Abstract
Down syndrome (DS), caused by trisomy of chromosome 21, is associated with immunological dysfunctions such as increased frequency of infections and autoimmune diseases. Patients with DS share clinical features, such as autoimmune manifestations and specific autoantibodies, with patients affected by autoimmune polyendocrine syndrome type 1. Autoimmune polyendocrine syndrome type 1 is caused by mutations in the autoimmune regulator (AIRE) gene, located on chromosome 21, which regulates the expression of tissue-restricted Ags (TRAs) in thymic epithelial cells. We investigated the expression of AIRE and TRAs in DS and control thymic tissue using quantitative PCR. AIRE mRNA levels were elevated in thymic tissue from DS patients, and trends toward increased expression of the AIRE-controlled genes INSULIN and CHRNA1 were found. Immunohistochemical stainings showed altered cell composition and architecture of the thymic medulla in DS individuals with increased frequencies of AIRE-positive medullary epithelial cells and CD11c-positive dendritic cells as well as enlarged Hassall’s corpuscles. In addition, we evaluated the proteomic profile of thymic exosomes in DS individuals and controls. DS exosomes carried a broader protein pool and also a larger pool of unique TRAs compared with control exosomes. In conclusion, the increased AIRE gene dose in DS could contribute to an autoimmune phenotype through multiple AIRE-mediated effects on homeostasis and function of thymic epithelial cells that affect thymic selection processes.
Introduction
Down syndrome (DS), or trisomy 21, is associated with an increased frequency of autoimmune disorders such as autoimmune hypothyroidism (1), hyperthyroidism (2), insulin-dependent diabetes mellitus (IDDM) (3), alopecia, and celiac disease (4). The mechanism underlying the high frequency of autoimmune manifestations in DS is not known. The human AIRE gene is located on chromosome 21 position q22.3 and codes for a transcription factor that in mice has been shown to control the expression of numerous TRAs in medullary thymic epithelial cells (mTEC) (5). The tissue-restricted Ag (TRA) expression is crucial for the negative selection of developing thymocytes in which self-reactive thymocytes with a high affinity for TRAs are eliminated within the thymus before they reach the periphery. Aire also has been shown to have other functions in mTECs such as controlling Ag presentation (6), chemokine production (7), and maturation (8–10). Thus, an altered expression of AIRE could have a complex influence on thymic selection including changes in the expression of TRAs, the presentation of TRAs, the movement of thymocytes, and the maturation of mTECs, possibly resulting in higher rates of autoimmune manifestations in DS patients. An additional link between DS and AIRE is that DS patients and patients with autoimmune polyendocrine syndrome type 1 (APS1), which is an autosomal recessive disease caused by mutations in AIRE, share specific autoantibodies. Autoantibodies against aromatic l-amino acid decarboxylase and cytochrome P4501A2 have not been found in other patient groups and are so far unique for DS and APS1 (11), except for aromatic l-amino acid decarboxylase autoantibodies found in a small subgroup of patients with isolated Addison’s disease (12). Previous investigations of DS thymic tissue have described a decrease in thymic size (13), altered thymocyte subpopulations (14), increased thymocyte depletion, markedly enlarged Hassall’s corpuscles (HCs) (15), decreased thymic output (16), and a decrease in the number of AIRE+ cells in the medullary region (17).
The thymic tissue also produce exosomes (18, 19), which are small membrane-bound vesicles that are released by various types of cells. Exosomes are important for intercellular communication and carry proteins, nucleic acids, and lipids (20). We have previously shown that exosomes containing TRAs are abundant in human thymic tissue (18), but exosomes from DS thymuses have previously not been studied.
On the basis of these observations, we analyzed thymic tissue from individuals with DS and controls without DS regarding the expression of AIRE/AIRE and selected TRAs, thymic cell subpopulations, and proteomic profile of thymic exosomes.
Materials and Methods
Collection of human thymic tissue
Human thymi removed during cardiac surgery of children 0–6 mo of age with DS and without DS at the Sahlgrenska University Hospital (Gothenburg, Sweden) were collected. The tissue was immediately put in 4% paraformaldehyde or RPMI 1640 medium (Invitrogen, Paisley, Scotland) on ice. Parents gave informed consent, and the study was approved by the local ethics committee (number 477-05, 2006-12-18). Thymic tissue for RNA extraction was frozen in 50-ml tubes with 50% RPMI 1640 medium (Invitrogen), 40% FBS, and 10% DMSO (both Sigma-Aldrich, St. Louis, MO) stepwise frozen from −20 to −85°C.
Thymic thawing RNA isolation and cDNA preparation
Thymic tissue was thawed in a 37°C water bath and RNA from each thymus was isolated from six different 20-mg pieces. The thymic tissue was pressed against a 100-μm mesh, followed by extensive washing with PBS to flush away thymocytes. To assess the enrichment of stromal cells in the pressed tissue, expression analysis of TEC markers FOXN1 and KERATIN 5 (K5) as well as the thymocyte marker CD3γ was performed using RT-quantitative PCR (qPCR) (see below). Remaining tissue was placed in RLT lysis buffer (1% 2-ME), followed by lysis in Qiagen TissueLyser (Qiagen, Hilden, Germany), two times for 2 min at 25 Hz. Samples were centrifuged to pellet debris, and RNA was isolated from the supernatant using RNeasy kit (Qiagen) on a Qiacube (Qiagen) with the following settings: RNeasy Mini, animal tissue and cells, standard, 50 μl eluate volume. cDNA was prepared from 1 μg RNA/sample with QuantiTect Rev. Transcription kit (Qiagen), according to the manufacturer’s instructions.
RT-qPCR
AIRE (Hs00230829_m1), INSULIN (Hs02741908_m1), GAPDH (Hs99999905_m1), THYROGLOBULIN (TG) (Hs00968042_m1), THYROID PEROXIDASE (TPO) (Hs00892519_m1), GAD1 (Hs01065893_m1), CHRNA1 (Hs00175578_m1), ATB4B (Hs01026288_m1), FOXN1 (Hs00186096_m1), K5 (Hs00361185_m1), CD3γ (Hs00962186_m1), and XCL1 (Hs00751481_s1) TaqMan assays 1 μl/reaction (Life Technologies, Foster City, CA), RNAse-free water 7 μl/reaction (Qiagen), and TaqMan Universal Master Mix II 10 μl/reaction (Life Technologies) were mixed and placed in a reaction plate, followed by 2 μl cDNA/20-μl reaction. Plates were run on a ViiA7 (Life Technologies) with the setting: Fast 96-well Block, Comparative CT, TaqMan Reagents, Standard. GAPDH was used as an endogenous control, and interplate variations were adjusted for using a calibrator sample. The samples were run in triplicates, and single wells with a divergent value resulting in a CT SD within triplicates exceeding 0.5 were omitted. For AIRE, INSULIN, TG, TPO, and GAD1 group comparisons, the median value from the six analyzed pieces was used. For the XCL1 group comparison, the six batches of cDNA were pooled prior to qPCR analysis. The expression of FOXN1, K5, and CD3γ in pressed versus untreated thymic tissues was investigated in four thymuses, from one piece of pressed tissue and on piece of untreated tissue per thymus.
Immunohistochemistry
Thymic tissue collected in the operating theater and directly submerged in 4% paraformaldehyde for 48 h were cut into smaller sections and embedded in paraffin. Four-micrometer-thick sections were placed on glass slides, followed by Ag retrieval in a 2100-Retriever (Electron Microscopy Sciences, Hatfield, PA) with Diva Decloaker and HotRinse (both Biocare Medical). Slides were rehydrated with TBS and blocked with serum-free protein block (DakoCytomation, Copenhagen, Denmark). The following primary anti-human Abs were added to different sections and incubated at 4°C overnight: AIRE (ab78065; Abcam, Cambridge, U.K.), FOXP3 (14-4777-82; eBioscience, San Diego, CA), INVOLUCRIN (IVL) (ab68; Abcam), CD11c (ab52632; Abcam), K5 (M7237; DakoCytomation), or caspase 3 (ab4051; Abcam). The slides were washed twice with TBS, followed by incubation with biotinylated secondary Ab for 30 min in room temperature and again washed twice with TBS. The slides were incubated 30 min at room temperature with streptavidin–alkaline phosphatase (BioLegend) and washed twice with TBS. Fast Red (Sigma-Aldrich) was added to the sections, and the reaction was stopped in dH2O. After two TBS washes, hematoxylin (Histolab) counterstaining was performed. Sections were mounted in aqueous mounting medium (DakoCytomation) and analyzed using a Lecia DMR microscope with ×10 or ×20 objective. The border between the thymic medulla and the cortex was identified, and stainings were quantified by counting positive cells or relative stained area within medullary regions, excluding HC areas, in a blinded fashion using Leica Qwin software.
Isolation of exosomes from thymic explants
Exosomes were isolated as previously described (18) from two DS thymi and two control thymi. Briefly, 1–2 g thymic tissue from each patient was cut into small pieces and incubated in RPMI 1640 medium (Invitrogen) with 5% exosome-depleted FBS (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), and penicillin/streptomycin (Sigma-Aldrich) for 8 h. The cultures were centrifuged for 10 min at 850 × g, and the supernatants were collected and centrifuged for another 15 min at 3000 × g to remove cell debris. Furthermore, the supernatants were spun for 30 min at 10,000 × g, followed by filtration through 0.2-μm filter. Finally the supernatants were ultracentrifuged for 70 min at 100,000 × g to pellet the exosomes. The pellets were washed in PBS and repelleted by an additional 100,000 × g centrifugation. Exosome concentration was determined with the Bradford protein concentration assay (Bio-Rad, Hercules, CA).
Proteomic analysis of isolated exosomes
Isolated exosomes, 50 μg/condition, were analyzed by one-dimensional SDS-PAGE (4–12% Bis-Tris Novex mini-gel; Invitrogen) and visualized using Coomassie blue staining (Novex; Invitrogen). The full gel lanes were excised and divided into equal slices and subjected to in-gel protein digestion with trypsin overnight at 37°C (21). Extracted peptides were dried under vacuum, reconstituted in 15 μl 0.2% formic acid, and 2-μl sample injections were made with an HTC-PAL autosampler (CTC Analytics, Zwingen, Switzerland) connected to an Agilent 1200 binary pump (Agilent Technologies, Palo Alto, CA). The peptides were trapped on a precolumn (45 × 0.075 mm i.d.) and separated on a 200 × 0.050-mm analytical column packed with 4-μm Reprosil-Pur C18-AQ particles (Dr. Maisch, Ammerbuch, Germany). Peptides were loaded in 0.2% formic acid and separated using a 40-min gradient 5–35% toward mobile phase B (acetonitrile). Mass spectrometry (MS) analysis was performed on an LTQ-Orbitrap operated in a data-dependent mode automatically switching between MS and MS/MS mode. Full MS scans were acquired in the orbitrap (from m/z 400 to 2000) with a resolution of 60,000 at m/z 400. The top six most intense double or triple protonated ions were selected for fragmentation in the linear ion trap using collision-induced dissociation fragmentation and afterward excluded for selection for 60 s. Spectral data were searched using MASCOT (version 2.3; Matrix Science) against the SwissProt database (release 2011_04, 20075 entries). Search parameters were set to taxonomy Human, MS mass tolerance 5 ppm, 0.5 Da MS/MS mass tolerance, trypsin allowing one missed cleavage, fixed modification of propionamide on cysteine and as variable modifications oxidized methionine and acetylation at protein N-terminal. The threshold for protein identification was set to <1% false discovery rate for both peptide and protein identification based on a minimum of one unique peptide. Overlap in protein identifications between the samples was determined by Venn diagrams generated using the gplots package in R (www.R-project.org). Expression of the identified proteins for 80 different tissue types was extracted from the human protein atlas (HPA) version 13 (22) when available, converted to numerical values (0–3 for negative, weak, moderate and strong expression respectively and missing observations as blanks), and evaluated by hierarchical cluster analysis, using “Euclidean distance” as similarity metric combined with complete linkage clustering. A TRA was defined as expressed in five tissue cell types or less according to the HPA. Known thymocyte proteins and proteins with an immune expression profile (bone marrow, lymph node, tonsil, or spleen expression) in the HPA were not considered TRAs.
Statistics
Values are presented as median with interquartile range. Data were statistically evaluated using an unpaired Mann–Whitney U test and a Spearman rank correlation coefficient. Values of p ≤ 0.05 were considered as statistically significant. All statistic tests were performed using GraphPad Prism software.
Results
AIRE, INSULIN, and CHRNA1 are overexpressed in DS thymic tissue
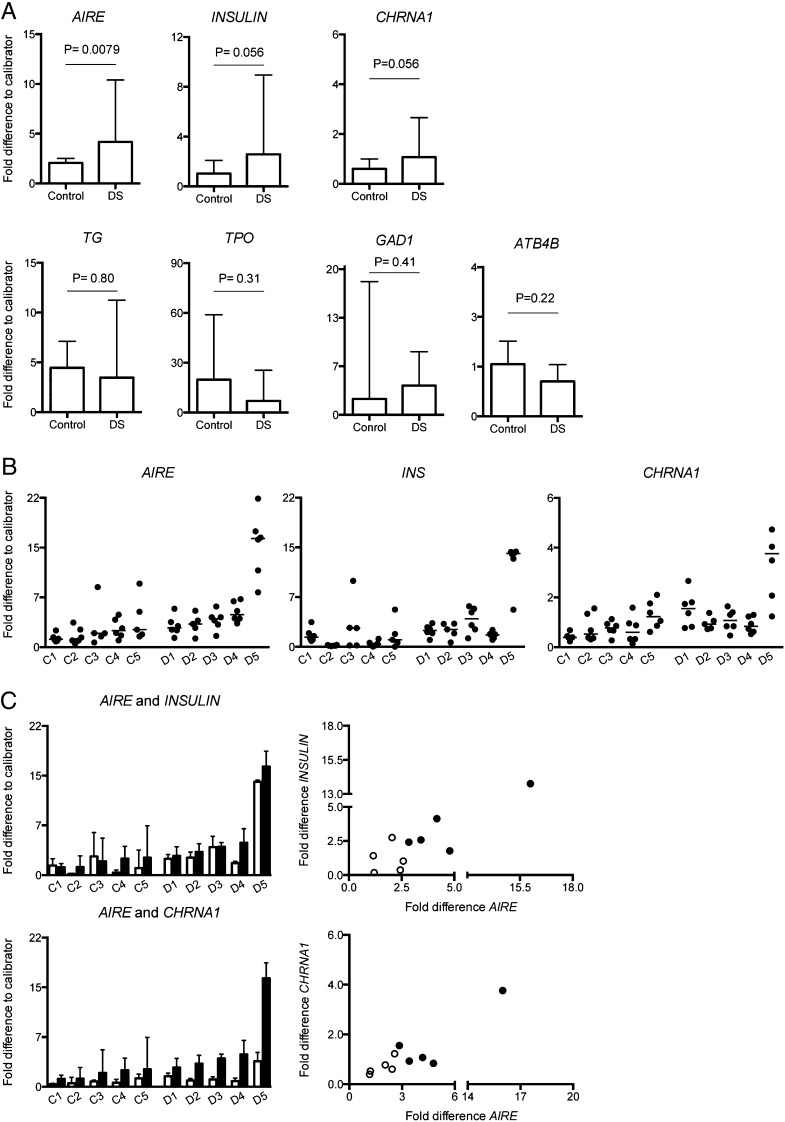
To assess the expression of AIRE and TRAs in the thymic tissue, we pressed and washed the thymi to remove thymocytes and thus enrich the thymic stroma. AIRE expression was significantly increased in DS patients compared with controls (p = 0.0079) (Fig. 1A). Trends toward increased expression also were found for INSULIN (p = 0.056) and CHRNA1 (p = 0.056) in DS patients compared with controls (Fig. 1A). For AIRE, INSULIN, and CHRNA1 mRNAs, the piece specific expression is presented in (Fig. 1B). No differences were found between the groups in expression of TG (p = 0.80), TPO (p = 0.31), GAD1 (p = 0.41), or ATB4B (p = 0.22) (Fig. 1A). XCL1 expression did not differ between the two groups (data not shown); however, the expression in the DS group was highly variable in comparison with the homogeneous control group. An overall positive correlation between the expression of AIRE and INSULIN (r = 0.64, p = 0.054) was found as well as between AIRE and CHRNA1 (r = 0.77, p = 0.0126) (Fig. 1C). Presence and enrichment of TECs in pressed thymic tissue was confirmed because the expression of epithelial cell markers FOXN1 and K5 seem higher, and CD3γ expression lower, in pressed compared with untreated tissues (Supplemental Fig. 1).
FIGURE 1.
Increased expression of AIRE in DS thymuses. Isolated RNA from DS and control thymuses analyzed by RT-qPCR. GAPDH was used as endogenous control and a calibrator sample was used for comparison between plates. (A) AIRE, INSULIN, CHRNA1, TG, TPO, GAD1, and ATB4B (n = 5+5). Data are presented as median with interquartile range per group based on the median value from each individual and were analyzed using Mann–Whitney U test. A value of p ≤ 0.05 was considered as statistically significant. (B) Data points for each piece of tissue in each individual for expression of AIRE (left panel) INS (middle panel), and CHRNA1 (right panel). (C) Individual specific expression of AIRE (▪) and INSULIN (□, top left panel) and CHRNA1 (□, bottom left panel), based on median values from the six pieces of thymic tissue presented with interquartile range. Top right panel, Correlation of INSULIN expression versus AIRE expression (r = 0.64, p = 0.054, n = 10). Bottom right panel, correlation of CHRNA1 expression versus AIRE expression (r = 0.77, p = 0.0126, n = 10) in DS individuals (●) and controls (○).
Increased frequency of AIRE+ mTECS, CD11c+ dendritic cells, and enlarged IVL+ HCs characterizes thymic medulla in DS
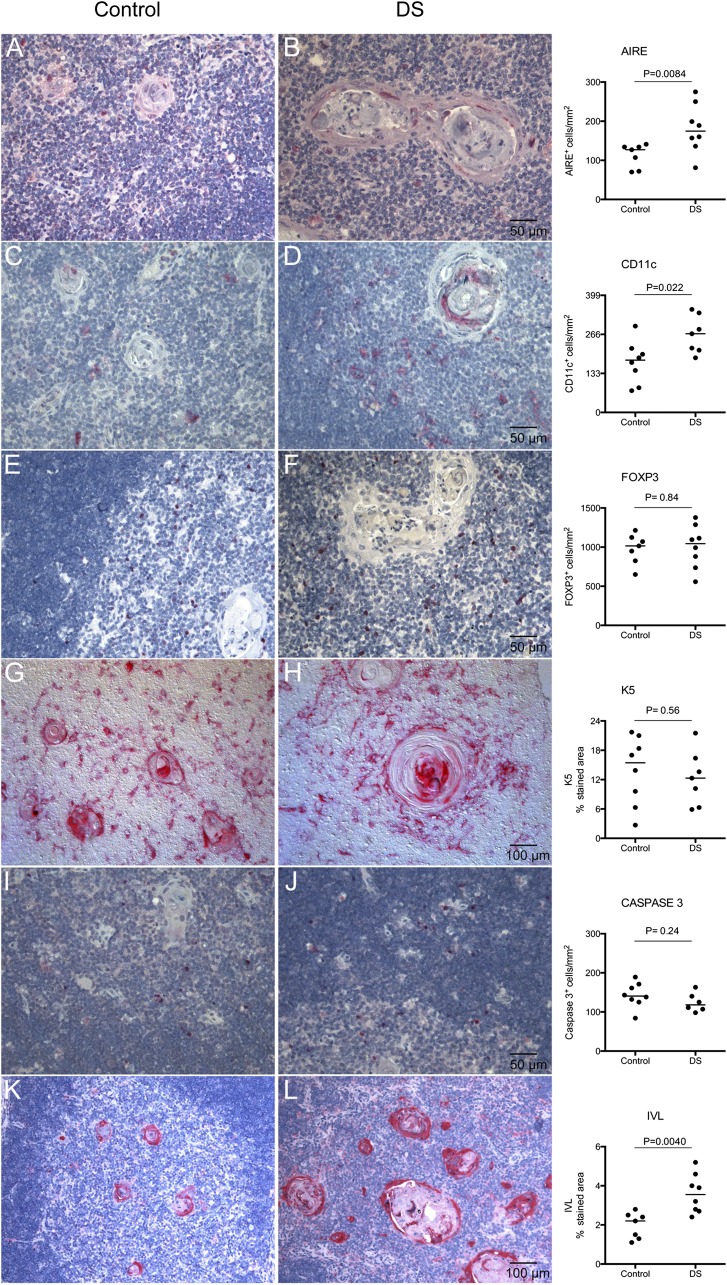
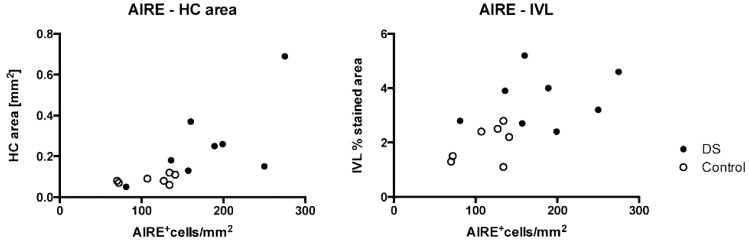
To investigate whether the overexpression of AIRE also resulted in more cells expressing the AIRE protein, we performed immunohistochemistry (IHC) on DS and control thymic tissue sections. The investigated area was medulla, excluding HC area, and for CASPASE3, the investigated area was cortex plus medulla excluding HC area. The number of AIRE+ cells pe square millimeter was significantly increased in DS thymic medullas compared with controls (DSmedian = 174.5, Controlmedian = 127, p = 0.0084) (Fig. 2A, 2B). Notable also was that AIRE expression adjacent to HCs was more frequently seen in DS patients. Furthermore, staining for the dendritic cell (DC) marker CD11c revealed higher number of CD11c+ cells per square millimeter medulla in the DS group (DSmedian = 268, Controlmedian = 178, p = 0.022) (Fig. 2C, 2D). No differences were found in the number of FOXP3+ cells per square millimeter (DSmedian = 1045, Controlmedian = 1015, p = 0.84), K5+ (percent stained area: DSmedian = 12.3, Controlmedian = 15.5, p = 0.56), or CASPASE 3+ cells per square millimeter (DSmedian = 118, Controlmedian = 140.5, p = 0.24) between DS and controls (Fig. 2E–J). Notable, however, was that DS K5+ mTECs were more spindly and orientated in a more distinct circular pattern around the HCs than corresponding cells in control stainings (Fig. 2G, 2H). In agreement with the previously reported striking enlargement of HCs in DS (15), we observed a markedly increased area stained for IVL in HCs (percent stained area: DSmedian = 3.6, Controlmedian = 2.2, p = 0.0040). Also, small non-HC areas within the medullas of DS thymus were positive for IVL while completely absent in controls (Fig. 2K, 2L). Furthermore, a positive correlation between AIRE and HC area was observed (r = 0.85, p = 0.0001) (Fig. 3, left panel) as well as between AIRE and IVL (r = 0.61, p = 0.02) (Fig. 3, right panel). K8 staining was present both in cortex and medulla. Some individuals displayed a network of small positive cells in cortex while they were completely absent in others regardless of group. In the medulla, both small and larger cells were positive to varying extent. Also, HCs stained positive to varying extent (data not shown).
FIGURE 2.
Increased AIRE, CD11c, and IVL protein expression in DS thymic tissue. Left panel, Representative IHC images of AIRE (A and B), CD11c (C and D), FOXP3 (E and F), K5 (G and H), CASPASE3 (I and J), and IVL (K and L) in thymic sections from controls respective DS individuals. Right panel, Quantification of IHC stainings. Note the enlarged HCs in DS tymic tissue. Data are presented with medians and were analyzed using Mann–Whitney U test (n = 8+8).
FIGURE 3.
AIRE expression correlates with epithelial maturation. Spearman rank correlation of AIRE+ cells per square millimeter to area of HCs in square millimeters (r = 0.85, p = 0.0001, n = 15) (left panel) and to percentage of area positive for IVL (r = 0.61, p = 0.02, n = 15) (right panel). DS individuals (●) and controls (○).
Broader total protein profile and TRA profile in DS thymic exosomes
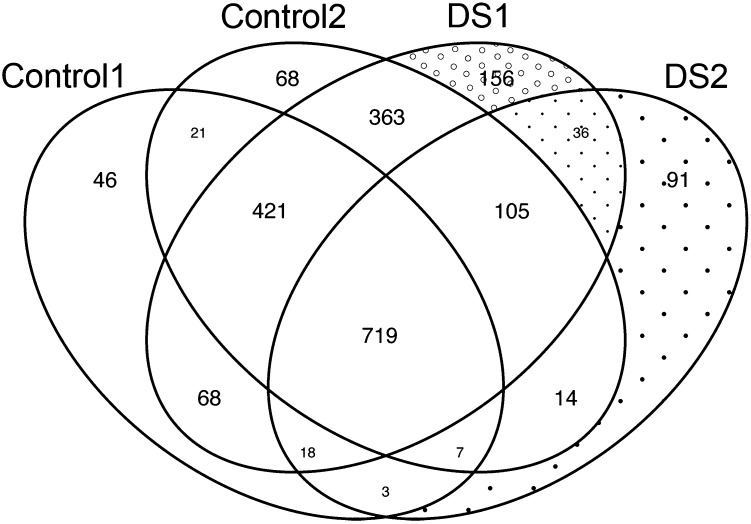
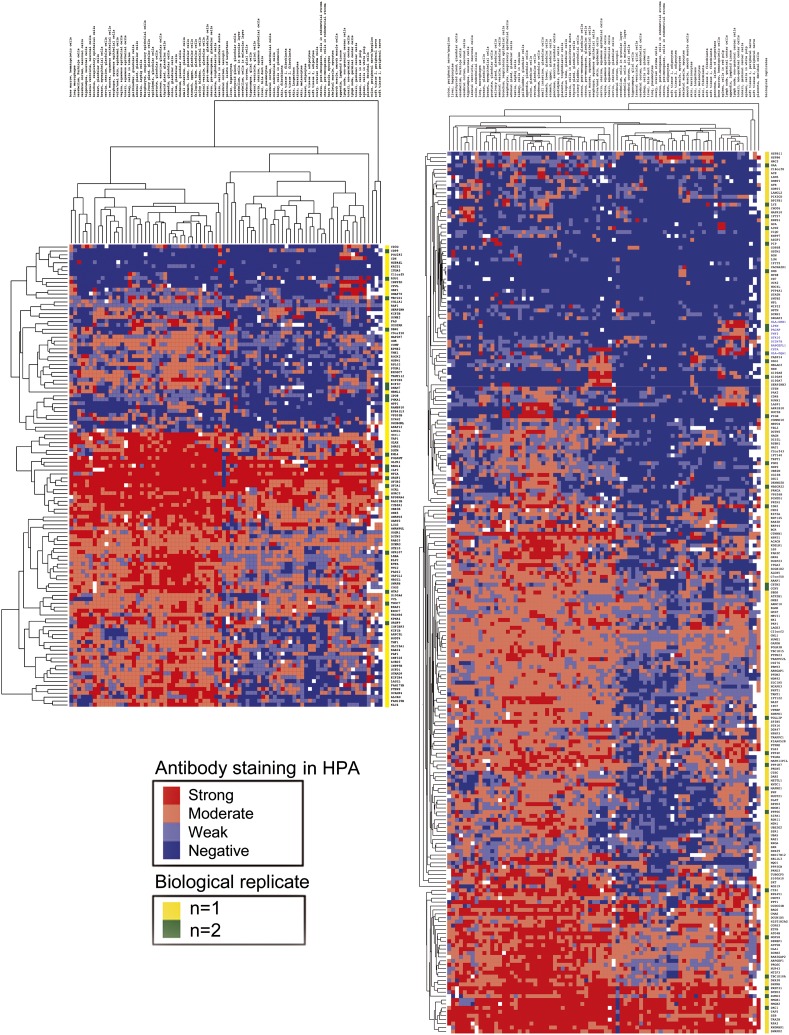
To further examine the possible effects of the abnormal AIRE levels on the thymic microenvironment, we analyzed the proteomic content of thymic exosomes in DS and control thymi. The proteomic data revealed a broader protein diversity in DS individuals compared with controls (i.e., the pool of unique proteins for a particular sample was larger in the two DS samples compared with the two control samples both in terms of absolute numbers [156 and 91 for DS exosomes respective 46 and 68 for control exosomes] and percentage [8.3 and 9.2% for DS exosomes respective 3.5 and 4.0% for control exosomes]) (Figs. 4, 5). Ten TRAs were restricted to DS thymic exosomes, whereas none was exclusive for control exosomes (Table I).
FIGURE 4.
Skewed proteomic profile in DS thymic exosomes. Venn diagram of protein content in thymic exosomes from two DS individuals and two controls show an increased number and percentage of proteins exclusively found in exosomes from DS individuals. DS individuals (DS1 and DS2) display the highest numbers (156 and 91 for DS versus 46 and 68 for controls) and percentages (8.3 and 9.2% for DS exosomes respective 3.5 and 4.0% for control exosomes) of proteins found exclusively in respective exosome sample (n = 2+2).
FIGURE 5.
A heat map illustrating a hierarchical cluster analysis of tissue expression of MS-identified proteins in thymic exosomes. The heat map is constructed from the unique proteins found only in control exosomes (left panel) or only in DS exosomes (right panel) using MS (proteins not yet investigated in the HPA database are not included). Proteins and tissues are hierarchally clustered according to biological function on the y- and x-axis, respectively. The expression level is graded as negative, weak, moderate, or strong, based on Ab staining of each protein in the HPA database, and illustrated by color shift from blue to red. Hence, a protein with a high number of blue fields is more tissue restricted than a protein with fewer blue fields and more red fields. Also note the difference in length between the two heat maps, which illustrates the difference in number of exclusive proteins between the two groups (n = 2+2).
Table I. TRAs (proteins expressed in five tissue cell types or less) found exclusively in DS exosomes.
| SwissProt | Chromosome | Tissue Expression in HPA |
|---|---|---|
| AZGP1 | 7q22.1 | Salivary gland, prostate, breast, kidney, cervix/uterine |
| PIP | 7q34 | Salivary gland, seminal vesicle, epididymis, breast |
| GSTM1 | 1p13.3 | Liver, testis, seminal vesicle, adrenal gland |
| DMD | Xp21.2 | Skeletal muscle, heart muscle, cerebral cortex |
| MYH8 | 17p13.1 | Skeletal muscle, heart muscle |
| CACNA2D1 | 7q21-q22 | Skeletal muscle, soft tissue, cerebral cortex, hippocampus, heart muscle |
| STATH | 4q13.3 | Salivary gland, gallbladder |
| UCK2 | 1q23 | Smooth muscle |
| NOC4L | 12q24.33 | Cerebral cortex, hippocampus, cerebellum |
| SNTB2 | 16q22.1 | Prostate, testis, cerebellum, cerebral cortex |
Five of 283 proteins (runt-related transcription factor 1, ubiquitin-conjugating enzyme E2G 2, lanosterol synthase, chromosome 21 open reading frame 2, and small ubiquitin-like modifier 3) uniquely found in the DS exosomes (three dotted fields in Fig. 4) have their corresponding coding gene on chromosome 21 (1.8%). Among proteins unique for control exosomes (three fields to upper left in Fig. 4), 1 of 135 (MORC Family CW-Type Zinc Finger 3, 21q22.13) has its coding gene placed on chromosome 21 (0.7%). This can be compared with the fact that the 230 genes located at chromosome 21 represents ∼1.2% of the 20,000 genes in the genome.
The Alzheimer’s disease associated protein DREBRIN1 (DBN1) (23) was found in thymic exosomes from both controls but in neither of the DS patients (Fig. 5).
Discussion
In this study, we found that patients with DS who have an extra copy of the AIRE gene also have an increased expression of AIRE at both mRNA and protein level in the thymus. This may alter thymic selection processes and affect the susceptibility for autoimmune diseases in DS patients. AIRE expression in mTECs is described to have multiple effects on various thymic functions such as TRA expression (5), mTEC differentiation (8–10), Ag presentation (6), chemokine production (7), and thymus-derived T regulatory cell (Treg) selection (24). Therefore, the net effect of an increased AIRE expression on the thymus is complex and not easily predicted.
The first aim of the current study was to investigate whether the increased copy number of AIRE in DS was accompanied by increased levels of AIRE mRNA in thymic tissue. Real-time PCR measurements showed that the AIRE mRNA level was increased in DS patients compared with controls (Fig. 1A). For these experiments, the study was designed to mimic the total thymic expression in DS and controls. Although thymocytes were washed away, the stromal compartment was kept intact, and information was obtained from six different parts of each thymus yielding information on a whole organ level rather than sorted cell level, which has been reported thoroughly elsewhere (25, 26). However, because many studies have shown that mTECs are the major cell type expressing AIRE in the thymus, our results still ought to reflect an increased expression of AIRE in the mTEC population.
Previous work by Peterson et al. (27) showed that overexpression of murine Aire in vitro results in an increased expression of TRAs. To examine whether the increased expression of AIRE in DS thymus leads to an altered expression of TRAs, the expression of both AIRE-dependent and AIRE-independent TRAs was analyzed. One predicted effect of AIRE overexpression is an increased expression of INSULIN, a known AIRE-dependent TRA, and in agreement with this a trend toward an increased expression of INSULIN in DS and an overall positive correlation between AIRE expression and INSULIN expression was seen (Fig. 1). This was also the case for AIRE-dependent CHRNA1, which codes for the α-subunit of the nicotinic acetylcholine repector (Fig. 1) (28). For other TRAs reported not to be Aire dependent in mice, such as tg (29) and gad67 (5), the overexpression of AIRE did not seem to affect the expression level of the corresponding human genes TG and GAD1 in DS thymic tissue (Fig. 1A). For TPO, a TRA with an enigmatic relation to Aire (30) and ATB4B, which is reported to be AIRE dependent (28), AIRE dependency was not confirmed in the DS phenotype (Fig. 1A). The reason could be that the effects of trisomy 21, other than the trisomy of AIRE, affect the expression levels of TPO and ATB4B, overcoming their possible AIRE dependency.
Immunohistochemical stainings confirmed the increased expression of AIRE on the mRNA level with more cells expressing AIRE protein in the thymic medulla in the DS group (Fig. 2A, 2B). At the same time, no difference was seen in the relative staining of K5 in the medullary regions, suggesting that the elevated number of AIRE+ cells per square millimeter in DS reflects a larger proportion of AIRE+ mTECs rather than an altered total number of mTECs. This could be explained by a general increase of AIRE expression in mTECs making a higher frequency of AIRE+ mTECs detectable by the IHC technique. An alternative explanation is that the mTEC turnover is affected so that a larger proportion of mTECs are in an AIRE expressing late phase of mTEC differentiation. The latter explanation is supported by studies showing that Aire promotes terminal differentiation of mTECs (31).
In a contrasting study, Lima et al. (17) demonstrated decreased protein expression of AIRE in thymic tissue from DS patients compared with controls. This discrepancy could be because of differences in the study design. In the current study, thymic tissue was collected from children of 0–6 mo of age, whereas Lima et al. (17) included thymic tissue from patients from 4 mo to 12 y of age, with the youngest group examined being 4 mo to 1 y of age. Differences in the IHC data could also in part be explained by the fact that in the current study the HC area was excluded from the medullary area when counting AIRE+ cells, whereas Lima et al. (17) use the whole “medullary region.” This could be of importance because, as shown in this and other studies, the HCs are massively enlarged in DS thymi compared with controls. Thus, if HCs are included in the medullary area, the number of AIRE+ cells per square millimeter will decrease in the DS group compared with the control group.
The results of the current study are strengthened by a study by Sabater et al. (32) in which they investigated the effect of IDDM2 alleles and AIRE levels on thymic INSULIN expression. In their material, the individual with the highest expression of both AIRE and INSULIN was a 6-y-old patient with DS, this despite the fact that the patient had a low classIII/classI IDDM2 ratio with classIII being the protective allele associated with higher INSULIN expression (this was the only DS patient in the study) (32).
The enlarged HCs seen in DS thymi also suggest that the increased AIRE expression in DS is affecting mTEC maturation (Fig. 2K, 2L). In addition, the increased staining of IVL seen in DS thymi points to an increased degree of epithelial cell turnover, because IVL is mainly present in terminally differentiated epithelial cells. Furthermore, as a comparison, Aire knockout mice have more mTECs, smaller HCs, and less IVL (33). Because Aire has been suggested to drive mTEC maturation into a post-TRA expression state characterized by formation of HCs (8), it is interesting to note that the increased AIRE expression and the elevated number of AIRE+ cells per square millimeter in DS thymi is accompanied by enlarged HCs and increased IVL expression. Hence, the enlarged HCs seen in DS thymi could be a result of an accelerated maturation of mTECs because of the overexpression of AIRE. These results strengthen the hypothesis that an altered AIRE expression affects the differentiation program of mTECs in DS. K8 expression was notoriously varying between individual sections and was therefore excluded from the analysis (data not shown).
Unexpectedly, the number of CD11c+ cells per square millimeter was elevated in DS thymic medulla compared with controls. Because it has been reported in mice that the production of the DC attracting chemokine XCL1 is Aire dependent (34), the level of XCL1 in DS and control tissue was measured. However, no difference between the two groups was found (data not shown). Possibly, elevated expression of AIRE directly influences CD11c+ DCs because thymic DCs in humans are known to express low levels of AIRE (26). At the same time, this CD11c+ population also could be a contributor to the higher levels of detected AIRE/AIRE in the DS thymus.
In agreement with previously published data that Aire does not influence the expression or function of Foxp3 (35–37), we found no difference in the frequency of FOXP3+ cells between DS and control thymus (Fig. 2E, 2F). However, a recent study of peripheral blood Tregs (CD4+CD25+FOXP3+) in DS patients reported an increased percentage of Tregs in DS compared with controls. Interestingly, at the same time, the inhibitory activity of DS CD4+CD25highCD127low was impaired in an in vitro suppression assay (38). The underlying reason for the overrepresented but less efficient Treg population is unknown.
Although the expression of CASPASE3 did not differ between DS and controls (Fig. 2I, 2J), this does not entirely rule out the possibility of altered apoptotic kinetics in negatively selected thymocytes in DS thymuses.
Because it has been speculated that exosomes may participate in the shuttling of TRAs from stromal cells to thymocytes or DCs (39, 40) and we and others have shown that thymic tissue is rich in exosomes (18, 19), we have taken interest in the characteristics and functions of thymic exosomes. When we investigated the protein profiles of thymic exosomes from DS and control patients, the proteins in exosomes from DS patients represent a greater diversity than the proteins from control exosomes (Fig. 4). This means that the group of proteins unique for each individual is larger in the DS patients compared with controls, both in terms of absolute number as well as percentage. A skewed TRA expression also is noted in the thymic exosomes because 10 TRAs are unique for DS exosomes and none for control exosomes, again indicating a broader protein expression within DS thymus (Table I). It cannot be ruled out that the increased AIRE expression in DS leads to the skewed proteomic profile, especially regarding TRA content, seen in DS thymic exosomes. The lack of DBN1 in DS exosomes in contrast to its presence in both control exosomal samples is an interesting observation because an accelerated development of Alzheimer’s disease is overrepresented among DS patients (Fig. 5). A lack of DBN1 has been observed in brain tissue from Alzheimer’s patients as well as from patients with DS (41).
Given the complexity of AIRE effects on thymic function, it is not surprising that the net outcome of AIRE overexpression does not lead to the opposite effects of AIRE deficiency. As illustrated in APS1, an AIRE-deficient situation, the efficacy of the negative selection is impaired by a deficiency of AIRE-driven TRAs. In DS in contrast, the negative selection does not seem to become more efficient, even though the increased AIRE gene dose is resulting in increased levels of AIRE protein and TRAs, but rather less efficient, possibly because of AIRE functions not committed to TRA expression or other effects of the trisomy 21. An additional important factor to bear in mind is the reduced thymic organ size in DS patients (13); hence, both size and function seem impaired in this group.
As illustrated by the presence of self-reactive T cell clones in healthy individuals (42), central tolerance is a very fine tuned process, which balances at the border of autoimmunity to achieve a highly effective T cell pool against pathogens. All alterations in the prerequisites for the negative selection process and/or the production of functional Tregs may result in escape of self-reactive uncontrolled T cells into the periphery. This study has shown a number of alterations in the thymus of DS patients that could be of importance for the development of an autoimmune phenotype: 1) an increased AIRE expression; 2) a skewed thymic Ag pool with overexpression of a subgroup of Ags, such as INSULIN, as well as a lowered expression of others leading to an altered stoichiometric milieu; 3) an increased frequency of AIRE expressing mTECs; 4) an accelerated mTEC maturation into apoptotic or post-AIRE stages; 5) massively enlarged, often centralized HCs, possibly affecting thymocyte movement in and into deep parts of the medulla; and 6) an accumulation of DCs, possibly altering the kinetics of Ag presentation. These alterations could all affect the selection and TCR repertoire of both conventional and regulatory T cells of importance for the development of an autoimmune phenotype in DS.
Supplementary Material
Acknowledgments
We thank Birgitta Romlin and Arvid Otterlind at the Queen Silvia Children’s Hospital (Gothenburg, Sweden) for assistance in collecting of thymic material, Ing-Marie Jonsson at the Department of Rheumatology and Inflammation Research for help with IHC tissue preparation, and the Proteomics Core Facility at Sahlgrenska Academy, Gothenburg University, for performing proteomic analyses.
This work was supported by the Swedish Research Council (Contract 80409601), the Marianne and Marcus Wallenberg Foundation, Region Västra Götaland (Grant ALFGBG-771712), Arbetsmarknadens Försäkringsaktiebolag (Contract 100258), the IngaBritt and Arne Lundbergs Research Foundation, the AnnMari and Per Ahlqvists Foundation, the Gothenburg Medical Society, and Wilhelm and Martina Lundgrens Research Foundation.
The online version of this article contains supplemental material.
- AIRE
- autoimmune regulator
- APS1
- autoimmune polyendocrine syndrome type 1, DC, dendritic cell
- DS
- Down syndrome
- HC
- Hassall’s corpuscle
- HPA
- human protein atlas
- IDDM
- insulin-dependent diabetes mellitus
- IHC
- immunohistochemistry
- IVL
- INVOLUCRIN
- K5,8
- keratin 5,8
- MS
- mass spectrometry, qPCR, quantitative PCR
- TEC
- medullary thymic epithelial cell
- Treg
- regulatory T cell
- TG
- thyroglobulin
- TPO
- thyroid peroxidase
- TRA
- tissue-restricted Ag.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Ivarsson S. A., Ericsson U. B., Gustafsson J., Forslund M., Vegfors P., Annerén G. 1997. The impact of thyroid autoimmunity in children and adolescents with Down syndrome. Acta Paediatr. 86: 1065–1067 [DOI] [PubMed] [Google Scholar]
- 2.Blumberg D., AvRuskin T. 1987. Down’s syndrome, autoimmune hyperthyroidism, and hypoparathyroidism: a unique triad. Am. J. Dis. Child. 141: 1149. [DOI] [PubMed] [Google Scholar]
- 3.Rabinowe S. L., Rubin I. L., George K. L., Adri M. N., Eisenbarth G. S. 1989. Trisomy 21 (Down’s syndrome): autoimmunity, aging and monoclonal antibody-defined T-cell abnormalities. J. Autoimmun. 2: 25–30 [DOI] [PubMed] [Google Scholar]
- 4.George E. K., Mearin M. L., Bouquet J., von Blomberg B. M., Stapel S. O., van Elburg R. M., de Graaf E. A. 1996. High frequency of celiac disease in Down syndrome. J. Pediatr. 128: 555–557 [DOI] [PubMed] [Google Scholar]
- 5.Anderson M. S., Venanzi E. S., Klein L., Chen Z., Berzins S. P., Turley S. J., von Boehmer H., Bronson R., Dierich A., Benoist C., Mathis D. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science 298: 1395–1401 [DOI] [PubMed] [Google Scholar]
- 6.Hubert F. X., Kinkel S. A., Davey G. M., Phipson B., Mueller S. N., Liston A., Proietto A. I., Cannon P. Z., Forehan S., Smyth G. K., et al. 2011. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood 118: 2462–2472 [DOI] [PubMed] [Google Scholar]
- 7.Laan M., Kisand K., Kont V., Möll K., Tserel L., Scott H. S., Peterson P. 2009. Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J. Immunol. 183: 7682–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Laan M., Bichele R., Kisand K., Scott H. S., Peterson P. 2012. Post-Aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front. Immunol. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillard G. O., Dooley J., Erickson M., Peltonen L., Farr A. G. 2007. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J. Immunol. 178: 3007–3015 [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto M. 2011. Contrasting models for the roles of Aire in the differentiation program of epithelial cells in the thymic medulla. Eur. J. Immunol. 41: 12–17 [DOI] [PubMed] [Google Scholar]
- 11.Söderbergh A., Gustafsson J., Ekwall O., Hallgren A., Nilsson T., Kämpe O., Rorsman F., Annerén G. 2006. Autoantibodies linked to autoimmune polyendocrine syndrome type I are prevalent in Down syndrome. Acta Paediatr. 95: 1657–1660 [DOI] [PubMed] [Google Scholar]
- 12.Erichsen M. M., Løvås K., Skinningsrud B., Wolff A. B., Undlien D. E., Svartberg J., Fougner K. J., Berg T. J., Bollerslev J., Mella B., et al. 2009. Clinical, immunological, and genetic features of autoimmune primary adrenal insufficiency: observations from a Norwegian registry. J. Clin. Endocrinol. Metab. 94: 4882–4890 [DOI] [PubMed] [Google Scholar]
- 13.Karl K., Heling K. S., Sarut Lopez A., Thiel G., Chaoui R. 2012. Thymic-thoracic ratio in fetuses with trisomy 21, 18 or 13. Ultrasound Obstet. Gynecol. 40: 412–417 [DOI] [PubMed] [Google Scholar]
- 14.Larocca L. M., Piantelli M., Valitutti S., Castellino F., Maggiano N., Musiani P. 1988. Alterations in thymocyte subpopulations in Down’s syndrome (trisomy 21). Clin. Immunol. Immunopathol. 49: 175–186 [DOI] [PubMed] [Google Scholar]
- 15.Levin S., Schlesinger M., Handzel Z., Hahn T., Altman Y., Czernobilsky B., Boss J. 1979. Thymic deficiency in Down’s syndrome. Pediatrics 63: 80–87 [PubMed] [Google Scholar]
- 16.Verstegen R. H., Borte S., Bok L. A., van Zwieten P. H., von Dobeln U., Hammarstrom L., de Vries E. 2013. Impact of Down syndrome on the performance of neonatal screening assays for severe primary immunodeficiency diseases. J. Allergy Clin. Immunol. 113: 1208–1211 [DOI] [PubMed] [Google Scholar]
- 17.Lima F. A., Moreira-Filho C. A., Ramos P. L., Brentani H., Lima Lde. A., Arrais M., Bento-de-Souza L. C., Bento-de-Souza L., Duarte M. I., Coutinho A., Carneiro-Sampaio M. 2011. Decreased AIRE expression and global thymic hypofunction in Down syndrome. J. Immunol. 187: 3422–3430 [DOI] [PubMed] [Google Scholar]
- 18.Skogberg G., Gudmundsdottir J., van der Post S., Sandström K., Bruhn S., Benson M., Mincheva-Nilsson L., Baranov V., Telemo E., Ekwall O. 2013. Characterization of human thymic exosomes. PLoS ONE 8: e67554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G. J., Liu Y., Qin A., Shah S. V., Deng Z. B., Xiang X., Cheng Z., Liu C., Wang J., Zhang L., et al. 2008. Thymus exosomes-like particles induce regulatory T cells. J. Immunol. 181: 5242–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons M., Raposo G. 2009. Exosomes—vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21: 575–581 [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- 22.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., et al. 2010. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28: 1248–1250 [DOI] [PubMed] [Google Scholar]
- 23.Harigaya Y., Shoji M., Shirao T., Hirai S. Disappearance of actin-binding protein, drebrin, from hippocampal synapses in Alzheimer’s disease. J. Neurosci. Res. 43: 87‑92 [DOI] [PubMed] [Google Scholar]
- 24.Kekäläinen E., Tuovinen H., Joensuu J., Gylling M., Franssila R., Pöntynen N., Talvensaari K., Perheentupa J., Miettinen A., Arstila T. P. 2007. A defect of regulatory T cells in patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J. Immunol. 178: 1208–1215 [DOI] [PubMed] [Google Scholar]
- 25.Derbinski J., Schulte A., Kyewski B., Klein L. 2001. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2: 1032–1039 [DOI] [PubMed] [Google Scholar]
- 26.Gotter J., Brors B., Hergenhahn M., Kyewski B. 2004. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J. Exp. Med. 199: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kont V., Laan M., Kisand K., Merits A., Scott H. S., Peterson P. 2008. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol. Immunol. 45: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taubert R., Schwendemann J., Kyewski B. 2007. Highly variable expression of tissue-restricted self-antigens in human thymus: implications for self-tolerance and autoimmunity. Eur. J. Immunol. 37: 838–848 [DOI] [PubMed] [Google Scholar]
- 29.Derbinski J., Gäbler J., Brors B., Tierling S., Jonnakuty S., Hergenhahn M., Peltonen L., Walter J., Kyewski B. 2005. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misharin A. V., Nagayama Y., Aliesky H. A., Rapoport B., McLachlan S. M. 2009. Studies in mice deficient for the autoimmune regulator (Aire) and transgenic for the thyrotropin receptor reveal a role for Aire in tolerance for thyroid autoantigens. Endocrinology 150: 2948–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray D., Abramson J., Benoist C., Mathis D. 2007. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204: 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabater L., Ferrer-Francesch X., Sospedra M., Caro P., Juan M., Pujol-Borrell R. 2005. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J. Autoimmun. 25: 312–318 [DOI] [PubMed] [Google Scholar]
- 33.Yano M., Kuroda N., Han H., Meguro-Horike M., Nishikawa Y., Kiyonari H., Maemura K., Yanagawa Y., Obata K., Takahashi S., et al. 2008. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J. Exp. Med. 205: 2827–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei Y., Ripen A. M., Ishimaru N., Ohigashi I., Nagasawa T., Jeker L. T., Bösl M. R., Holländer G. A., Hayashi Y., Malefyt Rde. W., et al. 2011. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J. Exp. Med. 208: 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liston A., Lesage S., Wilson J., Peltonen L., Goodnow C. C. 2003. Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4: 350–354 [DOI] [PubMed] [Google Scholar]
- 36.Anderson M. S., Venanzi E. S., Chen Z., Berzins S. P., Benoist C., Mathis D. 2005. The cellular mechanism of Aire control of T cell tolerance. Immunity 23: 227–239 [DOI] [PubMed] [Google Scholar]
- 37.Kuroda N., Mitani T., Takeda N., Ishimaru N., Arakaki R., Hayashi Y., Bando Y., Izumi K., Takahashi T., Nomura T., et al. 2005. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J. Immunol. 174: 1862–1870 [DOI] [PubMed] [Google Scholar]
- 38.Pellegrini F. P., Marinoni M., Frangione V., Tedeschi A., Gandini V., Ciglia F., Mortara L., Accolla R. S., Nespoli L. 2012. Down syndrome, autoimmunity and T regulatory cells. Clin. Exp. Immunol. 169: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collado J. A., Guitart C., Ciudad M. T., Alvarez I., Jaraquemada D. 2013. The repertoires of peptides presented by MHC-II in the thymus and in peripheral tissue: a clue for autoimmunity? Front. Immunol. 4: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyewski B., Klein L. 2006. A central role for central tolerance. Annu. Rev. Immunol. 24: 571–606 [DOI] [PubMed] [Google Scholar]
- 41.Shim K. S., Lubec G. 2002. Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer’s disease and Down syndrome. Neurosci. Lett. 324: 209–212 [DOI] [PubMed] [Google Scholar]
- 42.Lohse A. W., Dinkelmann M., Kimmig M., Herkel J., Meyer zum Büschenfelde K. H. 1996. Estimation of the frequency of self-reactive T cells in health and inflammatory diseases by limiting dilution analysis and single cell cloning. J. Autoimmun. 9: 667–675 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.