Abstract
Long noncoding RNAs (lncRNAs) have recently been associated with the development and progression of a variety of human cancers. However, to date, the interplay between known oncogenic or tumor suppressive events and lncRNAs has not been well described. Here the novel lncRNA, Prostate Cancer-Associated Transcript 29 (PCAT29), is characterized along with its relationship to the androgen receptor (AR). PCAT29 is suppressed by dihydrotestosterone (DHT) and upregulated upon castration therapy in a prostate cancer xenograft model. PCAT29 knockdown significantly increased proliferation and migration of prostate cancer cells, while PCAT29 overexpression conferred the opposite effect and suppressed growth and metastases of prostate tumors in chick chorioallantoic membrane (CAM) assays. Finally, in prostate cancer patient specimens, low PCAT29 expression correlated with poor prognostic outcomes. Taken together these data expose PCAT29 as an androgen-regulated tumor suppressor in prostate cancer.
Introduction
Recently, data from the ENCODE project has revealed that the majority of the transcriptome is comprised of non-coding RNAs (1). While the classification of these non-coding RNAs is still in development, long non-coding RNAs (lncRNAs) are of particular interest, given the similar features they share with protein-coding genes as well as recent evidence of their roles in cancer biology (2, 3). LncRNAs are polyadenylated RNA species that are more than 200 base pairs (bp) in length, transcribed by RNA polymerase II, and associated with common epigenetic signatures such as of histone 3 lysine 4 trimethylation (H3K4me3) at the transcriptional start site (TSS) and histone 3 lysine 36 trimethylation (H3K36me3) in the gene body (4). Several lncRNAs have been shown to play role in biological processes such as X-chromosomal inactivation, pluripoency (5) and gene regulation (6). Recently, several lncRNAs have been implicated in cancer initiation and progression (3, 7). Apart from their role in tumor initiation and progression, lncRNAs have been shown to be promising biomarkers. In prostate cancer, PCA3 is a well-studied prostate cancer biomarker that is now available for clinical use as a urine biomarker assay for diagnosis of prostate cancer (8, 9).
Despite their involvement in various cellular processes, the majority of lncRNAs are uncharacterized, and their role in cancer initiation and progression remains unclear. Using transcriptome sequencing, our group recently identified over 100 long-noncoding RNAs, named prostate cancer-associated transcripts (PCATs), which are differentially expressed or have outlier profiles in prostate cancer versus normal tissue (3). Here we find that one of these novel lncRNAs, PCAT29, exhibits cancer-suppressive phenotypes, including inhibition of cell proliferation, migration, tumor growth, and metastases. In accordance with this, PCAT29 is repressed by androgen signaling, and low PCAT29 expression associates with worse clinical outcomes.
Materials and Methods
Cell lines and Reagents
Prostate cancer cells were cultured as follows: VCaP cells in DMEM with Glutamax (Invitrogen), and LNCaP and DU145 cells in RPMI1640 (Invitrogen) in a 5% CO2 cell culture incubator. All the media were supplemented with 10% FBS (Invitrogen) and 1% penicillin-streptomycin (Invitrogen). All cell lines were purchased from ATCC and were authenticated.
For stable knockdown of PCAT29, LNCaP and VCaP cells were transfected with lentiviral constructs encoding 2 different PCAT29 shRNA or non-targeting shRNA in the presence of polybrene (8 μg/ml) (Supplementary Table S1A). After 48 hours, transduced cells were grown in culture media containing 3-5 μg/ml puromycin. For PCAT29 overexpression, two isoforms of PCAT29 were generated by subcloning the polymerase chain reaction (PCR) product into the CPO1 sites of the pCDH-CMV vector (System Biosciences). 500bp of the genomic region was attached at the 5’ end of each isoform. Lentiviral particles were made and DU145 cells were transduced as described above.
Gene Expression by Quantitative PCR
Total RNA was isolated using TRIzol (invotrogen) and an RNeasy kit (Qiagen) according to manufacturers’ instruction. Total RNA was reverse transcribed into cDNA using SuperScript III and random primers (Invitrogen). qPCR was performed using SYBR Geen Master Mix (Applied Biosystems) on a Applied Biosystems 7900HT Real-Time System. The relative quantity of the target gene was computed for each sample using the ΔΔCt method by comparing mean Ct of the gene to the mean Ct of the housekeeping gene, glyceraldehyde 3-phospate dehydrogenase (GAPDH). All the primers were obtained from Integrated DNA Technologies (IDT). Sequences of all the primers used is listed in supplementary table S1B
Rapid Amplification of cDNA ends (RACE)
5’ and 3’ RACE was performed using the GeneRacer RLM-RACE kit (Invitrogen) following manufacturers instruction. RACE PCR products were separated on a 1% agarose gel. Individual bands were gel purified, cloned in pcr4-TOPO vector and sequenced using M13 primers.
Expression of PCAT29 after castration in Prostate Tumor Xenograft Model
Five-week-old male nude athymic BALB/c nu/nu mice (Charles River Laboratory, Wilmington, MA) were used for xenograft studies. LNCaP cells were resuspended in 100 μl of phosphate-buffered saline with 20% Matrigel (BD Biosciences) and implanted subcutaneously into the left flank regions of the mice. Mice were castrated and euthanized 5 days after castration. RNA was extracted from the xenografts and expression of PCAT29 and FKBP5 was measured. All experimental procedures involving mice were approved by the University Committee on Use and Care of Animals at the University of Michigan and conform to their relevant regulatory standards.
Chromatin Immunoprecipitation
ChIP was performed with polyclonal Androgen Receptor (AR) antibody (Millipore PG21) using HiCell ChIP kit (Diagenode) following manufacturer’s instruction. Briefly, cells were treated with 10 μM MDV3100 or 10 μM bicalutamide 16 hours before the treatment with 10 nM DHT for 12 hours. Approximately 1 million cells were cross-linked per antibody with 1% formaldehyde. Chromatin was sonicated to an average length of 500 bp and centrifuged to remove debris. Magnetic protein-G Beads were coated with 6 μg of antibody and incubated with chromatin overnight at 4 °C. Protein-chromatin-antibody complees was washed thrice and cross-linking was reversed. ChIP products were cleaned using IPure kit (Diagenode). Eluted DNA was quantified by RT-PCR using primers described in Supplementary Table S1B.
Cell Proliferation and Migration assay
LNCaP and DU145 cells stably expressing PCAT29 shRNA-1 and 2 or PCAT29 isoform 1 and 2 were seeded in 24-well plates. Cells were trypsonized and counted by using Coulter Counter (Beckman Coulter, Fullerton, CA) at the indicated time points in triplicate. For migration assays, approximately 1 × 105 cells were seeded in the upper chamber of a boyden chamber. 500 μl of complete medium (10% FBS) was added to the lower chamber as a chemoattractant. 48 hours after seeding, cells on the upper surface were removed using a cotton swab. Inserts were fixed with 3.7% formaldehyde and migrated cells on the lower surface of the membrane were stained with crystal violet. The inserts were treated with 10% acetic acid, and absorbance was measured at 560 nm.
Gene Expression Microarray
Expression profiling of VCaP and LNCaP cells after PCAT29 knockdown was performed using the Agilent Whole Human Genome Oligo Microarray as described (7). GEO accession number: GSE58397.
Chicken Chorioallantoic Membrane (CAM) Assay
22RV1 cells were transduced with empty vector (pcDH) or PCAT29-isoform-1. 106 cells were inoculated on the CAM assay was done as described previously (13). For tumor growth and metastasis, the eggs were incubated for 18 days in total, after which the extra-embryonic tumor were exercised and weighed, and the embryonic livers were harvested and analyzed for the presence of tumor cells by quantitative human Alu-specific PCR. Quantification of human cells in the extracted DNA was performed as described (14). Fluorogenic TaqMan qPCR probes were applied as above and DNA copy numbers were quantified.
Kaplan-Meier analysis of PCAT29
For outcomes analysis, PCAT29 expression was determined on a cohort of 51 radical prostatectomy specimens from prostate cancer patients at the University of Michigan with long-term biochemical recurrence outcomes. Biochemical recurrence was defined by an increase of PSA of 0.2 ng/mL over the PSA nadir following prostatectomy. PCAT29 expression was determined by a SYBR-Green qPCR assay using the average of GAPDH + HMBS for data normalization using the delta-delta Ct method. Expression data was transformed using a z-score and patients were defined as high (top 33% of patients) or low (bottom 66% of patients) for PCAT29 expression. Kaplan-Meier curves for biochemical recurrence-free survival were generated for PCAT29-high and PCAT29-low patients using the GraphPad Prism program. Statistical significance was determined with a log rank test.
Results
PCAT29 is a novel long nuclear non-coding RNA
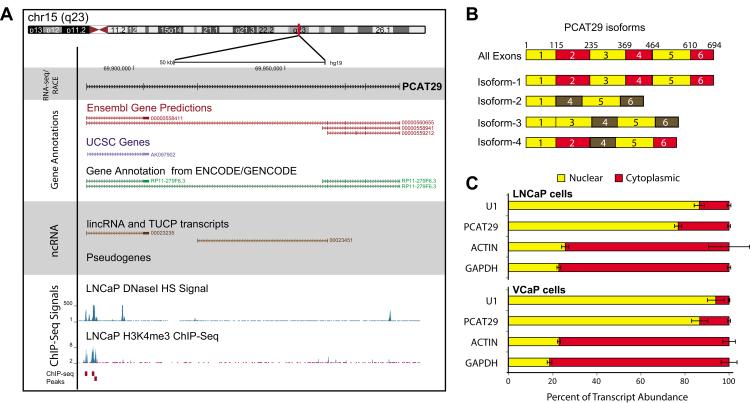
Using RNA-Seq data from prostate cancer tissues, we previously identified 121 lncRNAs, named prostate cancer-associated transripts (PCATs), which demonstrate differential expression or outlier profiles in prostate cancer compared to normal tissue (3). Here we characterize and functionally investigate one of the top outlier lncRNAs, PCAT29 (Ensembl ID ENSG00000259641). Using the predicted transcript structures, we designed exon spanning primers and performed rapid amplification of cDNA ends (RACE) to determine the full exon structure. As shown in a genome browser view, PCAT29 is a 694bp poly-adenylated transcript present on chr15(q23), and the PCAT29 gene spans over a 10kb stretch (Figure 1A; Supplementary Figure 1A). PCAT29 is comprised of 6 exons that are alternatively spliced to produce multiple isoforms (Figure 1B). To further characterize PCAT29, we interrogated recently published ENCODE data for H3K4 tri-methylation (H3K4me3) and DNase I hypersensitive sites (DNaseH), marks that predicts for open chromatin state and are commonly found near or at the transcription start sites (TSSs) sites, generated in the prostate cancer cell line LNCaP (4). We found several DNaseH and H3K4 tri-methylation peaks at the TSS of PCAT29, suggesting that PCAT29 is an actively transcribed gene (Figure 1A).
Figure 1. Characterization of PCAT29.
A. Genome Browser representation of PCAT29. Current gene annotations from Ensembl, ENCODE, UCSC genes and lncRNA datbases are shown. ChIP-Seq data for H3K4me3 and DNaseI HS signal in LNCaP cells obtained from ENCODE. B. Schematic representation of PCAT29 isoforms as determined by RACE analysis. C. Nuclear and cytoplasmic distribution of various non-coding and protein coding transcripts in LNCaP and VCaP cells. Error bar represents ± S.E.M. (standard error of mean).
To confirm that PCAT29 is indeed a non-coding RNA, we assessed the protein-coding potential of PCAT29 using the coding potential calculator (CPC) algorithm, which discriminates coding genes (positive score) from noncoding transcripts (negative score)(10). PCAT29 had a CPC score of −0.8921, while protein coding genes such as TP53 and beta-actin scored +8.25 and +3.70 respectively (Supplementary Figure 1B). Consistent with this finding, we found that in both LNCaP and VCaP cells, expression of PCAT29 was limited to nucleus, while other protein-coding mRNAs, such as GAPDH and beta-actin, were expressed in cytoplasm (Figure 1C). We then verified the expression of PCAT29 in various prostate cancer cell lines (LNCaP, VCaP, 22RV1, DU145, PC3) and immortalized or primary prostate epithelial cells (RPWE and PrEC). PCAT29 expression was highest in AR-dependent cell lines such as LNCaP, VCaP and 22RV1 (Supplementary Figure 1C). Next, we assessed the expression of PCAT29 in various tissues using transcriptome sequencing data. PCAT29 expression, although not limited to prostate, was enriched in prostate samples compared to other tissues (Supplementary Figure 1D).
Androgen receptor binds to the PCAT29 promoter and regulates PCAT29 expression
We next examined the effect of AR signaling on PCAT29 in LNCaP cells stimulated with 10nM dihydrotestosterone (DHT). As shown in Figure 2A, PCAT29 expression was suppressed upon stimulation with DHT in a time-dependent fashion both in LNCaP and VCaP cells. In contrast, expression of canonical AR target genes, such as FKBP5 and KLK3, were increased upon stimulation (Supplementary Figure 2A). To examine whether the suppression of PCAT29 was AR-specific, LNCaP cells were pre-treated with the AR antagonists MDV3100 or bicalutamide prior to treatment with DHT. As expected, DHT stimulation suppressed the expression of PCAT29 and pretreatment with MDV3100 or bicalutamide rescued this suppression. Similarly, expression of PCAT29 in LNCaP cells grown in charcoal-stripped media as well as in an AR-independent variant of LNCaP cells (C42) was higher compared to cells grown in serum-containing media and LNCaP cells respectively (Supplementary figure 2B, C). We next investigated whether AR suppresses the expression of PCAT29 in vivo. LNCaP xenografts were established in mice followed by physical castration to ablate AR signaling. As expected, 5 days of castration led to significant increase in the expression of PCAT29 in tumors (Figure 2C). In contrast, expression of FKBP5 was reduced in tumors from castrated mice. Taken together, our results suggest that stimulation of AR leads to suppression of PCAT29 expression.
Figure 2. Androgen receptor binds to the promoter of PCAT29 and suppresses its expression.
A. Expression of PCAT29 in LNCaP and VCaP cells treated with 10nM DHT for indicated time points. B. Expression of PCAT29 in LNCaP cells treated with 10nM DHT in the presence or absence of 10μM MDV3100 or bicalutamide C. Expression of PCAT29 and FKBP5 in LNCaP xenografts obtained from control mice and mice that were physically castrated for 5 days. D. Genome browser representation of AR binding on the promoter of PCAT29 before and after stimulation with 1nM R1881. Consensus androgen responsive elements (ARE) and ARE present in the PCAT29 promoter are shown. Inset: ChIP-PCR to confirm AR occupancy on TMPRSS2 and PCAT29 gene promoter. The y axis represents AR ChIP enrichment in VCaP cells treated with 10nM DHT normalized to ethanol (Ethl)-treated cells. Bars represent standard error of the mean (SEM).
To further study the association of PCAT29 expression with androgen signaling, we interrogated published ChIP-Seq data (ll) and found AR binding sites in the promoter region of PCAT29 (Figure 2D). These peaks were similar to those observed in other known AR-regulated genes (Supplementary Figure 2D). Upon closer inspection, we found a canonical AR binding site near the PCAT29 transcriptional start site in a putative enhancer region bounder by AR (Figure 2D). We confirmed our ChIP-Seq data by performing ChIP for AR followed by PCR for the PCAT29 promoter. As shown in Figure 2D, stimulation of VCaP cells with DHT led to an increase in association of AR with the PCAT29 promoter. This association was reduced in cells pretreated with bicalutamide and MDV3100. Taken together, our data suggests that AR can directly bind to the promoter of PCAT29 and leads to the suppression of gene expression.
PCAT29 regulates oncogenic phenotypes in vitro and in vivo
The androgen receptor drives oncogenesis in treatment-naïve prostate cancer as well as disease progression in castration-resistant prostate cancers. Since AR binds to the PCAT29 promoter and regulates gene expression, we investigated the functional role of PCAT29. Two independent shRNAs were designed to knock down the expression of PCAT29 in cells (Supplementary Figure 3A, B). VCaP and LNCaP cells were transfected with PCAT29 shRNAs following analysis using gene expression microarray. We found GO concepts enriched for cell cycle, proliferation, and migration related genes, suggesting a role of PCAT29 in proliferation and migration (Supplementary Figure 3D-G). Next, we defined a signature of genes positively and negatively correlated with PCAT29 expression from prostate cancer samples as described before (7). We checked the overlap of these genes with the top 1500 differentially expressed genes in PCAT29 knockdown samples of VCAP and LNCAP cells. As expected, the positively correlated genes show a significant overlap with genes downregulated with knockdown of PCAT29 and the negatively correlated genes show a significant overlap with genes upregulated by knockdown of PCAT29 in both VCAP and LNCAP (p<0.001 for all pairwise comparisons of overlapping genes, Supplementary figure 4A-D). For overlapping genes, we did see enrichment in pathways such as cell cycle, apoptosis, cell growth (Supplementary Figure 4A-D) Taken together this analysis suggested a role of PCAT29 in cell proliferation and migration.
To experimentally validate this observation, cell proliferation was assessed in LNCaP cells transfected with control versus PCAT29 shRNAs. To our surprise, knockdown of PCAT29 in LNCaP cells led to an increase in cell proliferation and migration (Figure 3A). To further validate this observation, we stably overexpressed the two most prevalent isoforms of PCAT29 in DU145 prostate cancer cells using a lentiviral vector (Supplementary Figure 3C). Consistent with the previous knockdown studies, overexpression of these two isoforms of PCAT29 in DU145 led to suppression of cell proliferation and migration (Figure 3B). We next assessed whether similar effects of PCAT29 could be achieved in vivo. 22RV1 prostate cancer cells overexpressing PCAT29 (isoform-1) were implanted on the chick chorioallantoic membrane (CAM) of a chicken egg. Compared to control cells, overexpression of PCAT29 significantly decreased the growth of tumor on the CAM as well as decreased liver metastases (Figure 3C).
Figure 3. PCAT29 suppresses oncogenic phenotypes.
A-B. Proliferation and migration of LNCaP cells stably expressing PCAT29 shRNA (B) and DU145 cells expressing PCAT29 expression constructs (C). Representative micrographs of crystal violet stained migrated cells are shown as insets. C. Quantification of tumor weight and metastasi to liver for 22Rv1 cells expressing PCAT29-isoform-1 or empty vector (pcDH) in the chick chorioallantoic membrane (CAM) assay. Data are represented as mean ± S.E.M. An asterisk (*) indicated p < 0.05 by Student’s T-test. D. Kaplan-Meier analyses of prostate cancer outcomes. PCAT29 expression was measured by qPCR and 51 patients were stratified according to their PCAT29 expression. Patient outcomes were analyzed for freedom from biochemical recurrence
Lastly, we measured the expression of PCAT29 in an independent cohort of 51 radical prostatectomy specimens from prostate cancer patients with localized disease and clinical follow-up. As shown in Kaplan-Meier analysis (Figure 3D), patients with lower PCAT29 expression had significantly higher rates of biochemical recurrence, consistent with our in vitro and in vivo findings.
Discussion
In this study, we characterize the novel lncRNA PCAT29. Our findings demonstrate that PCAT29 is directly regulated by the androgen receptor, which binds to the promoter of PCAT29 and suppresses its transcription. In vitro studies show that PCAT29 negatively regulates prostate cancer proliferation and migration, and CAM assays demonstrate that PCAT29 inhibits tumor growth and metastases. Low expression of PCAT29 is associated with higher rates of biochemical recurrence, suggesting that PCAT29 represses oncogenic phenotypes via a tumor suppressive role.
While previous studies have nominated and characterized lncRNAs that are dysregulated in cancer (3, 12), the majority of characterized lncRNAs, to date, have been associated with oncogenic roles instead of tumor suppressor functions. In fact, there have been only a handful of lncRNAs identified to date which function in repression of cancer phenotypes, and, to our knowledge, none of these are targets downregulated by known oncogenes (12). A recent study identifies a protein-coding gene, CCN3/NOV, as a tumor suppressor that is repressed by AR (15). Thus, our study represents the first identification of an AR-repressed lncRNA functioning as a tumor suppressor. While further studies will be required to identify the mechanism of PCAT29 and other tumor suppressor lncRNAs, it is clear that these lncRNAs represent an intriguing area for exploration in cancer biology.
In the context of prostate cancer, androgen-regulated lncRNAs are of fundamental importance, given that all stages of prostate cancer are exquisitely dependent on androgen receptor (AR) signaling for growth and survival. Since the majority of clinically relevant prostate cancer therapies target the androgen receptor, our studies would suggest that inhibition of androgen signaling will result in reactivation of PCAT29, providing another mechanism underlying the effectiveness of androgen deprivation therapy.
Clinically, there is a clear need for identification of prognostic biomarkers in prostate cancer to help guide decisions on treatment intensification. The association of high PCAT29 expression with good clinical prognosis and preclinical suppression of cell proliferation and tumor metastases suggests that decreased expression or loss of PCAT29 may identify subsets of patients who may require further intensification of therapy. As our clinical cohort was comprised of hormone-sensitive disease from prostatectomy patients, further stdies need to be performed to determine if PCAT29 can also serve as a prognostic biomarker in the context of more advanced, castration-resistant disease. Regardless, this study highlights the importance of lncRNAs in prostate cancer biology and prognosis, and suggests the need for further research in this relatively unexplored area.
Supplementary Material
Implications.
This study identifies PCAT29 as the first AR-repressed lncRNA that functions as a tumor suppressor and that its loss may identify a subset of patients at higher risk for disease recurrence.
Acknowledgements
We thank Xia Jiang and Vishal Kothari for helpful discussions and technical assistance, as well as Karen Giles for her review of the manuscript and submission of documents. We also thank the University of Michigan Vira1 vector core for generating the lentiviral constructs. This work was supported in part by the NIH Prostate Specialized Program of Research Excellence grant P50CA69568, the Early Detection Research Network grant UO1 CA111275, the US National Institutes of Health R01CA132874-0lAl, and the Department of Defense grant PCl00l7l (A.M.C.). A.M.C is supported by the Doris Duke Charitable Foundation Clinical Scientist Award, the Prostate Cancer Foundation, and the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor and a Taubman Scholar of the University of Michigan. F.Y.F. was supported by the Prostate Cancer Foundation and the Department of Defense grant PC094231. R.M. and J.R.P. were supported by a Prostate Cancer Foundation Young Investigator Award. R.M. was supported by the Department of Defense Post-doctoral Fellowship W81XWH-13-1-0284. A.S. was supported by the NIH Predoctoral Fellowship 1F30CA180376-01. M.K.I. was supported by the Department of Defense Predoctoral Fellowship BC100238.
Footnotes
Disclosures and Competing Financial Interests
The University of Michigan has filed a patent on lncRNAs in prostate cancer, including PCAT29, in which A.M.C., J.R.P. and M.K.I. are named as co-inventors. Wafergen, Inc. has a non-exclusive license for creating commercial research assays for lncRNAs in prostate cancer. A.M.C. serves on the Scientific Advisory Board of Wafergen; Wafergen had no role in the design or experimentation of this study, nor have they participated in the writing of the manuscript.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol. 2011;8:123–4. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- 10.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–9. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. American journal of translational research. 2012;4:127–50. [PMC free article] [PubMed] [Google Scholar]
- 13.Asangani IA, Ateeq B, Cao Q, Dodson L, Pandhi M, Kunju LP, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell. 2013;49:80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Horst EH, Leupold JH, Schubbert R, Ullrich A, Allgayer H. TaqMan-based quantification of invasive cells in the chick embryo metastasis assay. Biotechniques. 2004;37:940–2. doi: 10.2144/04376ST02. 4, 6. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Runkle C, Jin HJ, Yu J, Li J, Yang X, Kutzel T, Lee C, Yu J. CCN3/NOV gene expression in human prostate cancer is directly suppressed by the androgen receptor. Oncogene. 2014;33(4):504–13. doi: 10.1038/onc.2012.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.