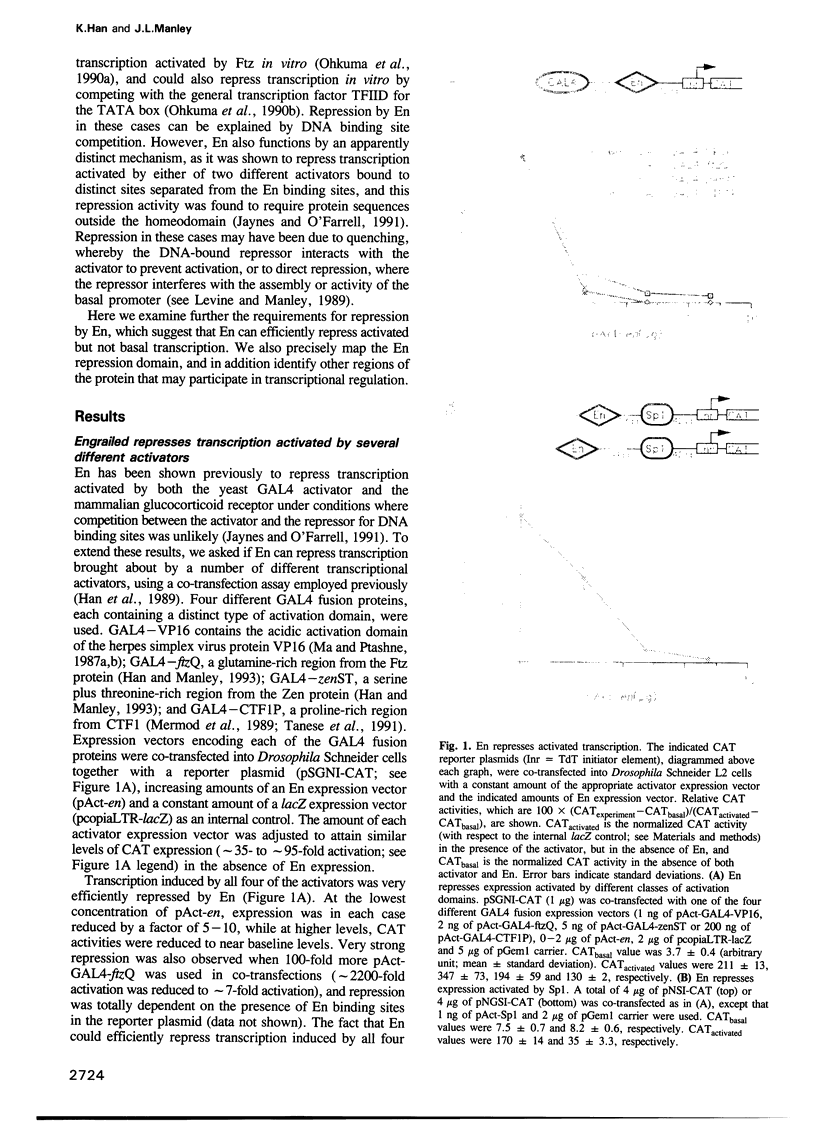
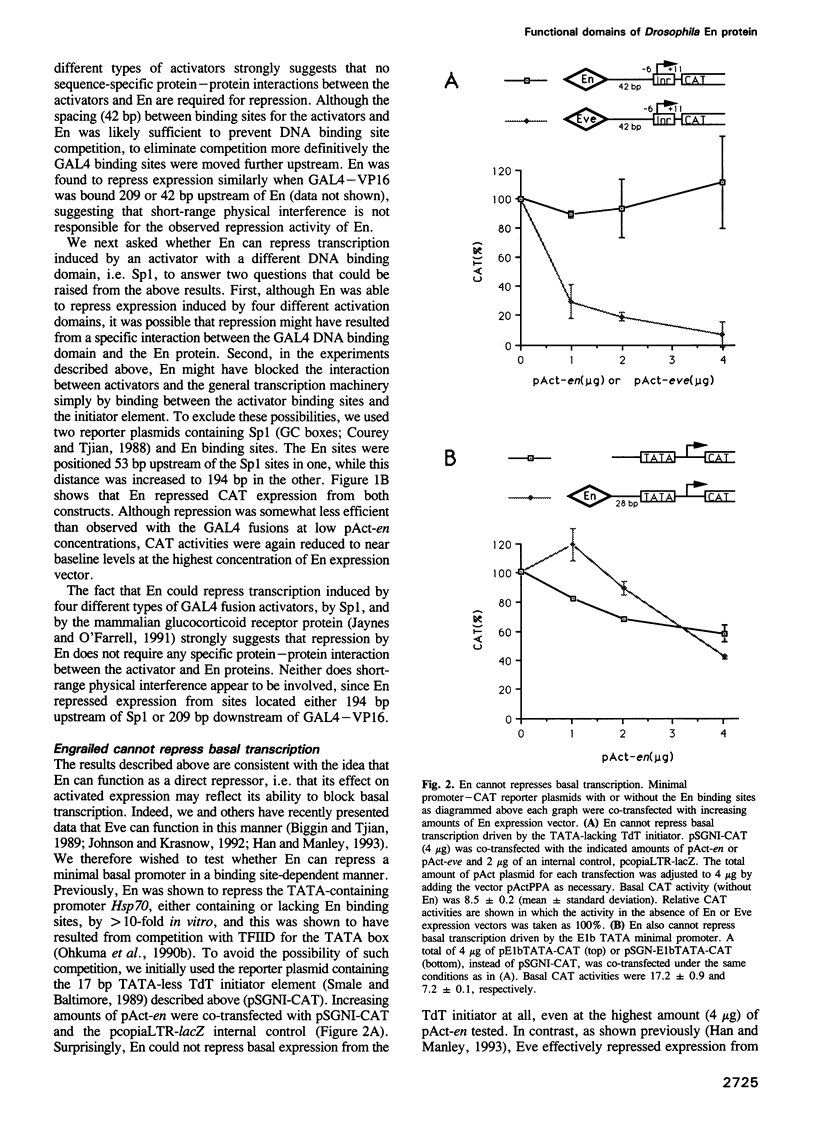
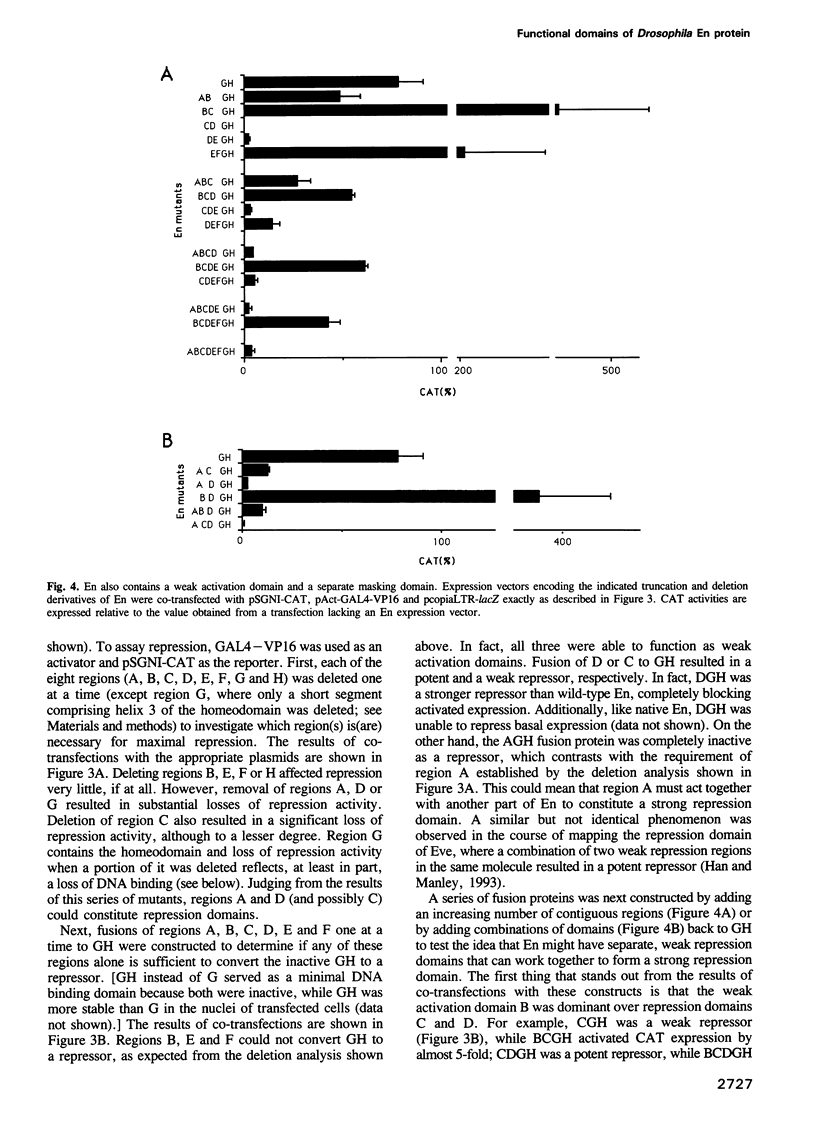
Abstract
We have studied the transcriptional activity of the Drosophila homeodomain protein Engrailed (En) by using a transient expression assay employing Schneider L2 cells. En was found to very strongly repress promoters activated by a variety of different activator proteins. However, unlike another Drosophila homeodomain-containing repressor, Even-skipped (Eve), En was unable to repress the activity of several basal promoters in the absence of activator expression. These findings indicate that En is a specific repressor of activated transcription, and suggest that En may repress transcription by a different mechanism than Eve, perhaps by interfering with interactions between transcriptional activators and the general transcription machinery. By analyzing the properties of a variety of En mutants, we identified a minimal repression domain composed of 55 residues, which can function when fused to a heterologous DNA binding domain. Like repression domains identified in the Drosophila repressors Eve and Krüppel, the En repression domain is rich in alanine residues (26%), but unlike these other domains, is moderately charged (six arginine and three glutamic acid residues). Separate regions of En that may in some circumstances function in transcriptional activation were also identified.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baniahmad A., Köhne A. C., Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 1992 Mar;11(3):1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989 Aug 11;58(3):433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Sonoda S., Ueda H., Scott M. P., Wu C. Repression of the Drosophila fushi tarazu (ftz) segmentation gene. EMBO J. 1991 Mar;10(3):665–674. doi: 10.1002/j.1460-2075.1991.tb07995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B. Zebra patterns in fly embryos: activation of stripes or repression of interstripes? Cell. 1990 Jan 12;60(1):9–16. doi: 10.1016/0092-8674(90)90711-m. [DOI] [PubMed] [Google Scholar]
- Colgan J., Manley J. L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992 Feb;6(2):304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- Colgan J., Wampler S., Manley J. L. Interaction between a transcriptional activator and transcription factor IIB in vivo. Nature. 1993 Apr 8;362(6420):549–553. doi: 10.1038/362549a0. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Desplan C., Theis J., O'Farrell P. H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988 Sep 23;54(7):1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eaton S., Kornberg T. B. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 1990 Jun;4(6):1068–1077. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J. A., Giniger E., Maniatis T., Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988 Apr 28;332(6167):853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Gay N. J., Poole S. J., Kornberg T. B. The Drosophila engrailed protein is phosphorylated by a serine-specific protein kinase. Nucleic Acids Res. 1988 Jul 25;16(14A):6637–6647. doi: 10.1093/nar/16.14.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Levine M. S., Manley J. L. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989 Feb 24;56(4):573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Han K., Manley J. L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993 Mar;7(3):491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Heemskerk J., DiNardo S., Kostriken R., O'Farrell P. H. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991 Aug 1;352(6334):404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., O'Farrell P. H. Activation and repression of transcription by homoeodomain-containing proteins that bind a common site. Nature. 1988 Dec 22;336(6201):744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes J. B., O'Farrell P. H. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991 Jun;10(6):1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. B., Krasnow M. A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992 Nov;6(11):2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Manley J. L. Transcriptional repression of eukaryotic promoters. Cell. 1989 Nov 3;59(3):405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Licht J. D., Grossel M. J., Figge J., Hansen U. M. Drosophila Krüppel protein is a transcriptional repressor. Nature. 1990 Jul 5;346(6279):76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Madden S. L., Cook D. M., Morris J. F., Gashler A., Sukhatme V. P., Rauscher F. J., 3rd Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991 Sep 27;253(5027):1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y., Horikoshi M., Roeder R. G., Desplan C. Binding site-dependent direct activation and repression of in vitro transcription by Drosophila homeodomain proteins. Cell. 1990 May 4;61(3):475–484. doi: 10.1016/0092-8674(90)90529-n. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y., Horikoshi M., Roeder R. G., Desplan C. Engrailed, a homeodomain protein, can repress in vitro transcription by competition with the TATA box-binding protein transcription factor IID. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2289–2293. doi: 10.1073/pnas.87.6.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz M. J., Jäckle H. Making stripes in the Drosophila embryo. Trends Genet. 1990 Sep;6(9):287–292. doi: 10.1016/0168-9525(90)90234-w. [DOI] [PubMed] [Google Scholar]
- Pankratz M. J., Seifert E., Gerwin N., Billi B., Nauber U., Jäckle H. Gradients of Krüppel and knirps gene products direct pair-rule gene stripe patterning in the posterior region of the Drosophila embryo. Cell. 1990 Apr 20;61(2):309–317. doi: 10.1016/0092-8674(90)90811-r. [DOI] [PubMed] [Google Scholar]
- Patel N. H., Martin-Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989 Sep 8;58(5):955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. A new bind for Myc. Trends Genet. 1992 Mar;8(3):91–96. doi: 10.1016/0168-9525(92)90196-b. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A. A. Activators and targets. Nature. 1990 Jul 26;346(6282):329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Read D., Levine M., Manley J. L. Ectopic expression of the Drosophila tramtrack gene results in multiple embryonic defects, including repression of even-skipped and fushi tarazu. Mech Dev. 1992 Sep;38(3):183–195. doi: 10.1016/0925-4773(92)90052-l. [DOI] [PubMed] [Google Scholar]
- Renkawitz R. Transcriptional repression in eukaryotes. Trends Genet. 1990 Jun;6(6):192–197. doi: 10.1016/0168-9525(90)90176-7. [DOI] [PubMed] [Google Scholar]
- Riddihough G., Ish-Horowicz D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 1991 May;5(5):840–854. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]
- Sauer F., Jäckle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991 Oct 10;353(6344):563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Carroll S. B. The segmentation and homeotic gene network in early Drosophila development. Cell. 1987 Dec 4;51(5):689–698. doi: 10.1016/0092-8674(87)90092-4. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Small S., Kraut R., Hoey T., Warrior R., Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991 May;5(5):827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Soeller W. C., Poole S. J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988 Jan;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Topol J., Dearolf C. R., Prakash K., Parker C. S. Synthetic oligonucleotides recreate Drosophila fushi tarazu zebra-stripe expression. Genes Dev. 1991 May;5(5):855–867. doi: 10.1101/gad.5.5.855. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992 Oct;6(10):1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- Zuo P., Stanojević D., Colgan J., Han K., Levine M., Manley J. L. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991 Feb;5(2):254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]