Abstract
The myocyte enhancer factor 2A-D (MEF2) proteins are members of the MCM1-agamous-deficiens-serum (MADS) response factor family of transcription factors. Various MEF2 isoform proteins are enriched in neurons and exhibit distinct patterns of expression in different regions of the brain. In neurons, MEF2 functions as a converging factor to regulate many neuronal functions including survival. MEF2 activities are tightly controlled in neurons in response to stress. Whether stress signal may differentially regulate MEF2s remains largely unknown. In this work, we showed that MEF2A but not MEF2C or MEF2D was modified by ubiquitination in dopaminergic neuronal cell line SN4741 cells. MEF2A was ubiquitinated at its N’-terminus, and the ubiquitination of MEF2A was first detectable in the nuclear compartment and later in the cytoplasm. Ubiquitination of MEF2A correlated with reduced DNA-binding activity and transcriptional activity. More importantly, disturbing the degradation of ubiquitinated MEF2A through proteasome pathway by neurotoxins known to induce Parkinson’s disease (PD) features in model animals caused accumulation of ubiquitinated MEF2A, reduced MEF2 activity, and impaired cellular viability. Our work thus provides the first evidence to demonstrate an isoforms specific regulation of MEF2s by ubiquitination-proteasome pathway in dopaminergic neuronal cell by neurotoxins, suggesting that stress signal and cellular context dependent dysregulation of MEF2s may underlie the loss of neuronal viability.
Keywords: myocyte enhancer factor 2, ubiquitination, proteasome, dopaminergic neurons, survival, death
MEF2 proteins are members of the MADS family of transcription factors (Naya & Olson 1999). Vertebrate MEF2 proteins are encoded by four different genes (MEF2A, B, C, and D), each of which gives rise to alternatively spliced transcripts. The first 56 amino acids at the N’-terminus of MEF2, termed MADS box, are the minimal region needed for DNA binding. Adjacent to the MADS box is a 29 amino acids MEF2 domain that mediates high affinity DNA binding and homo- and hetero-dimerization with other MEF2 proteins. The vertebrate MEF2 proteins share about 50% amino acid identity overall and about 95% similarity in the highly conserved MADS box and MEF2 domain (Heidenreich & Linseman 2004). The C’-terminal portion of MEF2 proteins contains the transcriptional activation domain as well as other regulatory minidomains including a nuclear localization sequence (Potthoff & Olson 2007). The C’-terminus is subject to complex patterns of alternative splicing and has relatively low amino acid homology between the various MEF2 isoforms.
Although MEF2s were initially identified in muscle cells due to their ability to bind an A/T rich consensus sequence [c/tTA(A/T)4TAg/a] found in the regulatory region of many muscle specific genes (Black & Olson 1998), all four MEF2 genes are expressed in neurons and exhibit distinct patterns of expression in different regions of the brain with highest levels in the cerebellum, cerebral cortex and hippocampus (Heidenreich & Linseman 2004). In neurons, MEF2 transcriptional activity is tightly regulated by extracellular stimuli. MEF2 can be activated by neurotrophin stimulation as well as calcium influx, and functions as a converging point to regulate neuronal proliferation, differentiation, survival, and synapse development (Shalizi & Bonni 2005). Deregulation of MEF2 activity had been associated with many neuronal stress conditions and degenerative diseases, including PD and Alzheimer's disease (Burton et al. 2002, Smith et al. 2006, Yang et al. 2009, She et al. 2011).
The structural organization of MEF2 proteins allows them to receive and respond to multiple inputs from various intracellular signaling pathways. Posttranslational modification is largely responsible for the function of MEF2 proteins (McKinsey et al. 2002). There are multiple phosphorylation motifs in MEF2 sequence (Potthoff & Olson 2007). Following our first demonstration that p38 MAPK directly phosphorylates and activates MEF2 to promote neuronal survival (Mao et al. 1999), calcineurin, protein kinase A (PKA), and extracellular signal regulated kinase 5 (ERK5) have been identified as positive MEF2 regulators (Liu et al. 2003, Mao & Wiedmann 1999, Wang et al. 2005). On the other hand, apoptotic signals inhibit MEF2 function in neurons. It was shown that nuclear cyclin dependent kinase 5 (Cdk5) and glycogen synthase kinase 3βb(GSK3β) in response to diverse toxic signals including oxidative stress, phosphorylated MEF2, inhibited its activity, and led to caspase- or calpain-dependent degradation and neuronal death (Gong et al. 2003, Tang et al. 2005, Wang et al. 2009, Wei et al. 2012). Recently, it was found that sumoylation of MEF2 inhibited its activity and modulated neuronal postsynaptic differentiation (Flavell et al. 2006, Kang et al. 2006, Riquelme et al. 2006, Shalizi et al. 2006). One key mechanism modulating MEF2 function is the control of its protein stability. In neurons, depending on the stress signals, MEF2s can turned over by caspase/calpain-mediated degradation (Tang et al. 2005) and by chaperone-mediated autophagy via lysosomes (Yang et al. 2009). However, what is not clear is whether different MEF2 isoforms may be differentially regulated in response to different stress signals.
Ubiquitination, another major type of posttranslational modification (Hershko 1983), regulates protein localization, degradation and function (Pickart 2004). Deregulation of ubiquitination has been associated with many human diseases including neurodegenerative disorders (Giasson & Lee 2003). We showed in this study that ubiquitination differentially regulates specific MEF2 isoforms to modulate MEF2 activity in response to neurotoxic stress.
Materials and methods
Antibodies and plasmids
The following antibodies used in this study were obtained commercially: anti-MEF2A rabbit polyclonal antibody, anti-ubiquitin rabbit polyclonal antibody (Santa Cruz); anti-MEF2D mouse monoclonal antibody, anti-c-Raf mouse monoclonal antibody (BD Bioscience); anti-MEF2C rabbit polyclonal antibody, anti-PARP rabbit polyclonal antibody (Cell Signaling); anti-Flag mouse monoclonal antibody, anti-β-actin rabbit polyclonal antibody (Sigma); anti-HA mouse monoclonal antibody (Roche). Flag-MEF2A(1-131aa) and Flag-MEF2A(132-507aa) were constructed by clone PCR fragments from Flag-MEF2A(human) into Nhe I and Xho I sites of pcDNA3.1(+). HA-ubiquitin (HA-Ub) was kindly provided by Dr. Junmin Peng (St. Jude Children’s Research Hospitol). MEF2 reporter plasmids (wild type and mutant type) were described previously (Wang et al. 2009).
Cell culture
SN4741 cells is a mouse midbrain dopaminergic cell line established and kindly provided by Dr. Jin H. Son at the Department of Neurology and Neuroscience, Cornell University. SN4741 cells were cultured at 33°C with 5% CO2 in RF medium (DMEM supplemented with 10% FBS, 1% D-glucose, 1% penicillin-streptomycin, and 140 mM L-glutamine). When cells reached 70% confluence (usually 3 days), split it into 3 plates. Experiments were usually done in 12-well or 96-well plate when cells reached 50–60% confluence.
Cytoplasmic and nuclear fractionation
Cytoplasmic and nuclear fractionation was performed using EZ nuclei isolation kit (Sigma, NUC-101) according to the manufacturer's procedures that involved three cycles of thorough cell lysis and washing.
Immunoprecipitation
Cells were sonicated in cell lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin). Primary antibody was added into 200 μg cell lysate and incubated overnight at 4°C with gentle rocking. Then, protein A/G agarose beads (20 μl of 50% bead slurry) were added and incubated for 1–3 hours at 4°C with gentle rocking. Then samples were microcentrifuged for 30 seconds at 4°C to pellet. The pellet was washed five times with 500 μl of 1× cell lysis buffer. Finally, the pellet was resuspended with 20 μl SDS sample buffer.
Immunoblotting
Cell lysates were suspended in the SDS sample buffer. For immunoblotting, the proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore) by electroblotting. The blots were blocked with 5% skim milk and 0.2% Tween 20 in PBS. After incubating them with a primary antibody, they were washed 3 times with PBS containing 0.2% Tween 20 and incubated with a peroxidase-conjugated secondary antibody. Immunoreactive bands were detected by enhanced chemiluminescence regents.
Luciferase reporter gene assay
MEF2 luciferase reporter assay was done as described previously (Mao & Wiedmann 1999, Wang et al. 2005). Briefly, cells were transiently transfected with various constructs with MEF2 luciferase reporter plasmid (WT, reporter with wild type MEF2 DNA binding sites; mt, reporter with the MEF2 DNA binding sites mutated) using Lipofectimane 2000 (Invitrogen) following procedures provided by the manufacturer. A β-galactosidase expression plasmid was used to determine the efficiency in each transfection. The total amount of DNA for each transfection was kept constant by using control vectors. Cell lysates were analyzed for luciferase activity (Roche Applied Science) following the manufacturer's instruction.
Electrophoretic mobility shift assay
DNA binding activity of MEF2 was studied by electrophoretic mobility shift assay (EMSA) with radiolabeled double-stranded oligonucleotides (She et al. 2011). Oligonucleotides corresponding to MEF2 binding sites (MEF2 probe: wile type, 5'-AGCTTCGCTCTAAAAATAACCCTGATC-3'; for the mutant probe, the three nucleotides in italics were mutated to GGC) were annealed and labeled with 32P using T4 polynucleotide kinase. Cell lysates (2 μg) were incubated at 4 °C for 10 minutes in binding buffer. Radiolabeled wild or mutant probe (1 pmol) was added to the reactions (total volume 20 μl) and incubated for an additional 20 minutes. Reaction mixtures were analyzed by electrophoresis at 4 °C on pre-run 5% native polyacrylamide gels. Images were captured after exposure the gels to autoradiography film (Denville Scientific).
Proteasome activity assay
The proteasome activity was determined using a kit from Millipore following the procedures provided. The assay is based on detection of the fluorophore 7-Amino-4-methylcoumarin (AMC) after cleavage from the labeled substrate LLVY-AMC. The free AMC fluorescence can be quantified using a 380/460 nm filter set in a fluorometer. Briefly, assay mixture were prepare in a 96-well fluorometer plate or standard microcentrifuge tubes. The samples were incubated for 1–2 hours at 37°C. Fluorescence was read using a 380/460 nm filter set in a fluorometer (Bio-Tek). The proteasome activity was calculated from the curve of 20S proteasome positive control contained in the kit.
Survival assays
WST-1 assay was done using a kit (cat.no.11644807001) from Roche and following procedures provided by the manufacturer. WST-1 (4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) is a water-soluble tetrazolium salt whose cleavage by cellular enzymes correlates with cell viability. Cells were seeded in 96-well plates and treated as indicated. Than 10 μl/well of premixed WST-1 Cell Proliferation Reagent was added, and the cells were incubated for an additional 4 hours under the same conditions. Absorbance at 450 nm was measured on a multi-well plate reader (Bio-Tek). The propidium iodide (PI) staining assay was carried out as described previously (She et al. 2011, Wang et al. 2009). Images were captured under an Olympus IX51 fluorescence microscope in a blind manner.
Statistical analyses
Data were presented as mean±SEM. The results were analyzed using one-way analysis of variance with Bonferroni multiple comparison to test significance between experimental groups. A p<0.05 was considered statistical significant.
Results
Ubiquitination of MEF2A
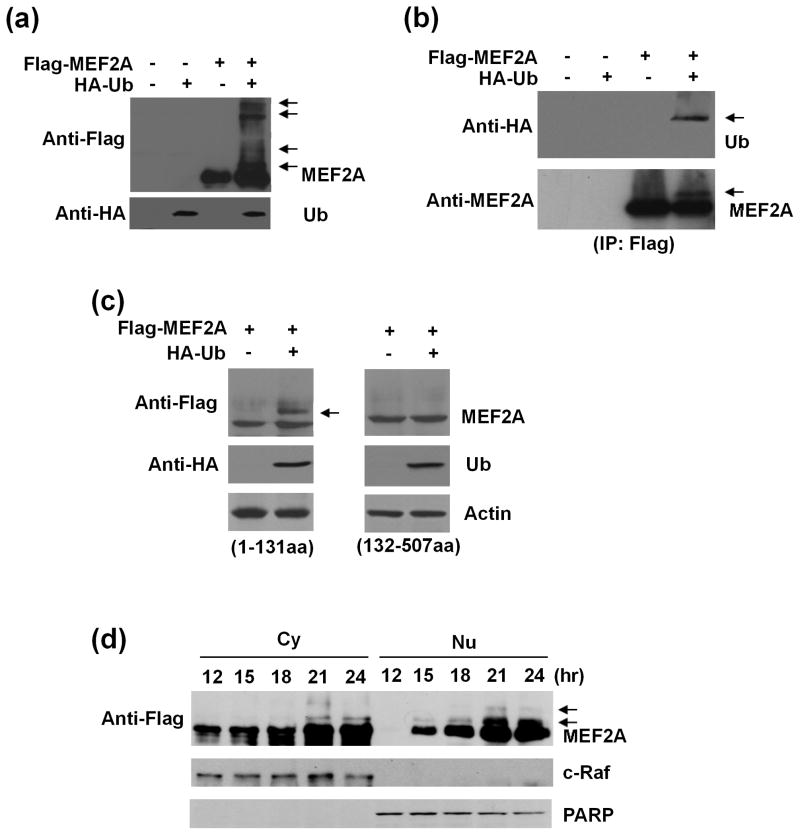
MEF2A activity is extensively controlled by posttranslational modifications (McKinsey et al. 2002). We tested whether MEF2A was regulated by ubiquitination in SN4741 cells, a mouse midbrain dopaminergic neuronal cell line. SN4741 cells were transfected with Flag-MEF2A alone or with HA-Ub simultaneously. Whole cell lysates were prepared 24 hours later and analyzed by western blot. Flag-MEF2A was well expressed in SN4741 cells (Fig. 1a). Co-expression of HA-Ub led to the appearance of several higher molecule weight bands which could be recognized by anti-Flag antibody (Fig. 1a), suggesting that MEF2A might be ubiquitinated. To confirm this, we transfected SN4741 cells as described above, immunoprecipitated Flag-MEF2A with anti-Flag antibody, and blotted the precipitates with anti-HA antibody and anti-MEF2A antibody sequentially. Our analysis showed that the minor band precipitated by anti-Flag antibody was recognized by anti-HA antibody, indicating clearly that Flag-MEF2A was indeed ubiquitinated (Fig. 1b). To identify the region of MEF2A which was responsible for ubiquitination, we over-expressed Flag-MEF2A-N’-terminus (1-131aa) and Flag-MEF2A-C’-terminus (132-507aa) fragments were transfected alone or with HA-Ub into SN4741 cells and analyzed for higher molecular band. The analysis showed that co-expression of HA-Ub with the N’-terminal but not the C’-terminal MEF2A fragment causes the appearance of the higher molecular MEF2A band (Fig. 1c). This higher molecular MEF2A band could be recognized by anti-HA antibody. Together, these findings indicate that over-expression of HA-Ub leads to the ubiquitination of the N’-terminal MEF2A. To determine the subcellular localization and kinetics of MEF2A ubiquitination, SN4741 cells were transfected with Flag-MEF2A and HA-Ub for various lengths of time and used to prepare cytoplasic (Cy) and nucleic (Nu) fractions. The analysis showed that the ubiquitinated MEF2A bands were detectable as early as 15 hours after transfection in the nucleus whereas its presence in the cytoplasm was evident only after 21 hours (Fig. 1d).
Fig. 1.
Ubiquitination of MEF2A in dopaminergic neuronal SN4741 cells. (a) SN4741 cells were transfected with Flag-MEF2A alone or together with HA-ubiquitin (HA-Ub) and analyzed by immunoblotting 24 hours later. Top panel: arrows indicated the higher molecular weight MEF2A. Bottom panel: blotting of HA-Ub. (b) After transfection as in (a), SN4741 cell lysates were immunoprecipitated using an anti-Flag antibody. The transferred membrane was immunoblotted with anti-HA and anti-MEF2A antibodies sequentially. Arrows indicated the ubiquitinated MEF2A. (c) SN4741 cells were transfected with constructs for either Flag-MEF2A-N’-terminus (1-131aa) or Flag-MEF2A-C’-terminus (132-507aa) alone or with HA-Ub simultaneously. After 24 hours, cell lysates were analyzed by sequential immunoblotting with anti-Flag and anti-HA antibodies. Arrows indicated the ubiquitinated N’-terminal MEF2A. (d) SN4741 cells were transfected with Flag-MEF2A and HA-Ub simultaneously. Cytoplasmic (Cy) and nuclear (Nu) lysates prepared at indicated time points after transfection were analyzed using anti-Flag antibody. c-Raf was used as cytoplasmic marker and PARP as nuclear marker. Arrows indicated the ubiquitinated MEF2A.
Degradation of MEF2A but not MEF2C or MEF2D through ubiquitin-proteasome pathway
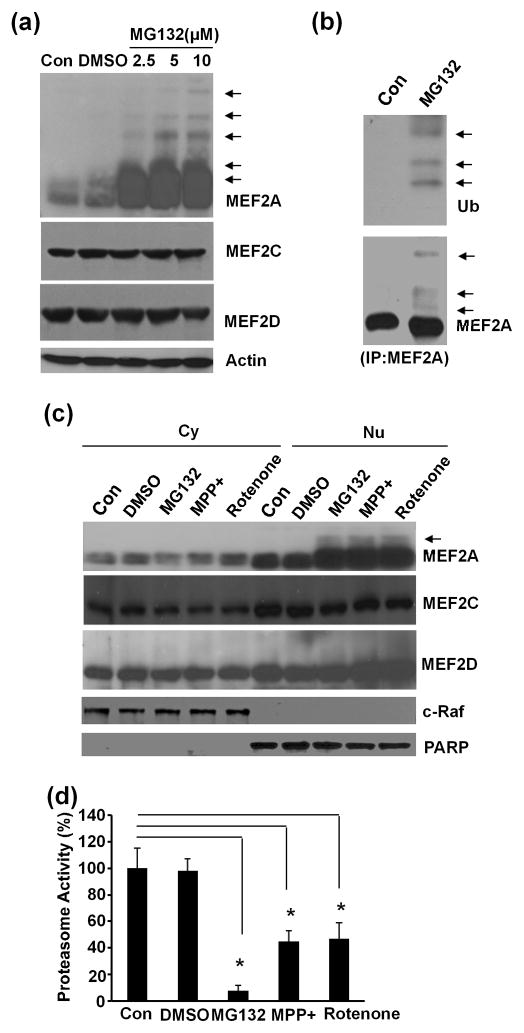
Many ubiquitinated proteins are degraded through proteasome pathway (Clague & Urbe 2010). To test if ubiquitinated MEF2A was degraded by proteasome, SN4741 cells were treated with proteasome inhibitor MG132 at different doses (2.5 μM, 5 μM, 10 μM) for 6 hours and analyzed for the levels and patterns of endogenous MEF2A, MEF2C, and MEF2D. Inhibition of proteasome activity led to a dose-dependent accumulation of MEF2A and appearance of higher molecular weight MEF2A bands (Fig. 2a). However, MG132 treatment did not significantly alter the levels and migration patterns of MEF2C and MEF2D. To further characterize these higher molecular weight MEF2As, we immunoprecipitated MEF2A with an anti-MEF2A antibody from SN4741 cells treated with MG132 and sequentially immunoblotted with an anti-ubiquitin antibody and then an anti-MEF2A antibody. This analysis showed that the higher molecular weight MEF2As were indeed ubiquitinated (Fig. 2b). To determine the subcellular localization of the ubiqitinated endogenous MEF2A, SN4741 cells were treated with MG132 (2.5 μM) for 6 hours and subjected to cytoplasmic and nuclear preparation. Immunoblotting these lysates showed that the ubiquitinated endogenous MEF2A was mainly present in the nucleus (Fig. 2c). Furthermore, 1-methyl-4-phenylpyridinium (MPP+) and rotenone, neurotoxins known to produce PD models in cells and animals, caused the accumulation of ubiquitinated MEF2A in the nucleus similar to MG132 treatment (Fig. 2c). Consistent with these, proteasome activity assay showed that MPP+ and rotenone inhibited proteasome activity (Fig. 2d). Unlike MEF2A, the levels of MEF2C and MEF2D in both cytoplasm and nucleus were not significantly affected by MG132 or MPP+ or rotenone treatment (Fig. 2c).
Fig. 2.
Selective degradation of endogenous MEF2A through ubiquitin-proteasome pathway. (a) SN4741 cells were treated with proteasome inhibitor MG132 at different doses (2.5 μM, 5 μM, 10 μM) for 6 hours. Whole cell lysates were analyzed using isoform specific MEF2 antibodies. Arrows indicated the higher molecule weight MEF2A bands. (b) Whole cell lysates prepared from SN4741 cells treated with MG132 (10 μM) or vehicle control for 6 hours were immunoprecipitated using anti-MEF2A antibody. The immunoprecipitates were immunoblotted with anti-ubiquitin antibody (top panel) and anti-MEF2A antibody (bottom panel). Arrows indicated the ubiquitinated MEF2A. (c) SN4741 cells were treated with MG132 (2.5 μM), MPP+ (25 μM), or rotenone (100 nM) for 6 hours. Cytoplasmic (Cy) and nuclear (Nu) lysates were prepared and subjected to immunoblotting. c-Raf was used as cytoplasmic marker and PARP as nuclear marker. Arrow indicated the ubiquitinated MEF2A. (d) The activities of proteasome pathway in SN4741 cells measured following treatment with MG132 (2.5 μM), MPP+ (25 μM) or rotenone (100 nM) for 6 hours. The experiments were repeated 4 times. *, p<0.01.
Inhibition of MEF2A activity by ubiquitination
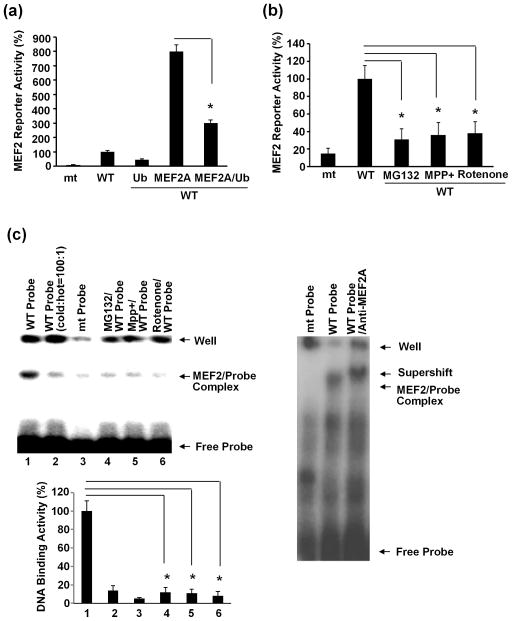
Posttranslational modifications affect transcriptional activity of MEF2. Phosphorylation of MEF2 could either enhance or inhibit its activity depending on the specific phosporylaytion sites whereas sumoylation of MEF2 had been shown to inhibit MEF2 function (Gong et al. 2003, Riquelme et al. 2006, Wang et al. 2005). Since our above data showed that MEF2A but not MEF2C and D was readily ubiquitinated, we tested the effect of ubiquitination on MEF2A function. SN4741 cells were transfected with MEF2 reporter and Flag-MEF2A alone, HA-Ub alone, or both. MEF2 reporter assay indicated that increasing the level of Ub reduces the transcriptional activity of the over-expressed MEF2A (Fig. 3a). Consistent with these findings, treating SN4741 cells with MG132 (2.5 μM) reduces the activity of endogenous MEF2A (Fig. 3b). Similarly, treatment SN4741 cells with MPP+ (25 μM) or rotenone (100 nM) for 6 hours, a condition which led to ubiquitinate MEF2A but not MEF2C or MEF2D (Fig. 2), also reduced MEF2 dependent reporter activity (Fig. 3b). Consistent with the reporter studies, EMSA analysis showed that exposure of SN4741 cells to MG132, MPP+, or rotenone also reduces MEF2 DNA-binding activity (Fig. 3c). Together, these results are consistent with the notion that ubiquitination inhibits MEF2A transcriptional activity.
Fig. 3.
Inhibition of MEF2A activity by ubiquitination. (a) SN4741 cells were transfected with constructs for MEF2 luciferase reporter (mt: MEF2 site mutated, wt: MEF2 site intact) alone or with Flag-MEF2A and HA-Ub as indicated. Luciferase activity was measured 24 hours later. The luciferase activity from cells transfected with wt MEF2 reporter alone was set as 100%. The experiments were repeated 5 times (n=5). (b) SN4741 cells were treated with MG132 (2.5 μM), MPP+ (25 μM) and rotenone (100 nM) for 6 hours. MEF2 reporter assay was carried out as in (a). The experiments were repeated 5 times (n=5). (c) SN4741 cells were treated as described in (b). MEF2 DNA-binding activity in whole cell lysates were tested by EMSA assay. The experiments were repeated 3 times. The quantification of DNA-binding activity was shown on the left-bottom graph. *, p<0.01.
Reduced neuronal viability following inhibition of MEF2A degradation pathway
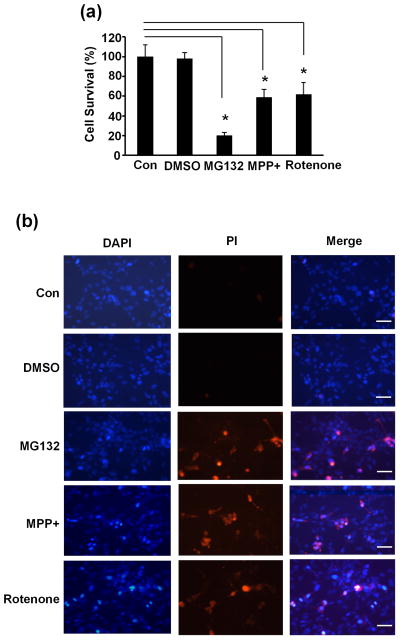
Our above results showed that MEF2A was uniquitinated and degradation of ubiquitinated MEF2A was blocked by proteasome inhibitor MG132 and by neurotoxins such as MPP+ and rotenone (Fig. 1 and Fig. 2). MEF2s have been shown to promote neuronal survival (She & Mao 2011). To test if neuronal viability was affected when the ubiquitin-proteasome pathway was inhibited, SN4741 cells were treated as described above and cell viability was analyzed with WST-1 assay and PI staining 24 hours later. These analyses showed that MG132 and neurotoxins MPP+ and rotenone all caused a significant increase in cell death (Fig. 4), which suggested that disturbing proteasome mediated degradation of ubiquitinated MEF2A may contribute to the reduced neuronal viability.
Fig. 4.
Impairment of neuronal viability following disruption of proteasome degradation pathway. (a) SN4741 cells were treated with MG132 (2.5 μM), MPP+ (25 μM) or rotenone (100 nM) treatment for 24 hours. Cell viability was measured by WST-1 assay. The experiments were repeated 4 times. (b) SN4741 cells were treated as in (a) and than stained with PI (Prodidium Iodide) and DAPI. The experiments were repeated 4 times. Bar=200 μm. *, p<0.01.
Discussion
MEF2s are shown to play an essential role in various neuronal functions including survival (Heidenreich & Linseman 2004). MEF2 activities are tightly regulated through multiple layers of mechanisms (McKinsey et al. 2002). Phosphorylation had been shown to either enhance or reduce MEF2 activity, whereas sumoylation inhibits its activity (Gong et al. 2003, Riquelme et al. 2006, Wang et al. 2005). Our current findings added ubiquitination as a new layer to the already sophisticated regulatory network. In the present study, we presented novel evidence that a member of MEF2 family, MEF2A, was specifically ubiquitinated and degraded through proteasome pathway under basal condition. Whether ubiquitination and other forms of posttranslational modifications regulate MEF2A in a coordinated manner remains to be determined. In addition to ubiquitination of MEF2A under basal condition, our data indicate that ubiquitination of MEF2A also constitutes a response to toxic stress induced by neurotoxins known to cause Parkinsonism in animals. Together with the findings that MEF2s are involved in neuronal survival (Mao et al. 1999, Gong et al. 2003) and loss of its function underlies PD pathogenesis (Yang et al. 2009, She et al. 2011), our new findings support the possibility that ubiquitination of MEF2A may also contribute the pathogenic process in PD.
A big puzzle in MEF2 remaining unresolved is whether and how different isoforms of MEF2 may be differentially regulated. Although neurons often express multiple MEF2 isoforms, it is not clear whether all MEF2s respond to and are regulated by the same stimuli. In vitro and in vivo studies have shown that MEF2A, MEF2C, and MEF2D are involved in neuronal survival (Tang et al. 2005, Mao et al. 1999, Gong et al. 2003, Akhtar et al. 2012), In addition, MEF2C appears to function in neuronal differentiation (Leifer et al. 1993). MEF2A is specifically involved in synapse development (Flavell et al. 2006, Shalizi et al. 2006). Both MEF2A and MEF2D have been shown to modulate mitochondrial function (Naya et al. 2002, She et al. 2011). There is some evidence that MEF2s may be differentially regulated. For example, although all four MEF2s could be sumoylated on the same consensus site (Gong et al. 2003, Riquelme et al. 2006, Wang et al. 2005), Cdk5 phosphorylates only MEF2A and MEF2D on their C’-termini but not MEF2C (Gong et al. 2003, Tang et al. 2005). In this work, we provided another line of evidence demonstrating differential regulation of MEF2 isoforms by ubiquitination in neuronal stress. MEF2A but not MEF2C or MEF2D is ubiquitinated in neuronal line SN4741 cells. Our data further suggest that ubiquitination of MEF2A is at the N’-terminal portion of MEF2A molecule. This is intriguing because the N’-termini of MEF2 are relatively conserved among different isforms. Therefore, it is not obvious why MEF2C and D are not readily ubiquitinated under neuronal stress in our model system. Further studies are needed to identify the molecular mechanisms underlying the specific ubiquitination of MEF2A.
Several studies have shown that the levels of MEF2 protein are a key point of regulation in neurons (Yang et al. 2009, Flavell et al. 2006). The expression levels of various MEF2s are developmentally controlled. In mature neurons, neuronal stress regulates MEF2 protein levels via several mechanisms. For example, neuronal stress causes caspases-dependent cleavage of MEF2D (Tang et al. 2005). Our more recent work revealed chaperone-mediated autophagy degrades MEF2D in dopaminergic neurons (Yang et al. 2009). Our new findings indicate that ubiquitination plays a central role in modulating the levels of MEF2A under basal and stress conditions. Together, these findings suggest that members of MEF2 family may be targeted by overlapping and distinct degradation mechanisms. Given the report that removal of depolarizing media triggers proteasome dependent degradation of MEF2A and D in cerebellar granule neurons (Butts et al. 2005), it raises an interesting possibility whether the selective usage of specific degradation pathways to remove MEF2s is both signal and cellular context dependent.
Acknowledgments
This work was supported in part by NIH grants ES015317 (Z.M.), AG023695 (Z.M.), NS048254 (Z.M.), ES016731-0002 (Z.M.) and Michael J. Fox Foundation (Z.M.).
Abbreviations used
- MEF2
myocyte enhancer factor 2
- PD
Parkinson’s disease
- MADS
MCM1-agamous-deficiens-serum
- Ub
ubiquitin
- EMSA
electrophoretic mobility shift assay
- WST-1
4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate
- Cy
Cytoplasmic
- Nu
nuclear
- MPP+
1-methyl-4-phenylpyridinium
Footnotes
Competing Interests
The authors declare that no competing interests exist.
References
- Akhtar MW, Kim MS, Adachi M, et al. In Vivo Analysis of MEF2 Transcription Factors in Synapse Regulation and Neuronal Survival. PLoS One. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Bryja V, Schulte G, Rawal N, Grahn A, Arenas E. Wnt-5a induces Dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J Cell Sci. 2007;120:586–595. doi: 10.1242/jcs.03368. [DOI] [PubMed] [Google Scholar]
- Burton TR, Dibrov A, Kashour T, Amara FM. Anti-apoptotic wild-type Alzheimer amyloid precursor protein signaling involves the p38 mitogen-activated protein kinase/MEF2 pathway. Brain Res Mol Brain Res. 2002;108:102–120. doi: 10.1016/s0169-328x(02)00519-3. [DOI] [PubMed] [Google Scholar]
- Butts BD, Hudson HR, Linseman DA, Le SS, Ryan KR, Bouchard RJ, Heidenreich KA. Proteasome inhibition elicits a biphasic effect on neuronal apoptosis via differential regulation of pro-survival and pro-apoptotic transcription factors. Mol Cell Neurosci. 2005;30:279–289. doi: 10.1016/j.mcn.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Chun HS, Gibson GE, DeGiorgio LA, Zhang H, Kidd VJ, Son JH. Dopaminergic cell death induced by MPP(+), oxidant and specific neurotoxicants shares the common molecular mechanism. J Neurochem. 2001;76:1010–1021. doi: 10.1046/j.1471-4159.2001.00096.x. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Lee VM. Are ubiquitination pathways central to Parkinson's disease? Cell. 2003;114:1–8. doi: 10.1016/s0092-8674(03)00509-9. [DOI] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Heidenreich KA, Linseman DA. Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Mol Neurobiol. 2004;29:155–166. doi: 10.1385/MN:29:2:155. [DOI] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983;34:11–12. doi: 10.1016/0092-8674(83)90131-9. [DOI] [PubMed] [Google Scholar]
- Kang J, Gocke CB, Yu H. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 2006;7:5. doi: 10.1186/1471-2091-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, et al. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci U S A. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cavanaugh JE, Wang Y, Sakagami H, Mao Z, Xia Z. ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc Natl Acad Sci U S A. 2003;100:8532–8537. doi: 10.1073/pnas.1332804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Mao Z, Wiedmann M. Calcineurin enhances MEF2 DNA binding activity in calcium-dependent survival of cerebellar granule neurons. J Biol Chem. 1999;274:31102–31107. doi: 10.1074/jbc.274.43.31102. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Riquelme C, Barthel KK, Liu X. SUMO–1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med. 2006;10:132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shalizi AK, Bonni A. brawn for brains: the role of MEF2 proteins in the developing nervous system. Curr Top Dev Biol. 2005;69:239–266. doi: 10.1016/S0070-2153(05)69009-6. [DOI] [PubMed] [Google Scholar]
- She H, Mao Z. Regulation of myocyte enhancer factor-2 transcription factors by neurotoxins. Neurotoxicology. 2011;32:563–566. doi: 10.1016/j.neuro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, Mao Z. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest. 2011;121:930–940. doi: 10.1172/JCI43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Mount MP, Shree R, et al. Calpain-regulated p35/cdk5 plays a central role in dopaminergic neuron death through modulation of the transcription factor myocyte enhancer factor 2. J Neurosci. 2006;26:440–447. doi: 10.1523/JNEUROSCI.2875-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JH, Chun HS, Joh TH, Cho S, Conti B, Lee JW. Neuroprotection and neuronal differentiation studies using substantia nigra dopaminergic cells derived from transgenic mouse embryos. J Neurosci. 1999;19:10–20. doi: 10.1523/JNEUROSCI.19-01-00010.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wang X, Gong X, Tong M, Park D, Xia Z, Mao Z. Cyclin-dependent kinase 5 mediates neurotoxin-induced degradation of the transcription factor myocyte enhancer factor 2. J Neurosci. 2005;25:4823–4834. doi: 10.1523/JNEUROSCI.1331-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, She H, Mao Z. Phosphorylation of neuronal survival factor MEF2D by glycogen synthase kinase 3beta in neuronal apoptosis. J Biol Chem. 2009;284:32619–32626. doi: 10.1074/jbc.M109.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang X, Li M, Marshall J, Mao Z. Regulation of neuroprotective activity of myocyte-enhancer factor 2 by cAMP-protein kinase A signaling pathway in neuronal survival. J Biol Chem. 2005;280:16705–16713. doi: 10.1074/jbc.M501819200. [DOI] [PubMed] [Google Scholar]
- Wei G, Yin Y, Li W, Bito H, She H, Mao Z. Calpain-mediated Degradation of Myocyte Enhancer Factor 2D Contributes to Excitotoxicity by Activation of Extrasynaptic N-Methyl-D-aspartate Receptors. J Biol Chem. 2012;287:5797–5805. doi: 10.1074/jbc.M111.260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]