. Dynamic pathways for viral capsid assembly. Biophys J. 2006;91(1):42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hagan MF, Elrad OM, Jack RL. Mechanisms of kinetic trapping in self-assembly and phase transformation. J Chem Phys. 2011;135:104115. doi: 10.1063/1.3635775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grant James, Jack Robert L, Whitelam Stephen. Analyzing mechanisms and microscopic reversibility of self-assembly. J Chem Phys. 2011;135(21) doi: 10.1063/1.3662140. [DOI] [PubMed] [Google Scholar]
- 83.Whitelam Stephen, Rogers Carl, Pasqua Andrea, Paavola Chad, Trent Jonathan, Geissler Phillip L. The Impact of Conformational Fluctuations on Self-Assembly: Cooperative Aggregation of Archaeal Chaperonin Proteins. Nano Lett. 2009;9:292–297. doi: 10.1021/nl8029306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bottcher B, Unseld S, Ceulemans H, Russell RB, Jeske H. Geminate structures of African cassava mosaic virus. J Virol. 2004;78(13):6758–6765. doi: 10.1128/JVI.78.13.6758-6765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katen SP, Chirapu SR, Finn MG, Zlotnick A. Trapping of Hepatitis B Virus Capsid Assembly Intermediates by Phenylpropenamide Assembly Accelerators. ACS Chemical Biology. 2010;5(12):1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katen Sarah P, Tan Zhenning, Chirapu Srinivas Reddy, Finn MG, Zlotnick Adam. Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure. Structure. 2013 Aug;21(8):1406–16. doi: 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elrad OM, Hagan MF. Nano Lett. 2008;8(11):3850–3857. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ortega A, Amorós D, de La Torre J García. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic-and residue-level models. Biophysical journal. 2011;101(4):892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Svergun D, Barberato C, Koch MHJ. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28(6):768–773. [Google Scholar]
- 90.Stockley Peter G, Twarock Reidun, Bakker Saskia E, Barker Amy M, Borodavka Alexander, Dykeman Eric, Ford Robert J, Pearson Arwen R, Phillips Simon EV, Ranson Neil A, et al. Packaging signals in single-stranded RNA viruses: nature's alternative to a purely electrostatic assembly mechanism. Journal of biological physics. 2013;39(2):277–287. doi: 10.1007/s10867-013-9313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Caspar Donald L. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophysical journal. 1980;32(1):103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc Natl Acad Sci U S A. 2008;105(42):16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Annamalai P, Rofail F, DeMason DA, Rao ALN. Replication-coupled packaging mechanism in positive-strand RNA viruses: Synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration. J Virol. 2008;82(3):1484–1495. doi: 10.1128/JVI.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rao ALN. Genome packaging by spherical plant RNA viruses. Annual Review of Phytopathology, volume 44 of Annual Review of Phytopathology. 2006:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 95.Bamunusinghe Devinka, Seo Jang-Kyun, Rao ALN. Subcellular Localization and Rearrangement of Endoplasmic Reticulum by Brome Mosaic Virus Capsid Protein. J Virol. 2011;85(6):2953–2963. doi: 10.1128/JVI.02020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Routh Andrew, Domitrovic Tatiana, Johnson John E. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proceedings of the National Academy of Sciences. 2012;109(6):1907–1912. doi: 10.1073/pnas.1116168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wales DJ. The energy landscape as a unifying theme in molecular science. Phil Trans R Soc A. 2005;363(1827):357–375. doi: 10.1098/rsta.2004.1497. [DOI] [PubMed] [Google Scholar]
- 98.Fejer Szilard N, James Tim R, Hernandez-Rojas Javier, Wales David J. Energy landscapes for shells assembled from pentagonal and hexagonal pyramids. Phys Chem Chem Phys. 2009;11(12):2098–2104. doi: 10.1039/b818062h. [DOI] [PubMed] [Google Scholar]
- 99.Johnston IG, Louis AA, Doye JPK. Modelling the self-assembly of virus capsids. J Phys : Condens Matter. 2010;22(10) doi: 10.1088/0953-8984/22/10/104101. [DOI] [PubMed] [Google Scholar]
- 100.Schwartz R, Shor PW, Prevelige PE, Berger B. Local rules simulation of the kinetics of virus capsid self-assembly. Biophys J. 1998;75(6):2626–2636. doi: 10.1016/S0006-3495(98)77708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hicks SD, Henley CL. Irreversible growth model for virus capsid assembly. Phys Rev E. 2006;74(3):031912. doi: 10.1103/PhysRevE.74.031912. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen HD, Reddy VS, Brooks CL. Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Lett. 2007;7(2):338–344. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- 103.Wilber AW, Doye JPK, Louis AA, Noya EG, Miller MA, Wong P. Reversible self-assembly of patchy particles into monodisperse icosahedral clusters. J Chem Phys. 2007;127(8) doi: 10.1063/1.2759922. [DOI] [PubMed] [Google Scholar]
- 104.Nguyen HD, Brooks CL. Generalized structural polymorphism in self-assembled viral particles. Nano Lett. 2008;8:4574. doi: 10.1021/nl802828v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nguyen Hung D, Reddy Vijay S, Brooks Charles L. Invariant polymorphism in virus capsid assembly. J Am Chem Soc. 2009;131(7):2606–14. doi: 10.1021/ja807730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilber AW, Doye JPK, Louis AA. Self-assembly of monodisperse clusters: Dependence on target geometry. J Chem Phys. 2009;131(17) doi: 10.1063/1.3243580. [DOI] [PubMed] [Google Scholar]
- 107.Wilber Alex W, Doye Jonathan PK, Louis Ard A, Lewis Anna CF. Monodisperse self-assembly in a model with protein-like interactions. J Chem Phys. 2009;131(17):175102. doi: 10.1063/1.3243581. [DOI] [PubMed] [Google Scholar]
- 108.Rapaport DC, Johnson JE, Skolnick J. Supramolecular self-assembly: molecular dynamics modeling of polyhedral shell formation. Comput Phys Commun. 1999;122:231–235. [Google Scholar]
- 109.Rapaport DC. Self-assembly of polyhedral shells: a molecular dynamics study. Phys. Rev. E. 2004;70:051905. doi: 10.1103/PhysRevE.70.051905. [DOI] [PubMed] [Google Scholar]
- 110.Rapaport DC. The role of reversibility in viral capsid growth: A paradigm for self-assembly. Phys Rev Lett. 2008;101:186101. doi: 10.1103/PhysRevLett.101.186101. [DOI] [PubMed] [Google Scholar]
- 111.Levandovsky Artem, Zandi Roya. Nonequilibirum assembly, retroviruses, and conical structures. Phys Rev Lett. 2009;102(19):198102–198102. doi: 10.1103/PhysRevLett.102.198102. [DOI] [PubMed] [Google Scholar]
- 112.Anderson Joshua A, Lorenz Chris D, Travesset A. General purpose molecular dynamics simulations fully implemented on graphics processing units. J Comput Phys. 2008;227(10):5342–5359. [Google Scholar]
- 113.Dac Nguyen Trung, Phillips Carolyn L, Anderson Joshua A, Glotzer Sharon C. Rigid body constraints realized in massively-parallel molecular dynamics on graphics processing units. Comput Phys Commun. 2011;182(11):2307–2313. [Google Scholar]
- 114.LeBard David N, Levine Benjamin G, Mertmann Philipp, Barr Stephen A, Jusufi Arben, Sanders Samantha, Klein Michael L, Panagiotopoulos Athanassios Z. Self-assembly of coarse-grained ionic surfactants accelerated by graphics processing units. Soft Matter. 2012;8(8):2385–2397. [Google Scholar]
- 115.Widom B. Some topics in the theory of fluids. J Chem Phys. 1963;39(11):2808–2812. [Google Scholar]
- 116.Kumar SK, Szleifer I, Panagiotopoulos AZ. Determination Of The Chemical-potentials Of Polymeric Systems From Monte-carlo Simulations. Phys Rev Lett. 1991 Jun 3;66(22):2935–2938. doi: 10.1103/PhysRevLett.66.2935. [DOI] [PubMed] [Google Scholar]
- 117.Stehle T, Gamblin SJ, Yan YW, Harrison SC. The structure of simian virus 40 refined at 3.1 angstrom resolution. Structure. 1996;4(2):165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 118.Amoros D, Ortega A, de la Torre J García. Hydrodynamic properties of wormlike macromolecules: Monte Carlo simulation and global analysis of experimental data. Macromolecules. 2011;44(14):5788–5797. [Google Scholar]
- 119.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 120.Mcghee JD, Hippel PHV. Theoretical Aspects Of Dna-protein Interactions - Cooperative And Non-cooperative Binding Of Large Ligands To A One-dimensional Homogeneous Lattice. J. Mol. BioEssays. 1974;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 121.Bowman GR, Voelz VA, Pande VS. Atomistic Folding Simulations of the Five-Helix Bundle Protein lambda(6–85) Am Chem Soc. 2011;133(4):664–667. doi: 10.1021/ja106936n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bowman GR, Ensign DL, Pande VS. Enhanced Modeling via Network Theory: Adaptive Sampling of Markov State Models. J Chem Theory Comput. 2010;6(3):787–794. doi: 10.1021/ct900620b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bowman GR, Beauchamp KA, Boxer G, Pande VS. Progress and challenges in the automated construction of Markov state models for full protein systems. J Chem Phys. 2009;131(12) doi: 10.1063/1.3216567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Swope WC, Pitera JW, Suits F, Pitman M, Eleftheriou M, Fitch BG, Germain RS, Rayshubski A, Ward TJC, Zhestkov Y, Zhou R. Describing protein folding kinetics by molecular dynamics simulations. 2. Example applications to alanine dipeptide and beta-hairpin peptide. J Phys Chem B. 2004;108(21):6582–6594. [Google Scholar]
- 125.Swope WC, Pitera JW, Suits F. Describing protein folding kinetics by molecular dynamics simulations. 1. Theory. J Phys Chem B. 2004;108(21):6571–6581. [Google Scholar]
- 126.Prinz Jan-Hendrik, Chodera John D, Pande Vijay S, Swope William C, Smith Jeremy C, Noe Frank. Optimal use of data in parallel tempering simulations for the construction of discrete-state Markov models of biomolecular dynamics. J Chem Phys. 2011;134(24) doi: 10.1063/1.3592153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Park S, Ensign DL, Pande VS. Bayesian update method for adaptive weighted sampling. Phys Rev E. 2006;74(6) doi: 10.1103/PhysRevE.74.066703. [DOI] [PubMed] [Google Scholar]
- 128.Pande VS, Beauchamp K, Bowman GR. Everything you wanted to know about Markov State Models but were afraid to ask. Methods. 2010;52(1):99–105. doi: 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci U S A. 2009;106(45):19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lane Thomas J, Bowman Gregory R, Beauchamp Kyle, Voelz Vincent A, Pande Vijay S. Markov State Model Reveals Folding and Functional Dynamics in Ultra-Long MD Trajectories. J Am Chem Soc. 2011;133(45):18413–18419. doi: 10.1021/ja207470h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jayachandran G, Vishal V, Pande VS. Using massively parallel simulation and Markovian models to study protein folding: Examining the dynamics of the villin headpiece. J Chem Phys. 2006;124(16) doi: 10.1063/1.2186317. [DOI] [PubMed] [Google Scholar]
- 132.Chodera John D, Swope William C, Noe Frank, Prinz Jan-Hendrik, Shirts Michael R, Pande Vijay S. Dynamical reweighting: improved estimates of dynamical properties from simulations at multiple temperatures. J Chem Phys. 2011;134(24):244107. doi: 10.1063/1.3592152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hinrichs NS, Pande VS. Calculation of the distribution of eigenvalues and eigenvectors in Markovian state models for molecular dynamics. J Chem Phys. 2007;126(24) doi: 10.1063/1.2740261. [DOI] [PubMed] [Google Scholar]
- 134.Chodera JD, Singhal N, Pande VS, Dill KA, Swope WC. Automatic discovery of metastable states for the construction of Markov models of macromolecular conformational dynamics. J Chem Phys. 2007;126(15) doi: 10.1063/1.2714538. [DOI] [PubMed] [Google Scholar]
- 135.Deuhard P, Weber M. Robust Perron cluster analysis in conformation dynamics. Linear Algebra and Its Applications. 2005;398:161–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.