Abstract
Dengue fever, a reemerging disease, is putting nearly 2.5 billion people at risk worldwide. The number of infections and the geographic extension of dengue fever infection have increased in the past decade. The disease is caused by the dengue virus, a flavivirus that uses mosquitos Aedes sp. as vectors. The disease has several clinical manifestations, from the mild cold-like illness to the more serious hemorrhagic dengue fever and dengue shock syndrome. Currently, there is no approved drug for the treatment of dengue disease or an effective vaccine to fight the virus. Therefore, the search for antivirals against dengue virus is an active field of research. As new possible receptors and biological pathways of the virus biology are discovered, new strategies are being undertaken to identify possible antiviral molecules. Several groups of researchers have targeted the initial step in the infection as a potential approach to interfere with the virus. The viral entry process is mediated by viral proteins and cellular receptor molecules that end up in the endocytosis of the virion, the fusion of both membranes, and the release of viral RNA in the cytoplasm. This review provides an overview of the targets and progress that has been made in the quest for dengue virus entry inhibitors.
1. Introduction
Dengue fever, dengue hemorrhagic fever, and dengue shock syndrome are caused by the dengue virus. According to the World Health Organization (WHO), dengue is one the most common mosquito borne diseases in the world [1]. It is estimated that up to 3.6 billion people live at risk of getting the disease [2]. Dengue virus is transmitted to humans by infected Aedes mosquitoes, A. aegypti and A. albopictus, which are distributed in tropical and subtropical areas and are widespread in urban and rural areas [1]. At present, A. albopictus can be found in temperate countries [3].
There is some uncertainty in the number and distribution of dengue cases due to the lack of reliable information and misdiagnosis and/or misreporting, thus emphasizing the importance of compiling more extensive records on dengue transmission [4]. It is estimated that dengue fever is present in 128 countries, including all continents, with figures differing from those reported by CDC and WHO [5, 6]. A recent study estimated that in 2010 there were 96 million apparent and 294 million unapparent dengue infections worldwide, with more infections in Asia (70%), followed by the Americas (14%) and Africa (16%) [7].
The pathogenic flaviviruses primarily include the four dengue serotypes, the Yellow fever virus, the West Nile virus, the Tick borne encephalitis virus, the Murray valley encephalitis virus, and the Japanese encephalitis virus. Dengue virus has an icosahedral symmetry, with diameter between 500 Å (mature virion) and 600 Å (spiky immature virion) [8]. The virus genome consists of a single stranded, positive, 11 Kb RNA coding for a single polyprotein. The polyprotein is cleaved in the cytoplasm into several structural and nonstructural polypeptides [9].
The structural proteins include the capsid (C), premembrane (PrM)/membrane (M), and envelope glycoprotein (E) that contains three main domains. These proteins are involved in the formation of the viral particle. The dengue virus membrane M protein has three portions, an extended N-terminal loop, an amphipatic perimembrane helix, and a pair of transmembrane helices [10]. The capsid protein of dengue consists of a dimer with four helices [11]. Ma et al. proposed that these helices may interact with the viral membrane or with viral RNA [12]. The nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) are responsible for the viral replication, assembly, and immune response escape [9].
The entry of the dengue virus into the host cell is a complex process, mediated mainly by E glycoprotein. The first step of dengue virus entry is the binding of the viral E glycoprotein to a cellular receptor and/or attachment factors (Figure 1). Several of these receptor/attachment factors have been identified and are considered important targets for the development of antivirals, as explained in detail later in the text. Following receptor binding, the virus gets internalized via clathrin-dependent endocytosis (reviewed in [13]).
Figure 1.
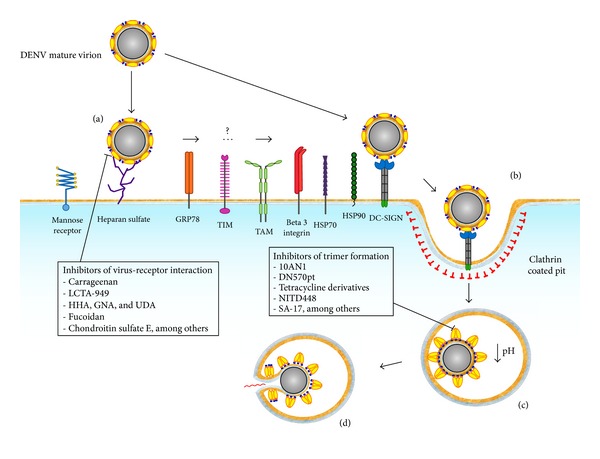
Schematic representation of the dengue virus entry process and possible antiviral targets. The dengue virus makes use of cellular membrane receptors and attachment factors to find its way to the cytoplasm. First, the mature virion gets attached to a cellular membrane receptor (a). It is not clear whether single interactions or sequential usage of several molecules is required to trigger the endocytic, clathrin-dependent pathway (b). The endocytic vesicle becomes a late endosome (c), where acidification triggers conformational changes on the E protein dimers to become fusogenic trimers. Finally, pores are formed and the genome of the virus is released into the cytoplasm (d). Possible antiviral targets are shown with examples of compounds inhibiting the step.
The dengue virus entry pathway is highly dependent on the cell type and viral strain. Although evidence suggests that the main way of entry for dengue virions is receptor initiated-clathrin mediated endocytosis [14], direct fusion through the plasmatic membrane has also been observed in particular cell lines [15, 16]. There is evidence that DENV-2 is able to use a clathrin-independent, noncaveolar, dynamin sensitive endocytic route which is also independent of macropinocytosis or phagocytosis [14]. The same virus, however, employs a classical clathrin-dependent pathway to enter human A549 cells. The “classical” postendocytosis fusion pathway depends on acidic pH to induce a conformational change in the viral envelope (E) protein to expose and insert the fusion loop into the endosomal membrane [17–19]. Once the endocytic and the viral membranes are bridged by the rearranged E glycoprotein, a final folding back movement of domain III brings the membrane closer to sequentially form apposing nipples, hemifusion stalk, and finally fusion pores. As a result, the membranes fuse and the viral RNA enters the cytoplasm [17].
Two broad directions have been undertaken to fight this disease. Most community level efforts are focused on reducing the mosquito populations, while at the patient level researchers are trying to develop vaccines and antivirals. The fight to reduce mosquito densities has been partially successful in some places, but the rapid development of resistance to chemicals and many other operative hurdles hamper the efficacy of these programs [20]. More sophisticated researches are trying to target the size of the mosquito population by releasing transgenic males carrying a dominant repressible lethal gene [21–23], but this solution might still have to overcome existing regulatory issues to be broadly applied.
The ideal solution for fighting dengue virus infection would be by developing a prophylactic vaccine. However, the fact is that current candidates have failed to give protection against all four serotypes, emphasizing the notion that a therapeutic agent—an antiviral drug—may be a key tool to fight the disease. An effective vaccine must protect against all four serotypes; otherwise, vaccinated individuals might be at risk of developing the severe form of dengue due to the phenomenon known as antibody-dependent enhancement (ADE). In this scenario, nonneutralizing antibodies opsonize the virions and mediate augmented entry and infection of cells expressing the Fc gamma receptor [24, 25]. This enhanced infection provokes proliferation of T cells, massive production of proinflammatory cytokines, and ultimately life-threatening systemic effects in infected patients [26–28].
Currently, there is no approved antiviral drug against the dengue virus. Several research groups have developed different approaches to identify molecules that inhibit dengue virus infection by targeting several structural and nonstructural viral proteins and its cellular receptors. One attractive approach could be blocking the virus before it enters the target cell. In this scenario, nature provides a proof of concept, as dengue virus generally induces the production of highly effective neutralizing antibodies in convalescent patients. The putative antiviral molecules targeting entry would not need to enter the cell, thereby relaxing structural and other constraints in the search and design of these molecules. Blocking the virus before entry could also reduce the hyperactivation of the immune system produced during the triggering of ADE. Dengue virus tropism in humans involves cells from liver and immune and endothelial systems. Other affected organs are lungs and brain (reviewed by [29]). Permissive cells include Langerhans cells, keratinocytes, monocytes, macrophages, and leukocytes. Also, bone marrow stromal cells are susceptible for dengue virus.
Several approved antiviral drugs show that entry inhibition is a feasible strategy to fight viruses. The anti-HIV drug maraviroc binds the virus coreceptor CCR5 [30], while enfuvirtide is able to block the viral protein GP41, reducing the fusion between viral and cellular membranes [31]. The drug Myrcludex-B, a lipopeptide based on the sequence of the preS1 domain of the HBV envelope protein, currently in clinical trials, is capable of stopping de novo viral infections [32]. This review provides an overview of the progress achieved in the search of dengue virus entry inhibitors, by targeting key molecules involved during this process.
2. Envelope Glycoprotein Structure Overview
The E glycoprotein is a class II fusion protein, mainly composed of β sheets and a short transmembrane domain for insertion into the viral membrane. This protein is highly flexible, undergoing significant conformational changes during the maturation and the fusogenic stages of the viral life cycle [17, 18, 33, 34]. In the mature virion, the E glycoprotein adopts a polygonal conformation, where dimers are arranged in polygons covering the surface of the viral particle [35].
Flavivirus virions structural studies show that the E glycoprotein is formed by three domains, in addition to a membrane proximal stem and a transmembrane anchor [17, 34, 36]. A central domain, the domain I, is formed by eight β-strand barrels containing two insertion loops. The dimerization domain, domain II, contains hydrophobic sequences that are conserved among all flaviviruses. These hydrophobic sequences, also known as fusion peptides, are responsible for the insertion of the rearranged E trimer into the cellular membrane during fusion [17, 33, 36–38]. Domain III is an immunoglobulin-like carboxy terminal domain, responsible for the initial cellular receptor binding [39]. The structure of this domain is arranged in a β barrel configuration, connecting with domain I [17].
Structural studies using X-ray crystallography and NMR solution structures of the domain III have shown that DENV-3 and DENV-4 contain nine β-strands, while DENV-2 has an extra β-strand [36, 40, 41]. The domain III can act as a virus fingerprint, due to the primary structure difference within the four dengue virus serotypes [42]. While the domain I and domain II are connected through four polypeptide strands, the domain I and domain III are connected by only one strand, working as a molecular hinge [18, 34]. The DI/DIII linker is a flexible region, allowing the interdomain rearrangement of E glycoprotein during the prefusion stage [43]. Several amino acids within this region have been identified as key for the viral replication and fusion [44, 45].
The E protein stem region is located at the C-terminus of the protein, from residues 395 to 450, and it is part of an outer lipid leaflet of the viral membrane [35]. This region consists of two α-helices (EH1 and EH2) connected by a conserved sequence [18, 38]. The EH1 and EH2 helices are amphipathic and slightly positively charged [35]. The arrangement of these helices differs depending on whether the virion is mature or immature [18]. The stem is important during the entry/fusion process because it promotes the E trimer assembly within the endosome [38].
The hydrophobic pocket is located in the domain I-domain II interface. This region plays an important role in E glycoprotein rearrangement, acting as a hinge that allows the exposure of the fusion peptide located in the domain II [34, 36, 46]. It has been proposed that the shift of two β-strands (β-hairpins) located at the hydrophobic pocket is the key structural element for the initiation of the E glycoprotein rearrangements, induced by a low pH environment in endosomes [36]. The hydrophobic pocket has been extensively used as a target for molecular docking approaches in order to identify molecules able to bind and ultimately show antiviral activity.
The E glycoprotein secondary and tertiary structure suffers significant rearrangements in order to adopt the fusogenic conformation [33]. This conformational change is triggered when pH is lowered inside the late endosome (Figure 1). These structural changes are involved in the dissociation of dimeric E glycoprotein and the formation of homotrimers, which point away from the virion surface and expose the fusion loops ready to be inserted into the endosomal membrane [19, 47–49]. These rearrangements are possible due to the linkers between domains [17]. During these changes, the domain II rotates approximately 30° with respect to domain I [17]. The hydrophobic core beneath this hairpin acts as a hinge, allowing the rotation between the domain I and domain II [17]. Domain III undergoes a significant rotation of about 70° toward domain II [17].
Dengue virus E glycoprotein adopts two main conformations: dimeric in the mature virion and trimeric in the immature virion and during the adoption of the fusogenic conformation inside the endosome [17, 33, 34, 36]. In the dimeric conformation, the antiparallel dimers lay flat on the virions surface, while trimers point away from the viral membrane with a conical rod shape to give the virion a spiky appearance. The fusion loop, which is exposed at the tip, contains three highly conserved amino acid residues, Trp101, Leu107, and Phe108 [17, 18, 33]. While the dimeric conformation is reversible, the transition to the trimeric conformation is irreversible.
The conformational changes leading to the formation of E glycoprotein trimer are thought to be initiated when low pH in the endosome is sensed by five histidine residues which are conserved among flaviviruses. These histidines are distributed in the three domains of the protein and in the stem region [10, 33, 50, 51]. Histidine 323, located at the DI–DIII interface, functions as a pH sensor and also stabilizes the trimer postfusion conformation. This stabilization might be due to the presence of a salt bridge between His323 and Glu373 [50]. When His323 is protonated, the salt bridge dissociates and destabilizes the DI–DIII interphase [33, 50]. Other two widely conserved histidines, His209 and His7, located in the E glycoprotein and the M protein, respectively, may also work as pH sensors. Hydrophobic interactions between E glycoprotein and M stabilize the E glycoprotein and are important for the conformational changes. When these histidines are in their protonated forms, the E glycoprotein dissociates from M protein, allowing the formation of the E trimer [10].
Other important structural characteristics of the E glycoprotein of DENV-2, identified by X-ray crystallography and cryo-EM, are the glycosylated Asn153 in domain I and Asn67 in domain II [10, 36]. The Asn153 residue is conserved among flaviviruses and interacts with the dimeric form of the protein, helping in the dengue virus's infectivity, whereas Asn67 is only found in dengue virus strains and it is thought that this glycosylation is important for the dengue virus assembly or exit [18, 52].
3. Possible Receptors for Dengue Virus
The most important target molecules on the cellular side of the dengue virus entry process are the attachment factors and receptors (Figure 1). Several molecules have been identified as possible receptors for the virus in mammalian cells, including the dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN; CD209) [53, 54], the heparan sulfate [39], the CD14 [55], the mannose receptor [56], the HSP90/HSP70 [57], the glucose regulated protein 78 (GRP78) [58], the laminin receptor [59], and the TIM and TAM proteins [60]. There is experimental evidence suggesting that dengue virus may also enter human cells by interacting with other molecules, including the vitronectin receptor [61], the scavenger receptor class B type I [62], the claudin 1 [63, 64], and the natural killer cell activating receptor [65]. Several of these receptors have been considered potential targets for the development of antivirals against the dengue virus.
It has been suggested that binding of the virus to the cell may require multiple, sequential interaction with several types of receptors. DC-SIGN and glycosaminoglycans (GAGs) are the first line of attachment factors (Figure 1). A second line of higher affinity receptors may then be recruited to allow viral entry, providing a possible explanation for the diverse tissue tropism of the virus [66].
3.1. DC-SIGN
The DC-SIGN is a type II transmembrane protein that is abundantly expressed in immature dendritic cells. It is a C-type lectin with an extracellular domain that binds mannose containing carbohydrates with high affinity. DC-SIGN is involved in dendritic cell migration, T-cell priming, and antigen recognition and presentation [67]. DC-SIGN also efficiently binds diverse viruses such as HIV-1 [68], Ebola [69], and CMV [70]. The DC-SIGN molecule was also identified as a cellular factor required for productive infection of immature dendritic cells by dengue virus [53, 54]. This interaction appears to be through the high mannose N-glycan groups located at the E glycoprotein [71]. Although this protein is glycosylated in two sites, Asn67 and Asn153, the former is believed to bind DC-SIGN [52].
The interaction DC-SIGN-DENV has been proposed as an attractive target to tackle the infection [72]. A recent study showed that the main interaction between E glycoprotein and DC-SIGN occurs by the formation of hydrogen bonds between the mannose molecule attached to Asn67 and Asn272 of DC-SIGN and six salt bridges present in different amino acid residues of both molecules. Blocking the E glycoprotein glycosylation site Asn67 and the conserved residues at the DC-SIGN-DENV interface might impair infectivity of all dengue serotypes [72]. Recently, it was proposed that the inhibition of the initial interaction between dengue virus and dendritic cells could prevent a deleterious immune response [73].
3.2. Glycosaminoglycans
Heparan sulfate (HS) belongs to the family of glycosaminoglycans (GAGs), which are very abundant in cell membrane proteoglycans. HS is composed of chains of disaccharides with uronic or L-iduronic acids and an O-sulfated glucosamine derivative [74]. It is well known that several virus families use the GAGs to enter the target cells. It is also known that HS is involved in the first interaction with the DENV, probably through GAG-binding sites on E glycoprotein [75–78]. These GAG-binding sites are located in the domain III of the protein [75]. Positively charged residues on E glycoprotein bind to the negatively charged HS [39, 75]. Recently, it was demonstrated that two conserved residues, Lys291 and Lys295, are critical for the interactions of GAG–E glycoprotein [45].
3.3. Other Possible Receptors
Besides DC-SIGN, dendritic cells (DCs) also express Fc gamma receptors (FcγRs). Evidence suggests that DENV uses two different ways to enter DCs, a primary infection through DC-SIGN into immature DCs and a secondary infection through FcγRs in mature DCs [79]. Although DENV particles should be previously opsonized with nonneutralizing antibodies to follow the FcγRs entry pathway, some authors consider these cellular molecules as receptors for the virus.
Recently, the TIM and TAM receptors were identified as new DENV cellular attachment molecules [60]. These proteins are involved in the engulfment and removal of apoptotic cells, recognizing the apoptotic marker phosphatidylserine (PtdSer). Since the viral membrane exposes PtdSer, the virus is able to enter cells by direct binding of TIM receptor or indirectly by the TAM receptor through the PtdSer binder molecule Gas6. TAM receptor activation may play an important role in dengue pathogenesis as it shuts down the expression of interferon pathway genes, a key cellular antiviral response [13, 80].
4. Targeting Envelope (E) Glycoprotein
The dengue E glycoprotein is by far the most important molecule during the viral entry process as it appears to be responsible for receptor recognition and attachment to the cell, the clathrin mediated endocytosis, and the subsequent fusion of viral and cellular membranes. This protein is able to interact with many diverse cellular molecules; thus, it is considered an ideal target to develop antivirals [33, 46, 81, 82] (Figure 1). Several parts of the dengue virus E glycoprotein might be suitable as drug targets, including the stem domain, the hydrophobic pocket, and the receptor binding domain III. Blocking any of these particular regions may interfere with the entry/fusion process of dengue virus (blocking compounds summarized in Table 1).
Table 1.
Inhibitors of dengue virus entry.
| Compound | Remarks | Cell line | Dengue virus serotype | IC50/EC50 # | Reference |
|---|---|---|---|---|---|
| 1OAN1 | Fusion inhibitor, peptide based on E protein domain II hinge | LLCMK2 | DENV-2 | IC50: 7 ± 1 μM3 | [96] |
|
| |||||
| DN57opt | Fusion inhibitor, peptide based on E glycoprotein DI/DII beta sheet connection | LLCMK2 | DENV-2 | IC50: 8 ± 1 μM3 | [96] |
|
| |||||
| DN59 | Fusion inhibitor, E glycoprotein stem and E trimer binder, peptide based on stem sequence | LLCMK2 | DENV-2 | IC50: 2–5 μM1 | [84, 85] |
|
| |||||
| Compound 6 | Fusion inhibitor, E glycoprotein hydrophobic pocket binder | A549/BHK21 | All | EC50: 0.068–0.49 μM1∗ | [91] |
|
| |||||
| Rolitetracycline | Fusion inhibitors, tetracycline derivative, and E glycoprotein hydrophobic pocket binder | BHK21 | DENV-2 | IC50: 67 μM2 | [88] |
|
| |||||
| Doxycycline | Fusion inhibitors, tetracycline derivative, and E glycoprotein hydrophobic pocket binder | BHK21 | DENV-2 | IC50: 55 μM2 | [87] |
|
| |||||
| NITD448 | Fusion inhibitor, E glycoprotein hydrophobic pocket binder | BHK21 | DENV-2 | EC50: 9.8 μM3
IC50: 6.8 μM |
[92] |
|
| |||||
| A5 | E glycoprotein hydrophobic pocket binder | Vero | DENV-2 | IC50: 1.2 μM2 | [93] |
|
| |||||
| 1662G07 and derivatives | Fusion inhibitor, E glycoprotein stem, and E trimer binders | BHK21 | DENV-2 | IC50: 8 μM1 | [87] |
|
| |||||
| SA-17 | Derivative of doxorubicin, possible E glycoprotein hydrophobic pocket binder | Vero/BHK21 | DENV-1 DENV-2 DENV-3 |
EC50: 12 μM4
EC50: 1.2 μM4 EC50: 1.7 μM4 |
[89] |
|
| |||||
| LCTA-949 | Analogue of the antibiotic teicoplanin, entry inhibitor | Vero | DENV-2 | EC50: 6.9 μM1 | [90] |
|
| |||||
| ST-148 | Inhibitor of capsid protein, effective in animal model | Vero | All | EC50: 0.016–2.8 μM4 | [109] |
|
| |||||
| E 419-447 peptides | Stem derived sequence, trimer binder | BHK | DENV-2 | IC90: 0.1–6 μM2 | [86] |
|
| |||||
| E 380-389 peptides | Domain III derived sequence, attachment inhibitor | LLCMK2 | DENV-2 | IC50: 35 μM2 | [98] |
|
| |||||
| P02 | Binds to DENV hydrophobic pocket | BHK | YFV-IRES-Luc | IC50: 13 ± 3 μM8 | [95] |
|
| |||||
| HHA, GNA, and UDA | Carbohydrate binding agent | Raji/DC-SIGN+ | All | EC50: 4.6; 3.8; 0.29 nM5 | [73] |
|
| |||||
| Pradimicin-S | Carbohydrate binding agent | Dendritic cells | DENV-2 | EC50: 11 μM5 | [73] |
|
| |||||
| PI-88, suramin, and pentosan polysulfate | Heparan mimetic | BHK | DENV-2 | EC50: 200 μg/mL9∗ | [105] |
|
| |||||
| Fucoidan | Heparan mimetic | BHK21 | DENV-2 | IC50: 4.7 μg/mL3 | [99] |
|
| |||||
| Sulfated galactomannan | Heparan mimetic | C6/36 | DENV-1 | EC50: 200 mg/L10∗ | [106] |
|
| |||||
| DL-galactan | Heparan mimetic | Vero | DENV-2 | IC50: 0.9–1 μg/mL1 | [103] |
|
| |||||
| Iota-carrageenan | Heparan mimetic | Vero | DENV-2 | EC50: 0.4 μg/mL1 | [103] |
|
| |||||
| Zosteric acid, CF-238 | Heparan mimetic | LLCMK2 | All | IC50: 14–47 μM3 | [110] |
|
| |||||
| Curdlan sulfate | Heparan mimetic | LLCMK2 | DENV-2 | EC50: 7 μg/mL6 | [101] |
|
| |||||
| Sulfated galactan, sulfated xylomannan | Heparan mimetic | Vero | All | IC50: 0.12–20 μg/mL1 | [102] |
|
| |||||
| Sulfated K5 polysaccharide | Heparan mimetic | HMEC-1 | DENV-2 | EC50: 111 nM9 | [104] |
|
| |||||
| Chebulagic acid, punicalagin | Hydrolysable tannins | Vero | DENV-2 | EC50: 13.1 and 7.8 µM7 | [108] |
|
| |||||
| Chondroitin sulfate E | Heparan mimetic | BHK21 | All | EC50: 0.3 µg/mL3 | [81] |
#IC50/EC50: in antiviral assays, half maximal effective concentration (EC50) refers to the concentration of compound causing 50% reduction of virus replication in cell based assays. Half maximal inhibitory concentration (IC50) is used when virus inhibition is estimated from in vitro inhibition assays, such as the inhibition of a viral enzyme. Sometimes both EC50 and IC50 values are used loosely to describe the same antiviral activity. In this review, we are following the original nomenclature used by each cited paper.
1Viral plaque reduction assay.
2Plaque formation assay.
3Focus forming assay.
4Virus induced CPEs.
5RNA quantitation.
6Cell viability.
7Viral EGFP (enhanced green fluorescent protein) expression.
8Reduction of luciferase activity.
9Flow cytometry.
10Immunofluorescens.
∗Tested in vivo in animal model.
4.1. The Stem Domain
Recent studies confirmed the viability of the stem region as an inhibition target. The disruption of the secondary structure of EH1 and EH2 by site-directed mutagenesis affected the viral entry, suggesting a key role of the stem region in the initial step of dengue infection [83]. Using the Wimley-White interfacial hydrophobicity (WWIH) scales algorithm, five possible inhibition sites of DENV were predicted. When peptides were designed, synthesized, and tested for antiviral activity, the best performing candidate corresponded to the stem region [84]. The peptide DN59, corresponding to amino acids 414 to 444, showed an IC50 of 2–5 μM against DENV-2 in LLC-MK2 cells. It was later found that this peptide is effective against all dengue serotypes through a virucidal mechanism. The peptide DN59 is able to destabilize the viral membranes causing the release of RNA and production of empty virions [85].
Peptides based on the sequences of the stem region (residues 419 to 447) of all dengue serotypes showed cross-reactive antiviral activity (IC90: 0.1 μM–>6 μM), although similar peptides from other flaviviruses are not effective against dengue viruses [86]. Notably, the highest inhibition levels on the dengue serotypes tested were not exerted by the homologous peptides. These authors found that the best correlate of inhibition related to the stem region was the hydrophobicity of residues 441–447. The peptides corresponding to the stem region are able to bind soluble E glycoprotein and block the viral fusion by binding a conformational intermediate of the envelope protein. These peptides are also able to bind mature virions, being transported into the endosome where they inhibit the fusion [86]. The same authors developed a screening system to look for small molecules able to interfere with the binding of stem based peptides and the E trimer [87]. Compound 1662G07 was identified as promising hit and several analogs were further developed and characterized (best IC50: 8 μM).
4.2. Hydrophobic Pocket
Several tetracycline derivatives have shown affinity for the hydrophobic pocket. These molecules were identified using GEMDOCK, a tool used to predict potential ligands for the active site of a target protein. The compounds doxycycline and rolitetracycline showed a significant in vitro inhibitory activity (IC50: 55 μM and 67 μM, resp.) against dengue virus type 2 in BHK-21 cells [88]. The binding of these molecules to the hydrophobic pocket halts the rearrangements of E glycoprotein domains I and II during pH induced rearrangement.
The compound SA-17, a derivative of the anthracycline antibiotic doxorubicin, showed antiviral activity against dengue virus serotypes 1, 2, and 3 (EC50: 12 μM, 1.2 μM, and 1.7 μM, resp.). As suggested by docking simulations, the SA-17 molecule appears to bind to the hydrophobic pocket, interacting with amino acid residues crucial for the fusion of the membranes (Ala50, Tyr137, and Gln200) [89, 90].
Using high throughput docking screening for hydrophobic pocket ligands, a thiophene-pyrimidine molecule (“compound 6”) was also identified as a potential dengue virus entry inhibitor. After optimization, one compound was able to inhibit all dengue serotypes in BHK cells (range EC50: 0.068 to 0.49 μM) and three additional flaviviruses [91]. However, testing in an in vivo dengue viremia animal model revealed that this compound precipitates in the gastrointestinal tract [91].
An in silico study using GLIDE docking software with a proprietary collection of compounds and natural products reported the finding of a molecule that potentially binds to the hydrophobic pocket [92]. The compound NITD448 effectively inhibited DENV-2 in BHK-21 cells (EC50: 9.8 μM). This molecule showed suppression of the fusion of membranes mediated by E glycoprotein, most likely through interaction with Lys128 and Gln52.
The compound A5 was also identified by an in silico docking screening, using the hydrophobic pocket as target [93]. This molecule showed strong antiviral activity against dengue 2, West Nile virus, and Yellow Fever virus (IC50: 1.2 μM, 3.8 μM, and 1.6 μM, resp.) and very low in vitro cytotoxicity (CC50: >100 μM). This and other studies suggest that compounds with central thiazole rings may be critical to the observed antiviral activity targeting the hydrophobic pocket [94]. Using a four-stage computational high throughput screening (HTS) of three National Cancer Institute compound libraries, P02 was identified as a potential β-OG pocket binder. Additional experiments demonstrated that P02 binds E glycoprotein and has antiviral activity (IC50: 13 ± 3 μM) [95].
A different approach for the in silico identification of potential drugs is the rational design of inhibitor molecules. Structural data from prefusion dengue virus E glycoprotein was used as a model for the design of new DENV entry peptide inhibitors [96]. The residue-specific all-atom probability discriminatory function (RAPDF) score was used for peptide design, allowing the identification of amino acid sequences that are likely to have structural and binding stability [96]. Two peptides were identified with this approach, 1OAN1 and DN57opt, with inhibitory activity against DENV-2 in LLC-MK2 cells (IC50: 7 μM and 8 μM, resp.). Both peptides were shown to bind E protein causing structural changes on the surface of DENV-2 virions.
4.3. Other Possible Targets on E Glycoprotein
E glycoprotein domain III was identified as a putative receptor binding domain that also stabilizes the E glycoprotein structure. Besides being the main target for neutralizing antibodies, several studies have shown that soluble domain III by itself may act as an antiviral through diverse mechanisms. Soluble domain III acts as a dominant negative inhibitor of class II fusion proteins, possibly by interacting with a core trimer intermediate and interfering with the folding back of the lipid bilayers [46]. Purified domain III from DENV-1 and DENV-2 inhibits entry of these serotypes into human HepG2 and mosquito C6/36 cells [97]. Also, a fusion protein containing maltose binding protein sequences, used as a solubility tag, and DENV-2 domain III showed antiviral activity in insect and human cells (IC50: 10 μM and 13 μM, resp.), probably due to competition for cellular receptors [45]. Further emphasizing the viability of domain III as a target, recent studies showed that small peptides designed from this region were active against DENV-2 in LLC-MK2 cells (IC50: 35 μM) [98].
Similarly, cellular receptor molecules that interact with domain III may also have antiviral activity. One such molecule, the sulfated polysaccharide fucoidan, blocks DENV-2 virus entry into target cells by binding to E glycoprotein domain III and competing with the cellular receptors (IC50: 4.7 μg/mL) [99]. The high molecular weight compound curdlan sulfate (CRDS) is a sulfated polysaccharide with branched β-d-(1→3) glucan backbone and piperidine-N-sulfonic acid groups. It was first identified as an inhibitor of HIV-1 entry and propagation [100]. Ichiyama and colleagues showed that CRDS binds E glycoprotein, probably near the fusion loop, and strongly and selectively inhibits DENV-2 in LLC-MK2 cells (EC50: 7 μg/mL) [101]. DENV-2 and DENV-3 were effectively inhibited, while DENV-1 and DENV-4 required a much higher concentration of the compound. The authors proposed that CRDS interacts with residues of the kl hairpin, obstructing the shifting motion required for the dimer-trimer transition.
Some studies have shown that carbohydrate binding agents (CBAs) possess antiviral activities against DENV, because its entry into human target cells is carbohydrate-dependent (Table 1). CBAs also inhibit the DC-SIGN independent entry pathway of DENV in monocyte derived dendritic cells, interrupting the interaction between DENV and DC-SIGN [73]. It was reported that plant lectins, which are an important source of CBAs, are also able to interact with DENV E glycoprotein. Several plant derived CBAs, such as the lectins HHA (Hippeastrum hybrid), GNA (Galanthus nivalis), and UDA (Urtica dioica), showed strong antiviral activity against all serotypes of DENV (lowest EC50: 4.6 nM, 3.8 nM, and 0.29 μM, resp.) [73]. Pradimicin-S, a small soluble nonpeptidic CBA, was also demonstrated to have an antiviral effect against DENV-2 in monocyte derived dendritic cells (EC50: 19 μM) [73]. This molecule is attractive because it may have better bioavailability than other CBAs. Evidence showed that the CBAs act at an early step of infection, most likely by binding mannose bearing E glycoprotein and blocking viral attachment.
A recent study has shown that the GAGs heparin and chondroitin sulfate E inhibited DENV in BHK cells, unlike the parent compounds chondroitin sulfate A and C (lowest EC50: 0.3 μg/mL) [81]. Heparin and chondroitin sulfate E inhibit the viral infection by directly interacting with E protein and blocking entry. On the other hand, it was found that sulfated polysaccharides isolated from different types of algae compete with dengue virus during interaction with target cell membrane components showing a significant antiviral activity for all DENV serotypes in Vero cells (IC50: 0.12–20 μg/mL) [102]. These compounds included sulfated galactan, fucan, and xylomannan-containing fractions from red seaweed and were particularly effective against DENV-2.
Other heparan sulfate-like molecules also interfere with the entry and multiplication of DENV. Carrageenans are molecules formed by linear chains of alternate (1–3)-β-galactopyranose and (1–4)-α-D-galactopyranose. The iota-carrageenan shows an inhibition of DENV-2 multiplication in infected Vero cells (EC50: 0.4 μg/mL) [78, 103], probably due to interference with the binding of virions to the HS receptor. Also, a sulfated K5 polysaccharide from Escherichia coli inhibits dengue infection in human dermal microvascular endothelial cells (EC50: 111 nM). This molecule interacts with E protein domain III, competing with heparan sulfate proteoglycans [104]. Lee et al. tested several HS-mimetics, suramin, pentosan polysulfate, and PI-88 showing relevant inhibition of virus attachment/entry against DENV, JEV, WNV, and MVEV. However, only PI-88 was active in an in vivo murine model of dengue viremia [105]. Two sulfated galactomannans extracted from the plants M. scabrella (BRS) and L. leucocephala (LLS) were found to be active against YFV and DENV-1, LLS being about ten times more active in C6/36 [106].
More recently, LCTA-949, an aglycon derivative molecule from the teicoplanin antibiotic, was found to possess antiviral activity against flaviviruses, including DENV-2 [90]. The antiviral activity of this molecule was evaluated using cytopathic effect (CPE) reduction assays, showing potent activity against several flaviviruses, including DENV-2 in Vero cells (EC50: 6.9 μM). It was reported that this molecule targets an early stage of infections and was able to reduce antibody-dependent enhancement (ADE) in vitro. In fact, in vitro inhibition of ADE has also been reported for several antiviral compounds [82, 101, 107].
In a recent study, Lin et al. tested two hydrolysable tannins, chebulagic acid and punicalagin, as possible dengue virus inhibitors [108]. The two compounds showed antiviral activity against several viruses including DENV-2 in Vero cells (EC50: 13.1 and 7.8 μM, resp.). It was suggested that these molecules can not only directly inactivate viral particles before attachment but also interfere with the entry process by binding the viral E glycoprotein and blocking interaction with GAGs.
The antiviral activity of sea grass derived natural product, zosteric acid, and several synthetic derivatives was tested by Rees and colleagues [110]. The derivative molecule CF 238 showed significant inhibition against all DENV serotypes in LLC-MK2 cells (range IC50: 14–47 μM). Notably, although the compound interferes with an early step during entry, it seems also to enhance the binding of the virions to the cell surface.
The E glycoprotein is the natural target of the humoral response against the virus. As naturally elicited antibodies do not provide protection across different serotypes, other approaches have been tried such as monoclonal antibodies (mAbs) and engineered derivatives like antibody fragments as another strategy for the blocking of dengue virus entry. It has identified three main epitope regions within dengue virus E glycoprotein: A (located at domain II), B (domain III), and C (domain I). The domains A and B epitopes allowed the generation of monoclonal antibodies with higher biological activity against dengue virus. Antibodies that were reactive to domain A epitopes neutralize the virus and may block virus cell membrane fusion. The domain B epitopes elicited mAbs that were potent neutralizers of virus infectivity and blocked hemagglutination; however, they did not block virus mediated cell membrane fusion [111].
Most anti-DENV mAbs neutralize at least in part, by disrupting the virus ability to bind to mammalian cellular receptors. The mAb 1A1D-2 recognizes a partially hidden epitope (antigenic domain B, E domain III) and inhibits dengue virus attachment. The possible inhibition mechanisms could be due to altering the distances between the glycans present in the E glycoprotein and impairing the binding to the DC-SIGN receptor [112]. Domain III is the E glycoprotein domain that elicits the strongest inhibiting mAb [113]. Shrestha et al. have found that there are differences in the neutralizing activity and protective potential of mAbs generated against several DENV-1 genotypes. However, they were able to identify two mAbs able to strongly bind and neutralize all genotypes [114]. Although promising, the potential use of these antibodies as therapeutic agents must take into account the possibility of generating ADE due to suboptimal neutralization in some patients. To tackle this limitation, some authors have engineered the antibodies to have deletions in the Fc region, avoiding the binding to FcγR [115].
5. Targeting Other Viral and Cellular Factors
Compound ST-148 was identified by using a high throughput screening for molecules able to block dengue cytopathic effect [109]. Notably, the compound was very potent against all dengue serotypes (range EC50: 0.016–2.8 μM) and against several other flaviviruses. The ST-149 binds to the virion capsid protein, being active also in a murine model of dengue viremia.
High levels of viremia correlate with severe dengue disease [116]; therefore, reducing uncontrolled virus production should be a priority of antiviral therapeutic interventions. Several research groups have looked into the possibility of using RNA interference (RNAi) technology to shut down key components of the entry machinery of the virus. In one study, silencing the expression of CD14 associated protein, clathrin heavy polypeptide, and dynamin 2 in human monocytes produced a significant reduction of infected cells and virus production [117]. Following the same logic of simultaneously inhibiting receptor- and clathrin-dependent entry pathways, this group also reported silencing the expression of GRP78, clathrin heavy polypeptide, and dynamin 2 in human hepatocytes [118]. These silenced genes allowed an important reduction of intra- and extracellular viral load and the number of infected cells. Although promising, the use of RNAi as therapeutic agent is still at the experimental stage, with many technical issues to resolve before becoming a real antiviral alternative. It is likely, for example, that silencing one or several host genes to reduce viral yield may also induce important off-target effects. Nevertheless, RNAi technology is still an invaluable analytical tool to understand the molecular biology of the virus and to identify new therapeutic targets.
6. Concluding Remarks
An extensive body of experimental evidence indicates that the DENV, while having a relatively simple structural organization, is able to exploit numerous molecules and pathways to infect cells and evade the immune response. Unfortunately, the virus is also able to cause severe, often deadly disease in a fraction of infected individuals. Therefore, the search for antiviral drugs is a top priority, as development of vaccines will also take some time to achieve a potent and balanced response against the four dengue serotypes.
Inhibiting the entry of the dengue virus to avoid the infection is an attractive approach to develop potent and specific antivirals. These molecules will exert their effects without having to enter the cells, thereby avoiding strict structural and chemical constraints. This kind of antiviral agents will have the added advantage of potentially limiting the immune system hyperactivation that leads to severe dengue.
Dengue viremia is acute during the first days of infection, decaying rapidly after 4-5 days [119]. Therefore, an antiviral drug could have a short early therapeutic window to fight the virus. However, as development of severe dengue is associated with uncontrolled viremia, the use of an antiviral may help stop further progression into this serious stage. To that purpose, early diagnosis may be crucial. Although an early diagnosis is not an easy task in routine clinical settings, considerable research has been done to incorporate early and robust biomarkers to the diagnosis, such as chemokines IP-10 and I-TAC expression in peripheral blood [120].
In general, antiviral development is focused on targeting viral components or cellular determinants of the infection. Targeting viral components has yielded the highest number of candidates, yet this strategy is prone to the rapid development of resistance. On the other hand, targeting cellular components reduces the possibility of developing resistance but may have a higher incidence of adverse effects. This approach may take into account cellular factors required for viral replication and maturation or pathogenic processes.
The growing body of knowledge about the biology of the virus tends to favor the approach of targeting the viral determinants of the entry process, instead of trying to block all possible cellular receptors and attachment molecules. Particularly promising as a target is the E glycoprotein because of its key role in viral attachment through receptor interaction and subsequent endosomal fusion. Several specific targets within the E glycoprotein are promising, including the stem domain, the hydrophobic pocket, and the domain III.
Another important challenge for the development of antivirals against DENV is the issue of bioavailability of the drugs. Particular drug families may have different chemical and biological properties that may dictate their fate in the organism. However, most of the molecules described here are small compounds in the proof of concept stage of research. Promising compounds may then soon move into preclinical studies where specific questions about in vivo efficacy and bioavailability are addressed.
Particularly useful for rapid development of antivirals are the animal models for viral diseases. In the past few years, there have been great efforts in the development of animal models for dengue virus infections. Currently, there are several dengue disease animal models. The AG129 mice deficient in type I and type II interferon receptors have been used to study dengue induced ADE (reviewed in [121]). This animal model was recently used to test the efficacy of an α-glucosides I and II inhibitor and an iminosugar as possible antiviral treatments [122, 123]. Another dengue animal model has been established by transplanting human CD34+ cells into NON/SCID (nonobese diabetic/severe combined immunodeficient) mice, showing clinical signs resembling human dengue fever disease [124, 125]. A recent investigation established a dengue animal model using a humanized NOD/SCID/interleukin 2 receptor gamma mouse strain. This animal model allows the evaluation human infection after the inoculation of dengue virus by infected mosquitoes [126]. However, the lack of an appropriate and convenient animal model for dengue infection is an important weakness for the rapid development and implementation of new antivirals [121].
Acknowledgments
The authors deeply thank Dr. Gabrielle Britton and Dr. Elaine Reynolds for reviewing the paper and Miss Rita Marissa Giovani for assistance in the preparation of Figure 1.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.World Health Organization. Treatment, prevention and control global strategy for dengue prevention and control. 2012.
- 2.Murray NEA, Quam MB, Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clinical Epidemiology. 2013;5(1):299–309. doi: 10.2147/CLEP.S34440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caminade C, Medlock JM, Ducheyne E, et al. Suitability of European climate for the Asian tiger mosquito Aedes albopictus: recent trends and future scenarios. Journal of the Royal Society Interface. 2012;9(75):2708–2717. doi: 10.1098/rsif.2012.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady OJ, Gething PW, Bhatt S, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Neglected Tropical Diseases. 2012;6(8) doi: 10.1371/journal.pntd.0001760.e1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. CDC Health Information for International Travel 2014. New York, NY, USA: Oxford University Press; 2014. Dengue—chapter 3—2014 Yellow Book|Travelers’ Health|CDC. [Google Scholar]
- 6.WHO. WHO: dengue and severe dengue. Fact Sheet No. 2012;(117) http://www.who.int/mediacentre/factsheets/fs117/en/
- 7.Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn RJ, Zhang W, Rossmann MG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindenbach B, Heinz-Jürgen T, Rice C. Flaviviridae: the viruses and their replication. In: Knipe D, Howley P, editors. Fields Virology. 5th edition. Philadelphia, Pa, USA: Lippincott-Raven; 2007. pp. 1103–1152. [Google Scholar]
- 10.Zhang X, Ge P, Yu X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nature Structural & Molecular Biology. 2012;20(1):105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein. Journal of Virology. 2003;77(12):7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(10):3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perera-Lecoin M, Meertens L, Carnec X, Amara A. Flavivirus entry receptors: an update. Viruses. 2014;6(1):69–88. doi: 10.3390/v6010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acosta EG, Castilla V, Damonte EB. Alternative infectious entry pathways for dengue virus serotypes into mammalian cells. Cellular Microbiology. 2009;11(10):1533–1549. doi: 10.1111/j.1462-5822.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hase T, Summers PL, Eckels KH. Flavivirus entry into cultured mosquito cells and human peripheral blood monocytes. Archives of Virology. 1989;104(1-2):129–143. doi: 10.1007/BF01313814. [DOI] [PubMed] [Google Scholar]
- 16.Lim HY, Ng ML. A different mode of entry by dengue-2 neutralisation escape mutant virus. Archives of Virology. 1999;144(5):989–995. doi: 10.1007/s007050050561. [DOI] [PubMed] [Google Scholar]
- 17.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhang W, Ogata S, et al. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12(9):1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Schaar HM, Rust MJ, Waarts B, et al. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. Journal of Virology. 2007;81(21):12019–12028. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui F, Raymond M, Qiao C-L. Insecticide resistance in vector mosquitoes in China. Pest Management Science. 2006;62(11):1013–1022. doi: 10.1002/ps.1288. [DOI] [PubMed] [Google Scholar]
- 21.Thomas DD, Donnelly CA, Wood RJ, Alphey LS. Insect population control using a dominant, repressible, lethal genetic system. Science. 2000;287(5462):2474–2476. doi: 10.1126/science.287.5462.2474. [DOI] [PubMed] [Google Scholar]
- 22.Alphey L, Andreasen M. Dominant lethality and insect population control. Molecular and Biochemical Parasitology. 2002;121(2):173–178. doi: 10.1016/s0166-6851(02)00040-3. [DOI] [PubMed] [Google Scholar]
- 23.Phuc H, Andreasen MH, Burton RS, et al. Late-acting dominant lethal genetic systems and mosquito control. BMC Biology. 2007;5, article 11 doi: 10.1186/1741-7007-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takada A, Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Reviews in Medical Virology. 2003;13(6):387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- 25.Kliks SC, Nisalak A, Brandt WE, Wahl L, Burke DS. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. The American Journal of Tropical Medicine and Hygiene. 1989;40(4):444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 26.Rigau-Pérez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue haemorrhagic fever. The Lancet. 1998;352(9132):971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 27.Halstead SB. Dengue. The Lancet. 2007;370(9599):1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 28.Mairuhu ATA, Wagenaar J, Brandjes DPM, Van Gorp ECM. Dengue: an arthropod-borne disease of global importance. European Journal of Clinical Microbiology and Infectious Diseases. 2004;23(6):425–433. doi: 10.1007/s10096-004-1145-1. [DOI] [PubMed] [Google Scholar]
- 29.Martina BEE, Koraka P, Osterhaus ADME. Dengue virus pathogenesis: an integrated view. Clinical Microbiology Reviews. 2009;22(4):564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugach P, Ketas TJ, Michael E, Moore JP. Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology. 2008;377(2):401–407. doi: 10.1016/j.virol.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lalezari JP, Henry K, O'Hearn M, et al. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. The New England Journal of Medicine. 2003;348(22):2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 32.Volz T, Allweiss L, Barek MB, et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. Journal of Hepatology. 2013;58(5):861–867. doi: 10.1016/j.jhep.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Bressanelli S, Stiasny K, Allison SL, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. The EMBO Journal. 2004;23(4):728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Chipman PR, Corver J, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nature Structural Biology. 2003;10(11):907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(12):6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. Journal of Virology. 2001;75(9):4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. Journal of Virology. 1999;73(7):5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung J, Hsieh M, Young M, Kao C, King C, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. Journal of Virology. 2004;78(1):378–388. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volk DE, Lee Y-C, Li X, et al. Solution structure of the envelope protein domain III of dengue-4 virus. Virology. 2007;364(1):147–154. doi: 10.1016/j.virol.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang K, Lee M, Wu C, Lei H, Cheng J. Solution structure and neutralizing antibody binding studies of domain III of the dengue-2 virus envelope protein. Proteins: Structure, Function and Genetics. 2008;70(3):1116–1119. doi: 10.1002/prot.21806. [DOI] [PubMed] [Google Scholar]
- 42.Soares ROS, Caliri A. Stereochemical features of the envelope protein Domain III of dengue virus reveals putative antigenic site in the five-fold symmetry axis. Biochimica et Biophysica Acta: Proteins and Proteomics. 2013;1834(1):221–230. doi: 10.1016/j.bbapap.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 43.de Wispelaere M, Yang PL. Mutagenesis of the DI/DIII linker in dengue virus envelope protein impairs viral particle assembly. Journal of Virology. 2012;86(13):7072–7083. doi: 10.1128/JVI.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butrapet S, Childers T, Moss KJ, et al. Amino acid changes within the E protein hinge region that affect dengue virus type 2 infectivity and fusion. Virology. 2011;413(1):118–127. doi: 10.1016/j.virol.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 45.Watterson D, Kobe B, Young PR. Residues in domain III of the dengue virus envelope glycoprotein involved in cell-surface glycosaminoglycan binding. Journal of General Virology. 2012;93(1):72–82. doi: 10.1099/vir.0.037317-0. [DOI] [PubMed] [Google Scholar]
- 46.Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. Journal of Cell Biology. 2005;171(1):111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. Journal of Virology. 1995;69(2):695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. Journal of Virology. 1996;70(11):8142–8147. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Schaar HM, Rust MJ, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathogens. 2008;4(12) doi: 10.1371/journal.ppat.1000244.e1000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. Journal of Cell Biology. 2008;183(2):353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. Journal of Virology. 2009;83(9):4338–4344. doi: 10.1128/JVI.02574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mondotte JA, Lozach P-Y, Amara A, Gamarnik AV. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. Journal of Virology. 2007;81(13):7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. The Journal of Experimental Medicine. 2003;197(7):823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro-Sanchez E, Altmeyer R, Amara A, et al. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Reports. 2003;4(7):723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Wang S, King C. Bacterial lipopolysaccharide inhibits dengue virus infection of primary human monocytes/macrophages by blockade of virus entry via a CD14-dependent mechanism. Journal of Virology. 1999;73(4):2650–2657. doi: 10.1128/jvi.73.4.2650-2657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JL, de Wet BJM, Martinez-Pomares L, et al. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathogens. 2008;4(2, article e17) doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reyes-del Valle J, Chávez-Salinas S, Medina F, del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. Journal of Virology. 2005;79(8):4557–4567. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jindadamrongwech S, Thepparit C, Smith DR. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Archives of Virology. 2004;149(5):915–927. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- 59.Thepparit C, Smith DR. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. Journal of Virology. 2004;78(22):12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meertens L, Carnec X, Lecoin MP, et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host and Microbe. 2012;12(4):544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Wang J, Gao N, Chen Z, Tian Y, An J. Up-regulated expression of β3 integrin induced by dengue virus serotype 2 infection associated with virus entry into human dermal microvascular endothelial cells. Biochemical and Biophysical Research Communications. 2007;356(3):763–768. doi: 10.1016/j.bbrc.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Kakinami C, Li Q, Yang B, Li H. Human apolipoprotein A-I is associated with dengue virus and enhances virus infection through SR-BI. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0070390.e70390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao F, Duan X, Lu X, et al. Novel binding between pre-membrane protein and claudin-1 is required for efficient dengue virus entry. Biochemical and Biophysical Research Communications. 2010;391(1):952–957. doi: 10.1016/j.bbrc.2009.11.172. [DOI] [PubMed] [Google Scholar]
- 64.Che P, Tang H, Li Q. The interaction between claudin-1 and dengue viral prM/M protein for its entry. Virology. 2013;446(1-2):303–313. doi: 10.1016/j.virol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 65.Hershkovitz O, Rosental B, Rosenberg LA, et al. NKp44 receptor mediates interaction of the envelope glycoproteins from the West Nile and dengue viruses with NK cells. The Journal of Immunology. 2009;183(4):2610–2621. doi: 10.4049/jimmunol.0802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Jesús Martínez-Barragán J, del Angel RM. Identification of a putative coreceptor on Vero cells that participates in dengue 4 virus infection. Journal of Virology. 2001;75(17):7818–7827. doi: 10.1128/JVI.75.17.7818-7827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geijtenbeek TBH, Van Kooyk Y. Pathogens target DC-SIGN to influence their fate: DC-SIGN functions as a pathogen receptor with broad specificity. APMIS. 2003;111(7-8):698–714. doi: 10.1034/j.1600-0463.2003.11107803.x. [DOI] [PubMed] [Google Scholar]
- 68.Geijtenbeek TBH, Kwon DS, Torensma R, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 69.Alvarez CP, Lasala F, Carrillo J, Muñiz O, Corbí AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola Virus in cis and in trans. Journal of Virology. 2002;76(13):6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halary F, Amara A, Lortat-Jacob H, et al. Human Cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity. 2002;17(5):653–664. doi: 10.1016/s1074-7613(02)00447-8. [DOI] [PubMed] [Google Scholar]
- 71.Lozach P-Y, Burleigh L, Staropoli I, et al. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. The Journal of Biological Chemistry. 2005;280(25):23698–23708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- 72.Shah M, Wadood A, Rahman Z, Husnain T. Interaction and inhibition of dengue envelope glycoprotein with mammalian receptor DC-Sign, an in-silico approach. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0059211.e59211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alen MMF, de Burghgraeve T, Kaptein SJF, Balzarini J, Neyts J, Schols D. Broad Antiviral activity of Carbohydrate-binding agents against the four serotypes of dengue virus in monocyte-derived dendritic cells. PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0021658.e21658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annual Review of Biochemistry. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 75.Chen Y, Maguire T, Hileman RE, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nature Medicine. 1997;3(8):866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 76.Hilgard P, Stockert R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology. 2000;32(5):1069–1077. doi: 10.1053/jhep.2000.18713. [DOI] [PubMed] [Google Scholar]
- 77.Hung SL, Lee PL, Chen HW, Chen LK, Kao CC, King C. Analysis of the steps involved in dengue virus entry into host cells. Virology. 1999;257(1):156–167. doi: 10.1006/viro.1999.9633. [DOI] [PubMed] [Google Scholar]
- 78.Talarico LB, Noseda MD, Ducatti DRB, Duarte MER, Damonte EB. Differential inhibition of dengue virus infection in mammalian and mosquito cells by iota-carrageenan. Journal of General Virology. 2011;92(6):1332–1342. doi: 10.1099/vir.0.028522-0. [DOI] [PubMed] [Google Scholar]
- 79.Boonnak K, Slike BM, Burgess TH, et al. Role of dendritic cells in antibody-dependent enhancement of dengue virus infection. Journal of Virology. 2008;82(8):3939–3951. doi: 10.1128/JVI.02484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhattacharyya S, Zagórska A, Lew ED, et al. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host and Microbe. 2013;14(2):136–147. doi: 10.1016/j.chom.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kato D, Era S, Watanabe I, et al. Antiviral activity of chondroitin sulphate E targeting dengue virus envelope protein. Antiviral Research. 2010;88(2):236–243. doi: 10.1016/j.antiviral.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 82.Nicholson CO, Costin JM, Rowe DK, et al. Viral entry inhibitors block dengue antibody-dependent enhancement in vitro. Antiviral Research. 2011;89(1):71–74. doi: 10.1016/j.antiviral.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 83.Lin S, Zou G, Hsieh S, et al. The helical domains of the stem region of dengue virus envelope protein are involved in both virus assembly and entry. Journal of Virology. 2011;85(10):5159–5171. doi: 10.1128/JVI.02099-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virology Journal. 2005;2(article 49) doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lok S, Costin JM, Hrobowski YM, et al. Release of dengue virus genome induced by a peptide inhibitor. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0050995.e50995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of flavivirus entry derived from the E protein stem. Journal of Virology. 2010;84(24):12549–12554. doi: 10.1128/JVI.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt AG, Lee K, Yang PL, Harrison SC. Small-molecule inhibitors of dengue-virus entry. PLoS Pathogens. 2012;8(4) doi: 10.1371/journal.ppat.1002627.e1002627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang J-M, Chen Y-F, Tu Y-Y, Yen K-R, Yang Y-L. Combinatorial computational approaches to identify tetracycline derivatives as flavivirus inhibitors. PLoS ONE. 2007;2(5, article e428) doi: 10.1371/journal.pone.0000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaptein SJF, De Burghgraeve T, Froeyen M, et al. A derivate of the antibiotic doxorubicin is a selective inhibitor of dengue and yellow fever virus replication in vitro. Antimicrobial Agents and Chemotherapy. 2010;54(12):5269–5280. doi: 10.1128/AAC.00686-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Burghgraeve T, Kaptein SJF, Ayala-Nunez NV, et al. An analogue of the antibiotic teicoplanin prevents flavivirus entry in vitro. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0037244.e37244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Q-Y, Patel SJ, Vangrevelinghe E, et al. A small-molecule dengue virus entry inhibitor. Antimicrobial Agents and Chemotherapy. 2009;53(5):1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poh MK, Yip A, Zhang S, et al. A small molecule fusion inhibitor of dengue virus. Antiviral Research. 2009;84(3):260–266. doi: 10.1016/j.antiviral.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 93.Kampmann T, Yennamalli R, Campbell P, et al. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antiviral Research. 2009;84(3):234–241. doi: 10.1016/j.antiviral.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 94.Li Z, Khaliq M, Zhou Z, Post CB, Kuhn RJ, Cushman M. Design, synthesis, and biological evaluation of antiviral agents targeting flavivirus envelope proteins. Journal of Medicinal Chemistry. 2008;51(15):4660–4671. doi: 10.1021/jm800412d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou Z, Khaliq M, Suk J, et al. Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein. ACS Chemical Biology. 2008;3(12):765–775. doi: 10.1021/cb800176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Costin JM, Jenwitheesuk E, Lok S, et al. Structural optimization and de novo design of dengue virus entry inhibitory peptides. PLoS Neglected Tropical Diseases. 2010;4(6, article e721) doi: 10.1371/journal.pntd.0000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chin JFL, Chu JJH, Ng ML. The envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microbes and Infection. 2007;9(1):1–6. doi: 10.1016/j.micinf.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 98.Alhoot MA, Rathinam AK, Wang SM, Manikam R, Sekaran SD. Inhibition of dengue virus entry into target cells using synthetic antiviral peptides. International Journal of Medical Sciences. 2013;10(6):719–729. doi: 10.7150/ijms.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hidari KIPJ, Takahashi N, Arihara M, Nagaoka M, Morita K, Suzuki T. Structure and anti-dengue virus activity of sulfated polysaccharide from a marine alga. Biochemical and Biophysical Research Communications. 2008;376(1):91–95. doi: 10.1016/j.bbrc.2008.08.100. [DOI] [PubMed] [Google Scholar]
- 100.Kaneko Y, Yoshida O, Nakagawa R, et al. Inhibition of HIV-1 infectivity with curdlan sulfate in vitro . Biochemical Pharmacology. 1990;39(4):793–797. doi: 10.1016/0006-2952(90)90161-d. [DOI] [PubMed] [Google Scholar]
- 101.Ichiyama K, Gopala Reddy SB, Zhang LF, et al. Sulfated polysaccharide, curdlan sulfate, efficiently prevents entry/fusion and restricts antibody-dependent enhancement of dengue virus infection in vitro: a possible candidate for clinical application. PLoS Neglected Tropical Diseases. 2013;7(4) doi: 10.1371/journal.pntd.0002188.e2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pujol CA, Ray S, Ray B, Damonte EB. Antiviral activity against dengue virus of diverse classes of algal sulfated polysaccharides. International Journal of Biological Macromolecules. 2012;51(4):412–416. doi: 10.1016/j.ijbiomac.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 103.Talarico LB, Pujol CA, Zibetti RGM, et al. The antiviral activity of sulfated polysaccharides against dengue virus is dependent on virus serotype and host cell. Antiviral Research. 2005;66(2-3):103–110. doi: 10.1016/j.antiviral.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 104.Vervaeke P, Alen M, Noppen S, Schols D, Oreste P, Liekens S. Sulfated escherichia coli K5 polysaccharide derivatives inhibit dengue virus infection of human microvascular endothelial cells by interacting with the viral envelope protein E domain III. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0074035.e74035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee E, Pavy M, Young N, Freeman C, Lobigs M. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antiviral Research. 2006;69(1):31–38. doi: 10.1016/j.antiviral.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Ono L, Wollinger W, Rocco IM, Coimbra TLM, Gorin PAJ, Sierakowski M. In vitro and in vivo antiviral properties of sulfated galactomannans against yellow fever virus (BeH111 strain) and dengue 1 virus (Hawaii strain) Antiviral Research. 2003;60(3):201–208. doi: 10.1016/s0166-3542(03)00175-x. [DOI] [PubMed] [Google Scholar]
- 107.Ayala-Nuñez NV, Jarupathirun P, Kaptein SJF, Neyts J, Smit JM. Antibody-dependent enhancement of dengue virus infection is inhibited by SA-17, a doxorubicin derivative. Antiviral Research. 2013;100(1):238–245. doi: 10.1016/j.antiviral.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 108.Lin L-T, Chen T-Y, Lin S-C, et al. Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiology. 2013;13(1, article 187) doi: 10.1186/1471-2180-13-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Byrd CM, Dai D, Grosenbach DW, et al. A novel inhibitor of dengue virus replication that targets the capsid protein. Antimicrobial Agents and Chemotherapy. 2013;57(1):15–25. doi: 10.1128/AAC.01429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rees CR, Costin JM, Fink RC, et al. In vitro inhibition of dengue virus entry by p-sulfoxy-cinnamic acid and structurally related combinatorial chemistries. Antiviral Research. 2008;80(2):135–142. doi: 10.1016/j.antiviral.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 111.Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246(2):317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- 112.Lok S, Kostyuchenko V, Nybakken GE, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nature Structural and Molecular Biology. 2008;15(3):312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 113.Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to vero cells. Journal of Virology. 2001;75(16):7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shrestha B, Brien JD, Sukupolvi-Petty S, et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathogens. 2010;6(4):1–18. doi: 10.1371/journal.ppat.1000823.e1000823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goncalvez AP, Engle RE, St. Claire M, Purcell RH, Lai C. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(22):9422–9427. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang W-K, Chao D-Y, Kao C-L, et al. High levels of plasma dengue viral load during defervescence in patients with dengue hemorrhagic fever: implications for pathogenesis. Virology. 2003;305(2):330–338. doi: 10.1006/viro.2002.1704. [DOI] [PubMed] [Google Scholar]
- 117.Alhoot MA, Wang SM, Sekaran SD. Inhibition of dengue virus entry and multiplication into monocytes using RNA interference. PLoS Neglected Tropical Diseases. 2011;5(11) doi: 10.1371/journal.pntd.0001410.e1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alhoot MA, Wang SM, Sekaran SD. RNA interference mediated inhibition of dengue virus multiplication and entry in HepG2 cells. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0034060.e34060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Libraty DH, Endy TP, Houng H-SH, et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. Journal of Infectious Diseases. 2002;185(9):1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 120.Fink J, Gu F, Ling L, et al. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Neglected Tropical Diseases. 2007;1(2, article e86) doi: 10.1371/journal.pntd.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zompi S, Harris E. Animal models of dengue virus infection. Viruses. 2012;4(1):62–82. doi: 10.3390/v4010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watanabe S, Rathore APS, Sung C, et al. Dose- and schedule-dependent protective efficacy of celgosivir in a lethal mouse model for dengue virus infection informs dosing regimen for a proof of concept clinical trial. Antiviral Research. 2012;96(1):32–35. doi: 10.1016/j.antiviral.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 123.Perry ST, Buck MD, Plummer EM, et al. An iminosugar with potent inhibition of dengue virus infection in vivo . Antiviral Research. 2013;98(1):35–43. doi: 10.1016/j.antiviral.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 124.Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. Journal of Virology. 2005;79(21):13797–13799. doi: 10.1128/JVI.79.21.13797-13799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mota J, Rico-Hesse R. Humanized mice show clinical signs of dengue fever according to infecting virus genotype. Journal of Virology. 2009;83(17):8638–8645. doi: 10.1128/JVI.00581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cox J, Mota J, Sukupolvi-Petty S, Diamond MS, Rico-Hesse R. Mosquito bite delivery of dengue virus enhances immunogenicity and pathogenesis in humanized mice. Journal of Virology. 2012;86(14):7637–7649. doi: 10.1128/JVI.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


