Abstract
Polyvalent vaccines use a mixture of Ags representing distinct pathogen strains to induce an immune response that is cross-reactive and protective. However, such approaches often have mixed results, and it is unclear how polyvalency alters the fine specificity of the Ab response and what those consequences might be for protection. In this article, we present a coarse-grain theoretical model of B cell affinity maturation during monovalent and polyvalent vaccinations that predicts the fine specificity and cross-reactivity of the Ab response. We stochastically simulate affinity maturation using a population dynamics approach in which the host B cell repertoire is represented explicitly, and individual B cell subpopulations undergo rounds of stimulation, mutation, and differentiation. Ags contain multiple epitopes and are present in subpopulations of distinct pathogen strains, each with varying degrees of cross-reactivity at the epitope level. This epitope- and strain-specific model of affinity maturation enables us to study the composition of the polyclonal response in granular detail and identify the mechanisms driving serum specificity and cross-reactivity. We applied this approach to predict the Ab response to a polyvalent vaccine based on the highly polymorphic malaria Ag apical membrane antigen-1. Our simulations show how polyvalent apical membrane Ag-1 vaccination alters the selection pressure during affinity maturation to favor cross-reactive B cells to both conserved and strain-specific epitopes and demonstrate how a polyvalent vaccine with a small number of strains and only moderate allelic coverage may be broadly neutralizing. Our findings suggest that altered fine specificity and enhanced cross-reactivity may be a universal feature of polyvalent vaccines.
Introduction
The humoral or Ab response to a vaccine is often a key component in its ability to induce protection against a targeted pathogen. This Ab response is polyclonal in nature, arising from multiple clonal B cell populations, each producing unique Abs with respect to their binding affinity and Ag epitope. Although this complex polyclonal response can be measured in the aggregate, it has only recently become possible to quantitatively assess the individual contributions of the clonal subpopulations. The fine specificity of the Ab response can play a major role in vaccine efficacy, because distinct Ag epitopes can vary significantly in terms of their neutralization and their degree of conservation across pathogen strains. Although polyvalent vaccines, which use a mixture of Ags representing multiple pathogen strains, have been used to broaden the efficacy of a vaccine Ag, it is still unclear how such formulations alter the fine specificity of the Ab response and what those implications might be for protection.
The polyclonal response is an aggregate of individual monoclonal responses, each with unique properties with respect to binding epitope, binding affinity, and neutralization, and the fine specificity of this polyclonal response can be a critical determinant of efficacy. Sera with similar overall Ab titers to a given Ag can vary significantly in neutralization or in cross-reactivity to alternate pathogen strains. Recently, there have been a number of efforts to rationally design vaccine Ags that exploit fine specificity to target highly neutralizing or highly conserved epitopes that are poorly immunogenic in natural infections, as in the case of HIV-1 (1, 2) and respiratory syncytial virus (3).
The serum Ab response is the result of affinity maturation within the germinal centers (GCs) of lymph nodes in the host lymphatic system. The host immune system is thought to contain ≥107–108 naive B cells (4), each expressing a unique BCR created through the somatic recombination of several BCR gene segments. During a primary infection or vaccination, a subset of B cells that express BCRs with some threshold Ag-binding affinity (Ag-specific B cells) bind to the Ag and undergo activation. Within the GC, these B cells undergo repeated rounds of stimulation, mutation, and replication to selectively expand B cell clonal lines with increasing Ag-binding affinity (5). In the latter stages of affinity maturation, GC B cells undergo differentiation into plasma cells and memory cells. Plasma cells secrete a soluble form of the BCR as Abs that make up the serum Ab response, whereas memory cells remain dormant until reactivation during secondary exposure to the Ag months or years after the initial infection.
Mathematical modeling of affinity maturation relies on a mechanistic “first principles” approach to immunology; theories and hypotheses describing the underlying immune processes are applied in an in silico manner to explain experimental results and clinical observations. Key components of the immune system, such as lymphocytes (B and T cells), Abs, cytokines, and Ags, are modeled dynamically, and their levels grow or shrink as they interact with each other throughout a simulated immunological event, such as an infection or vaccination. Such modeling efforts have made substantial contributions to our understanding of immunology. Seminal studies in 1970 and 1971 by G. I. Bell (6–8) led to the first computer simulations of affinity maturation and demonstrated the theoretical basis for clonal selection in the Ab immune response. Subsequent work by Oprea, Perelson, and Kepler (9–11) developed simulation approaches to study somatic hypermutation and GC dynamics. Contributions by later modeling and simulation efforts included the elucidation of the role of key immune system properties, such as repertoire size, diversity, and somatic mutation rates (12, 13), and the development of detailed kinetic models of B cell GC dynamics and morphologies (9, 14, 15). Pioneering research by Perelson and other investigators (12, 16, 17) helped to develop an immunological “shape space” model for representing complex shape-based Ab–Ag interactions. Subsequent work by Smith et al. established parameters for a shape space model using immunological data (18) and later adapted it for use in a stochastic simulation of affinity maturation to accurately predict the efficacy of repeated influenza vaccinations from clinical data (19). Beyond studying affinity maturation, computational immunology approaches been used to study T cell activation (20, 21) and specificity (22), viral infection and adaptation (23–27), and innate immunity (28).
In this article, we present a novel coarse-grain theoretical model of B cell affinity maturation to simulate the progression of an Ab response against a monovalent or polyvalent Ag. We model affinity maturation using a B cell population dynamics approach. The host B cell repertoire is represented explicitly, and individual B cell subpopulations undergo rounds of stimulation, mutation, and differentiation. Ags in this model contain multiple epitopes and are present as distinct strains, each with specified degrees of cross-reactivity at the epitope level. During the simulation, B cells bind with Ag and become stimulated, mutate, and proliferate. GC B cells differentiate into memory cells, which have long half-lives, and plasma cells, which produce Abs. Abs, in turn, bind with Ag and neutralize or clear it. BCR/Ab–Ag interactions are modeled using the immunological shape space model developed by Smith et al. (18), and the simulation is carried out using the Gillespie algorithm (26, 29). Our approach is distinct from previous B cell repertoire models (12, 13) in its use of a multiple-strain and multiple-epitope representation of the Ag. This allows us to simulate the Ab response with respect to epitope-level and strain-level specificity, which is critical to modeling vaccine efficacy for multiple-strain or multiple-serotype pathogens, such as Plasmodium falciparum or dengue virus. Furthermore, to our knowledge, this model represents the first computational simulation of affinity maturation for a real-world polyvalent vaccine candidate, enabling us to identify potential immune mechanisms driving serum specificity and cross-reactivity in polyvalent vaccine formulations.
Antigenic variation between pathogen strains remains one of the greatest challenges in vaccine design. This variation can be the result of antigenic drift, such as in the case of the influenza virus (30, 31), or the result of pathogen evolution in response to immune-selective pressure, such as in the case of HIV (32–34) and both hepatitis B (35) and C (36). Antigenic variation can be addressed through the use of polyvalent vaccines, and understanding the mechanisms that guide the polyvalent immune response is critical to vaccine design. We hypothesize that the complex polyclonal Ab response to a polyvalent vaccine can be accurately simulated using a model of affinity maturation that takes into account multiple epitopes and strains, along with experimental information regarding the relative immunogenicity and antigenic distance of various epitopes on the Ag. We applied our model to examine the mechanisms guiding the specificity and cross-reactivity of the Ab response to a polyvalent vaccine based on the highly polymorphic malaria Ag apical membrane antigen-1 (AMA1).
The malaria vaccine candidate AMA1 is a subunit-based vaccine against P. falciparum that induces protection (37), but only against parasite strains that are closely related to the vaccine (38–40). A recent phase 2b AMA1 vaccine trial in 1–6-y-old children in Mali found 64% efficacy against malaria caused by vaccine-like strains, underscoring its potential as a malaria vaccine (41). Unfortunately, AMA1 displays immense antigenic variation: >10% of its residues are polymorphic, and >200 unique haplotypes have been identified (42, 43). A crystal structure of AMA1 reveals that the protein consists of three domains that present two immunologically distinct regions: a conserved face, consisting of a cluster of largely conserved epitopes along domain I and III, and a polymorphic face, consisting of a cluster of highly polymorphic epitopes along domain II (44, 45). Previous studies suggested that no fewer than 6–10 strains of AMA1, and possibly many more, must be included in a polyvalent vaccine to provide sufficient allelic coverage for broad protection (42, 43, 46). However, several studies developed polyvalent formulations of AMA1, using two to six strains, and demonstrated significant cross-reactivity and protection relative to the monovalent vaccine of any single strain (47–50).
In a recent study, Dutta et al. (47) showed that the tetravalent formulation had surprisingly high efficacy against a diverse panel of 22 non–vaccine strains of P. falciparum. Extensive epitope mapping of the Ab response revealed that the polyvalent vaccine induced an enhanced response toward the conserved face of AMA1, but this enhancement alone was insufficient to explain the broadly neutralizing humoral response. Furthermore, it was experimentally difficult to determine to what degree the cross-reactivity of the polyvalent response was a result of strain-transcending Abs that neutralize all strains or the result of overlapping partial cross-reactivity of strain-specific Abs. We applied our stochastic model of affinity maturation using a two-epitope model of AMA1 to identify possible answers to these questions and provide a mechanistic explanation underlying this broadly cross-reactive Ab response.
Materials and Methods
Stochastic model for affinity maturation
In this study, we used the immunological shape space model developed by Smith et al. (18) to model multiple epitopes for multiple strains of the polymorphic AMA1 Ag. This allowed us to describe the antigenic relationships between each strain, at the epitope level, with respect to B cell specificity and cross-reactivity. We used a B cell population dynamics approach, in conjunction with this immunological shape space model, to simulate how a B cell repertoire evolves in response to exposure to these epitopes during affinity maturation in the context of a specified vaccination schedule.
We modeled B cell affinity maturation as a set of rate reactions, similar to chemical reactions, that describe the underlying immunological processes, such as Ag binding or B cell replication. We carried out stochastic simulations of affinity maturation by applying the Gillespie algorithm to this set of rate equations outlined below. The Gillespie algorithm was adapted for modeling the immune system, as described by Woo and Reifman (26). The main parameters that were set to model AMA1 in this simulation were the number of epitopes in the AMA1 Ag, the relative immunogenicity of each epitope, and the antigenic distance of each epitope between different AMA1 strains. These parameters were determined using previous experimental data from monovalent AMA1 vaccination studies (47, 48, 51). We coded the algorithm in Python, and the source code is freely available from the authors by request.
Immunological shape space
Abs and BCRs interact with Ags through the shape complementarity between the Ab/BCR paratope and the Ag epitope. We used the immunological space model developed by Smith et al. to describe the BCR/Ab and Ag shape-based interactions (18, 19). Briefly, the shape of the paratope (on the BCR or Ab) and epitope (on the Ag) is symbolically represented as a 20-character string made up of 4 unique characters that defines a space of 1012 distinct epitope and paratope shapes. In our model, this string serves as the genotype sequence defining the paratope or epitope. Any mutations during affinity maturation are direct mutations of this genotype sequence and can be thought of as nonsilent mutations that alter the shape of the paratope. Likewise, any differences in this genotype sequence between epitopes reflect polymorphisms that alter the epitope structure. The number of phenotypically distinct BCRs is thought to be ≥1010 (52) and when including effects, such as somatic mutations, as high as 1016 (53). The simplified representation of Ab Ag “shape space” in our model allows us to represent 1012 possible BCR sequences (18); increasing the length or number of unique characters that makes up the paratope sequence increases the number of possible BCR sequences that can be represented.
The binding affinity Qij between the paratope i and the epitope j is a function of the Hamming distance between the paratope and epitope genotype sequences, d(i,j), and is described by Eq. 1. The epitope and paratope sequences have equal lengths, and the Hamming distance was calculated following a simple linear comparison of the sequences without any further sequence alignment.
| (1) |
In this approach, the parameter ε reflects the fold increase in apparent binding affinity following a single beneficial mutation for BCR and Ab binding (εB and εAb, respectively). In the model, εB and εAb, are set to values of 10 and 2.5, respectively. The εB parameter for B cells is larger than εAb for Abs, because unlike Abs, which bind to Ag through a single Ab–Ag–binding event, BCR activation is a cooperative process that requires multiple individual BCR–Ag–binding events. In our model, this cooperativity is represented by a larger fold change in apparent binding affinity of BCR–Ag interactions compared with Ab–Ag interactions. The parameter α is the minimum Hamming distance that defines maximum binding affinity (19). This value was set to 4 and reflects the degeneracy in paratope sequences that can achieve maximum binding affinity to the epitope. Finally, λ is the maximum Hamming distance that defines the threshold binding affinity, where the binding affinity is nonzero and is set to 7 based on parameterization of immunological data by Smith et al. (18). These values of α and λ result in four BCR binding-affinity levels that span a 104-fold difference in binding affinity between naive and “fully” matured B cells. Unless otherwise specified, all immunological shape space parameters were set to the values defined by Smith et al. (19).
The antigenic distance between strains for the same epitope is equivalent to the Hamming distance between the genotype sequences that define each epitope. This antigenic distance can be adjusted based on the cross-reactivity between those epitopes. Conserved epitopes have low antigenic distances between them, whereas polymorphic or diverse epitopes have higher antigenic distances between them. Values for antigenic distance can range from 0 (for identical epitopes) to >10 (for significantly dissimilar epitopes).
Immune system components
In this model, each Ag has its own genotype, as described above, denoted by the subscript i. The Ag genotype contains the epitope genotypes for two epitopes. All B cells and Abs in the system are likewise denoted by a paratope genotype, subscript j. There are four types of B cells: naive N, GC B, plasma P, and memory M, along with Abs, all of which have paratope genotypes.
Rate equations
We modeled affinity maturation using a set of equations that describe B cell stimulation and proliferation, memory and plasma cell differentiation, Ab production, and Ag clearance. In the system, the genotype of every B cell and Ag epitope was explicitly described (by a subscript in the following equations). All parameters used below are summarized in Table I.
Table I. Parameter values for the immune system model.
| Parameter | Symbol | Value |
|---|---|---|
| Simulation conditions | ||
| No. of naive B cells | 5 × 107 cells | |
| Ag dosea | 360 units | |
| GC carrying capacity | κ | 5000 cells |
| Ag parameters | ||
| Serotypesa | 1–4 | |
| Epitopes | 2 | |
| Epitope 1 | ||
| Immunogenicitya | γ | 0.8 |
| Clearancea | ρ | 1.0 × 10-4 |
| Antigenic distancea | 0 | |
| Epitope 2 | ||
| Immunogenicitya | γ | 1.2 |
| Clearancea | ρ | 1.0 × 10-4 |
| Antigenic distancea | 4 | |
| Intrinsic decay | gAg | (12 h)−1 |
| B cell parameters | ||
| B cell enhancement factor | εB | 10 |
| Ab enhancement factor | εAb | 2.5 |
| Naive B cell formation rate | kN | (4.6 × 105 h)−1 |
| Naive B cell stimulation | σN | (1 d)−1 |
| GC B cell stimulation (base) | σbase | (8 h)−1 |
| GC B cell stimulation (maximum) | σmax | (15 min)−1 |
| GC B cell replication rate | r | (8 h)−1 |
| Mutation probability | μ | 0.10 |
| Differentiation probability | δ | 0.10 |
| Memory cell stimulation | σM | (1 d)−1 |
| Ab production | kAb | 1.0 |
| Naive B cell decay rate | gN | (4.5 d)−1 |
| GC B cell decay rate (base) | gB | (4.5 d)−1 |
| Plasma cell decay rate | gP | (3 d)−1 |
| Ab decay rate | gAb | (10 d)−1 |
Adjusted parameter.
Throughout the simulation, each B cell clonal line is represented as a “subpopulation” and followed individually. As new BCR genotypes are introduced, either through the formation of naive B cells or through somatic mutation of existing B cells, a new clonal line corresponding to that genotype is added to the system, with an initial population of 1, and it increases or decreases as the simulation progresses. The overall populations of naive B cells, GC B cells, memory cells, plasma cells, and Abs are each represented as sets of clonal subpopulations.
In the model, there was a revolving population of 5 × 107 naive B cells (N) that was continuously replenished and whose formation and decay were described as zero-order and first-order reactions, respectively. The decay rate gN of naive B cells was set based on an estimated naive B cell half-life of 4.5 d (Eq. 2a). The B cell formation rate (kN) was set such that a steady-state population size of 5 × 107 naive B cells was maintained for the given decay rate (Eq. 2b). A newly formed B cell was assigned a random paratope genotype (denoted as j).
| (2a) |
| (2b) |
Two adjustments were made to modeling the naive B cell population. First, because the vast majority of naive B cells in the host repertoire have no binding affinity to a given set of Ags, for reasons of computational efficiency, only naive B cells with a nonzero binding affinity to any Ag in the system were explicitly modeled using Eq. 2a and 2b. This was done by modifying the decay and formation rates with the probability that a random naive B cell would be specific to a given Ag in the system (PAg), which was set to 10−5 (18). Second, naive B cells were limited to paratope genotypes with a minimum Hamming distance of 7 to any one epitope. This reflects the more limited diversity and low binding affinity of germ line BCRs expressed on naive B cells.
A starting naive B cell repertoire was generated with two criteria: that there be PAg ⋅ 5 × 107 Ag-specific naive B cells for each Ag (based on the Ag-specific B cell frequency PAg = 10−5) and that naive B cells be evenly distributed between each epitope. First, for each Ag, PAg ⋅ 5 × 107 B cells specific to either epitope 1 or epitope 2 were randomly generated to create an initial B cell repertoire. Second, this initial B cell repertoire was “rebalanced” to ensure that the appropriate number of B cells (PAg ⋅ 5 × 107) was present for any single Ag. This was done by randomly removing excess B cells for a given Ag and epitope until an equal number of B cells were present for all epitopes. Without rebalancing the initial B cell repertoire, B cells specific for cross-reactive epitopes shared between two Ags would be overrepresented because the B cells generated to be specific for one Ag would also be specific for a second Ag. This would increase the total number of Ag-specific B cells over the intended value (PAg ⋅ 5 × 107) and increase the proportion of Ag-specific B cells that target the cross-reactive epitope.
We modeled B cell stimulation as a second-order reaction between the Ag and the B cell. The rate of this reaction for a B cell with a BCR paratope of j (Nj for naive B cells, Bj for GC B cells) was determined by the base stimulation rate (σN and σB for naive and GC B cells, respectively), the immunogenicity of Ag epitope i (γi), and the binding affinity between the paratope j and epitope i (Qij [Eq. 1]). The immunogenicity parameter, γi, of an epitope (i), reflects that epitope’s ability to elicit an immune response, which, biologically, can be the result of a multitude of factors from epitope accessibility or arrangement to residue composition and similarity to self-Ags. We set values for immunogenicity for each epitope based on the relative Ab titers for each epitope determined from experimental data (see Results). The stimulation rate for both naive and memory B cells was set to (3 d)−1, which reflects experimental data that suggest that 50–80% of these cells begin division ∼75 h following Ag exposure (54). The B cell stimulation reaction has a maximum rate of σmaxBj, which reflects the maximum rate of GC B cell stimulation in the presence of excess Ag. As shown below, we modeled activation and migration of naive B cells into GC B cells and stimulation of GC B cells into stimulated GC B cells (denoted as B*) following Ag binding (Eq. 3a, 3b).
| (3a) |
| (3b) |
We modeled B cell proliferation as a first-order reaction (Eq. 4a), the products of which were the original B cell and a single daughter B cell of genotype k, which had, at most, one mutation from the parent genotype j. The rate of this reaction was a function of the B cell replication rate (r) and the probability of mutation from genotype j to genotype k (Rjk), which itself is a function of the mutation rate (μ) and is given by Rjk = (1 − μ)19(μ/3). Experimental studies suggested that B cell proliferation may have doubling times as high as 6–7 h; we set the value of r based on a more conservative doubling time of 8 h (55). Although in reality B cells consume Ag during activation, in our model they did not. We made the simplifying assumption that relative to nonspecific clearance processes in the body (Eq. 8) and Ab-based clearance (Eq. 7a), the overall rate of Ag clearance was not measurably affected by B cell consumption.
B cell differentiation was modeled as a symmetric process in which stimulated B cells have an equal probability of differentiating into plasma and memory cells at the rate of δ (Eq. 4b, 4c). Based on this model, the probability of a given activated B cell to differentiate is constant, but the overall rate of differentiation changes as the pool of activated GC B cells increases or decreases in size. This simple one-step model of B cell differentiation was necessary to efficiently model the B cell population at a repertoire level.
| (4a) |
| (4b) |
| (4c) |
| (4d) |
Affinity maturation in the GC occurs under high apoptotic pressure that drives the selection of higher-affinity BCRs. In this model, we applied a carrying capacity approach to apoptosis, where the apoptosis rate η was a function of the B cell replication rate (r) and the total GC B cell population (B) relative to the GC carrying capacity (κ) shown in Eq. 4d. At low GC B cell populations (B << κ), the GC B cell decay rate reflects the base B cell decay rate gB (half-life of 4.5 d). However, as the GC B cell population reaches carrying capacity (B ≈ κ), the GC B cell decay rate approaches the B cell replication rate (half-life of 8 h), halting further net GC B cell population growth. This reflects the high apoptotic pressure within the GC as the B cell population reaches carrying capacity, which is limited by both physiological constraints, such as the physical space within the GC, and biological constraints, such as the availability of T cell help. This high rate of apoptosis provides a selection pressure that favors B cells that are stimulated faster over those that are stimulated more slowly.
Plasma cells produce serum Abs (Eq. 5a) and decay at a rate (gP) based on a half-life of 3 d (Eq. 5b). Memory cells act identically to naive B cells (Eq. 6) but do not decay. Because the time scale of the simulation is from days to weeks, we only modeled shorter-lifespan plasma cells descended from either naive B cells or memory cells, which are thought to dominate the acute Ab response and have half-lives of less than a week (19, 56, 57). Similarly, memory cells, which are estimated to have lifespans of months to years (58, 59), do not decay in the simulation.
| (5a) |
| (5b) |
| (6) |
Abs produced by plasma cells bind to the Ag and clear it through a second-order reaction (Eq. 7a), with a reaction rate that is a function of the binding affinity between the Ab paratope j and the Ag epitope i (Qij), as well as the clearance or neutralization parameter of epitope i (ρi). This neutralization parameter reflects the fact that different epitopes can have different neutralization characteristics, including some epitopes that might be entirely nonneutralizing. Abs decay with a rate gAb based on a half-life of 10 d (Eq. 7b), which is an approximation within the range of serum IgG Ab half-lives for mice (6–8 d) (60) and humans (7–21 d) (61).
| (7a) |
| (7b) |
Finally, there is an intrinsic rate of Ag decay (gAg) due to nonspecific clearance processes in the body (Eq. 8). For the purpose of this work, the AMA1 Ag was assigned a half-life of 12 h. Although there is no measured value for the half-life for recombinant AMA1, this number is well within a range of normal serum half-life values for a recombinant protein in the human body (62).
| (8) |
Simulation conditions
The simulation conditions were set up to reflect the vaccination strategy used by Dutta et al. (47) for the tetravalent AMA1 vaccine. For the monovalent vaccine, 360 Ag units was inserted into the system at days 1, 28, and 56. For the polyvalent vaccine, 90 units of each of the four Ags were inserted into the system at the same intervals. Each Ag was represented as two epitopes: a conserved epitope, which had an antigenic distance of 0 between all serotypes, and a polymorphic epitope, which had an antigenic distance of 4 between each serotype (see Results).
In this stochastic model, each simulation produced a unique trajectory that represented the outcome of a single individual. We carried out 50 runs for both monovalent and polyvalent vaccinations. Each simulation was run out to 63 d, 1 wk following the final immunization, and lasted ∼10 min on a desktop computer. We report the median values, as well as the overall distribution of outcomes.
Simulated in vitro results
To compare the simulation results with experimental data, we used the output from the simulations to predict experimental in vitro results for two types of assays: Ab titer assays based on ELISA and serum-depletion and -neutralization assays. In the simulated ELISA assay, the total Ab concentration that has nonzero binding to a specified target Ag is shown on a log scale. We used a model non–vaccine strain, S5, as the heterologous Ag in the simulated in vitro results. Strain S5 is a strain that was not found in the monovalent (S1) or polyvalent (S1, S2, S3, and S4) vaccines and had pairwise antigenic distances of 0 and 4 for epitopes 1 and 2, respectively, to all other strains. An Ab or B cell is described as cross-reactive with respect to two Ags if it has nonzero binding affinity to both Ags, as defined by Eq. 1.
The simulated neutralization and depletion assay was carried out in two steps. First, the Abs that were specific for a given depleting Ag epitope k (Abi, for which Qik > 0) are removed. Then, the neutralization capacity of the remaining Abs to the target Ag epitope j was calculated as a function of Ab concentration Abi, the neutralization rate of epitope j (ρj), and binding affinity Qij for all remaining Ab genotypes (Eq. 9).
| (9) |
During the depletion step, Abs specific to any of the epitopes in the depleting Ag are removed. Neutralization is reported as a percentage of neutralization relative to undepleted serum. This simplified model for neutralization was used to represent the Ab response as a function of both Ab titer and avidity.
Results
Overall immune system model
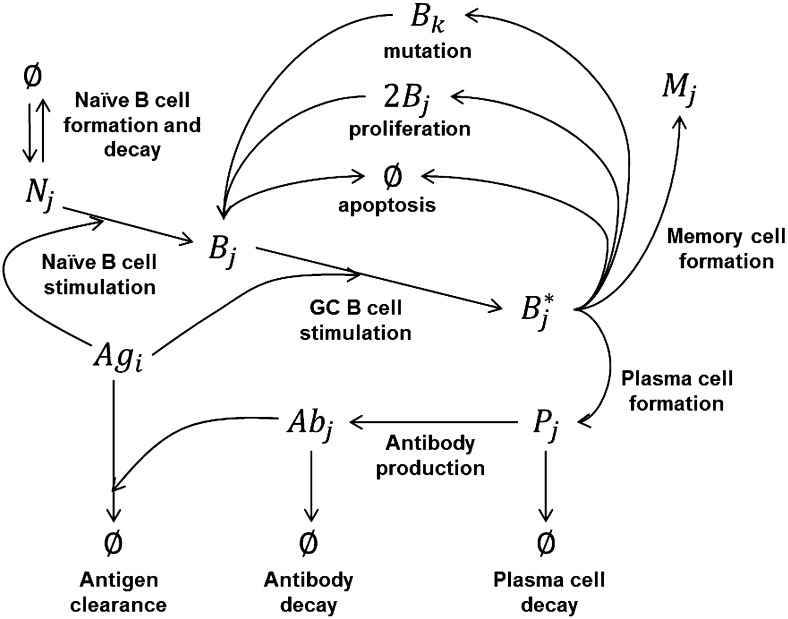
Fig. 1 provides a summary of the immune system model used in this study to simulate affinity maturation. Briefly, Ag-specific naive B cells (N) in the host B cell repertoire bind to Ag and migrate to the GC and become GC B cells (B). Within the GC, these B cells continue to bind Ag and become stimulated GC B cells (B*). Stimulated GC B cells proliferate and mutate into unstimulated daughter cells (B) and differentiate into mature plasma (P) and memory (M) B cells. The GC B cell population is under increasing apoptotic pressure as it approaches the GC carrying capacity. Plasma cells produce Abs that bind to the Ag, leading to Ag clearance.
FIGURE 1.
Immune system model for affinity maturation. A summary of the immune system model that is used to simulate affinity maturation, including B cell stimulation, mutation and proliferation, differentiation, Ab production, and Ag clearance. The components in the model include Ags, naive B cells (N), GC B cells (B), stimulated GC B cells (B*), memory cells (M), plasma cells (P), and Abs. Subscripts denote epitope and paratope genotypes.
We designed this affinity-maturation model to be minimally dependent on extensive fitting and parameterization. Table I defines all major model parameters and their corresponding symbols used in Materials and Methods. Many parameters, such as the B cell replication rate of 8 h−1, reflect standard established literature values (63). Immunological shape space parameters, which define the BCR and Ab–Ag interactions, were kept at the values defined by Smith et al. (18). Likewise, B cell parameters (such as B cell formation and decay rates) and mutation and differentiation probabilities were set to values defined by Smith et al. (18, 19) or Celada and Seiden (13). We adjusted the B cell stimulation rates to roughly approximate affinity-maturation kinetics with respect to the number and frequency of somatic mutations.
We created a simplified model of the polymorphic AMA1 Ag. The polyvalent vaccine candidate tested by Dutta et al. was a subunit-based vaccine that consisted of a mixture of recombinant AMA1 proteins from four strains of P. falciparum (3D7, FVO, HB3, and W2mef) (47). In our model, each AMA1 Ag consisted of two epitopes: a conserved epitope (epitope I) and a polymorphic epitope (epitope II). We modeled four strains of AMA1, termed S1, S2, S3, and S4. The four strains shared an identical epitope I, but each had a unique epitope II. In the immunological shape space model, a paratope has maximum complementarity to an epitope if the Hamming distance between their sequences is ≤4. Likewise, a Hamming distance of 7 defines the maximum distance with the minimum nonzero binding affinity.
We fitted two parameters for each epitope: immunogenicity and antigenic distance, based on experimental data from monovalent AMA1 vaccination. The immunogenicity of the conserved face (γ1) was set to 0.8 and that of the polymorphic face (γ2) was set to 1.2, to reflect that ∼80% of the Ab response in a monovalent AMA1 vaccination was toward epitopes on the polymorphic face. We set pairwise antigenic distance values between each of the four strains for each epitope. For the four conserved epitope I sequences, this distance was set to 0, meaning that all four strains had the same sequence for epitope I. For the four polymorphic epitope II sequences, this distance was set to 4, meaning that all four epitope II sequences differed from each other by a Hamming distance of 4. We selected these antigenic distance values to reflect ∼100% cross-reactivity of Abs to the conserved face epitope and ∼20% cross-reactivity of Abs to the polymorphic face epitope suggested in the monovalent vaccination results (47). In cases in which experimental data from a vaccination study are unavailable, the antigenic distance can be inferred from in vitro testing of serum samples (31) or estimated from epitope sequence identity (64, 65). A third epitope-specific parameter, neutralization (ρ1 and ρ2), was set to 1.0 for both epitopes to reflect experimental data, which suggested that both epitopes have comparable neutralization characteristics (47).
Affinity maturation kinetics
The affinity maturation model is stochastic, meaning that each simulation represents a unique trajectory of a single vaccination event. For each condition, we ran 50 simulations and present the average results. Fig. 2 (upper panels) shows the overall output of the monovalent vaccine simulation in terms of Ab and Ag levels, B cell subpopulations, and Ab and BCR affinities. In Dutta et al. (47), rabbits were immunized on days 0, 28, and 56 and sacrificed on day 63 for serum analysis. The same vaccination schedule was used in the simulation. Each subsequent boost led to an increase in the Ag-specific Ab levels. Fig. 2A shows the underlying B cell populations. On day 0, naive B cells were stimulated, leading to a growing population of stimulated GC B cells, which slowly differentiated into memory and plasma B cells. Subsequent boosts were carried out well beyond the half-life of naive and GC B cells (4.5 d) and were triggered primarily through stimulation of memory B cells. These memory B cells re-entered the affinity maturation process, eventually differentiating into mature memory and plasma cell populations.
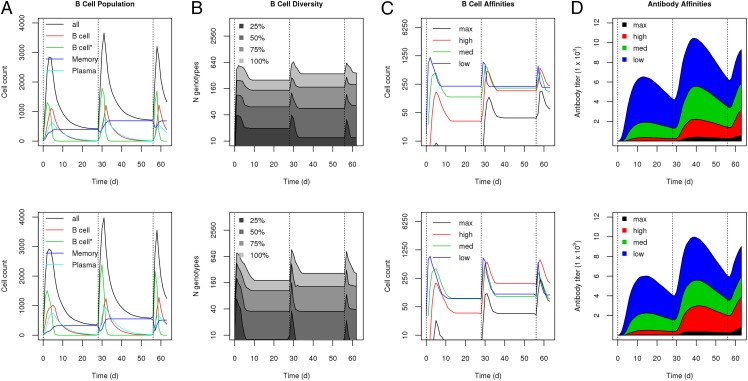
FIGURE 2.
Affinity maturation simulation results for 50 independent trajectories for monovalent (upper panel) and polyvalent (lower panel) vaccinations. Median values are shown for each panel. (A) B cell populations, including GC B cells, stimulated GC B cells, memory cells, and plasma cells. (B) B cell diversity, with the number of unique genotypes that make up 25, 50, 75, and 100% of the total B cell population. B cell populations (C) and Ab levels (D) with respect to affinity to the Ag strain S1 for different levels of affinity, from low affinity (Hamming distance = 7) to high affinity (Hamming distance = 4).
In the simulation, we explicitly modeled individual B cell clonal lines, or genotypes. Fig. 2B illustrates the diversity of the B cell population. Overall, for monovalent vaccination, ∼20 clonal lines made up 25% of the B cell population, and ∼40 clonal lines made up >50% of the B cell population, whereas the entire population consisted of >330 clonal lines. This shows that, although the overall response is highly diverse, there is significant enrichment of ∼10% of the B cell clonal lines in the immune response. Fig. 2C shows a breakdown of the B cell population in terms of subpopulations by binding affinity to the Ag, from low affinity (Hamming distance = 7) to maximum affinity (Hamming distance = 4). The results show that, over time and through subsequent boost immunizations, higher-affinity B cells are significantly enriched for. This enrichment of higher-affinity B cells is also reflected in the Ab response (Fig. 2D): medium- and high-affinity Abs begin to dominate the total Ab response by the third boost immunization. Overall, Fig. 2 illustrates the various stages of affinity maturation modeled in this simulation: Ag presentation leads to stimulation, proliferation, mutation, and differentiation of B cells, which leads to increased B cell diversity, enrichment of higher-affinity B cells, and, finally, maturation of a high-affinity Ab response.
Polyvalent vaccination
In the polyvalent vaccine simulation, immunization consisted of 90 units of each of four strains, instead of 360 units of a single strain, to ensure that total Ag dose was held constant while Ag composition was altered. Fig. 2 (lower panels) shows the results of 50 simulations run under the polyvalent vaccine condition. Overall, the results were very similar to the monovalent vaccine condition. Subsequent boost immunizations led to higher Ab titers and higher Ab affinities. Higher-affinity B cells made up a significant portion of the GC B cell population by the second boost and dominated the B cell population by the third boost. Similarly, following the third boost, >50% of the Abs in the simulation consisted of high-affinity Abs. There were marginally higher Ab titers and avidities in the polyvalent vaccination case but no other significant differences from the monovalent vaccination. Interestingly, polyvalent vaccination led to a greater enrichment of B cell clonal lines; ∼10 clonal lines made up 25% of the B cell population, and 30 clonal lines made up 50% of the population, despite the fact that total B cell diversity was approximately the same as in the monovalent case, with ∼330 genotypes represented in the entire population.
Simulated in vitro experiments
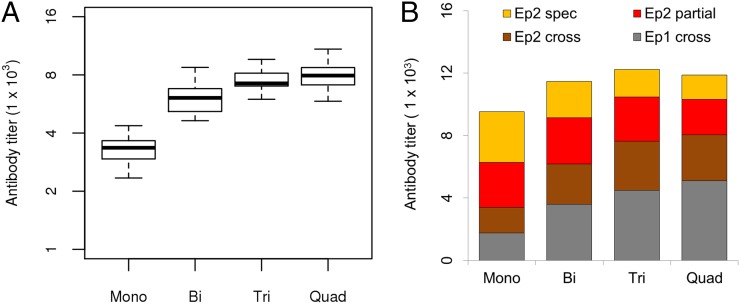
The affinity maturation simulation yielded a profile of the Ab response at day 63, 1 wk after the third immunization. We sought to determine whether the polyvalent vaccine simulation successfully predicted the specificity, cross-reactivity, and neutralization of the Ab response in experimental data. Recall that only monovalent vaccine experimental data were used to determine parameters for the simulation. We compared our simulation results to the results of two experimental assays carried out by Dutta et al. (47): indirect ELISA, which measures the total amount of Ab that binds to a given target Ag, and growth inhibition reversal assays (GIA reversal), which measure the amount of neutralization by an Ab sample that is reversed following depletion using a depleting Ag. The first assay measures the overall Ab response, whereas the second assay quantifies the degree of cross-reactivity of that response. When referring to prior experimental work by Dutta et al. (47), monovalent vaccination refers to vaccination using 3D7 AMA1, and polyvalent vaccination refers to their tetravalent QV AMA1 vaccine. Because the experimental data from Dutta et al. (47) are derived from pooled sera (n = 3), we used average results from the simulations for comparison. However, we present the SDs of the simulation results to illustrate the variability in the simulation data, which reflects the intersubject variability in the model.
Fig. 3A shows the simulated and experimental ELISAs to four target Ags. The data are shown as a percentage of the total response to the homologous strain (strain S1 in the simulation, strain 3D7 in the experimental data), defined as the strain used in the monovalent vaccination. S5 represents a heterologous strain that is not included among the four strains in the polyvalent vaccine. The heterologous strain experimental results shown reflect the median value for three heterologous strains 7G3, M24, and 102-1 from Dutta et al. (47) POLY refers to a recombinant chimera of AMA1 that includes only the polymorphic face, whereas CONS refers to a recombinant chimera that includes only the conserved face (47).
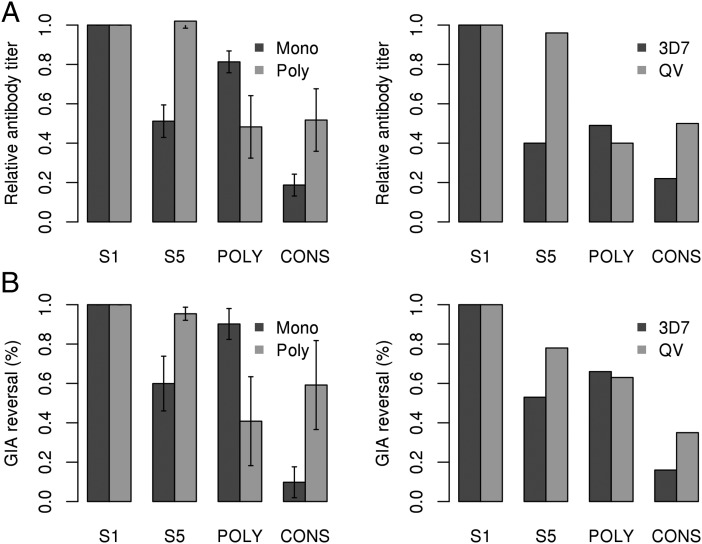
FIGURE 3.
Simulated in vitro results compared with prior experimental data. (A) The median Ab titers from the monovalent and polyvalent simulations are reported for the homologous (strain S1) and heterologous (strain S5) Ags, as well as a model chimera containing only the polymorphic (POLY) or conserved (CONS) epitope. Experimental results for monovalent (3D7) and polyvalent (QV) vaccination against homologous (strain 3D7) and heterologous (strains 7G3, M24, and 102-1) Ags, as well as recombinant POLY and CONS AMA1 chimeras. (B) Median simulated GIA reversal and experimental GIA reversal assays for the same Ags as above used for depletions. SDs are given for the simulated results; all experimental data are from Dutta et al. (47) and are derived from pooled serum samples. All differences in the simulation results between monovalent and polyvalent vaccinations were significant based on a Welch t test (p < 10−5).
The simulation results show that the Ab titers to the heterologous strain S5 were 50% of the titer to the homologous strain S1 for the monovalent vaccine. In the polyvalent case, the titers to both strains were the same. This was similar to the experimental data: the polyvalent vaccine showed equal Ab titers to all four vaccine strains, whereas the monovalent vaccine showed ∼40% of the response to heterologous strains. The simulated ELISA results using the POLY and CONS chimera revealed that 80% of the Ab response in the monovalent vaccination was directed toward the polymorphic face, and 20% was directed to the conserved face, whereas the polyvalent vaccine showed ∼50% response to each face. This also agrees well with the prior experimental data, which showed that the monovalent 3D7 vaccine showed a significantly higher response to the polymorphic face than to the conserved face (50% versus 20%), whereas the polyvalent QV vaccine showed an approximately equal response to both faces (40% versus 50%). One difference in the experimental data lies in the relative response of the monovalent vaccine to the polymorphic face: 80% in the simulation compared with 50% in the experiment. Interestingly, in the experimental data, the monovalent response to the POLY and CONS chimeras do not add up to the total response to AMA1, indicating that this discrepancy might be a limitation of the two-epitope model used in the simulation. Overall, these results show that the simulation successfully recapitulated the trends observed in the experimental data with respect to the overall Ab response for both homologous versus heterologous strains, as well as to the polymorphic versus conserved face.
Fig. 3B shows the simulated and experimental GIA reversal assay results using the same four Ags described above (S1, S2, POLY, and CONS), as depleting Ags. In this assay, one of the four Ags was used to deplete the serum, followed by testing for neutralization using the S1 strain. As expected, in both the simulation results and the experimental data, depletion using the homologous Ag (S1 for simulation, 3D7 for experimental) resulted in complete reversal of inhibition. In the simulation results, depletion using the heterologous S5 strain resulted in only a 60% reversal of inhibition for the monovalent response and a 95% reversal for the polyvalent response, indicating that the polyvalent response is highly cross-reactive and that the same Abs that bind to the heterologous Ag bind to the homologous Ag. These results agree with the prior experimental work, in which depletion with a heterologous Ag resulted in an average 50% reversal of inhibition in the monovalent responses and an average 80% reversal in the polyvalent QV vaccine. In the simulation results, depletion using POLY and CONS resulted in a 90 and 10% reversal of inhibition in the monovalent response, respectively, compared with a 40 and 60% reversal in the polyvalent response. These results agree to some degree with experimental work by Dutta et al. (47), which showed that POLY and CONS depletions led to 70 and 15% reversal in the monovalent response and 60 and 40% reversal in the polyvalent response. The lack of quantitative agreement may reflect the limitations of modeling a complex phenomenon like Ab-dependent inhibition in an in vitro growth assay using simple characteristics, such as Ab titer, binding affinity, and specificity. Overall, however, the simulation results reproduced the experimental finding that the polyvalent response is highly cross-reactive and well-balanced between both the polymorphic and conserved face, whereas the monovalent vaccine was largely strain specific and biased toward the polymorphic face.
Both our simulation results and prior experimental work identified three key outcomes. First, the polyvalent response had equal neutralization to homologous and heterologous/non–vaccine strains, whereas the monovalent response had ∼50% neutralization to heterologous strains. Second, the polyvalent vaccination showed an ∼2-fold increase in the Ab response toward the conserved face. Third, polymorphic face Abs were responsible for at least half of the neutralization of the polyvalent response. These results present a conundrum: if conserved face Abs represent only 40% of the polyvalent response, and polymorphic face Abs contribute a significant portion of the neutralization, how does the polyvalent Ab response achieve 80–100% neutralization of heterologous nonvaccine strains? To address this issue, we analyzed the fine specificity of the polyclonal Ab response modeled in the simulation.
Ab fine specificity
In this affinity maturation model, B cells and Abs were represented at the genotype level, enabling us to analyze both the fine specificity of the Ab response with respect to each epitope and the degree of cross-reactivity to heterologous strains. Fig. 4A shows the overall Ab response for both the monovalent and polyvalent vaccine simulations. As was seen earlier, subsequent boosts resulted in increased Ag-specific Ab levels. We used the end point values for the Ab response at day 63 to determine the Ab titer results in Fig. 3A, which showed that the monovalent response was largely biased toward the conserved face epitope, whereas the polyvalent response was evenly balanced between the conserved and polymorphic epitopes. Fig. 4A shows that, through subsequent boosts, the relative Ab response to the polymorphic face increased in both the monovalent and polyvalent vaccine simulations. This was especially striking in the polyvalent case: the polymorphic face response was 20% of the total strain S1-specific response after the first immunization and 55% of the response after the third.
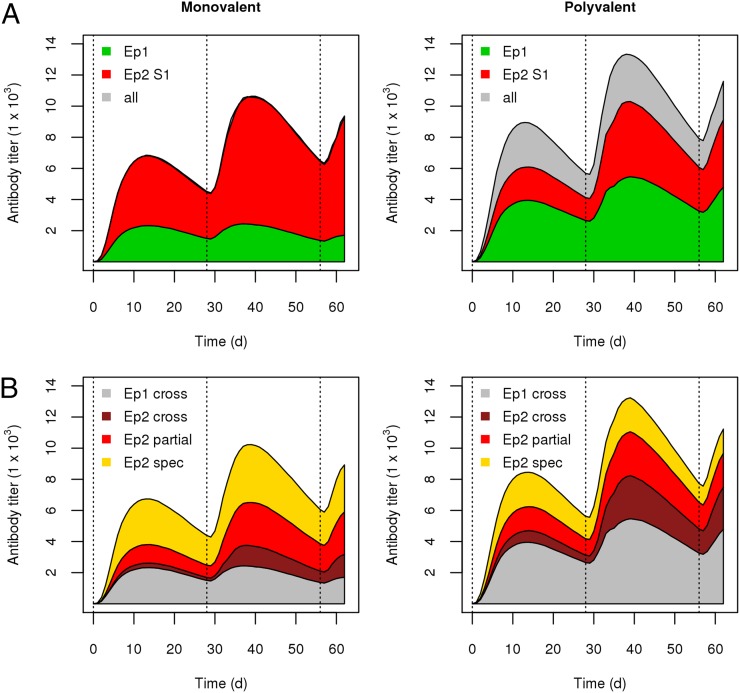
FIGURE 4.
Fine specificity and cross-reactivity of the Ab response. (A) The Ab response toward the conserved (Ep 1) and polymorphic (Ep 2) epitopes for strain S1 in the monovalent and polyvalent vaccine simulations. (B) The Ab response is further broken down with respect to reactivity across multiple Ag alleles: fully cross-reactive (cross), partially cross-reactive (partial), and strain-specific (spec) for both epitopes.
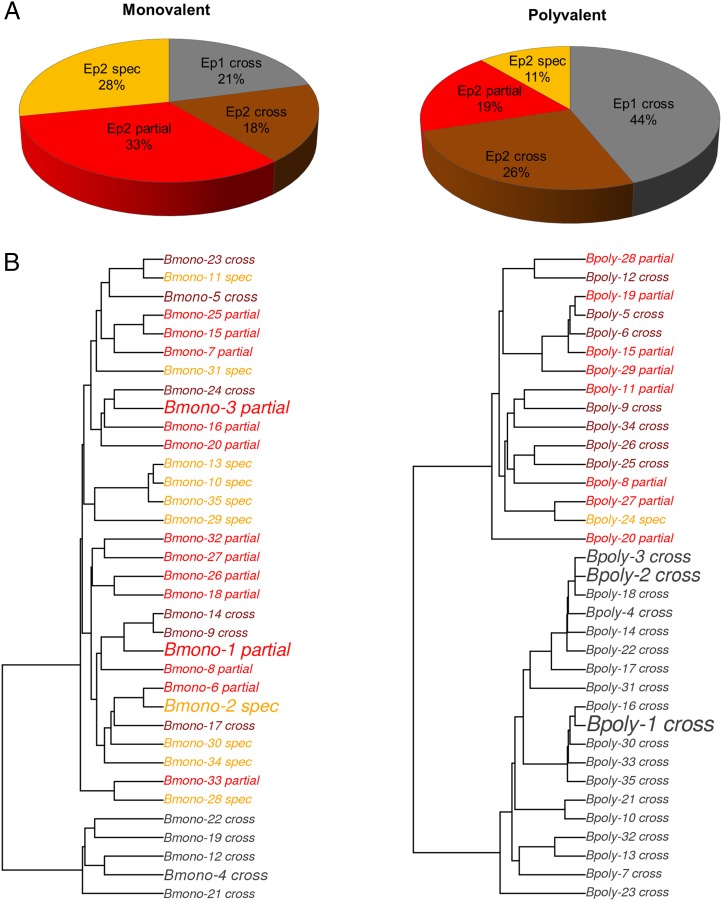
Next, we assessed the degree of cross-reactivity of the simulated Ab response. In Fig. 4B, the Ab response is broken down by both epitope and the degree of cross-reactivity from cross-reactive to all four strains, cross-reactive to more than one strain but not all four, and specific to only one strain. For the monovalent vaccine simulation, 32% of the Ab response was fully cross-reactive, and 35% was strain-specific, whereas in the polyvalent vaccine simulation 67% of the response was fully cross-reactive, and 11% was strain specific. In addition to the shift previously observed for the polyvalent vaccine—that there was an ∼2-fold enhancement of the conserved-face Ab response—a second, related shift was present. In the monovalent vaccine simulation, 19% of the polymorphic-face Ab response was fully cross-reactive, and 42% was strain specific. By contrast, in the polyvalent vaccine simulation, 39% of this response was fully cross-reactive, whereas 21% was strain specific. Finally, the epitope-level cross-reactivity of the response varied with time, and subsequent immunization boosts led to an increase in the proportion of the polymorphic-face response that was partially or fully cross-reactive. Interestingly, the overall proportion of the Ab response that was fully cross-reactive seemed to be largely constant, at ∼30% in the monovalent simulation and ∼70% in the polyvalent simulation.
These results show that, despite the same overall Ag dose, there was a substantial shift in the fine specificity and cross-reactivity of the Ab response during polyvalent vaccination. This was reflected in an enhancement of the conserved face Abs, as well as the cross-reactive polymorphic face Abs. This shift suggests that the polymorphic face Ab response was qualitatively different in the polyvalent vaccine than in the monovalent vaccine and provides a possible explanation for the broad neutralization observed in the polyvalent response. Furthermore, it suggests that a large percentage of the Abs in the polyvalent response (>66%) was truly strain transcending.
Number of strains and strain dilution
The simulations presented in this article demonstrate that a polyvalent vaccine consisting of four strains showed an apparent enhancement of cross-reactive Abs toward both the conserved and polymorphic faces of AMA1. This was most clearly reflected in the high Ab titers to non–vaccine heterologous strains. We ran the polyvalent simulation again, this time including two (bivalent) and three (trivalent) strains instead of four, to explore how this enhancement effect is related to the number of strains used in the polyvalent vaccine. In Fig. 5A, we show the Ab titers of each of these polyvalent vaccines to a heterologous non–vaccine strain, in which the polymorphic epitope had an antigenic distance of 4.
FIGURE 5.
Monovalent, bivalent, trivalent, and tetravalent vaccine responses. (A) The Ab response toward a heterologous, non–vaccine strain is shown from simulations of monovalent, bivalent, trivalent, and tetravalent vaccinations. (B) The cross-reactive, partially cross-reactive, and strain-specific Ab response to the conserved (Ep 1) and polymorphic (Ep 2) epitopes is shown for monovalent, bivalent, trivalent, and quadvalent vaccine conditions.
The results show that there was substantial enhancement of the heterologous response in the bivalent and trivalent conditions compared with the monovalent condition. Overall, the bivalent and trivalent formulations resulted in an ∼60 and ∼80% increase, respectively, in Ab titers against a non–vaccine strain relative to the monovalent formulation compared with an ∼100% increase for the tetravalent formulation. Furthermore, the degree of variation in the titers of the monovalent and bivalent simulations was much higher than in the trivalent and tetravalent simulations. These findings agree with the experimental results of Dutta et al. (47), which show, for a variety of inhibition assays and heterologous strains, that the trivalent vaccine performs almost as well as the tetravalent vaccine, whereas the bivalent vaccine performs measurably worse.
We next analyzed the fine specificity and cross-reactivity of the Ab response for all four vaccination conditions. In Fig. 5B, the Ab titers to each epitope are plotted, along with their cross-reactivity. The results show two trends as the number of strains in the vaccine increased: the percentage of the Ab response directed toward the conserved face increased, in a largely linear fashion, and the percentage of the polymorphic face Ab response that was fully cross-reactive increased, reaching a maximum of ∼40% of the total polymorphic face response in the trivalent and tetravalent conditions. These results highlight the strain-dilution effect observed in this work: the introduction of additional strains diluted the relative response of strain-specific Abs and enhanced the response of partial and fully cross-reactive Abs to both polymorphic and conserved face epitopes. We next sought to identify the mechanism for this strain-dilution effect by analyzing the B cell response underlying the Ab response in the simulation.
B cell specificity in a polyvalent vaccine
Like Abs, B cell genotypes are explicitly modeled in our simulation. Fig. 6A shows the profile of the B cell population 1 wk after the third immunization. As expected, the B cell population largely reflects the Ab response with respect to both epitope specificity and cross-reactivity. We found that 21% of the B cell population in the monovalent simulation was specific to the conserved face epitope compared with 44% in the polyvalent simulation, and a total of 39% of the B cell population was fully cross-reactive in the monovalent simulation compared with 70% in the polyvalent simulation. By contrast, the monovalent B cell response was 28% strain specific, whereas the polyvalent response was only 11% strain specific.
FIGURE 6.
Memory B cell response. (A) A breakdown of the memory B cell response in terms of specificity for the conserved (Ep 1) and polymorphic (Ep 2) epitopes in the monovalent (left) and polyvalent (right) vaccine simulations. (B) A sequence phylogeny tree of the 35 largest B cell clonal populations, which represent ∼30–40% of the total B cell population, colored with respect to epitope and cross-reactivity in the same scheme as in (A). The font size reflects the relative population size of that clonal line.
Recent studies using high-throughput sequencing of B cell repertoires were able to determine phylogenetic maps of the B cell population that provided the first detailed glimpses of a polyclonal B cell population’s makeup (66, 67). We provide a similar analysis of our simulated B cell repertoires with a phylogenetic tree for 35 clonal lines with the largest populations, from a representative case for a monovalent and polyvalent vaccination simulation (Fig. 6B). The phylogenetic trees of these high-frequency B cell clones reflect the overall B cell population results in Fig. 6A. B cell clones from the polyvalent simulation were much more likely to be specific to the conserved face or specific to the polymorphic face and cross-reactive. These results show that, in the polyvalent simulation, the broadly neutralizing cross-reactive Ab response resulted from a highly cross-reactive B cell response.
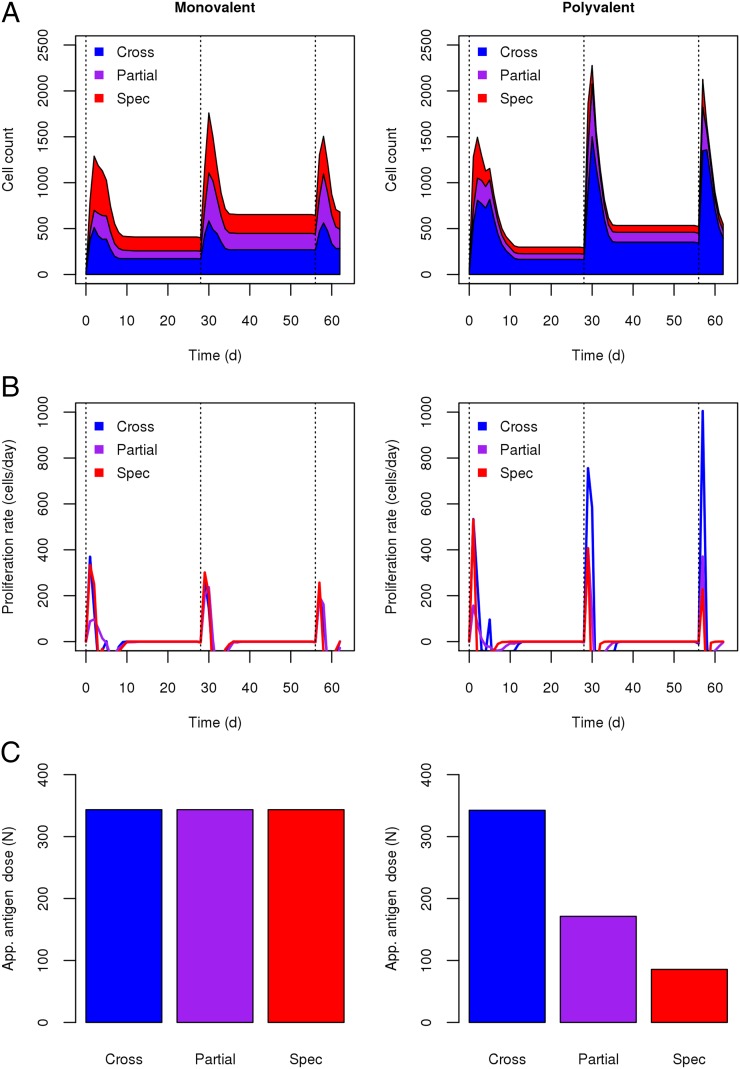
To determine the underlying mechanisms for the enhancement of this cross-reactive B cell response in the polyvalent simulation, we analyzed the dynamics of the B cell population with respect to epitope specificity and cross-reactivity. Fig. 7A shows a profile of the B cell response as a function of time in monovalent and polyvalent simulations. As was seen in the Ab response, subsequent boost immunizations led to an increased B cell response. Furthermore, it is clear that, even at a very early stage, the polyvalent simulation had much higher levels of cross-reactive B cells. Fig. 7B plots the growth rate of the B cell populations with respect to their epitope specificity and their cross-reactivity. These results show that cross-reactive B cells grew at a significantly higher rate (approximately 2–3-fold) than did the strain-specific B cells in the polyvalent simulation but not in the monovalent simulation.
FIGURE 7.
B cell cross-reactivity and proliferation rate. (A) The number of B cells with fully cross-reactive, partially cross-reactive, and strain-specific Ag specificity for monovalent (left panel) and polyvalent (right panel) vaccination conditions. (B) The growth rate of B cell populations (dB/dT) corresponding to cross-reactive, partially cross-reactive, and strain-specific B cells for monovalent (left panel) and polyvalent (right panel) conditions. (C) Epitope dose for fully conserved (Cross), partially conserved (Partial), and strain-specific (Spec) epitopes in monovalent (left panel) and polyvalent (right panel) vaccine formulations. Total Ag dose was 360 units.
B cells not only bind to Ag, they bind to a particular epitope on the Ag. As a result, the overall Ag dose does not necessarily reflect the apparent Ag dose for a given B cell line. This apparent Ag dose is a reflection of the total number of epitopes in the system for which that B cell is specific. Although the overall Ag dose was the same in both the monovalent and polyvalent cases, the relative abundance of each epitope in the system varied between the two. In the monovalent case, cross-reactive, partially cross-reactive, and strain-specific B cell lines were exposed to the same apparent Ag dose (Fig. 7B). However, in the polyvalent case, B cell lines that were cross-reactive to all four strains were exposed to an apparent Ag dose that was four times higher than the dose to which the strain-specific B cell lines were exposed. This apparent Ag dose was the key mechanism underlying the strain dilution effect seen in the polyvalent vaccine simulation. As the number of strains in the polyvalent vaccine increased, the selective advantage of cross-reactive B cells over strain-specific B cells increased, because the relative Ag dose for cross-reactive B cells increased. By contrast, in a monovalent vaccine, cross-reactive B cells had no selective advantage over strain-specific B cells because both were exposed to the same apparent Ag dose.
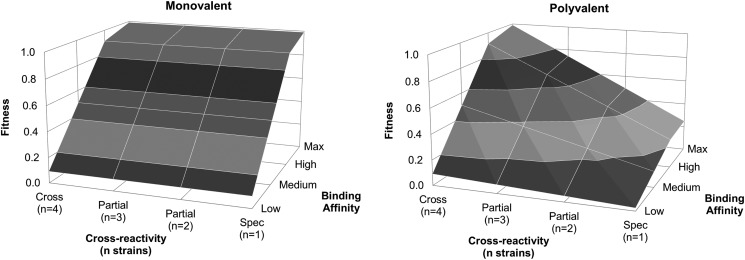
The selective advantage of cross-reactive B cells in affinity maturation during a polyvalent vaccination can most clearly be represented as a “fitness landscape” (Fig. 8), which plots the fitness of a B cell line as a function of two attributes: its binding affinity and its cross-reactivity. “Fitness” refers to the proliferation rate of a B cell line (based on Eqs. 5 and 6 in the simulation), with a value of 1 indicating maximum proliferation. As is the case in classical affinity maturation models, B cells with higher binding affinities show higher fitness, which is a reflection of the faster stimulation and proliferation rates; as affinity increases from low to max, B cell fitness increases monotonically in both monovalent and polyvalent conditions. Because our model explicitly included multiple Ag strains, we can analyze the fitness landscape with regard to cross-reactivity. We showed earlier that, in monovalent conditions, B cells were exposed to the same apparent Ag dose regardless of cross-reactivity, whereas in the polyvalent case, cross-reactive B cells were exposed to higher apparent Ag levels (Fig. 7C). We see those effects on B cell fitness. In the monovalent case, B cells had the same fitness regardless of cross-reactivity, whereas in the polyvalent case, a B cell’s fitness increases as the number of strains to which it is reactive increases. In monovalent simulations, the fitness peak corresponded to all high-affinity B cells, whereas in polyvalent simulations, the fitness peak corresponded to fully cross-reactive, high-affinity B cells. These fitness landscapes guide the evolutionary behavior of B cells during affinity maturation, suggesting that, in polyvalent vaccination, B cell lines that are initially more cross-reactive are favored over those that are not and that mutations during affinity maturation that increase cross-reactivity are favored as well.
FIGURE 8.
B cell fitness landscape during affinity maturation. The fitness landscape for B cells during affinity maturation in the monovalent (left panel) and polyvalent (right panel) simulations, as a function of cross-reactivity and binding affinity. A fitness of 1 represents the highest fitness; cross-reactivity is listed as the number of strains that the B cell line is specific to, and the binding affinity is shown as low, medium, high, and max, corresponding to Hamming distances of 4, 5, 6, and 7.
Discussion
In this study, we sought to apply a first-principles approach to simulating the progression of B cell affinity maturation to predict the fine specificity of an Ab response in the case of a real-world polyvalent vaccine candidate. We combined the theoretical model of immunological shape space, developed by Smith et al. (18) to describe BCR and Ab interactions, with Ags, using the stochastic approach to immune system dynamics developed by Woo and Reifman (26) to model B cell population dynamics during affinity maturation. The model was designed to have a minimal amount of adjustable parameters: the Ag dose, the number of Ag strains, the number of epitopes/Ag, the immunogenicity of each epitope, and the antigenic distance between each epitope among the different strains. To our knowledge, this work represents the first computational simulation of affinity maturation that incorporates both multiple strains and multiple epitopes, which are key requirements to modeling a polyvalent immune response. We applied this approach to study the immune response to the polyvalent AMA1 Ag and used the simulation results to recapitulate experimentally verifiable characteristics of the immune response in terms of specificity and cross-reactivity.
Our theoretical results provide a mechanistic explanation for the experimental studies carried out by Dutta et al. (47) and other investigators (48) that directly compared monovalent and polyvalent vaccine Ab responses. We show qualitative, and sometimes quantitative, agreement with the Ab titers and efficacies in the monovalent and polyvalent conditions, as well as in the depletion studies that determined the degree to which the responses are strain specific and cross-reactive. We also show good agreement with their findings on the relative response to conserved and polymorphic face epitopes, as well as the degree to which the enhancement of cross-reactivity is observed in bivalent, trivalent, and tetravalent formulations. Overall, with minimal parameterization, we reproduced a number of the trends observed in prior experimental studies with respect to fine specificity, cross-reactivity, and efficacy.
We successfully recapitulated the experimental observations that the polyvalent AMA1 vaccine enhances the immunogenicity of cross-reactive epitopes (47). In our simulations, this cross-reactive response in polyvalent vaccination is a result of an enhanced response to conserved face epitopes, as well as greater cross-reactivity to polymorphic face epitopes. Furthermore, our model suggests that this cross-reactivity is largely (but not entirely) a result of strain-transcending Abs to both conserved and polymorphic face epitopes. Although further experimental studies are needed to confirm this hypothesis, several findings from previous studies (46–49), such as a largely balanced response across vaccine strains, comparable efficacy to non–vaccine strains, similar results following heterologous depletion, and increased cross-reactivity relative to pooled monovalent sera, are all consistent with the theory that the broadly neutralizing activity of the polyvalent vaccine is largely a result of strain-transcending Abs.
Our model attributes the mechanism underlying the enhanced cross-reactivity in the polyvalent response to a strain dilution effect, in which the introduction of additional polymorphic strains results in an increased selection of cross-reactive B cells over strain-specific B cells during affinity maturation. This effect occurs because, as the number of strains in the polyvalent vaccine increases, the apparent Ag dose available for cross-reactive B cells increases relative to the Ag dose for strain-specific B cells. This increase results in a selective pressure that favors increasing binding affinity (as is the case in classical models of affinity maturation), as well as increasing cross-reactivity. The effects of this selective pressure can be described as follows: in the GC environment in which B cells must compete with each other for survival and proliferation, the B cells that are specific to more Ags have higher growth rates and dominate the population. In a polyvalent vaccine, as the number of strains increases, the selective advantage of cross-reactive B cells increases, resulting in an increasingly cross-reactive Ab response.
There is a subtle difference between our proposed mechanism, which is based on B cell selective fitness, and the “epitope-dilution” concept proposed by Dutta et al. (47). As would be predicted by epitope dilution, our simulations show an enhanced response to the conserved epitope, but they also show an enhanced cross-reactivity to the polymorphic epitope. Because of the degeneracy of BCR-epitope binding, many distinct B cell clonotypes can bind to the same epitope, and not all clonotypes that bind to the polymorphic epitope are the same. Those that happen to be able to bind to more than one strain have a selective advantage over those that do not in the polyvalent vaccine, regardless of the overall “dilution” of the polymorphic epitope, compared with the conserved epitope. The distinction between our mechanism and that proposed by Dutta et al. (47) is subtle, but significant; it suggests that broadly conserved epitopes need not be a prerequisite for enhanced cross-reactivity in the polyvalent vaccine and that even disparate strain-specific epitopes may induce increased cross-reactivity in polyvalent vaccines.
AMA1 is a leading candidate for a subunit-based malaria vaccine; however, the high degree of polymorphic variation in this Ag among different strains of P. falciparum has been a major impediment to efficacy in both preclinical and clinical testing. Researchers have explored two approaches to tackle the issue of antigenic variation in AMA1: polyvalent formulations using multiple strains of AMA1 (47, 48) and chimeric variants of AMA1 (51, 68), which incorporate a subset of high-frequency polymorphisms into a small set of constructs. In both cases, allelic coverage is the guiding principle, meaning that the vaccine should incorporate as broad a range of polymorphic variation as is possible (43). The results of this theoretical study, along with the experimental results of Dutta et al. (47), present an alternate approach: to directly enhance the cross-reactivity of the Ab response through the use of a polyvalent formulation that biases affinity maturation toward shared or cross-reactive epitopes. The computational work presented in this article provides a theoretical basis for experimental findings observed in a number of polyvalent AMA1 vaccine studies: a polyvalent AMA1 vaccine is capable of inducing a robust cross-reactive Ab response despite limited allelic coverage.
There are a number of future experiments that could validate this mechanism for enhanced cross-reactivity. First, ELISAs on sufficiently large panels of mAbs derived from hybridized B cells from rabbits immunized with the monovalent and polyvalent vaccines against the POLY or CONS recombinant chimeras, as well as recombinant non–vaccine AMA1 strains, could directly measure whether the polyvalent response shows enhanced cross-reactivity on both the conserved and polymorphic face and whether that response is truly strain transcending. Second, serum-depletion and -neutralization assays could assess the degree to which the polymorphic face response in the polyvalent vaccine is systematically different from the polymorphic face response in the monovalent vaccine with respect to cross-reactivity. Third, serial vaccinations first with the polyvalent vaccine, followed by the monovalent vaccine, should show an immune response that remains highly cross-reactive.
Our findings have implications for polyvalent vaccine research, as well as general vaccine design. First, a number of studies use DNA vaccines, virus-like particles, nanoparticles, or protein scaffold–based approaches to engineer Ags that isolate and present a conserved and/or neutralizing epitope. Our results suggest a novel alternative, not unlike heterologous priming (69), by using a heterogeneous mixture in which the targeted epitope is shared among otherwise diverse Ags. Second, our findings suggest that an enhanced cross-reactive Ab response may be a universal feature of polyvalent vaccines in cases in which the component Ags contain shared epitopes. Although polyvalent vaccines are used to accommodate Ag diversity, our results suggest, somewhat counterintuitively, that the broadening effect of some polyvalent vaccines may not come from pooled strain-specific responses toward the polymorphic epitopes but, instead, from an enhanced cross-reactive response toward shared epitopes. In some cases, these polymorphic regions are thought to arise from positive immune-selective pressure, where mutations within neutralizing epitopes confer a selective advantage, whereas conserved regions may be indicative of nonneutralizing or subneutralizing epitopes. In such cases, a polyvalent vaccine may enhance immunogenicity of conserved, nonneutralizing epitopes at the expense of polymorphic neutralizing epitopes.
In the case of AMA1, both conserved and polymorphic epitopes are known to be neutralizing, and the polyvalent AMA1 vaccine shows robust efficacy against a wide range of P. falciparum strains (47). With other Ags this may not be the case. For example, in dengue virus, subneutralizing cross-reactive Abs and cross-reactive memory B cells are thought to play a major pathogenic role in dengue hemorrhagic fever during secondary infections (70–72). Our findings suggest that polyvalent vaccines may be a double-edged sword: effective in cases in which there are no shared epitopes or if conserved epitopes are neutralizing and detrimental in cases in which conserved epitopes are present, but polymorphic or strain-specific epitopes are critical for neutralization.
We developed this stochastic model of B cell affinity maturation to serve as a platform from which to carry out theoretical and experimental studies of the Ab response to complex multiepitope Ags. We are currently exploring a number of directions in which to extend this model, such as the incorporation of T cells and cell-mediated immunity, as well as the simulation of viral kinetics and pathogenesis. We hope that the work presented in this article highlights the potential for theoretical biology to investigate the mechanisms underlying experimental observations in immunology, and we will continue to explore the application of computational methods in advancing our understanding of basic immunological principles.
Acknowledgments
We thank Dr. Sheetij Dutta and Dr. J. David Haynes (Walter Reed Army Institute of Research, Silver Spring, MD) for invaluable discussions about AMA1 and Dr. Hyung Jun Woo (Department of Defense Biotechnology High Performance Computing Software Applications Institute) for contributions in developing the stochastic simulation method.
This work was supported by the U.S. Army’s Network Science Initiative, the U.S. Army Military Infectious Disease Research Program, and the U.S. Army Medical Research and Materiel Command (Ft. Detrick, MD).
The opinions and assertions contained in this article are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or the U.S. Department of Defense. This article has been approved for public release with unlimited distribution.
- AMA1
- apical membrane antigen-1
- CONS
- chimeric AMA1 containing only the conserved face
- GC
- germinal center
- GIA
- growth inhibition assay
- POLY
- chimeric AMA1 containing only the polymorphic face.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Jardine J., Julien J. P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P. S., MacPherson S., Jones M., et al. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Correia B. E., Ban Y. E., Holmes M. A., Xu H., Ellingson K., Kraft Z., Carrico C., Boni E., Sather D. N., Zenobia C., et al. 2010. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18: 1116–1126 [DOI] [PubMed] [Google Scholar]
- 3.McLellan J. S., Correia B. E., Chen M., Yang Y., Graham B. S., Schief W. R., Kwong P. D. 2011. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J. Mol. Biol. 409: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klinman N. R. 1976. The acquisition of b-cell competence and diversity. Am. J. Pathol. 85: 693–704 [PMC free article] [PubMed] [Google Scholar]
- 5.Küppers R., Zhao M., Hansmann M. L., Rajewsky K. 1993. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J. 12: 4955–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell G. I. 1970. Mathematical model of clonal selection and antibody production. J. Theor. Biol. 29: 191–232 [DOI] [PubMed] [Google Scholar]
- 7.Bell G. I. 1970. Mathematical model of clonal selection and antibody production. Nature 228: 739–744 [DOI] [PubMed] [Google Scholar]
- 8.Bell G. I. 1971. Mathematical model of clonal selection and antibody production. II. J. Theor. Biol. 33: 339–378 [DOI] [PubMed] [Google Scholar]
- 9.Oprea M., Perelson A. S. 1997. Somatic mutation leads to efficient affinity maturation when centrocytes recycle back to centroblasts. J. Immunol. 158: 5155–5162 [PubMed] [Google Scholar]
- 10.Kepler T. B., Perelson A. S. 1993. Somatic hypermutation in B cells: an optimal control treatment. J. Theor. Biol. 164: 37–64 [DOI] [PubMed] [Google Scholar]
- 11.Kepler T. B., Perelson A. S. 1993. Cyclic re-entry of germinal center B cells and the efficiency of affinity maturation. Immunol. Today 14: 412–415 [DOI] [PubMed] [Google Scholar]
- 12.Seiden P. E., Celada F. 1992. A model for simulating cognate recognition and response in the immune system. J. Theor. Biol. 158: 329–357 [DOI] [PubMed] [Google Scholar]
- 13.Celada F., Seiden P. E. 1996. Affinity maturation and hypermutation in a simulation of the humoral immune response. Eur. J. Immunol. 26: 1350–1358 [DOI] [PubMed] [Google Scholar]
- 14.Meyer-Hermann M. 2002. A mathematical model for the germinal center morphology and affinity maturation. J. Theor. Biol. 216: 273–300 [DOI] [PubMed] [Google Scholar]
- 15.Meyer-Hermann M., Deutsch A., Or-Guil M. 2001. Recycling probability and dynamical properties of germinal center reactions. J. Theor. Biol. 210: 265–285 [DOI] [PubMed] [Google Scholar]
- 16.Farmer J. D., Packard N. H., Perelson A. S. 1986. The immune system, adaptation, and machine learning. Physica D 22: 187–204 [Google Scholar]
- 17.Perelson A. S., Oster G. F. 1979. Theoretical studies of clonal selection: minimal antibody repertoire size and reliability of self-non-self discrimination. J. Theor. Biol. 81: 645–670 [DOI] [PubMed] [Google Scholar]
- 18.Smith D. J., Forrest S., Hightower R. R., Perelson A. S. 1997. Deriving shape space parameters from immunological data. J. Theor. Biol. 189: 141–150 [DOI] [PubMed] [Google Scholar]
- 19.Smith D. J., Forrest S., Ackley D. H., Perelson A. S. 1999. Variable efficacy of repeated annual influenza vaccination. Proc. Natl. Acad. Sci. USA 96: 14001–14006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrickson S. E., Mempel T. R., Mazo I. B., Liu B., Artyomov M. N., Zheng H., Peixoto A., Flynn M. P., Senman B., Junt T., et al. 2008. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol. 9: 282–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H., Jin B., Henrickson S. E., Perelson A. S., von Andrian U. H., Chakraborty A. K. 2008. How antigen quantity and quality determine T-cell decisions in lymphoid tissue. Mol. Cell. Biol. 28: 4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosmrlj A., Jha A. K., Huseby E. S., Kardar M., Chakraborty A. K. 2008. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc. Natl. Acad. Sci. USA 105: 16671–16676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H. Y., Topham D. J., Park S. Y., Hollenbaugh J., Treanor J., Mosmann T. R., Jin X., Ward B. M., Miao H., Holden-Wiltse J., et al. 2009. Simulation and prediction of the adaptive immune response to influenza A virus infection. J. Virol. 83: 7151–7165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y., Ghersi D., Calcagno C., Selin L. K., Puzone R., Celada F. 2009. A discrete computer model of the immune system reveals competitive interactions between the humoral and cellular branch and between cross-reacting memory and naïve responses. Vaccine 27: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancioglu B., Swigon D., Clermont G. 2007. A dynamical model of human immune response to influenza A virus infection. J. Theor. Biol. 246: 70–86 [DOI] [PubMed] [Google Scholar]
- 26.Woo H. J., Reifman J. 2012. A quantitative quasispecies theory-based model of virus escape mutation under immune selection. Proc. Natl. Acad. Sci. USA 109: 12980–12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Deutekom H. W., Wijnker G., de Boer R. J. 2013. The rate of immune escape vanishes when multiple immune responses control an HIV infection. J. Immunol. 191: 3277–3286 [DOI] [PubMed] [Google Scholar]
- 28.Nagaraja S., Wallqvist A., Reifman J., Mitrophanov A. Y. 2014. Computational approach to characterize causative factors and molecular indicators of chronic wound inflammation. J. Immunol. 192: 1824–1834 [DOI] [PubMed] [Google Scholar]
- 29.Gillespie D. T. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81: 2340–2361 [Google Scholar]
- 30.Huang J. W., Yang J. M. 2011. Changed epitopes drive the antigenic drift for influenza A (H3N2) viruses. BMC Bioinformatics 12(Suppl. 1): S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith D. J., Lapedes A. S., de Jong J. C., Bestebroer T. M., Rimmelzwaan G. F., Osterhaus A. D., Fouchier R. A. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305: 371–376 [DOI] [PubMed] [Google Scholar]
- 32.Crawford H., Matthews P. C., Schaefer M., Carlson J. M., Leslie A., Kilembe W., Allen S., Ndung’u T., Heckerman D., Hunter E., Goulder P. J. 2011. The hypervariable HIV-1 capsid protein residues comprise HLA-driven CD8+ T-cell escape mutations and covarying HLA-independent polymorphisms. J. Virol. 85: 1384–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer W., Ganusov V. V., Giorgi E. E., Hraber P. T., Keele B. F., Leitner T., Han C. S., Gleasner C. D., Green L., Lo C. C., et al. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS ONE 5: e12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gils M. J., Bunnik E. M., Burger J. A., Jacob Y., Schweighardt B., Wrin T., Schuitemaker H. 2010. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. J. Virol. 84: 3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowd K. A., Netski D. M., Wang X. H., Cox A. L., Ray S. C. 2009. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology 136: 2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cento V., Mirabelli C., Dimonte S., Salpini R., Han Y., Trimoulet P., Bertoli A., Micheli V., Gubertini G., Cappiello G., et al. 2013. Overlapping structure of hepatitis B virus (HBV) genome and immune selection pressure are critical forces modulating HBV evolution. J. Gen. Virol. 94: 143–149 [DOI] [PubMed] [Google Scholar]
- 37.Remarque E. J., Faber B. W., Kocken C. H., Thomas A. W. 2008. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 24: 74–84 [DOI] [PubMed] [Google Scholar]
- 38.Coley A. M., Parisi K., Masciantonio R., Hoeck J., Casey J. L., Murphy V. J., Harris K. S., Batchelor A. H., Anders R. F., Foley M. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect. Immun. 74: 2628–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohler B., Puzone R., Seiden P. E., Celada F. 2000. A systematic approach to vaccine complexity using an automaton model of the cellular and humoral immune system. I. Viral characteristics and polarized responses. Vaccine 19: 862–876 [DOI] [PubMed] [Google Scholar]
- 40.Stanisic D. I., Richards J. S., McCallum F. J., Michon P., King C. L., Schoepflin S., Gilson P. R., Murphy V. J., Anders R. F., Mueller I., Beeson J. G. 2009. Immunoglobulin G subclass-specific responses against Plasmodium falciparum merozoite antigens are associated with control of parasitemia and protection from symptomatic illness. Infect. Immun. 77: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thera M. A., Doumbo O. K., Coulibaly D., Laurens M. B., Ouattara A., Kone A. K., Guindo A. B., Traore K., Traore I., Kouriba B., et al. 2011. A field trial to assess a blood-stage malaria vaccine. N. Engl. J. Med. 365: 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duan J., Mu J., Thera M. A., Joy D., Kosakovsky Pond S. L., Diemert D., Long C., Zhou H., Miura K., Ouattara A., et al. 2008. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc. Natl. Acad. Sci. USA 105: 7857–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takala, S. L., D. Coulibaly, M. A. Thera, A. H. Batchelor, M. P. Cummings, A. A. Escalante, A. Ouattara, K. Traore, A. Niangaly, A. A. Djimde, et al. 2009. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci. Transl. Med. 1: 2ra5. [DOI] [PMC free article] [PubMed]
- 44.Dutta S., Lee S. Y., Batchelor A. H., Lanar D. E. 2007. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. USA 104: 12488–12493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coley A. M., Gupta A., Murphy V. J., Bai T., Kim H., Foley M., Anders R. F., Batchelor A. H. 2007. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. 3: 1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drew D. R., Hodder A. N., Wilson D. W., Foley M., Mueller I., Siba P. M., Dent A. E., Cowman A. F., Beeson J. G. 2012. Defining the antigenic diversity of Plasmodium falciparum apical membrane antigen 1 and the requirements for a multi-allele vaccine against malaria. PLoS ONE 7: e51023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutta S., Dlugosz L. S., Drew D. R., Ge X., Ababacar D., Rovira Y. I., Moch J. K., Shi M., Long C. A., Foley M., et al. 2013. Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1. [Published erratum appears in 2014 PLos Pathog. 10(1).] PLoS Pathog. 9: e1003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kusi K. A., Faber B. W., Thomas A. W., Remarque E. J. 2009. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS ONE 4: e8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miura K., Herrera R., Diouf A., Zhou H., Mu J., Hu Z., MacDonald N. J., Reiter K., Nguyen V., Shimp R. L., Jr., et al. 2013. Overcoming allelic specificity by immunization with five allelic forms of Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 81: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy M. C., Wang J., Zhang Y., Miles A. P., Chitsaz F., Saul A., Long C. A., Miller L. H., Stowers A. W. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70: 6948–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kusi K. A., Faber B. W., Riasat V., Thomas A. W., Kocken C. H., Remarque E. J. 2010. Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response. PLoS ONE 5: e15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berek C., Milstein C. 1988. The dynamic nature of the antibody repertoire. Immunol. Rev. 105: 5–26 [DOI] [PubMed] [Google Scholar]
- 53.Lodish H., Baltimore D., Berk A., Zipursky S. L., Matsudaira P., Darnell J. 1999. Molecular Cell Biology. Scientific American Books, New York [Google Scholar]
- 54.Tangye S. G., Avery D. T., Deenick E. K., Hodgkin P. D. 2003. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J. Immunol. 170: 686–694 [DOI] [PubMed] [Google Scholar]
- 55.Liu Y. J., Zhang J., Lane P. J., Chan E. Y., MacLennan I. C. 1991. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur. J. Immunol. 21: 2951–2962 [DOI] [PubMed] [Google Scholar]
- 56.Smith K. G., Hewitson T. D., Nossal G. J., Tarlinton D. M. 1996. The phenotype and fate of the antibody-forming cells of the splenic foci. Eur. J. Immunol. 26: 444–448 [DOI] [PubMed] [Google Scholar]
- 57.Driver D. J., McHeyzer-Williams L. J., Cool M., Stetson D. B., McHeyzer-Williams M. G. 2001. Development and maintenance of a B220− memory B cell compartment. J. Immunol. 167: 1393–1405 [DOI] [PubMed] [Google Scholar]
- 58.Liu X. Q., Hart D. N., MacPherson G. G., Good M. F., Wykes M. N. 2008. Soluble CD38 significantly prolongs the lifespan of memory B-cell responses. Immunology 125: 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dogan I., Bertocci B., Vilmont V., Delbos F., Mégret J., Storck S., Reynaud C. A., Weill J. C. 2009. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 10: 1292–1299 [DOI] [PubMed] [Google Scholar]
- 60.Vieira P., Rajewsky K. 1988. The half-lives of serum immunoglobulins in adult mice. Eur. J. Immunol. 18: 313–316 [DOI] [PubMed] [Google Scholar]
- 61.Morell A., Terry W. D., Waldmann T. A. 1970. Metabolic properties of IgG subclasses in man. J. Clin. Invest. 49: 673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konterman, R. 2012. Therapeutic Proteins: Strategies to Modulate Their Plasma Half-lives. Wiley-Blackwell, Weinham, Germany.
- 63.Zhang J., MacLennan I. C., Liu Y. J., Lane P. J. 1988. Is rapid proliferation in B centroblasts linked to somatic mutation in memory B cell clones? Immunol. Lett. 18: 297–299 [DOI] [PubMed] [Google Scholar]
- 64.Liao Y. C., Lee M. S., Ko C. Y., Hsiung C. A. 2008. Bioinformatics models for predicting antigenic variants of influenza A/H3N2 virus. Bioinformatics 24: 505–512 [DOI] [PubMed] [Google Scholar]
- 65.Pan K., Subieta K. C., Deem M. W. 2011. A novel sequence-based antigenic distance measure for H1N1, with application to vaccine effectiveness and the selection of vaccine strains. Protein Eng. Des. Sel. 24: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeKosky B. J., Ippolito G. C., Deschner R. P., Lavinder J. J., Wine Y., Rawlings B. M., Varadarajan N., Giesecke C., Dörner T., Andrews S. F., et al. 2013. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parameswaran P., Liu Y., Roskin K. M., Jackson K. K., Dixit V. P., Lee J. Y., Artiles K. L., Zompi S., Vargas M. J., Simen B. B., et al. 2013. Convergent antibody signatures in human dengue. Cell Host Microbe 13: 691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Remarque E. J., Faber B. W., Kocken C. H., Thomas A. W. 2008. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect. Immun. 76: 2660–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guenaga J., Dosenovic P., Ofek G., Baker D., Schief W. R., Kwong P. D., Karlsson Hedestam G. B., Wyatt R. T. 2011. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS ONE 6: e16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beltramello M., Williams K. L., Simmons C. P., Macagno A., Simonelli L., Quyen N. T., Sukupolvi-Petty S., Navarro-Sanchez E., Young P. R., de Silva A. M., et al. 2010. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8: 271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dejnirattisai W., Jumnainsong A., Onsirisakul N., Fitton P., Vasanawathana S., Limpitikul W., Puttikhunt C., Edwards C., Duangchinda T., Supasa S., et al. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328: 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith S. A., Zhou Y., Olivarez N. P., Broadwater A. H., de Silva A. M., Crowe J. E., Jr. 2012. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J. Virol. 86: 2665–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]