Abstract
Toll-like receptors (TLRs) play an important role in innate immunity to virus infections. We investigated the role of TLR3 in the pathogenesis of H5N1 and pandemic H1N1 (pH1N1) influenza virus infections in mice. Wild-type mice and those defective in TLR3 were infected with influenza A/HK/486/97 (H5N1) or A/HK/415742/09 (pH1N1) virus. For comparison, mice defective in the gene for myeloid differential factor 88 (MyD88) were also infected with the viruses, because MyD88 signals through a TLR pathway different from TLR3. Survival and body weight loss were monitored for 14 days, and lung pathology, the lung immune-cell profile, viral load and cytokine responses were studied. H5N1-infected TLR3−/− mice had better survival than H5N1-infected WT mice, evident by significantly faster regain of body weight, lower viral titre in the lung and fewer pathological changes in the lung. However, this improved survival was not seen upon pH1N1 infection of TLR3−/− mice. In contrast, MyD88−/− mice had an increased viral titre and decreased leukocyte infiltration in the lungs after infection with H5N1 virus and poorer survival after pH1N1 infection. In conclusion, TLR3 worsens the pathogenesis of H5N1 infection but not of pH1N1 infection, highlighting the differences in the pathogenesis of these two viruses and the different roles of TLR3 in their pathogenesis.
Introduction
Both pandemic H1N1 (pH1N1) and highly pathogenic avian influenza (HPAI) H5N1 viruses pose a significant threat to public health. The pH1N1 virus has spread globally, but pH1N1 infection is clinically relatively mild (Presanis et al., 2009). However, pH1N1 can cause severe disease and death, especially in pregnant women or people with underlying diseases. HPAI H5N1 continues to zoonotically infect humans and causes severe disease with an apparently high mortality rate, but has not acquired efficient human-to-human transmissibility. Innate immunity dysregulation is believed to play a role in the pathogenesis of severe influenza infection (Peiris et al., 2009).
Toll-like receptors (TLRs) play an important role in induction of innate immunity after influenza and other virus infections, but their biological role remains controversial (Daffis et al., 2008; Le Goffic et al., 2006; Gowen et al., 2006, 2007; Majde et al., 2010). On one hand, TLR3-deficient mice were reported to have higher survival (Le Goffic et al., 2006) and milder behavioural changes associated with influenza virus infection (Majde et al., 2010) after challenge with mouse-adapted, low-pathogenic influenza A viruses. Infection of TLR knockout (TLR3−/−) mice with the phlebovirus Punta Toro is associated with reduced disease severity (Gowen et al., 2006). On the other hand, TLR3−/− mice have been found to be more susceptible to encephalomyocarditis virus (EMCV) infection (Hardarson et al., 2007) and to have increased mortality and neuroinvasion after West Nile virus infection (Daffis et al., 2008). Here, we use TLR3−/− mice to investigate the pathogenesis of HPAI A/HK/486/97 (H5N1) and A/HK/415742/09 (pH1N1) infection. As a comparison, we also challenged MyD88 gene knockout mice with the two viruses.
Influenza virus H5N1 (HPAI) strains commonly used in mouse experimental studies are those with high virulence (e.g. A/HK/483/97, A/VN/1194/2004 and A/VN/1203/2004) (Perrone et al., 2010; Salomon et al., 2007; Zheng et al., 2008) and are typically neurotropic. This makes it difficult to evaluate these strains in mouse models because the severe disease and death seen in human H5N1 disease is due to respiratory distress rather than encephalitis in most patients (Uiprasertkul et al., 2005; Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A (H5N1) Virus, 2008). Thus, we chose A/HK/486/97 for our study as this virus in mice produces lung pathology with no virus dissemination to the brain, reflecting the extent of disease observed in humans (Tumpey et al., 2000). Furthermore, for our study to be physiologically relevant, we used viruses that have not been adapted in mice or in embryonated eggs.
We demonstrated that TLR3 signalling is detrimental in H5N1 virus infection, but not in pH1N1 virus infection. These findings have relevance to the pathogenesis of H5N1 and pH1N1 influenza, and in the understanding of the role of TLR receptors in the pathogenesis of severe influenza infection.
Results
TLR3−/− mice show improved survival and less body weight loss after challenge with H5N1 but not with pH1N1 influenza virus
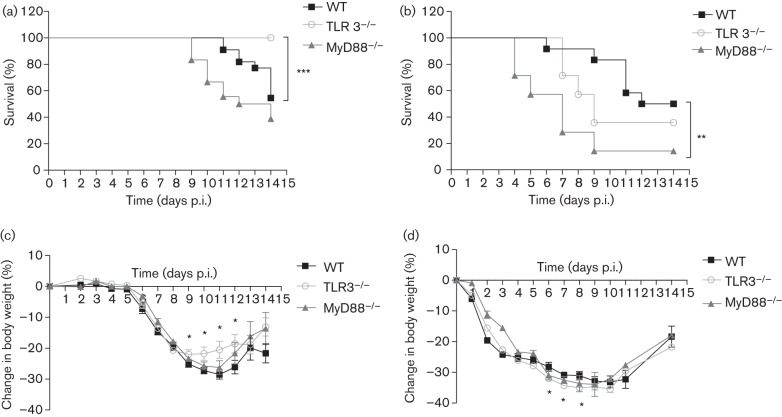
TLR3−/− and WT mice were challenged with 1 LD50 of HPAI H5N1 or pH1N1. While 50 % of the WT mice died after H5N1 challenge, none of the TLR3−/− mice died (P<0.001) (Fig. 1a). However, increased survival was not observed in TLR3−/− mice challenged with 1 LD50 of pH1N1 (Fig. 1b). Comparing the survival of MyD88−/− and WT mice, there was no significant difference upon HPAI H5N1 infection (Fig. 1a), but MyD88−/− mice had significantly worse (P = 0.01) survival after pH1N1 challenge (Fig. 1b).
Fig. 1.
Survival and weight loss in mice after challenge with 1 LD50 HPAI H5N1 or pH1N1 virus. Survival of WT, TLR3−/− and MyD88−/− mice after challenge with HPAI H5N1 (a) or pH1N1 (b) virus. TLR3−/− mice had a significantly improved survival compared with WT mice after challenge with H5N1, but not with pH1N1. MyD88−/− mice had a poorer survival after challenge with pH1N1, but there was no significant difference between H5N1-infected MyD88−/− and WT mice. Percentage weight loss of WT, TLR3−/− and MyD88−/− mice after challenge with H5N1 (c) or pH1N1 (d) virus; asterisks denote statistically significant differences between WT and TLR3−/− mice. The data represent means±sem (TLR3−/−, n = 20; WT, n = 22; MyD88−/−, n = 18 for H5N1 infection. TLR3−/−, n = 14; WT, n = 12; and MyD88−/−, n = 7 for pH1N1 infection). Statistical significance of differences is denoted as: *, P≤0.05; **, P≤0.01; ***, P≤0.001.
Upon H5N1 challenge, body weight loss in TLR3−/− and WT mice was comparable until day 8 post-infection (p.i.), after which TLR3−/− mice regained body weight faster from day 9 until day 12 p.i. than WT mice (P<0.05) (Fig. 1c). However, TLR3−/− mice had significantly greater weight loss than WT mice from day 6 to 8 after pH1N1 challenge (Fig. 1d). In comparison, MyD88−/− mice did not differ in weight loss from WT mice upon challenge with H5N1 or pH1N1 virus. Although the same challenge dose of virus (1 LD50) was used for both H5N1 and pH1N1, the loss in body weight of mice in the first 5 days after the challenge was minimal in H5N1 infection regardless of the mouse strain used. In contrast, pH1N1 infection decreased body weight in all mouse strains from day 1 p.i. (Fig. 1c, d).
TLR3−/− mice clear viruses significantly faster in the lungs
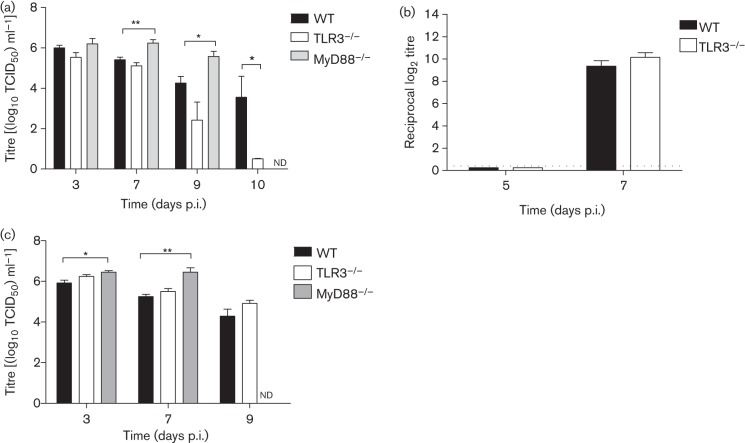
The viral titre in the lungs of H5N1-infected TLR3−/− mice was significantly lower than in WT mice at day 10 p.i. (P = 0.01), while at the other time points (days 3, 7 and 9 p.i) titres were not significantly different between TLR3−/− and WT mice (Fig. 2a). No virus was isolated from the brain of TLR3−/− and WT mice at any time point. Furthermore, there was no difference in antibody titre in serum at days 5 and 7 p.i. between TLR3−/− and WT mice (Fig. 2b). In contrast, in comparison with WT mice, MyD88−/− mice had a significantly higher viral titre in the lung on days 7 (P = 0.002) and 9 p.i. (P = 0.04) (Fig. 2a). H5N1 virus was detectable at a very low titre in the brain of one of five MyD88−/− mice sacrificed at day 3 p.i. and in five out of six mice sacrificed at day 7 p.i. (mean TCID50 of 102 ml−1).
Fig. 2.
Viral titres and serum antibodies in mice infected with H5N1 or pH1N1. (a) Viral titre in lungs of TLR3−/−, MyD88−/− and WT mice after challenge with 1 LD50 of H5N1 at days 3, 7, 9 and 10 p.i. The viral titre in TLR3−/− mice was significantly lower than in WT mice at day 10 p.i. Viral titre in MyD88−/− was significantly higher than in WT mice at days 7 and 9 p.i. (b) Serum antibody against A/HK/486/1999 H5N1 virus in TLR3−/− and WT mice at days 5 and 7 p.i. The dotted line indicates the detection limit. (c) Viral titres in lungs of pH1N1-infected TLR3−/−, MyD88−/− and WT mice. At days 3 and 7 p.i., the viral titre in MyD88−/− mice was significantly higher than in WT mice. Viral titre in MyD88−/− mice at day 9 p.i. was not determined as most of these mice had succumbed to the infection by this time. The data represent means±sem (n = 3–7 per group). Viral titre was determined in MDCK cells. Statistical significance of differences is denoted as: *, P≤0.05; **, P≤0.01. nd, Not determined.
Lungs of TLR3−/− mice challenged with pH1N1 at days 3, 7 and 9 p.i. had a viral titre similar to that in lungs of WT mice, while MyD88−/− mice had a significantly higher virus titre compared with WT mice at days 3 p.i. (P = 0.03) and 7 p.i. (P = 0.008) (Fig. 2c). Infectious virus was detected in the brain in one of three MyD88−/− mice (TCID50 of 102.7 ml−1) sacrificed at day 3 p.i. and in two of four mice dying 4–7 days p.i. (mean TCID50 of 101.2 ml−1). None of the WT or TLR3−/− mice had detectable pH1N1 in the brain.
Lung infiltration of macrophages, neutrophils and T cells in TLR3−/− and MyD88−/− mice challenged with H5N1 or pH1N1
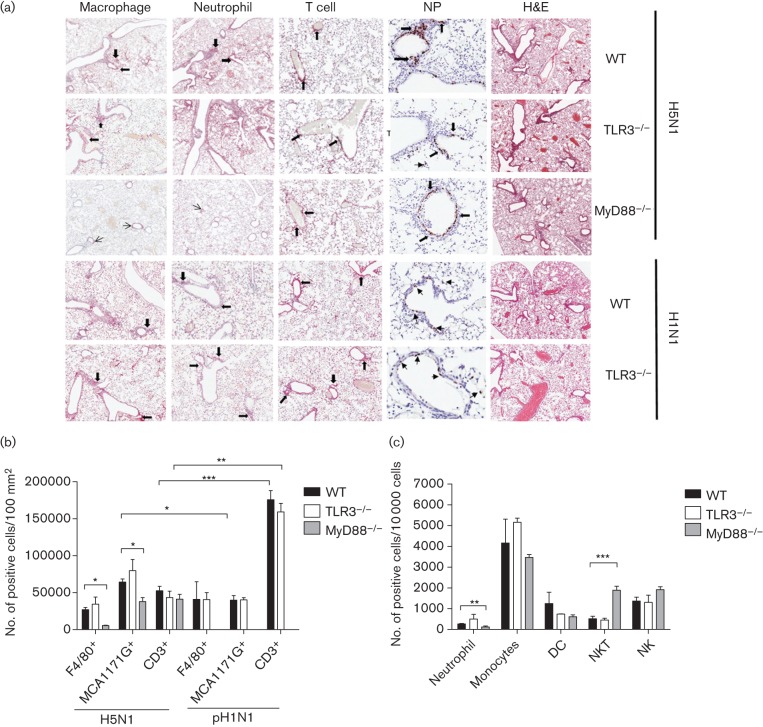
The lung pathology and infiltration by macrophages, neutrophils and T cells of H5N1-infected TLR3−/−, MyD88−/− and WT mice, and of pH1N1-infected TLR3−/− and WT mice, were studied at days 3 and 10 p.i. At day 3 p.i, H5N1-infected TLR3−/−, MyD88−/− and WT mice had comparable lung pathologies (determined by grading of the haematoxylin and eosin staining of lung sections) (Fig. 3a), as did pH1N1-infected TLR3−/− and WT mice. The numbers of macrophages, neutrophils and CD3+ T cells, and of influenza A nucleoprotein+ cells in lung sections of TLR3−/− and WT mice were comparable after infection with H5N1 and pH1N1. In contrast, H5N1-infected MyD88−/− mice had significantly lower numbers of macrophages (P = 0.003) and neutrophils (P = 0.008) in the lungs compared to H5N1-infected WT mice (Fig. 3b). Comparing the number of lung neutrophils and CD3+ cells at day 3 p.i. in H5N1- and pH1N1-infected WT mice, H5N1-infected mice had significantly higher numbers of neutrophils (P = 0.013), but lower numbers of CD3+ cells in the lungs (P<0.001). Similarly, numbers of CD3+ cells in H5N1-infected TLR3−/− mice were lower than in pH1N1-infected mice (P = 0.004) (Fig. 3b). Further immune profiling by flow cytometry showed that the percentages of neutrophils, monocytes, dendritic, NK cells or NK T cells in the lungs of TLR3−/− mice were not significantly different from those in WT mice. In contrast, MyD88−/− mice had a lower percentage of neutrophils (P = 0.005) and a higher percentage of NK T cells (P = 0.002) in the lungs at day 3 p.i. (Fig. 3c).
Fig. 3.
Lung histology and immunohistochemistry of H5N1- or pH1N1-infected mice at day 3 p.i. (a) Immunohistochemistry of lung sections stained with F4/80 (macrophage, ×80), MCA1171G (neutrophil, ×80), CD3 (mainly T cells, ×80), Hb65 (NP, ×200) and haematoxylin and eosin (H&E, ×80) from TLR3−/− and MyD88−/− mice compared with wild-type mice after H5N1 infection (first to third row of panel) and from TLR3−/− compared with WT mice after pH1N1 infection (fourth and fifth row of panel). Pictures are representatives of three mice per group. Arrows indicate cells with positive staining. (b) Quantification of macrophage (F4/80+), neutrophil (MCA1171G+) and T cell (CD3+) infiltration per 100 mm2 of cross-sectional area of lung sections counted by Aperigo Image Scope after H5N1 and pH1N1 infection at day 3 p.i. (c) The number of neutrophils, monocyte, dendritic, NK T and NK cells per 10 000 cells, after infection of H5N1 at day 3 p.i. in WT, TLR3−/− and MyD88−/− mice. The data were analysed by flow cytometry on a FACS LSRII. The data represent means±sem (n = 3). Statistical significance of differences is denoted as: *, P≤0.05; **, P≤0.01; ***, P≤0.001.
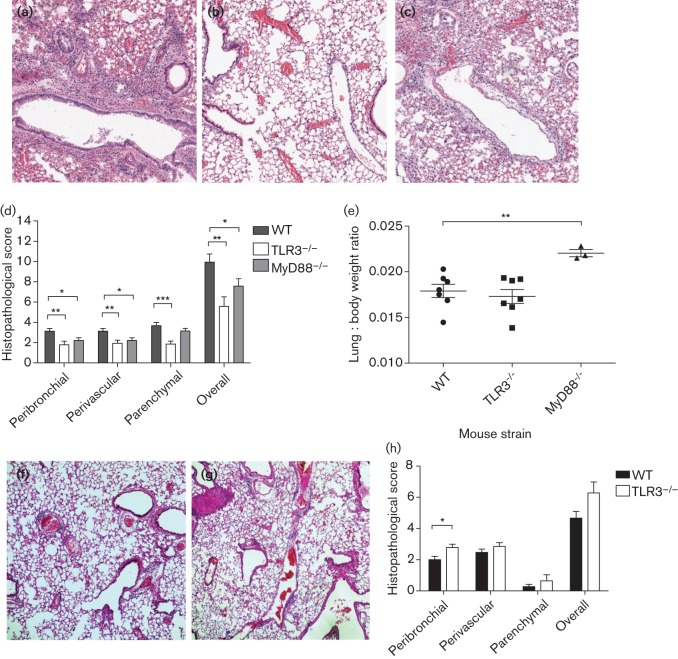
The extent of overall lung pathology in WT, TLR3−/− and MyD88−/− mice was compared using a scoring system of lung pathology at day 10 p.i. H5N1-infected lungs of WT mice exhibited severe peribronchial and perivascular inflammation with massive cell infiltrates; lungs of MyD88−/− mice exhibited less inflammation, while those from TLR3−/− mice had the least peribronchial and perivascular inflammation (Fig. 4a–c). The mean histopathological scores in H5N1-infected WT, MyD88−/− and TLR3−/− mice were 9.93, 7.07 and 5.57, respectively, out of a total point score of 13, indicating that lungs of TLR3−/− (P = 0.002) and MyD88−/− (P = 0.039) mice had significantly fewer pathological lesions than those in WT mice, indicating an overall hierarchy of lung immunopathology. When the pathologies of the peribronchial and perivascular lesions of the lungs were considered separately, both TLR3−/− (peribronchial, 1.79, and perivascular, 1.93) and MyD88−/− mice (peribronchial, 2.21, and perivascular, 2.21) had significantly lower scores (peribronchial, P = 0.004 and P = 0.02, respectively; perivascular, P = 0.007 and P = 0.02, respectively) than WT mice (peribronchial, 3.13, and perivascular, 3.13). The parenchymal histopathological score, however, was significantly lower in TLR3−/− mice (1.86, P<0.001) than in WT mice (3.67), but in MyD88−/− mice (3.14, P = 0.21) this score was not different from that in WT mice (Fig. 4d). Compared with H5N1-infected WT mice at day 10 p.i., H5N1-infected MyD88−/− mice had a significantly (P = 0.007) higher lung : body weight ratio while that of TLR3−/− mice was not different from WT (Fig. 4e). Upon challenge with pH1N1, the severity of lung inflammation was not much different in TLR3−/− and WT mice (Fig. 4f, g). Although the peribronchial lung histopathology score was higher in TLR3−/− than in WT mice (P = 0.016), the overall lung pathology score was comparable (Fig. 4h). Comparing H5N1 and pH1N1 infection for both TLR3−/− and WT mice at day 10 p.i, a relatively lower lung-pathological score at the parenchyma was observed in pH1N1-infected mice.
Fig. 4.
Lung histology, histological score and lung-to-body weight ratio of H5N1- and pH1N1-infected mice. Haematoxylin and eosin staining of lung sections from WT (a), TLR3−/− (b) and MyD88−/− (c) mice after infection of H5N1 on day 10 p.i. (×80). (d) The histopathology score of the peribronchial, perivascular and lung parenchyma, and the overall lung histopathology score of WT, TLR3−/− and MyD88−/− mice at day 10 p.i. after H5N1 influenza virus infection. The overall lung histopathology score of both TLR3−/− and MyD88−/− mice was significantly lower than that of WT mice. TLR3−/− mice had less parenchymal, perivascular and peribronchial inflammation compared with WT mice; while MyD88−/− mice had less perivascular and peribronchial inflammation, parenchymal inflammation was comparable to WT mice. (e) Lung-to-body weight ratio of H5N1-infected TLR3−/− and MyD88−/− mice compared with H5N1-infected WT mice at day 10 p.i. The lung-to-body weight ratios of MyD88−/− mice were significantly higher than those of WT mice. Haematoxylin and eosin staining of lung sections (×80) from WT (f), and TLR3−/− mice (g) and the histopathology scores of the peribronchial, perivascular and lung parenchyma, and overall lung histopathology scores of WT and TLR3−/− mice at day 10 p.i. after infection with pH1N1 (h). Pictures a, b, c, f and g are representatives of three mice per group. The data represent means±sem (n = 3 per group). Statistical significance of differences is denoted as: *, P≤0.05; **, P≤0.01; ***, P≤0.001.
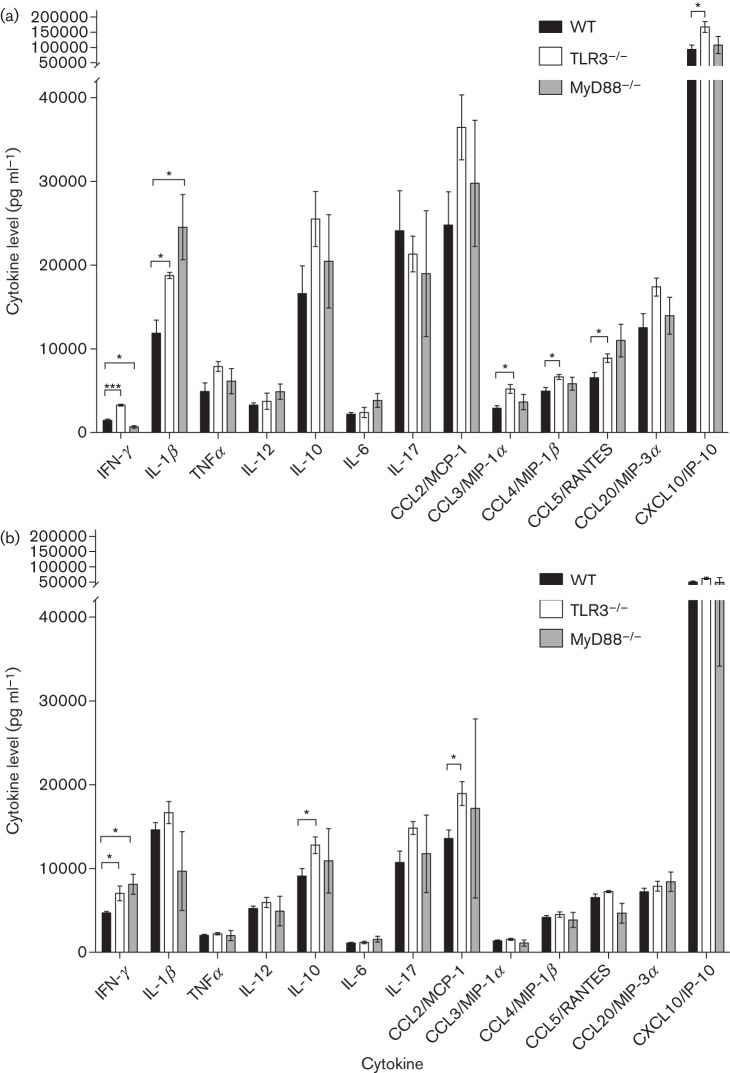
Cytokine expression in lungs of TLR3−/−, MyD88−/− and WT mice after infection with H5N1 or pH1N1
Prior to infection, there was no significant difference in any of the levels of cytokines examined in the lungs of TLR3−/− and WT mice. However, pre-infection levels of IFN-γ, IL-1β, TNF-α, IL-12, IL-10, IL-6, chemokine (C-C motif) ligand (CCL)2, CCL3, CCL4, CCL5 CCL20 and chemokine (C-X-C motif) ligand (CXCL)10 were all significantly higher in MyD88−/− mice compared with WT mice (data not shown). Compared with WT mice, TLR3−/− mice had higher IFN-γ (P<0.001), Il-1β (P = 0.013), CCL3 (P = 0.018), CCL4 (P = 0.033), CCL5 (P = 0.045) and CXCL10 (P = 0.032) at day 3 p.i., and MyD88−/− mice had lower IFN-γ (P = 0.02) but higher IL-1β (P = 0.039) after H5N1 challenge. Upon challenge with pH1N1, TLR3−/− mice also exhibited higher lung IFN-γ (P = 0.028) but with a different overall cytokine profile of significantly higher levels of IL-10 (P = 0.042) and CCL2 (P = 0.025) levels than in WT mice. IFN-γ levels in pH1N1-infected MyD88−/− mice were also significantly higher (P = 0.02) than in WT mice (Fig. 5a, b).
Fig. 5.
Lung cytokine and chemokine levels in mice infected with H5N1 (a) or pH1N1 (b) at day 3 p.i. Cytokines and chemokines were measured from lung homogenates using R&D Systems DuoSet ELISAs. The data represent means±SEM (n = 3 per group). Asterisks indicate statistically significant differences between groups. *, P≤0.05; **, P≤0.01; ***, P ≤ 0.001.
Overall, HPAI H5N1- and pH1N1-infected TLR3−/− mice produced different phenotypes, including survival, loss of body weight, rate of virus clearance, lung histopathology and immune-cell and cytokine profiles.
Discussion
The discovery of the TLR family has led to a revolutionary change in our understanding of how immune systems recognize pathogens, and of the initiation of the innate and adaptive immune responses (Bowie & Haga, 2005; Finberg & Kurt-Jones, 2004). TLR3 is known to recognize dsRNA and polyinosinic/polycytidylic acid (polyI : C) (Choe et al., 2005). TLR3 uses TIR-domain-containing adaptor protein-inducing IFN-β (TRIF) to induce signalling pathways, whereas all other TLRs use MyD88 as the adaptor protein (Akira et al., 2006). The MyD88-dependent pathway activates the transcription factor NF-κB and mitogen-activated protein kinases (MAPKs) for downstream signalling of inflammatory cytokines. The MyD88-independent pathway, however, uses TRIF, leading to the activation of transcription factor IFN receptor factor 3 (IRF3) and induction of type I IFN and NF-κB, which leads to the induction of inflammatory cytokines. (Kawai & Akira, 2010).
This study reveals that TLR3 signalling is detrimental in infections with H5N1 but not in infections with pH1N1, both of which were lethal to WT mice, and TLR3 could thus be associated with the severe immunopathology of HPAI H5N1. When challenged with H5N1 virus, TLR3−/− mice had better survival and regained body weight faster than WT mice. At day 3 p.i., infiltration of lung macrophages, neutrophils and T cells, and the overall histopathologies, were similar in TLR3−/− and WT mice. The viral titres in the lung were comparable between the two groups of mice up to day 7 p.i. But by day 10 p.i., TLR3−/− mice had significantly fewer lesions in the lung and a significantly lower lung virus titre than WT mice. In contrast, pH1N1-infected TLR3−/− mice did not show improved survival, and the percentage of body weight loss was even greater at day 6 to day 9 p.i than in WT mice. The lung viral titre in pH1N1-infected TLR3−/− mice was still higher than that in WT mice at day 9 p.i., and the lung pathology was worse than that in WT mice at day 10 p.i. The role of TLR3 in pathogenesis is supported by the observation that a polymorphism in TLR3, rs5743313/CT, is associated with increased risk of influenza pneumonia in children (Esposito et al., 2012). Whether this polymorphism is associated with an increase or decrease in TLR3 function is unclear, however.
The challenge of WT mice with 1 LD50 of H5H1 or pH1N1 revealed that mice showed a different pattern of body weight loss after infection with the two viruses. H5N1-infected WT mice had a significantly higher number of lung neutrophils and a lower number of T cells at day 3 p.i. than pH1N1-infected mice. There are marked differences in disease severity of H5N1 and pH1N1 infections in humans and in the profile of host responses induced in primary human respiratory epithelium (Chan et al., 2010; Lee et al., 2010). These differences may also account for the contrasting outcomes of these viruses in TLR3−/− mice. Infections with human H5N1 and pH1N1 in BALB/c and C57BL/6J mouse models lead to different disease outcomes in terms of survival, weight loss and lung-pathological features (Otte et al., 2011). This highlights that the viral and host determinants responsible for the pathogenicity of H5N1 are different from those determining the pathogenicity of pH1N1 influenza infections.
A previous study using a lethal mouse-adapted, seasonal H3N2 influenza virus also found that TLR3−/− mice had improved survival (Le Goffic et al., 2006). It may be possible that the mouse-adapted lethal H3N2 virus model is more akin to our (non-mouse-adapted) H5N1 infection of mice. TLR3−/− mice challenged with the phlebovirus Punta Toro also showed faster clearance of the systemic virus and an improved survival rate (Gowen et al., 2006). Similarly, West Nile virus was more efficiently cleared from the spleen of TLR3−/− mice compared with WT mice (Daffis et al., 2008). Conversely, TLR3−/− mice were more susceptible to EMCV infection (Hardarson et al., 2007). It is interesting to note that enteroviruses such as EMCV do not activate RIG-I, and the host defence depends on the TLR3 and IFN-γ pathway. Influenza, however, is a potent activator of RIG-I, and thus this may be the key difference between these two opposite phenotypes associated with infection of TLR3−/− mice. TLR3−/− mice were protected in an experimental model of polyI : C-induced shock and TLR3−/− mice responded with attenuated IL6, IL12p40 and IFN-γ profiles (Zhang et al., 2008). But this profile of cytokine induction was not observed in H5N1-infected TLR3−/− mice.
TLR3−/− mice appeared to show higher lung IFN-γ levels after challenge with influenza viruses. The mechanism by which loss of TLR3 signalling may contribute to enhanced cytokine responses is intriguing. TLR3-Tg/IFNAR1−/− mice have been shown to have a higher expression of TLR3, but a lower expression of RIG-I and MDA-5 (Negishi et al., 2008). Thus, it is possible that TLR3−/− mice have a compensatory increase in baseline expression of alternative innate-sensing receptors such as RIG-I and MDA-5, resulting in a more potent innate immune response to infection. While there is no evidence for such a mechanism at present, it deserves further investigation. Studies with Sendai virus have suggested that there could be a novel intracellular pathway that is independent of the TLR signalling pathway in triggering of DC maturation subsequently leading to antiviral response (López et al., 2004). In addition, a study on the MyD88 adaptor-like (Mal) adaptor has demonstrated that this adaptor protein inhibits the polyI : C activation of IRF7, concomitant with IFN-β production, and it also negatively regulates TLR3 signalling. Whether TLR3−/− could conversely inhibit IRF7 activation and IFN-β production via Mal or other pathways requires further study (Siednienko et al., 2010).
Previous studies have shown that mice deficient in IL-6 or MIP-1 did not differ in their response to H5N1 challenge. Mice defective in TNF-receptor 1 or those treated with antibody against TNF-α appear to have less weight loss after H5N1 virus challenge, although overall mortality is not affected (Szretter et al., 2007). Others have shown that mice lacking both TNF and IL1 receptors exhibited reduced lung inflammation and delayed mortality after challenge with H5N1. Reminiscent of our own observations with TLR3-defective mice, the beneficial effect of the lack of TNF and IL1 signalling was observed only after challenge with H5N1 and not with PR8 (H1N1) virus (Perrone et al., 2010).
We observed that MyD88−/− mice have poorer survival when challenged with low-pathogenic influenza viruses such as pH1N1, and the lungs of pH1N1-infected MyD88−/− mice have a higher viral titre. Comparable results have been reported in MyD88−/− mice challenged with PR8 (H1N1) (Seo et al., 2010). Interestingly, H5N1-infected MyD88−/− and WT mice had comparable survival, even though MyD88−/− mice had increased levels of virus in the lung. Lungs of MyD88−/− mice had decreased infiltration of macrophages and neutrophils, but increased NK T cells. These observations reinforce the fundamental differences in pathogenesis and the importance of innate immune activation between H5N1 virus and low-pathogenic influenza viruses such as pH1N1.
Conclusion
We have demonstrated that TLR3 signalling is detrimental to survival and pathology after H5N1 infection, but not after pH1N1 infection. These findings highlight the differences in pathogenic mechanisms between H5N1 and pH1N1 infections. Depending on the virus, different sensing pathways in innate immunity may be beneficial or detrimental to the overall outcome of infection, and this is key to understanding the pathogenesis of H5N1 influenza and the development of future therapeutic options.
Methods
Virus preparation and quantification.
Influenza viruses A/HK/486/97 (H5N1) and A/HK/415742/09 (pH1N1) grown in Madin–Darby canine kidney (MDCK) cells were used. The LD50 of the viruses for C57BL/6N mice was determined by challenging groups of eight mice with 25 µl of 10−1, 10−2, 10−3, 10−4 and 10−5 dilutions of each virus. The LD50 was calculated using the Spearman–Karber method. One LD50 (equivalent to a dose of 106 TCID50 for both viruses) was used in all challenge experiments to determine the pathogenesis of H5N1 and pH1N1 in knockout and WT mice.
Mouse strains.
Mice with genetic defects in the innate-sensing receptors TLR3 and MyD88 were provided by Dr S. Akira at Osaka University, Japan. They were back-crossed with the same genetic background C57BL/6N mice provided by the Animal Laboratory Unit of the University of Hong Kong, accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The genotype was confirmed by extracting DNA from the ear flap using a Qiagen DNA kit, followed by PCR using the primers 5′-CCAGAGCCTGGGTAAGTTATTGTGCTG-3′ and 5′-TCCAGACAATTGGCAAGTTATTCGCCC-3′ for the WT allele, and 5′-TCCAGACAATTGGCAAGTTATTCGCCC-3′ and 5′-ATCGCCTTCTATCGCCTTCTTGACGAG-3′ for the mutated allele in TLR3-knockout (TLR3−/−) mice. The primers for genotyping MyD88 were 5′-AGACAGGCTGAGTGCAAACTTGTGCTG-3′ and 5′-AGCCTCTACACCCTTCTCTTCTCCACA-3′ for the WT allele, and 5′-AGCCTCTACACCCTTCTCTTCTCCACA-3′ and 5′-ATCGCCTTCTATCGCCTTCTTGACGAG-3′ for the mutated allele of the MyD88-knockout (MyD88−/−) mice.
Animal challenge.
The animal infection experiments were approved by the Committee of the Use of Live Animals for Teaching and Research of the University of Hong Kong (CULATR no. 2637-12) and were performed in the biosafety level 3 containment laboratory at the University of Hong Kong. Female mice (WT, TLR3−/− and MyD88−/−) aged 6–9 weeks were anaesthetized using a mixture of ketamine and xylazine (1.88 and 0.11 mg per mouse, respectively), followed by inoculation with 1 LD50 of H5N1 or pH1N1 virus. Mice were monitored daily for 14 days, and clinical symptoms and weight recorded daily. At days 5 and 7, four mice from each group were sedated using ketamine and xylazine, and blood was collected via the intracardial route, followed by euthanasia of the mice. Sera obtained were tested for antibody against the virus using a haemagglutination inhibition test.
Titration of infectious virus and studying cytokine induction in lungs.
At days 3, 7, 9 and 10 p.i., mice were sacrificed with a dose of pentobarbital. Lungs and brain were dissected from three mice of each group, weighed and further homogenized with 1 ml PBS using a homogenizer (Omni tissue homogenizer; Omni International). The supernatant was aliquoted and stored at −80 °C for later titration of virus load and cytokines or chemokines. Titration of infectious virus in lung homogenates was carried out in MDCK cells, and the TCID50 was calculated using the Spearman–Karber method. TNF-α, IFN-γ, IL-1β, IL-12, IL-10, IL-6, IL-17, CCL2 (MCP-1) CCL3 (MIP-1α), CCL4 (MIP-1β), CCL5 (RANTES), CCL20 (MIP-3α) and CXCL10 (IP-10) were measured in lung homogenates using DuoSet ELISAs (R&D Systems).
Study of lung histology and immunohistochemistry.
At days 3 and 10 p.i., lungs from another three mice were inflated with 1 ml of 10 % formalin/saline, and the whole organ was dissected and placed in 10 % formalin/saline for histology and immunohistochemistry. Tissues in 10 % formalin/PBS were processed and embedded in paraffin. mAbs to mouse macrophage F4/80 antibody (eBioscience), mouse neutrophils MCA1171G (Serotec), mouse CD3 (Abcam) and anti-influenza A nucleoprotein antibody Hb65 (European Veterinary Laboratories) were used for immunohistochemistry. The slides were scanned using a scanscope CS scanner (Aperigo Technologies), and the cells were counted using the software Aperigo Image Scope version 10.0.36.1805 (Aperigo Technologies). The results were expressed as the number of cells (100 mm2 of cross-sectional area)−1. Histopathological scoring was performed by a pathologist and a researcher blinded to the experimental details of the infecting viruses and mouse strains; the scoring is based on a 13-point scale, which evaluates the airway, vascular and parenchymal inflammation as described elsewhere (Horvat et al., 2007).
Isolation of mouse tissue and immune-cell profiling.
At day 3 p.i., three mice were sacrificed, and their lungs were dissected and digested to study the immune-cell profile. The lungs were cut into small pieces and digested with collagenase II (1000 U) and DNase I (250 U) (Worthington) in Dulbecco's Modified Eagle's Medium (with 10 % FBS and 1 % penicillin/streptomycin) for 60 min at 37 °C. Digested lungs were homogenized and red blood cells were lysed. Cells were subjected to Fc receptor blocking (anti-CD16/CD32; eBioscience), and further stained with a panel of mAbs for innate and adaptive immune cells (all from Biolegend, unless indicated otherwise), for 30 min on ice. mAbs included: mouse CD11b, CD11c, Ly6G/C, Dx-5 and CD3 (eBioscience). Cells were fixed in 4 % para-formaldehyde for 30 min, and samples were acquired by flow cytometry on a FACS LSRII and analysed with FlowJo software.
Statistical analysis.
Statistical significance of differences between any two groups compared was analysed using the unpaired t-test with a threshold of P<0.05 using GraphPad version 5.0 (GraphPad Software). Survival of mice was compared using Kaplan–Meier analysis and log-rank test.
Acknowledgements
We acknowledge research funding from the National Institutes of Health (NIAID contract HHSN272201400006C) and the Area of Excellence Scheme of the University Grants Committee, Hong Kong SAR Government (AoE/M-12/06). We thank Kelvin Fung and Ji-Chun Chu, Department of Pathology, University of Hong Kong, for processing the histology slides, and Yu-On Wu, Kai-Chi Chow, Shing-Chun Tang, Isaac Chow and Ching-Lam Yu of the School of Public Health, University of Hong Kong, for technical support.
References
- Akira S., Uematsu S., Takeuchi O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Bowie A. G., Haga I. R. (2005). The role of Toll-like receptors in the host response to viruses. Mol Immunol 42, 859–867 10.1016/j.molimm.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Chan R. W., Yuen K. M., Yu W. C., Ho C. C., Nicholls J. M., Peiris J. S., Chan M. C. (2010). Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS ONE 5, e8713 10.1371/journal.pone.0008713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Kelker M. S., Wilson I. A. (2005). Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309, 581–585 10.1126/science.1115253 [DOI] [PubMed] [Google Scholar]
- Daffis S., Samuel M. A., Suthar M. S., Gale M., Jr, Diamond M. S. (2008). Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol 82, 10349–10358 10.1128/JVI.00935-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., Molteni C. G., Giliani S., Mazza C., Scala A., Tagliaferri L., Pelucchi C., Fossali E., Plebani A., Principi N. (2012). Toll-like receptor 3 gene polymorphisms and severity of pandemic A/H1N1/2009 influenza in otherwise healthy children. Virol J 9, 270 10.1186/1743-422X-9-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg R. W., Kurt-Jones E. A. (2004). Viruses and Toll-like receptors. Microbes Infect 6, 1356–1360 10.1016/j.micinf.2004.08.013 [DOI] [PubMed] [Google Scholar]
- Gowen B. B., Hoopes J. D., Wong M.-H., Jung K.-H., Isakson K. C., Alexopoulou L., Flavell R. A., Sidwell R. W. (2006). TLR3 deletion limits mortality and disease severity due to phlebovirus infection. J Immunol 177, 6301–6307 10.4049/jimmunol.177.9.6301 [DOI] [PubMed] [Google Scholar]
- Gowen B. B., Wong M.-H., Jung K.-H., Sanders A. B., Mitchell W. M., Alexopoulou L., Flavell R. A., Sidwell R. W. (2007). TLR3 is essential for the induction of protective immunity against Punta Toro virus infection by the double-stranded RNA (dsRNA), poly(I:C12U), but not poly(I:C): differential recognition of synthetic dsRNA molecules. J Immunol 178, 5200–5208 10.4049/jimmunol.178.8.5200 [DOI] [PubMed] [Google Scholar]
- Hardarson H. S., Baker J. S., Yang Z., Purevjav E., Huang C.-H., Alexopoulou L., Li N., Flavell R. A., Bowles N. E., Vallejo J. G. (2007). Toll-like receptor 3 is an essential component of the innate stress response in virus-induced cardiac injury. Am J Physiol Heart Circ Physiol 292, H251–H258 10.1152/ajpheart.00398.2006 [DOI] [PubMed] [Google Scholar]
- Horvat J. C., Beagley K. W., Wade M. A., Preston J. A., Hansbro N. G., Hickey D. K., Kaiko G. E., Gibson P. G., Foster P. S., Hansbro P. M. (2007). Neonatal chlamydial infection induces mixed T-cell responses that drive allergic airway disease. Am J Respir Crit Care Med 176, 556–564 10.1164/rccm.200607-1005OC [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11, 373–384 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- Le Goffic R., Balloy V., Lagranderie M., Alexopoulou L., Escriou N., Flavell R., Chignard M., Si-Tahar M. (2006). Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog 2, e53 10.1371/journal.ppat.0020053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M., Chan R. W., Gardy J. L., Lo C. K., Sihoe A. D., Kang S. S., Cheung T. K., Guan Y. I., Chan M. C. & other authors (2010). Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I-like alveolar epithelial cells in vitro. Respir Res 11, 147 10.1186/1465-9921-11-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López C. B., Moltedo B., Alexopoulou L., Bonifaz L., Flavell R. A., Moran T. M. (2004). TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J Immunol 173, 6882–6889 10.4049/jimmunol.173.11.6882 [DOI] [PubMed] [Google Scholar]
- Majde J. A., Kapás L., Bohnet S. G., De A., Krueger J. M. (2010). Attenuation of the influenza virus sickness behavior in mice deficient in Toll-like receptor 3. Brain Behav Immun 24, 306–315 10.1016/j.bbi.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi H., Osawa T., Ogami K., Ouyang X., Sakaguchi S., Koshiba R., Yanai H., Seko Y., Shitara H. & other authors (2008). A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci U S A 105, 20446–20451 10.1073/pnas.0810372105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte A., Sauter M., Alleva L., Baumgarte S., Klingel K., Gabriel G. (2011). Differential host determinants contribute to the pathogenesis of 2009 pandemic H1N1 and human H5N1 influenza A viruses in experimental mouse models. Am J Pathol 179, 230–239 10.1016/j.ajpath.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J. S. M., Cheung C. Y., Leung C. Y. H., Nicholls J. M. (2009). Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol 30, 574–584 10.1016/j.it.2009.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone L. A., Szretter K. J., Katz J. M., Mizgerd J. P., Tumpey T. M. (2010). Mice lacking both TNF and IL-1 receptors exhibit reduced lung inflammation and delay in onset of death following infection with a highly virulent H5N1 virus. J Infect Dis 202, 1161–1170 10.1086/656365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presanis A. M., De Angelis D., Hagy A., Reed C., Riley S., Cooper B. S., Finelli L., Biedrzycki P., Lipsitch M., New York City Swine Flu Investigation Team (2009). The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med 6, e1000207 10.1371/journal.pmed.1000207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R., Hoffmann E., Webster R. G. (2007). Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc Natl Acad Sci U S A 104, 12479–12481 10.1073/pnas.0705289104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.-U., Kwon H.-J., Song J.-H., Byun Y.-H., Seong B. L., Kawai T., Akira S., Kweon M.-N. (2010). MyD88 signaling is indispensable for primary influenza A virus infection but dispensable for secondary infection. J Virol 84, 12713–12722 10.1128/JVI.01675-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siednienko J., Halle A., Nagpal K., Golenbock D. T., Miggin S. M. (2010). TLR3-mediated IFN-β gene induction is negatively regulated by the TLR adaptor MyD88 adaptor-like. Eur J Immunol 40, 3150–3160 10.1002/eji.201040547 [DOI] [PubMed] [Google Scholar]
- Szretter K. J., Gangappa S., Lu X., Smith C., Shieh W.-J., Zaki S. R., Sambhara S., Tumpey T. M., Katz J. M. (2007). Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol 81, 2736–2744 10.1128/JVI.02336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey T. M., Lu X., Morken T., Zaki S. R., Katz J. M. (2000). Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J Virol 74, 6105–6116 10.1128/JVI.74.13.6105-6116.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uiprasertkul M., Puthavathana P., Sangsiriwut K., Pooruk P., Srisook K., Peiris M., Nicholls J. M., Chokephaibulkit K., Vanprapar N., Auewarakul P. (2005). Influenza A H5N1 replication sites in humans. Emerg Infect Dis 11, 1036–1041 10.3201/eid1107.041313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A (H5N1) Virus (2008). Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med 358, 261–273 10.1056/NEJMra0707279 [DOI] [PubMed] [Google Scholar]
- Zhang S. Y., Boisson-Dupuis S., Chapgier A., Yang K., Bustamante J., Puel A., Picard C., Abel L., Jouanguy E., Casanova J. L. (2008). Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev 226, 29–40 10.1111/j.1600-065X.2008.00698.x [DOI] [PubMed] [Google Scholar]
- Zheng B.-J., Chan K.-W., Lin Y.-P., Zhao G.-Y., Chan C., Zhang H.-J., Chen H.-L., Wong S. S. Y., Lau S. K. P. & other authors (2008). Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci U S A 105, 8091–8096 10.1073/pnas.0711942105 [DOI] [PMC free article] [PubMed] [Google Scholar]