Abstract
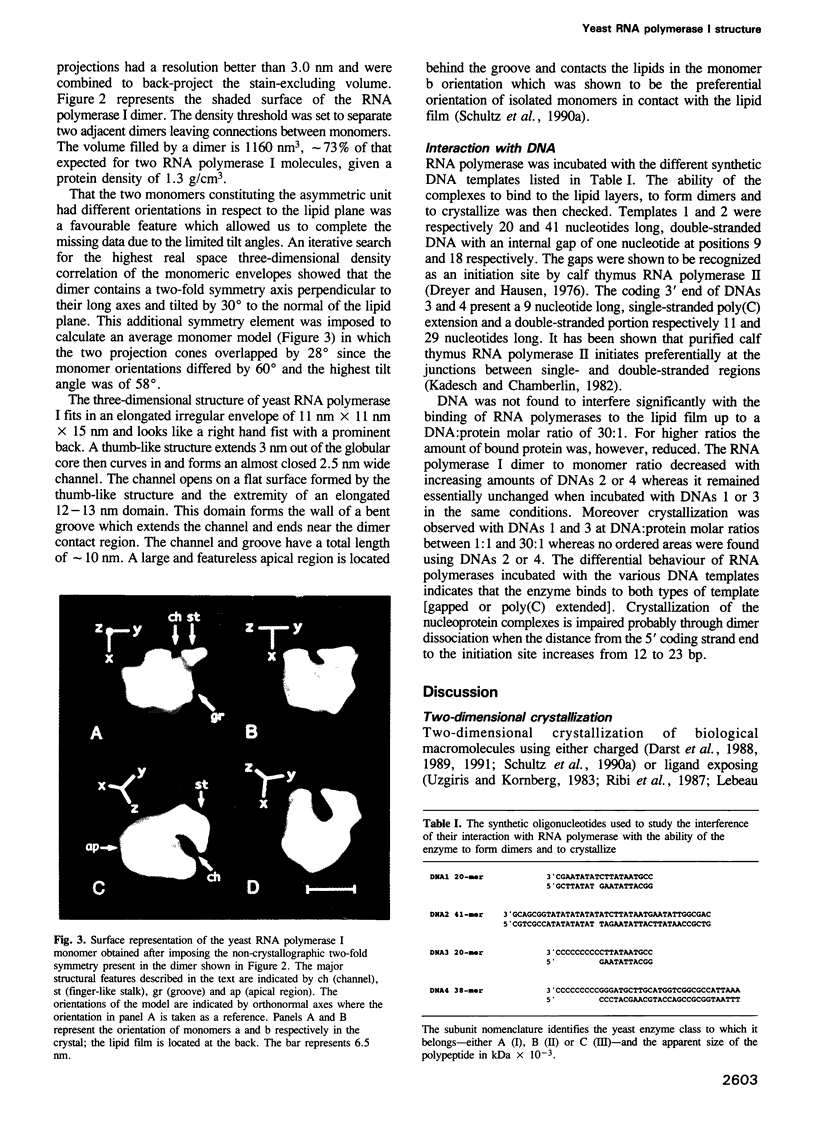
Two-dimensional crystals of yeast RNA polymerase I dimers were obtained upon interaction with positively charged lipid layers. A three-dimensional surface model of the enzyme was determined by analyzing tilted crystalline areas and by taking advantage of the non-crystallographic internal symmetry of the dimer to correct for the missing viewing directions. The structure shows, at approximately 3 nm resolution, an irregularly shaped molecule 11 nm x 11 nm x 15 nm in size characterized by a 3 nm wide and 10 nm long groove which constitutes a putative DNA binding site. The overall structure is similar to the Escherichia coli holo enzyme and the yeast RNA polymerase II delta 4/7 structures. The most remarkable structural feature is a finger-shaped stalk which partially occludes the entrance of the groove and forms a 2.5 nm wide channel. We discuss the possible location of the catalytic centre and of the carboxy-terminal region of the beta-like subunit in the channel. The interference of different DNA fragments with RNA polymerase dimerization and crystallization indicates the orientation of the template in the putative DNA binding groove.
Full text
PDF


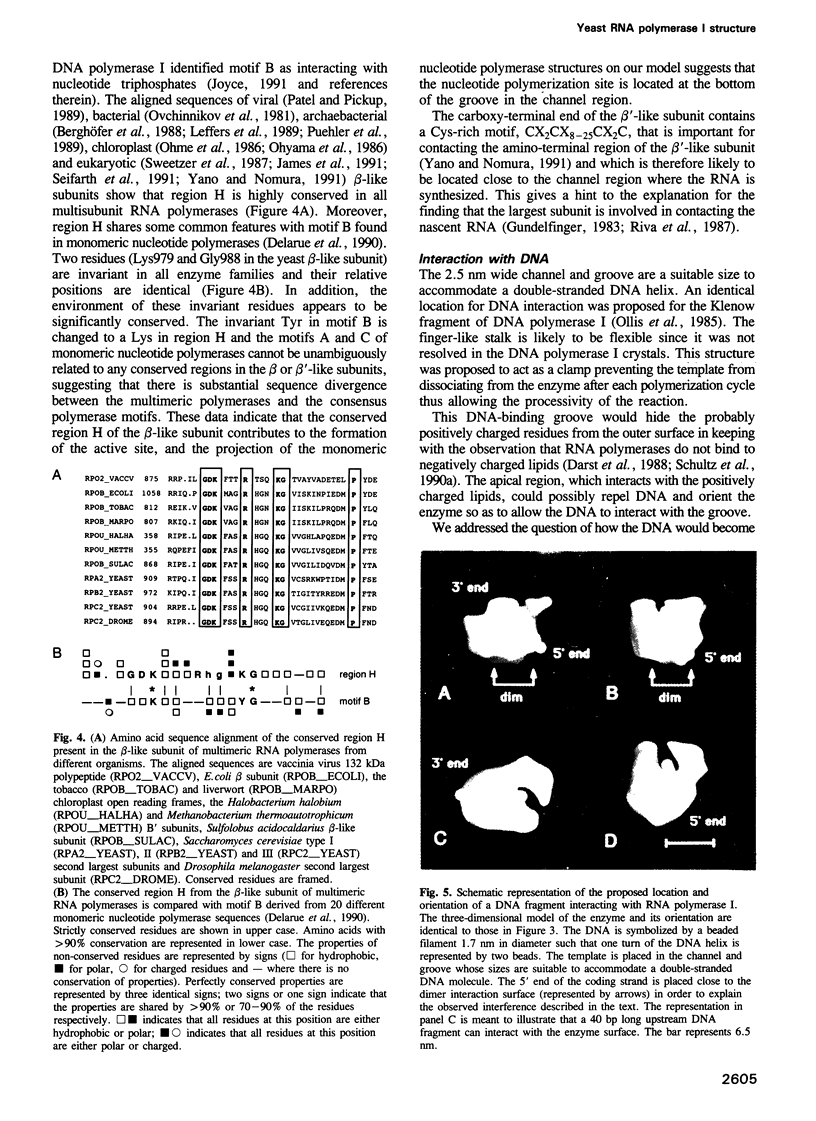


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahearn J. M., Jr, Bartolomei M. S., West M. L., Cisek L. J., Corden J. L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987 Aug 5;262(22):10695–10705. [PubMed] [Google Scholar]
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991 Jul 12;66(1):1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréant B., Huet J., Sentenac A., Fromageot P. Analysis of yeast RNA polymerases with subunit-specific antibodies. J Biol Chem. 1983 Oct 10;258(19):11968–11973. [PubMed] [Google Scholar]
- Buhler J. M., Sentenac A., Fromageot P. Isolation, structure, and general properties of yeast ribonucleic acid polymerase A (or I). J Biol Chem. 1974 Sep 25;249(18):5963–5970. [PubMed] [Google Scholar]
- Darst S. A., Kubalek E. W., Kornberg R. D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989 Aug 31;340(6236):730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- Darst S. A., Ribi H. O., Pierce D. W., Kornberg R. D. Two-dimensional crystals of Escherichia coli RNA polymerase holoenzyme on positively charged lipid layers. J Mol Biol. 1988 Sep 5;203(1):269–273. doi: 10.1016/0022-2836(88)90107-6. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Dequard-Chablat M., Riva M., Carles C., Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J Biol Chem. 1991 Aug 15;266(23):15300–15307. [PubMed] [Google Scholar]
- Dreyer C., Hausen P. On the initiation of mammalian RNA polymerase at single-strand breaks in DNA. Eur J Biochem. 1976 Nov 1;70(1):63–74. doi: 10.1111/j.1432-1033.1976.tb10956.x. [DOI] [PubMed] [Google Scholar]
- Edwards A. M., Darst S. A., Feaver W. J., Thompson N. E., Burgess R. R., Kornberg R. D. Purification and lipid-layer crystallization of yeast RNA polymerase II. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2122–2126. doi: 10.1073/pnas.87.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Kolocheva T. I., Lukhtanov E. A., Mustaev A. A. Studies on the functional topography of Escherichia coli RNA polymerase. Highly selective affinity labelling by analogues of initiating substrates. Eur J Biochem. 1987 Feb 16;163(1):113–121. doi: 10.1111/j.1432-1033.1987.tb10743.x. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Zaychikov E. F., Abdukayumov M. N., Rabinov I. V., Richter V. I., Skoblov Y. S., Chistyakov P. G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989 Apr 1;180(3):577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- Gundelfinger E. D. Interaction of nucleic acids with the DNA-dependent RNA polymerases of Drosophila. FEBS Lett. 1983 Jun 27;157(1):133–138. doi: 10.1016/0014-5793(83)81131-4. [DOI] [PubMed] [Google Scholar]
- Heumann H., Ricchetti M., Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988 Dec 20;7(13):4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Buhler J. M., Sentenac A., Fromageot P. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3034–3038. doi: 10.1073/pnas.72.8.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Phalente L., Buttin G., Sentenac A., Fromageot P. Probing yeast RNA polymerase A subunits with monospecific antibodies. EMBO J. 1982;1(10):1193–1198. doi: 10.1002/j.1460-2075.1982.tb00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Whelen S., Hall B. D. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. Structural analysis of the wild-type and ret1-1 mutant alleles. J Biol Chem. 1991 Mar 25;266(9):5616–5624. [PubMed] [Google Scholar]
- Jokerst R. S., Weeks J. R., Zehring W. A., Greenleaf A. L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989 Jan;215(2):266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- Kadesch T. R., Chamberlin M. J. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem. 1982 May 10;257(9):5286–5295. [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Lebeau L., Regnier E., Schultz P., Wang J. C., Mioskowski C., Oudet P. Two-dimensional crystallization of DNA gyrase B subunit on specifically designed lipid monolayers. FEBS Lett. 1990 Jul 2;267(1):38–42. doi: 10.1016/0014-5793(90)80282-n. [DOI] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Liljelund P., Mariotte S., Buhler J. M., Sentenac A. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9302–9305. doi: 10.1073/pnas.89.19.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Metzger W., Schickor P., Heumann H. A cinematographic view of Escherichia coli RNA polymerase translocation. EMBO J. 1989 Sep;8(9):2745–2754. doi: 10.1002/j.1460-2075.1989.tb08416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser G., Ravanat C., Freyssinet J. M., Brisson A. Sub-domain structure of lipid-bound annexin-V resolved by electron image analysis. J Mol Biol. 1991 Jan 20;217(2):241–245. doi: 10.1016/0022-2836(91)90538-h. [DOI] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Mémet S., Saurin W., Sentenac A. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. J Biol Chem. 1988 Jul 25;263(21):10048–10051. [PubMed] [Google Scholar]
- Ohme M., Tanaka M., Chunwongse J., Shinozaki K., Sugiura M. A tobacco chloroplast DNA sequence possibly coding for a polypeptide similar to E. coli RNA polymerase beta-subunit. FEBS Lett. 1986 May 5;200(1):87–90. doi: 10.1016/0014-5793(86)80516-6. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Chertov OYu, Modyanov N. N., Grinkevich V. A., Makarova I. A., Marchenko T. V., Polovnikova I. N. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the beta-subunit. Eur J Biochem. 1981 Jun 1;116(3):621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- Patel D. D., Pickup D. J. The second-largest subunit of the poxvirus RNA polymerase is similar to the corresponding subunits of procaryotic and eucaryotic RNA polymerases. J Virol. 1989 Mar;63(3):1076–1086. doi: 10.1128/jvi.63.3.1076-1086.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pühler G., Lottspeich F., Zillig W. Organization and nucleotide sequence of the genes encoding the large subunits A, B and C of the DNA-dependent RNA polymerase of the archaebacterium Sulfolobus acidocaldarius. Nucleic Acids Res. 1989 Jun 26;17(12):4517–4534. doi: 10.1093/nar/17.12.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribi H. O., Reichard P., Kornberg R. D. Two-dimensional crystals of enzyme-effector complexes: ribonucleotide reductase at 18-A resolution. Biochemistry. 1987 Dec 1;26(24):7974–7979. doi: 10.1021/bi00398a064. [DOI] [PubMed] [Google Scholar]
- Riva M., Carles C., Sentenac A., Grachev M. A., Mustaev A. A., Zaychikov E. F. Mapping the active site of yeast RNA polymerase B (II). J Biol Chem. 1990 Sep 25;265(27):16498–16503. [PubMed] [Google Scholar]
- Riva M., Schäffner A. R., Sentenac A., Hartmann G. R., Mustaev A. A., Zaychikov E. F., Grachev M. A. Active site labeling of the RNA polymerases A, B, and C from yeast. J Biol Chem. 1987 Oct 25;262(30):14377–14380. [PubMed] [Google Scholar]
- Saxton W. O., Baumeister W., Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13(1-2):57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Schultz P., Célia H., Riva M., Darst S. A., Colin P., Kornberg R. D., Sentenac A., Oudet P. Structural study of the yeast RNA polymerase A. Electron microscopy of lipid-bound molecules and two-dimensional crystals. J Mol Biol. 1990 Nov 20;216(2):353–362. doi: 10.1016/S0022-2836(05)80326-2. [DOI] [PubMed] [Google Scholar]
- Schultz P., Nobelis P., Colin P., Louys M., Huet J., Sentenac A., Oudet P. Electron microscopic study of yeast RNA polymerase A: analysis of single molecular images. Chromosoma. 1990 Jul;99(3):196–204. doi: 10.1007/BF01731130. [DOI] [PubMed] [Google Scholar]
- Seifarth W., Petersen G., Kontermann R., Riva M., Huet J., Bautz E. K. Identification of the genes coding for the second-largest subunits of RNA polymerases I and III of Drosophila melanogaster. Mol Gen Genet. 1991 Sep;228(3):424–432. doi: 10.1007/BF00260636. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I., Carles C., Sentenac A., Riva M. Determination of lysine residues affinity labeled in the active site of yeast RNA polymerase II(B) by mutagenesis. Nucleic Acids Res. 1992 Sep 25;20(18):4721–4725. doi: 10.1093/nar/20.18.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II: subunit structure and function. Trends Biochem Sci. 1990 Sep;15(9):347–351. doi: 10.1016/0968-0004(90)90074-l. [DOI] [PubMed] [Google Scholar]
- Yano R., Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991 Feb;11(2):754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Heggeler-Bordier B., Wahli W., Adrian M., Stasiak A., Dubochet J. The apical localization of transcribing RNA polymerases on supercoiled DNA prevents their rotation around the template. EMBO J. 1992 Feb;11(2):667–672. doi: 10.1002/j.1460-2075.1992.tb05098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]