Abstract
RNA-binding proteins (RBPs) can act as stem cell modulators and oncogenic drivers, but have been largely ignored by the pharmaceutical industry as potential therapeutic targets for cancer. The MUSASHI (MSI) family has recently been demonstrated to be an attractive clinical target in the most aggressive cancers. Therefore, the discovery and development of small molecule inhibitors could provide a novel therapeutic strategy. In order to find novel compounds with MSI RNA binding inhibitory activity, we have developed a fluorescence polarization (FP) assay and optimized it for high throughput screening (HTS) in a 1536-well microtiter plate format. Using a chemical library of 6,208 compounds, we performed pilot screens, against both MSI1 and MSI2, leading to the identification of 7 molecules for MSI1, 15 for MSI2 and 5 that inhibited both. A secondary FP dose-response screen validated 3 MSI inhibitors with IC50 below 10μM. Out of the 25 compounds retested in the secondary screen only 8 demonstrated optical interference due to high fluorescence. Utilizing a SYBR-based RNA electrophoresis mobility shift assay (EMSA), we further verified MSI inhibition of the top 3 compounds. Surprisingly, even though several aminoglycosides were present in the library, they failed to demonstrate MSI inhibitor activity challenging the concept that these compounds are pan-active against RBPs. In summary, we have developed an in vitro strategy to identify MSI specific inhibitors using an FP HTS platform, which will facilitate novel drug discovery for this class of RBPs.
Keywords: RNA-binding protein, MUSASHI, HTS, fluorescence polarization, cancer, small-molecule inhibitors
INTRODUCTION
RNA-binding proteins (RBPs) are key components of RNA metabolism as they regulate RNA biogenesis, maturation, stability, subcellular localization, translation and RNA degradation [1]. Dysregulation of RBPs expression or activity has been reported in several diseases and a few have been found to play relevant roles in the development and maintenance of cancer, due to their oncogenic or tumor-suppressive function [2–4]. The MSI gene family, consisting of the two closely related RBPs MUSASHI-1 (MSI1) and MUSASHI-2 (MSI2), has been identified as translational regulators that are highly expressed in the most aggressive solid cancers and hematopoietic malignancies [5]. Genetic and functional studies have shown that the expression of MSI1 is upregulated in pediatric brain tumors and metastatic breast cancer; and, accordingly, knockdown of MSI1 in an adenocarcinoma cell line reduces tumor burden in vivo [6, 7]. Additionally, MSI2 is highly expressed in gliomas and medulloblastoma [8]. The MSI2 gene has also been found amplified and overexpressed by deep sequencing of an aggressive prostate adenocarcinoma and in metastatic prostate cancer [9]. In addition to its role in aggressive solid tumors [5], MSI2 fusions have been found in several patients with blast crisis Chronic Myeloid Leukemia (CML-BC), where chromosomal translocations fused MSI2 and HOXA9 [10]. Recent studies have reported that MSI2 overexpression occurs in a variety of hematopoietic malignancies including CML-BC, AML and B-Cell Acute Lymphoblastic Leukemia, and can contribute as a negative prognostic marker [3, 11, 12]. Moreover, recent studies have demonstrated a functional role in which MSI2 can maintain self-renewal and control of hematopoietic differentiation in human myeloid leukemia cell lines [3].
The MSI gene family is normally expressed in stem and progenitor cells by regulating the switch between symmetric and asymmetric cell division and altering cellular fate [13]. Consistent with its role as a modulator of self-renewal, our laboratory has determined that MSI2 maintains hematopoietic stem cells [14]. Furthermore, the aberrant expression of the MSI family in aggressive cancers results in a gain of self-renewal properties [3, 15]. MSI1 and MSI2 are characterized by the presence of two tandem RNA recognition motifs (RRMs) [13, 16]. Mechanistically, MSI1 has been shown to interact with the 3′UTRs of target mRNAs and block translation initiation by interfering with the poly A binding protein (PABP) and its association with the elongation initiation complex [16]. The minimal binding sequence of mammalian MSI1 has been identified and corresponds to [(G/A) Un AGU, n=1–3] [17]. Although the specific targets for human MSI proteins remain to be fully characterized, studies from our laboratory and others have demonstrated that they control many essential oncogenic pathways including cell cycle, proliferation, metabolism, c-MYC and TGF-b signaling [3, 14, 15]. Thus, we reasoned that blocking MSI function with small molecule inhibitors would have a great therapeutic potential in a variety of tumor settings and hematological malignancies, and will represent a proof of concept for targeting RBPs for cancer therapeutics.
In this study, we have developed, optimized and miniaturized into a1536-well format an FP assay to identify novel small molecules inhibitors of MSI RNA binding activity. With a total assay volume of 10μL, a pilot HTS assay was run with a 6,208 compound library obtaining an optimal Z′ factor of 0.6 and a very low overall percentage of dual MSI positive hits (0.08%). We further validated the list of initial hits by performing dose-response studies; and for those hits with an IC50 value less than 10 μM, we performed an orthogonal assay using an EMSA approach to confirm their activity. Of note, this effective and reliable strategy provides the tools to identify specific MSI inhibitors. It represents the first steps toward obtaining novel chemical species for targeting RNA binding proteins.
MATERIALS AND METHODS
RNA oligos and chemicals
The RNAse free HPLC purified single-stranded RNA (ssRNA) oligos were purchased from Integrated DNA Technologies (Coralville, IA). The optimal ssRNA oligo [8 nucleotides, r(GUAGUAGU)] for the FP assay, determined by SYBR-based RNA EMSA, was obtained Cy3-labelled with a 9 carbon (C9) spacer between the RNA and the fluorophore (Integrated DNA Technologies). Other chemical reagents were purchased from Fisher Scientific (Pittsburgh, PA).
Cloning of MSI1, MSI2, LIN28A and p53 into protein expression vectors
The ORF mRNA sequences of human MSI1 and MSI2 (accession numbers NM_002442.3 and NM_138962.2, respectively) were subcloned into pGEX6P-3 (GE Healthcare, Port Washington, NY) from pcDNA3.1-MSI1 and -MSI2 (as previously reported [3]), by introducing a 5′FLAG sequence (5′-ATGGATTACAAGGATGACGACGATAAG-3′) and using BamHI and NotI (MSI1) or two EcoRI (MSI2) restriction sites. Similarly, human LIN28A mRNA full-length (accession number NM_024674.4)and human P53 mRNA (accession number NM_000546.5) were subcloned into pGEX6P-3 from pBABE-LIN28A and pGEX2TK-P53 introducing a 5′FLAG sequence and using two EcoRI restriction sites. The resulting plasmids (pGEX6P-3-MSI1, pGEX6P-3-MSI2, pGEX6P-3-LIN28A, pGEX6P-3-P53) were sequence verified anda plasmid DNA stock was prepared by MaxiPrep (QIAgen, Germantown, MD) and quantified by NanoDrop 8000 (Thermo Scientific, Waltham, MA).
Expression and purification of GST tagged MSI1, MSI2, LIN28A and p53 recombinant proteins
To produce the GST tagged recombinant proteins, 2μL of pGEX6P-3-MSI1, -MSI2,-LIN28A or -p53 were transformed into BL21 (DE3) competent cells (Agilent Technologies, Santa Clara, CA). Transformed bacteria were inoculated in 100 mL LB Broth Miller media containing 100 μg/mL ampicillin and grown overnight at 225 rpm and 37°C. The overnight culture was diluted 1:40 in 4L of fresh LB (100 μg/mL ampicillin) and grown until the optical density measured at 600 nm reached 0.4–0.7. Induction was then started by addition of 1mM IPTG followed by an overnight incubation at 20°C. Cells were harvested by centrifugation at 5000 rpm for 30 min and the obtained cell pellet was kept at −80°C until use. Each cell pellet (obtained from 2L culture) was re-suspended in 25 mL final volume of PBS containing 2.5 mL of 10X protease inhibitor cocktail prepared from FAST protease inhibitor tablets (Sigma Aldrich) and lysed by passing 2–3 times through a French Press. The resultant cell lysate was centrifuged at 15,000 rpm for 1 hour and the supernatant was applied to a XK16/20 column pre-packed with Glutathione Sepharose 4 Fast Flow (GE Healthcare) connected to an AKTA Prime FPLC (GE Healthcare). The collected fractions, eluted in a 50 mM Tris-HCl, 20 mM reduced L-glutathione buffer, were analyzed by gel electrophoresis and the ones containing the GST-tagged proteins were pooled and dialyzed at 4°C against PBS (pH 7.4) (for EMSA analysis) or 100 mM sodium phosphate, 0.001% BSA (w/v), 0.01% glycerol (w/v), 1 mM TCEP (pH 8.0) (for FP assay). After dialysis, the protein concentration was measured by BCA assay (Pierce, Rockford, IL). Up to 12.5mg of GST-protein was obtained per liter of bacterial cell culture.
SYBR-based RNA Electrophoresis Mobility Shift Assay
EMSAs were performed to measure the RNA-binding ability of GST-MSI1, GST-MSI2 and GST-LIN28A to different RNA oligos (in length and number of binding consensus sequence repeats). The ssRNA oligos characteristics and binding affinities of the three proteins tested are listed in Table 1. The RNA-binding consensus repeats were previously determined for MSI1 as [(G/A) Un AGU, n=1–3] [17]. In short, different quantities of GST-proteins (500 to 2500 ng) were incubated in EMSA buffer (10 mM Tris-HCl, 150 mM KCl, 0.1 mM EDTA, 0.5 mM DTT, pH 7.4) with a fixed ssRNA oligo quantity (200 pmols) in a final volume of 20μL for 30 min at room temperature. When confirming activity of compounds identified in the pilot screen using the EMSA assay, we made use of a non-specific inhibitor oleic acid to abrogate all RNA binding activity to achieve total inhibition instead of 1% sodium dodecyl sulfate (SDS) to avoid bubbles generation during pipetting as we had used in other projects performed at the HTS Core Facility. As such, the GST-protein was pre-incubated either in 1% DMSO (v/v) to achieve 100% binding activity or 10 μM oleic acid in 1% DMSO (v/v) to abolish binding activity for 30 min at room temperature. After this period, the ssRNAoligo was added and incubated during 30 additional min. Then, the EMSA reaction was loaded in ssRNA running buffer (New England BioLabs, Ipswich, MA), onto a native 10% polyacrylamide gel (in 1X Tris-Borate-EDTA, TBE) (BioRad, Hercules, CA) and electrophoresis was carried out in chilled 0.5X TBE buffer at 4°C for up to 2.5 hours at 50–75V (until the bromophenol blue dye migrated approximately 3/4 down the length of the gel). The gel was then washed twice in MilliQ H2O and incubated for 30 min in 2X SYBR safe solution (Invitrogen, Grand Island, NY) in 1X TBE and the results were visualized and analyzed using a BioRad Gel DocTM XR+ imager (BioRad). RNA band intensity was quantified using ImageJ (v10.2) software.
Table 1.
Characteristics of ssRNAoligo sequences tested in EMSA assay against GST-proteins.
|
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Binding affinity(EMSA) | ||||||||
|
| ||||||||
| # | Motif(s) | ssRNAoligo sequence | #binding motifs | Length (nt) | MSI1 | MSI2 | LIN28A | P53 |
| 1 | MSI | UAGAAGUAGUUUUAA | 1 | 15 | + | + | + | − |
| 2 | MSI | UAGUAGUAAGUAGUA | 2 | 15 | ++ | ++ | ++ | − |
| 3 | MSI | AGUUAGUAGUUAGUA | 3 | 15 | ++ | ++ | ++ | − |
| 4 | MSI | GUAGUAGUAGUAGUA | 4 | 15 | +++ | +++ | +++ | − |
| 5 | MSI | GUAGUAGU | 2 | 8 | ++ | ++ | n.d. | − |
|
| ||||||||
| 6 | LIN28 | GAGGAGGAGCUG | 2 | 12 | − | − | ++ | n.d. |
| 7 | LIN28 | GGGAGGCUGAGG | 3 | 12 | − | −/+ | ++ | n.d. |
|
| ||||||||
| 8 | - | AGGGAUCGAAAAUUG | 0 | 15 | − | − | +++ | − |
Chemical Libraries
The library screened combines 6,208 chemicals obtained from MicroSource, Prestwick, Tocris, and other commercial sources as previously described [18]. The MicroSource library contains 2,000 known drugs, natural products, and other bioactive components, such as enzyme inhibitors, receptor blockers, membrane-active compounds, and cellular toxins. The Prestwick chemical library is a collection of 1,119 clinically approved drugs, selected for their high chemical and pharmacological diversity, as well as known bioavailability and safety in humans. The Tocris library represents a unique and diverse collection of high purity compounds with known activity in kinases, ion channels, nuclear receptors, and transporter assays.
Fluorescence Polarization (FP) control run and pilot screen
For the control run and pilot screen, the assay was conducted in 1536-well format (black polystyrene, Corning #3724) and automated on a previously described linear track robotic platform [19] (CRS F3 Robot System, Thermo Scientific) with integrated Flexdrop and LEADseeker, according to the following protocol. The control run consisted of two 1536-well microtiter plates assayed in the presence of 1% of DMSO (v/v) vehicle only as High Control (HC) or 1 mM oleic acid in 1% DMSO (v/v) as Low Control (LC). Tested compounds or controls were added to the wells at a volume of 1 μL using a custom designed 384 head on a PP-384-M Personal Pipettor (Apricot Designs, Covina, CA). The pilot screen was performed in duplicate and compounds were tested at a final concentration of 10 μM in 1% DMSO (v/v). GST-MSI1 or GST-MSI2 recombinant protein preparations were diluted in the assay buffer (10 mM Tris-HCl, 150 mM KCl, 0.1 mM EDTA, 0.005% Tween 20 (v/v), 1 mM TCEP, pH 7.4, vacuum-degassed for 2 hours) were dispensed at a volume of 4 μL for a final concentration of 1 μM using a FlexDrop IV (Perkin Elmer, Waltham, MA). After 1 hour pre-incubation, 5 μL probe in solution in assay buffer were added to the wells for a final concentration of 10 nM. After additional 1 hour incubation at room temperature, the FP was read using the GE Healthcare LEADseeker Multimodality Imaging System equipped with Cy3 excitation/emission filters and Cy3 FP epi-mirror as previously reported [20]. Screening data files from LEADseeker were loaded onto the HTS Core Screening Data Management System, a custom built suite of modules for compound registration, plating and data management powered by ChemAxon Cheminformatic tools (ChemAxon, Hungary). The percentage inhibition for the tested compounds was calculated as follows: % inhibition = (high control average − read value)/(high control average − low control average) × 100 based on the average of the high and low controls. Z′ values and FP measurements subtracting the buffer background were performed as previously described [20].
Dose-Response Studies
To assess compound potency and confirm activity of identified positives, compounds were assessed in dose-response study using 12 doubling dilutions in duplicate with 10 μM concentration as the upper limit, unless stated otherwise. Control and compound dilutions were made in an intermediate 384-well polypropylene plate (ABgene, Thermo Fisher Scientific), and 1μL were transferred to the assay plates (Low volume, black polystyrene, round bottom 384-well, Corning #3676) with a custom-designed 384 head on a PP-384-M Personal Pipettor (Apricot Designs). The enzyme and the probe were subsequently dispensed in 5 μL assay buffer using FlexDrop and the plates were read using LEADseeker as described above. Dose-response data files were uploaded onto the HTS Core Screening Data Management System for curve fitting and IC50 calculation. Curves presented in this article were fitted as a logistic 4-parameter sigmoid using SigmaPlot 9.0 (Systat Software, Inc.).
Optical interference and solubility assessment
To assess any potential compound optical interference, resupplied positives were tested in dose response as described above, but in absence of protein. Compounds affecting fluorescence polarization and/or fluorescence intensity (FI) in a dose-dependent manner in these conditions were flagged as optically interfering in the assay. To identify any non-specific activity due to low solubility in the assay conditions, compound solubility limit was assessed in dose response in triplicate using laser nephelometry in 384-well format as previously described [20].
RESULTS
MSI specific RNA probe and EMSA assay optimization
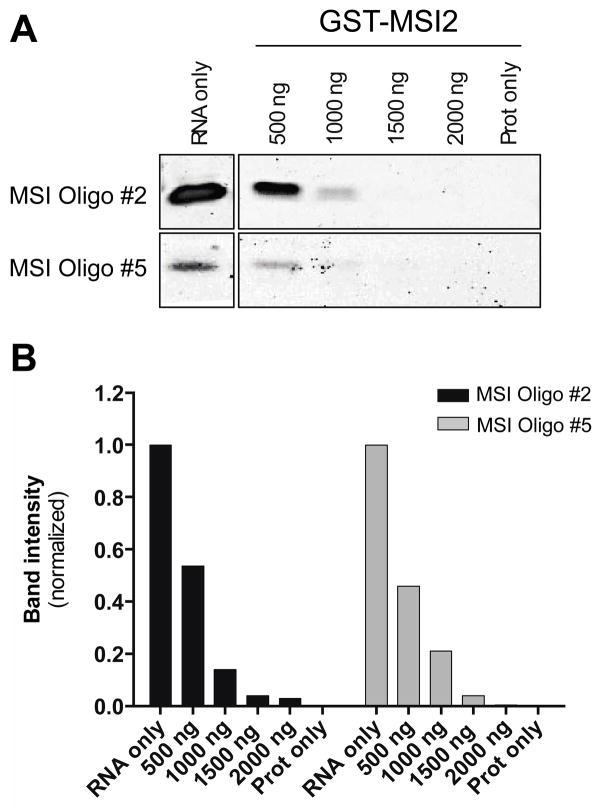
In order to develop an in vitro binding assay to be used for chemical screening, we first assessed the best sequence and length of the ssRNA probe using different oligos (5–15 nt length) containing different repeats of the previously determined MSI1 consensus RNA binding site [(G/A) Un AGU, n=1–3] [17] (Table 1 and Fig 1). We tested binding activities of each oligo using the RNA SYBR-based EMSA assay against the following GST-tagged recombinant proteins: MSI1, MSI2, LIN28A and p53. LIN28A and p53 were used as orthogonal controls for non-specific RNA and DNA oligo binding, respectively. The binding affinity of MSI1, MSI2 and LIN28A increased in accordance with the number of consensus RNA binding site [considering r(GUAGU) as the shortest sequence for binding]. Thus, the 15-nt oligo containing 4 MSI binding motifs (MSI Oligo #4) displayed the highest affinity. However, a 15-nt oligo containing 2 MSI motifs (MSI Oligo #2) had very similar binding affinity compared to the 2 MSI motifs and 8-nt oligo (MSI Oligo #5) (Fig 1 and Table 1). As the longer oligos could form secondary structures reducing the robustness of our screen, we selected the MSI Oligo #5 [r(GUAGUAGU), 8-nt, 2 overlapping binding motifs] as the optimal shortest oligo to be used in the FP assay. Most importantly, LIN28A lacked specificity to the different ssRNA oligos tested (binding with high affinity to the non-specific ssRNA Oligo #8, Table 1) suggesting that it would allow us to distinguish from identifying general RNA-protein inhibitors from MSI sequence specific inhibitors.
Figure 1. SYBR-based Electrophoresis Mobility Shift Assay (EMSA) optimization of ssRNA oligos for fluorescence polarization.
(A)The GST-tagged protein MSI2 at the indicated concentrations (500 to 2000 ng) was incubated for 30 min at room temperature in EMSA buffer with MSI Oligo #2 [r(UAGUAGUAAGUAGUA), 15 nucleotides, 2 MSI motifs] or MSI Oligo #5 [r(GUAGUAGUA), 8 nucleotides, 2 MSI overlapping motifs] at 200 pmols. After the incubation, the reaction was run onto a native 10% polyacrylamide TBE gel in chilled 0.5X TBE at 4°C at 100V for 2–2.5h. The controls consisted of RNA only and protein only incubations and were run in the same gel. The gel was stained with 2X SYBR Safe and the results were imaged by BioRadGel DocTM XR+ imager. (B) Quantification of SYBR RNA bands intensity. Results are shown normalized to RNA only intensity band for both MSI Oligo #2 and #5. Representative data from at least two independent experiments for each oligo is shown.
Development of a specific MSI FP RNA binding assay
The FP assay provides a quantitative and scalable in vitro system to identify small molecules with the ability to block RNA binding activity. To avoid typical interference (either direct or indirect) of many compounds with tracers such as fluorescein [21], we labeled our RNA probe with Cy3 (red-shifted dye with an excitation wavelength of 550 nm and emission wavelength of 570 nm). Additionally, Cy3 labeling provides a photostable, pH insensitive and generally results in less background than most other fluorophores [22]. The optimal probe for the FP assay was generated from the selected ssRNA oligo (MSI Oligo #5) from the SYBR-based EMSA that was then chemically coupled with Cy3 through a 9-carbon (C9) linker in the 5′ end to form the MSI FP oligo [Cy3-labelled ssRNA probe; Cy3-C9-r(GUAGUAGU), 8-nt, 2 overlapping MSI motifs].
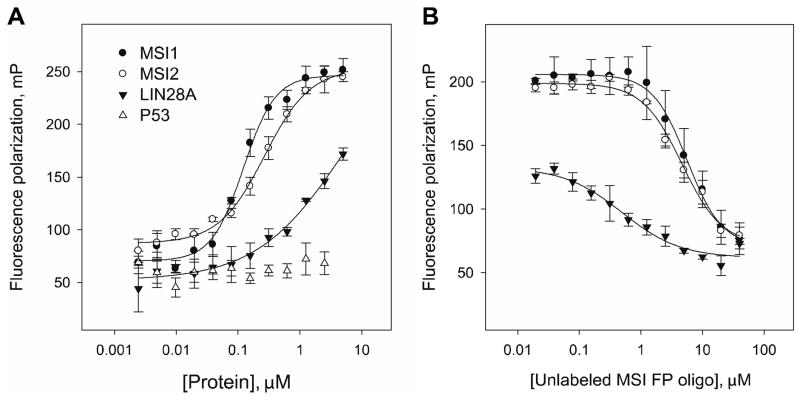
Using recombinant MSI1, MSI2 and LIN28A, we generated plots that measured the binding affinity for the MSI specific FP probe in a 384-well format (Fig 2A). By plating increasing concentrations of GST-protein with a fixed concentration of 10 nM MSI FP oligo we confirmed a high-affinity binding for MSI1 and MSI2 (~1 μM) compared to LIN28A (~2.5 μM). p53 lacked any measurable binding suggesting our FP probe was not binding non-specifically to a DNA binding protein. To understand the reversibility of this RNA binding activity and feasibility for inhibitor screens, we performed a probe displacement assay with MSI1, MSI2 (at 1 μM, fixed concentration) and LIN28A (at 2.5 μM). Interestingly, the non-labeled MSI FP oligo successfully competed off the fluorescent labeled one in a concentration dependent manner with IC50 values of ~5 μM (5.6 μM for MSI1 and 4.6 μM for MSI2) (Fig 2B). LIN28A also showed a reversible binding to the MSI RNA oligo with a lower IC50 (0.5 μM) due to a weaker binding affinity. Overall, these experiments suggested that we had successfully developed a MSI specific FP based assay.
Figure 2. Fluorescence polarization assay in 384-well plate format.
(A) Binding of the MSI FP oligo [Cy3-C9-r(GUAGUAGU) at 10 nM] to increasing concentrations of GST-tagged proteins MSI1, MSI2, LIN28A and p53. The error bars represent plus or minus one standard deviation from triplicate measurements; the protein concentration chosen for following experiments was 1 μM (MSI1 or MSI2) and 2.5 μM (LIN28A); (B) Displacement curves of the Cy3-MSI FP oligo (10 nM) from MSI1, MSI2 and LIN28A with the unlabeled MSI FP oligo; the IC50 values were: MSI1 (5.6 μM), MSI2 (4.6 μM) and LIN28A (0.5 μM).
Miniaturization and quality control for a 1536-well plate MSI FP RNA binding assay
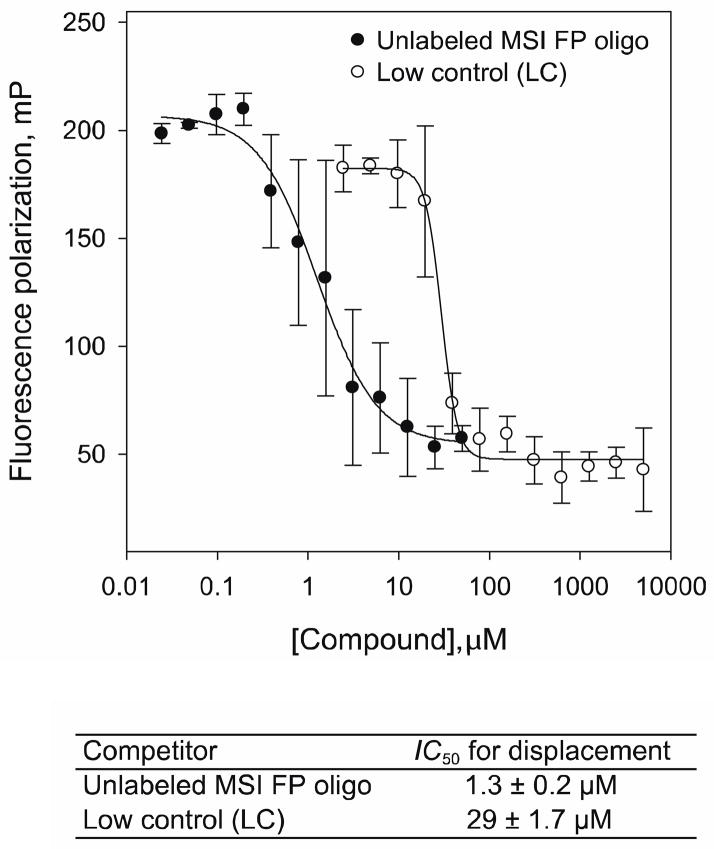
To provide a platform for screening of large compound libraries, we wanted to determine if our MSI FP RNA binding assay could be miniaturized down to a 1536-well format (Fig 3). Additionally, we wanted to identify a non-specific control for the assay other than SDS, since the latter tends to generate bubbles rendering the FP measurements problematic. We opted to use an alternative non-specific compound like oleic acid to abolish all binding activity of the probe to the proteins. We thus compared the oleic acid potency to the unlabeled MSI FP oligo in a dose response utilizing a 1536-well plate format. This resulted in a displacement of MSI2 binding at an IC50value of 1.3 ± 0.2 μM and IC50 value of 29 ± 1.7 μM with the unlabeled probe and oleic acid, respectively; suggesting that oleic acid could indeed be used as a low control for our screening efforts (Fig 3). Taken together, our data demonstrated the development of a scalable and robust assay amenable for further high throughput screening.
Figure 3. Displacement of MSI2 binding in 1536-well plate format fluorescent polarization assay.
Displacement curves of the MSI FP oligo from MSI2, with the unlabeled MSI FP oligo and oleic acid (Low Control, LC); IC50 values for displacement of two independent experiments performed in triplicate are shown in the bottom table.
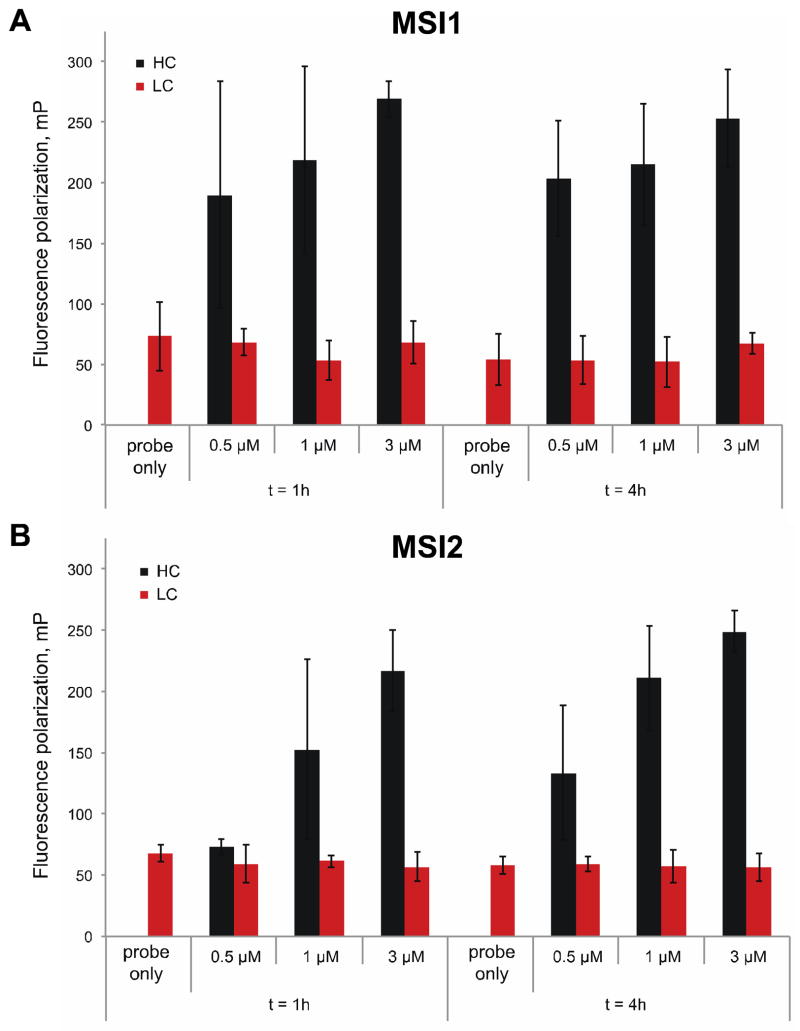
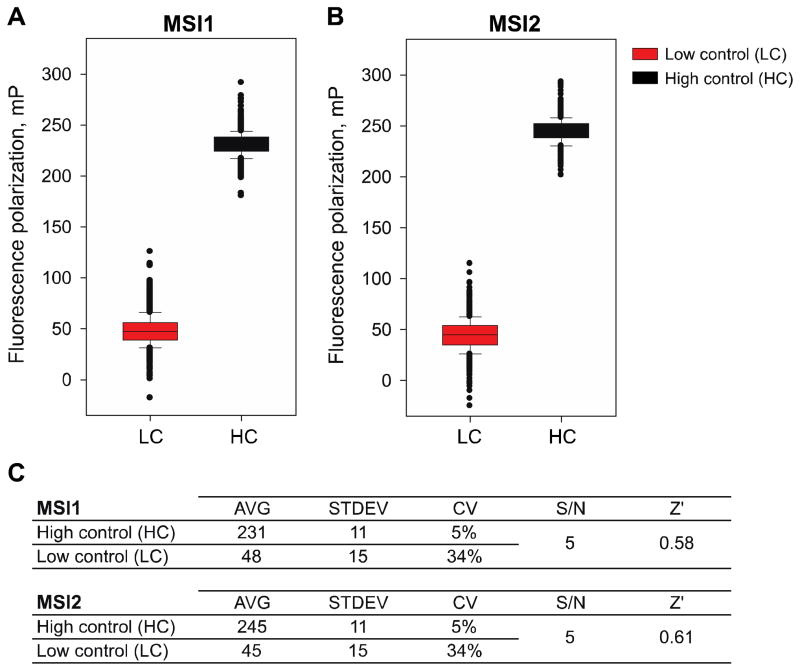
To assess that the recombinant MSI proteins were stable and maintained their activity throughout the duration of the screen (~4 hours), we tested three different protein concentrations (0.5, 1 and 3 μM) of MSI1 and MSI2 in the presence of 1% DMSO (v/v) (HC) or 1 mM oleic acid in 1% DMSO (v/v) (LC) and pre-incubated them at room temperature for 1 or 4 hours. After this period, the MSI FP oligo, at a concentration of 10 nM, was added and then further incubated for an additional hour. Importantly, the incubations at room temperature did not significantly alter the dose dependence or the differential polarization values (ΔmP = 150–200) (Fig 4). We then evaluated the miniaturized assay performance under screening conditions by automated plating of two 1536-well plates with the MSI proteins and probe in the presence of HC or LC (Fig 5). The dynamic range (differential between FP values for HC and LC) was calculated at ~180 mP and the Z′ values were greater than 0.5 (0.58 for MSI1 -Fig 5A, 0.61 for MSI2 -Fig 5B) suggesting that our automated screening window would allow us identify inhibitors for MSI RNA binding activity (Fig 5C).
Figure 4. Stability of MSI proteins in fluorescent polarization assay.
(A) Stability of GST-MSI1 protein at three different concentrations (0.5, 1, and 3 μM) was tested during binding to MSI FP oligo during1h and 4h incubation at room temperature in 1% DMSO (v/v) (High Control, HC; black bars) or 1 mM oleic acid in 1% DMSO (v/v) (LC; grey bars); (B) Same as (A) with GST-MSI2 protein. In both (A) and (B), a control with MSI FP oligo only is shown as a measure of FP (mP) background. The dynamic range (differential between HC and LC) at 1h and 4h did not show significant differences and was demonstrated to be optimal at 4h incubation (ΔmP > 150), the approximate time of the pilot screen. Values represent the average and standard deviation of two independent experiments performed in triplicate.
Figure 5. Control run plates of MSI1 and MSI2 in the pilot screen.
MSI (1 μM) High Control wells were seeded with 1% DMSO (HC) and Low Control wells (LC) with 1 mM oleic acid in 1% DMSO (v/v); (A) Box plot comparing MSI1 HC and LC demonstrated an optimal dynamic range of 4.8-fold (ΔmP=183); (B) Box plot for MSI2 showing a dynamic range of 5.4-fold (ΔmP=200); (C) Fluorescence polarization (mP) average (AVG) values, standard deviation (STDEV), coefficient of variation (CV)and Z′ factor of each control plate. Z′ values for both MSI1 and MSI2 were >0.5.
Pilot screen using the MSI FP RNA binding assay
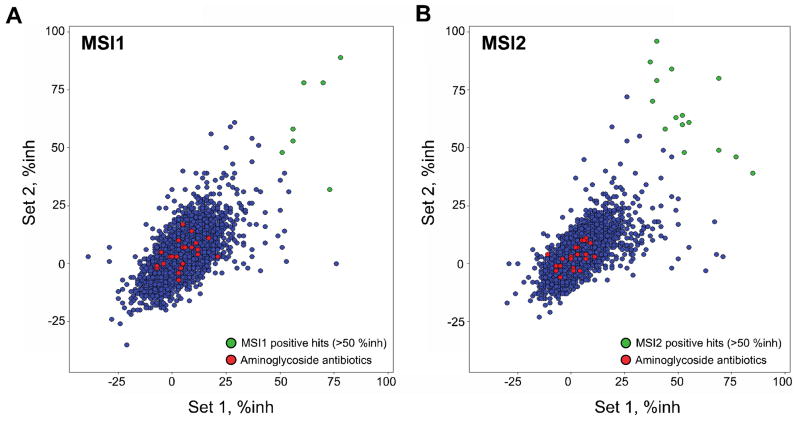
Based on the performance from our feasibility studies, we performed a pilot screen against a library of 6,208 compounds, which included known drugs, experimental bio-actives and natural products. Each compound was screened in duplicate across two separate sets of plates (Set 1 and Set 2) at a final concentration of 10μM in 1% DMSO (v/v). HC and LC were also included in each plate as internal references for the assay. Both data sets exhibited Z′ values greater than 0.5 suggesting a robust and reproducible assay. We then compared the percentage inhibition (%inh) values from all the pilot screen compounds in Set 1 and Set 2 for both MSI1 and MSI2 and found a linear correlation between the replicates suggesting that the screen demonstrated reproducibility (Fig 6). We defined positives as compounds with an average inhibition greater than 50%. We identified a total of 17 compounds with an initial hit rate of 0.27%, five of which inhibited both MSI1 and MSI2 (Table 2 and Fig 6). MSI1 screening resulted in 7 compounds (0.11%) and 15 compounds were identified as MSI2 inhibitors (0.24%), with an overall initial hit rate of 0.08% from the total number of compounds screened that scored as dual MSI inhibitors (Table 2). We flagged 37 compounds due to high FI and discarded 4 small molecules known as commonly identified false positives in FP assay (internal screening data; e.g. brazilein). Among the list of compounds that were inhibiting MSI binding activity, 5 were anthracyclines used in cancer chemotherapy such as idarubicin, daunorubicin, rutilantinone, aklavine or doxorubicin and two more were nucleotide analogs. Most interestingly, the classical RNA-protein disruptive molecules (23 compounds from different vendors including 12 unique aminoglycoside antibiotics, e.g. streptomycin) lacked significant MSI inhibition activity (see red circles, Fig 6A–B and Table 4).
Figure 6. High-throughput fluorescence polarization pilot screen for MSI1 and MSI2 inhibitors.
A library of 6,208 compounds was screened in duplicate assay plates (Set 1 and Set2) to determine the reproducibility of the HTS for both MSI1 (A) and MSI2 (B). The % inhibition of each compound was determined and shown in blue circles. Signals generated from the aminoglycoside antibiotics (known RNA disruptive molecules) present in the library are depicted as red circles and positive hits (>50% average inhibition for Set1 and Set2) are depicted as green circles [for either MSI1 (A) or MSI2 (B)]. The hit rate for MSI1 was 0.11%, for MSI2 0.24% and for dual MSI inhibition 0.08% (see Table 2).
Table 4.
Percentage inhibition (%inh) of MSI1 and MSI2 binding of the 12 unique aminoglycoside antibiotics (23 total compounds from different sources) present in the FP pilot screen.
| # | SKI ID | Compound name | MSI1 positives |
MSI1 %inh (Set1) |
MSI1 %inh (Set2) |
MSI1 AVG %inh |
MSI2 positives |
MSI2 %inh (Set1) |
MSI2 %inh (Set2) |
MSI2 AVG %inh |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 217717 | Amikacin | No | 21 | 3 | 12 | No | 5 | 10 | 8 |
| 2 | 209802 | Amikacinsulfate | No | −5 | 5 | 0 | No | −7 | −1 | −4 |
| 3 | 218382 | Dibekacin | No | 17 | 11 | 14 | No | 9 | 9 | 9 |
| 4 | 417942 | Dihydrostreptomycinsulfate #1 | No | 9 | 14 | 12 | No | 7 | 11 | 9 |
| 5 | 417942 | Dihydrostreptomycinsulfate #2 | No | 12 | 6 | 9 | No | 6 | 10 | 8 |
| 6 | 210040 | Gentamicinesulfate #1 | No | −4 | 0 | −2 | No | 1 | −2 | −1 |
| 7 | 217582 | Gentamicinesulfate #2 | No | 12 | 4 | 8 | No | 2 | 7 | 5 |
| 8 | 217702 | Kanamycin A sulfate | No | 3 | −7 | −2 | No | −5 | −6 | −6 |
| 9 | 210095 | Kanamycin sulfate | No | 3 | 10 | 7 | No | 11 | 3 | 7 |
| 10 | 274387 | Netilmicinsulfate | No | 4 | −2 | 1 | No | 1 | −3 | −1 |
| 11 | 211005 | Paromomycinsulfate #1 | No | 2 | 3 | 2 | No | 7 | 4 | 6 |
| 12 | 218166 | Paromomycinsulfate #2 | No | 15 | 32 | 24 | Yes | 99 | 90 | 94 |
| 13 | 417074 | Paromomycinsulfate #3 | No | 5 | 0 | 2 | No | −4 | 2 | −1 |
| 14 | 211215 | Ribostamycinsulfate | No | 6 | 7 | 7 | No | 0 | 2 | 1 |
| 15 | 217950 | Ribostamycinsulfate salt | No | 9 | 7 | 8 | No | 7 | 2 | 4 |
| 16 | 210292 | Sisomicinsulfate #1 | No | −7 | −2 | −5 | No | 0 | 3 | 1 |
| 17 | 218057 | Sisomicinsulfate #2 | No | 0 | 3 | 2 | No | 4 | −3 | 0 |
| 18 | 210294 | Spectinomycindihydrochloride | No | 3 | −4 | 0 | No | −7 | −3 | −5 |
| 19 | 210294 | Spectinomycin hydrochloride | No | 5 | 17 | 11 | No | 3 | 4 | 3 |
| 20 | 210297 | Spectinomycinsulfate | No | 5 | 7 | 6 | No | 7 | 10 | 9 |
| 21 | 217689 | Streptomycin sulfate | No | −7 | −1 | −4 | No | −11 | 4 | −4 |
| 22 | 210332 | Tobramycin #1 | No | −1 | 3 | 1 | No | −5 | −1 | −3 |
| 23 | 218059 | Tobramycin #2 | No | 11 | 9 | 10 | No | 3 | 7 | 5 |
Compound validation and secondary screening
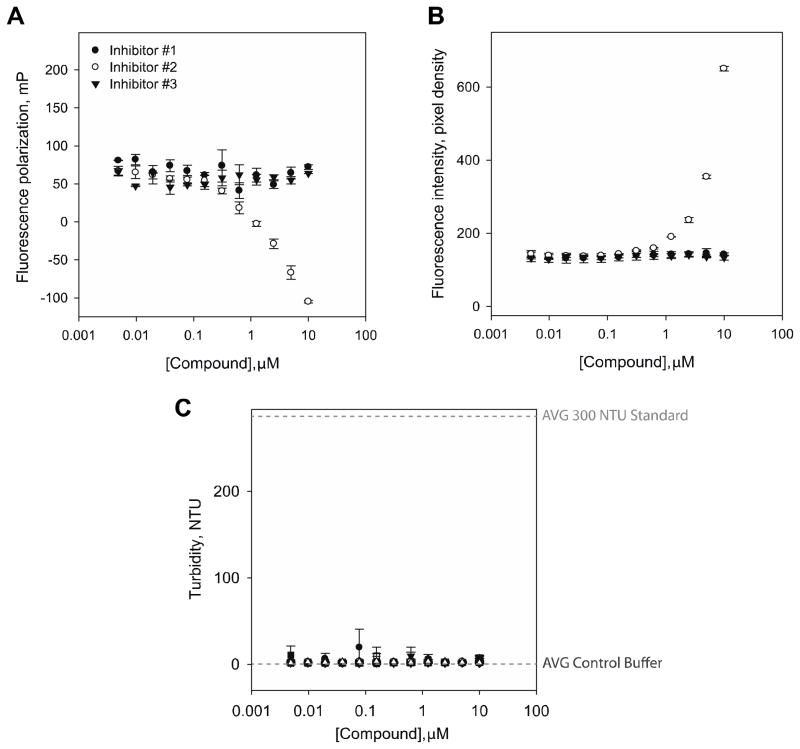
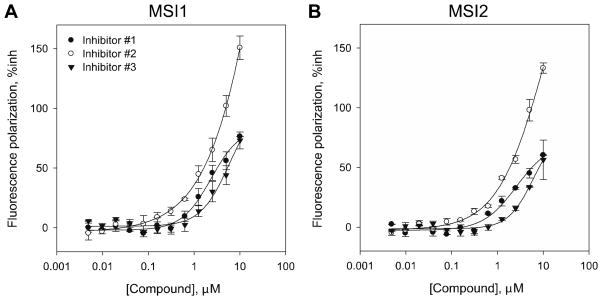
We determined the optical interference with the top 28 compounds that were identified for secondary screening (inhibitors #1–3 are shown in Fig 7A–B and data not shown) by adding the compounds alone at different concentrations in a 384-well plate and assessing the optical interference with the FP and the FI values (Fig 7A–B). Inhibitors #2 and #18–25 decreased the FP values and increased the FI when the concentration of the molecule was increased (Table 3 and Fig 7A–B). Furthermore, we confirmed the solubility of the compounds by measuring the turbidity of the solution up to 10 μM (Fig 7C). Of note, none of the tested compounds showed increased turbidity, confirming an optimal solubility at the concentrations used in our assay. We then performed dose-response curves with 12 serial dilutions up to a concentration of 10 μM with 28 compounds. Of the 25 compounds tested, only 3 validated in the dose-response secondary screen with IC50 values below 10 μM (Fig 8 and Table 3). Of note, MSI1 demonstrated a slightly higher potency than MSI2 for these three validated hits.
Figure 7. Fluorescence polarization, intensity and turbidity assessment.
For the three top hits, the optical interference in the FP values (mP) (A) and fluorescence intensity (pixel density) (B) were assessed up to 10μM concentration of the drugs. Of note, inhibitor #2 interfered with the assay due to high fluorescence intensity of the compound significantly decreasing mP values at concentrations >1 μM; (C) the 3 small molecules were also tested for solubility by analysis of their turbidity in solution at different concentrations up to 10 μM. Values represent the average and standard deviation of two independent experiments performed in triplicate.
Table 3.
Percentage inhibition from pilot screen run and IC50 values from dose-response validation of the 28 hits from the FP screen.
| Pilot screen | Secondary screen | ||||||
|---|---|---|---|---|---|---|---|
| # | SKI ID | Compound name | MSI1 HTS %inh | MSI2 HTS %inh | High FI | MSI1 AVG IC50 | MSI2 AVG IC50 |
| 1 | 210998 | Inhibitor #1 | 55 | 59 | no | 3.1 | 5.5 |
| 2 | 398565 | Inhibitor #2 | 83 | 75 | no | 3.6 | 4.6 |
| 3 | 417128 | Inhibitor #3 | 70 | 56 | no | 5.3 | 6.6 |
| 4 | 209954 | Inhibitor #4 | 22 | 60 | no | >10 | >10 |
| 5 | 210920 | Inhibitor #5 | 23 | 62 | no | >10 | >10 |
| 6 | 211201 | Inhibitor #6 | 50 | 36 | no | >10 | >10 |
| 7 | 211551 | Inhibitor #7 | 52 | 31 | no | >10 | >10 |
| 8 | 217615 | Inhibitor #8 | 28 | 54 | no | >10 | >10 |
| 9 | 397311 | Inhibitor #9 | 31 | 62 | no | >10 | >10 |
| 10 | 397438 | Inhibitor #10 | 46 | 62 | no | >10 | >10 |
| 11 | 398321 | Inhibitor #11 | 32 | 66 | no | >10 | >10 |
| 12 | 416737 | Inhibitor #12 | 43 | 51 | no | >10 | >10 |
| 13 | 417049 | Inhibitor #13 | 45 | 56 | no | >10 | >10 |
| 14 | 417182 | Inhibitor #14 | 34 | 68 | no | >10 | >10 |
| 15 | 417183 | Inhibitor #15 | 17 | 58 | no | >10 | >10 |
| 16 | 398034 | Inhibitor #16 | 63 | 70 | no | >10 | >10 |
| 17 | 398563 | Inhibitor #17 | 52 | 73 | no | >10 | >10 |
| 18 | 209480 | Inhibitor #18 | 72 | 80 | yes | >10 | >10 |
| 19 | 209995 | Inhibitor #19 | 149 | 138 | yes | >10 | >10 |
| 20 | 274191 | Inhibitor #20 | 41 | 54 | yes | >10 | >10 |
| 21 | 397366 | Inhibitor #21 | 55 | 38 | yes | >10 | >10 |
| 22 | 416584 | Inhibitor #22 | 141 | 155 | yes | >10 | >10 |
| 23 | 416735 | Inhibitor #23 | 50 | 47 | yes | >10 | >10 |
| 24 | 416876 | Inhibitor #24 | 45 | 58 | yes | >10 | >10 |
| 25 | 416984 | Inhibitor #25 | 63 | 78 | yes | >10 | >10 |
Figure 8. Dose-response curves of the three top hits in HTS for MSI inhibition.
(A) MSI1 binding curves with % inhibition (%inh) against a dose response of the inhibitors #1–#3 up to 10μM; (B) same as (A) with MSI2. The IC50 values found are shown in Table 3. Inhibitor #2 showed a potential optical interference with the assay as the % inhibition of FP was greater than 100% at concentrations >5 μM. Values represent the average and standard deviation of two independent experiments performed in triplicate.
Orthogonal screen by SYBR-based EMSA analysis
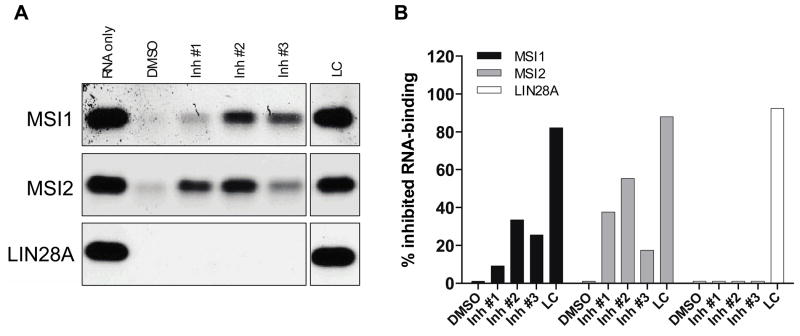
To further confirm the identified 3 MSI inhibitors, we performed our previously described orthogonal SYBR-based EMSA with MSI1, MSI2 and LIN28A. As expected, oleic acid at 1 mM (LC) inhibited the binding of all 3 RBPs significantly (>80 % inhibition, Fig 9A–B). Most importantly, all three MSI inhibitors demonstrated activity against both MSI1 and MSI2, but not against LIN28A. Inhibitor #1 demonstrated modest inhibition for MSI1 compared with its activity against MSI2, while Inhibitor #3 demonstrated increased potency with MSI1 compared to MSI2. Although Inhibitor #2 scored with high FI in the optical interference assay (Fig 7A–B), it retained inhibitory activity in the RNA EMSA and was the molecule demonstrating the best inhibition for both MSI. Altogether we have performed a 6,208 compounds pilot screen, verified the top hits with a dose-response secondary screen and then validated the 3 high potency (italic>10 μM) hits using an EMSA orthogonal assay simultaneously testing their optical interference and solubility. We have identified 3 putative MSI-specific inhibitors using the strategy outlined in Fig 10. These results will be further validated using in vitro cellular systems and MSI-dependent systems.
Figure 9. SYBR-based Electrophoresis Mobility Shift Assay (EMSA) orthogonal validation of MSI inhibitors.
(A) The GST-tagged proteins MSI1, MSI2 and LIN28A (1000 ng) were incubated for 30 min at room temperature in the absence of 1% DMSO (v/v) or in the presence of 10 μM inhibitors #1–#3 or 1 mM oleic acid) in EMSA buffer. After this period, 200 pmols of ssRNA MSI Oligo #2 [r(UAGUAGUAAGUAGUA)] were added and incubated for an additional 30 min. A final volume of 20μL reaction was run onto a native 10% polyacrylamide TBE gel in chilled 0.5X TBE at 4°C at 100V for 2–2.5h. The gel was then stained with 2X SYBR Safe and results were obtained by BioRadGel DocTM XR+ imager. (B) Percentage inhibition (%) of RNA-binding activity with inhibitors same as in (A) against MSI1, MSI2 and LIN28A. After quantification of RNA band intensity, results were graphed as % of inhibited RNA-binding for MSI1 (black bars), MSI2 (grey bars) or LIN28A (white bars) considering the RNA only band as the control (highest signal) and the DMSO control as the highest binding (lowest signal). One representative gel of at least two repeats per oligo and recombinant protein is shown.
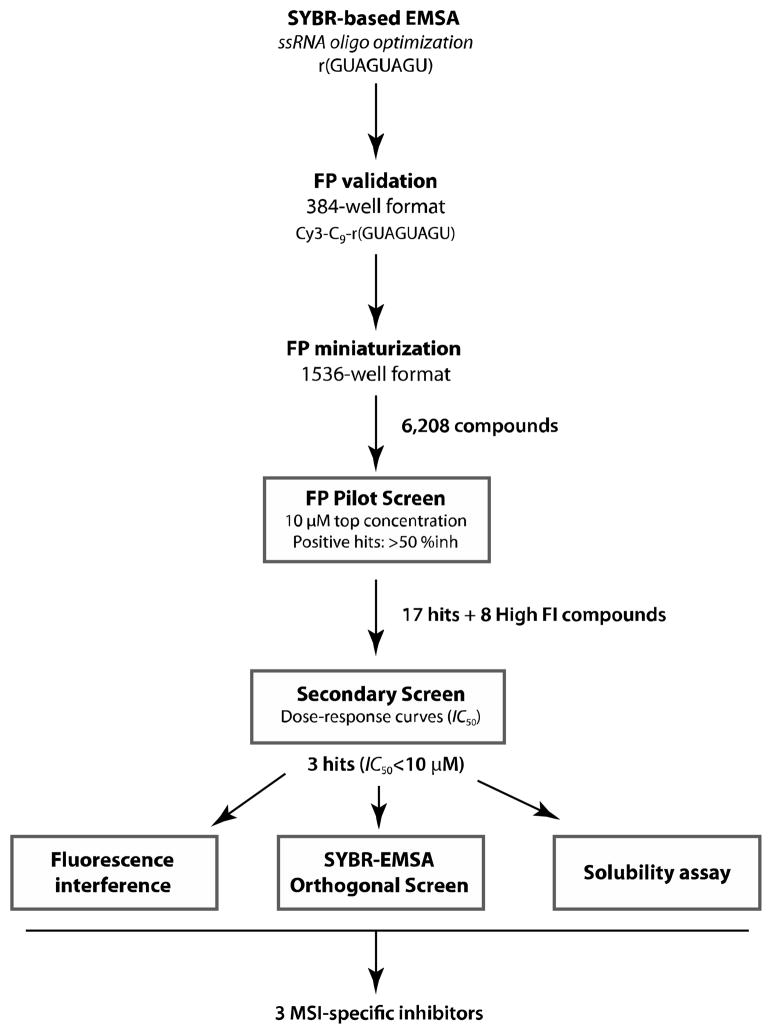
Figure 10. Strategy for the development of the fluorescence polarization (FP) assay to screen for inhibitors of MSI RNA-binding proteins.
The optimal ssRNA oligo for FP was obtained by testing GST-MSI1 and -MSI2 binding in SYBR-based EMSA. FP validation was done in 384-well plates and then miniaturized and optimized in 1536-well plates. A 6,208 chemical library was run as a pilot screen to validate the assay and 17 total positive hits were found (5 common to both MSI). A secondary screen by dose-response curves (IC50) was performed and 3 final chemical candidates, with <10μM, were obtained. Finally, optical interference and solubility assays were simultaneously performed with the orthogonal screen against both MSI by SYBR-EMSA. The three top hits were validated as putative MSI selective inhibitors.
DISCUSSION
Oncogenic drivers that regulate the translational machinery can drive cancer pathogenesis. The MSI gene family has been identified as a translational regulator that is highly expressed and associated with some of the most aggressive solid cancers and hematopoietic malignancies [5]. MSI1 has been mainly studied in the neural system and it is known to be a key regulator in the maintenance of multi-potential neural progenitors in their proliferative stem-like state and to influence cellular differentiation [13, 14, 22]. The MSI family has been demonstrated to be dysregulated in cancer in a variety of tumors including in the brain, prostate, breast, and colon [7, 23–25]. MSI2 is mainly expressed in the hematopoietic system and it is an important modulator of proliferation and differentiation in normal hematopoietic stem cells whereas its overexpression has been related to myeloid [3, 11] and lymphoid malignancies [12].
Due to the inherent challenges of deciphering RBPs biology, pharmaceutical companies have largely ignored them as putative therapeutic targets. Moreover, one of the major challenges in developing selective inhibitors for RBPs is the lack of high-throughput in vitro assays in which to rapidly screen small molecules targeting the interaction between RBP and mRNA targets. Traditional methods such as EMSA assays cannot easily be adapted for high throughput. Other biophysical methods such as surface plasmon resonance or circular dichroism are most commonly used as validation methods to obtain information on protein-RNA binding kinetics and the molecular mechanism of action [26, 27] rather than for screening large chemical libraries.
Despite recent studies that have highlighted the importance of the MSI family as a therapeutic target, there are currently no drugs or tool compounds available for this class of RBPs. Other groups have previously reported the optimization of the protein production and the development of similar assays for the eukaryotic translation initiation factor 4E (eIF4E) binding to mRNA 5′ cap sequence [28] or the eukaryotic cap-binding complex CBP80/CBP20 [29]. An FP assay has been developed to study the molecular mechanisms of RNA binding to AU-rich elements with specificity RRMs found in Hu antigen R (HuR) protein [30]. More recently, similar structure-binding studies using FP have shown conformational changes in these RRMs during RNA binding [31]. Nevertheless, our study provides the first developed platform for the identification of small molecules that block MSI in vitro binding activity.
We have developed and optimized an FP based HTS strategy to target MSI interaction with its RNA consensus binding sequence. We screened a 6,208 compound library obtaining 17 initial hits and then further validating 3 MSI-specific inhibitors. Our assay showed a great reproducibility, robustness (Z′ factor = 0.6) and specificity against MSI proteins due to the use of a very short MSI probe (8 nucleotides) containing 2 overlapping specific MSI motifs. Of note, the low overall hit rate of our assay (0.27%, Table 2) and the lower hit rate for dual MSI inhibitors (0.08%) confirms it as a suitable experimental approach for large screens with libraries of thousands of compounds to obtain more specific and potent MSI inhibitors.
Table 2.
Summary of statistical data from the Pilot Screen and Validation of Hits.
| Characteristic | Value |
|---|---|
| Total compounds from Pilot Screen | 6,208 |
| Total high confidence hits | 17 |
| MSI1 | 7 |
| MSI2 | 15 |
| Both MSI | 5 |
| Overall hit rate, % | 0.27 |
| MSI1 | 0.11 |
| MSI2 | 0.24 |
| Both MSI | 0.08 |
| Compounds tested in Secondary Screen | 25 |
| Validated MSI inhibitors (IC50<10 μM) | 3 |
| Z′ factor, mean | 0.60 |
Although our screen initially identified compounds that were specific to either MSI1 or MSI2, the validated hits ended up inhibiting both. Moreover, the closely conserved RRMs of both MSI might explain why this might be challenging. Nevertheless, it remains necessary to screen a larger library to determine if MSI isoform specific inhibitors can be identified. We found that the orthogonal assay for MSI is necessary to verify the hits in our screen as Inhibitor #2 demonstrated bold>10μM activity and high FI in our validation screen. Previous studies have identified inhibitors of protein RNA-binding activity with IC50 values in the low micromolar range (with the exception of one compound inhibiting influenza NS1 binding to dsRNA at 300 nM) suggesting that the potency of our compounds were comparable. By utilizing an additional RBP (LIN28A), we found that these inhibitors retained their selectivity against MSI. However, to assess the specificity of these inhibitors for MSI proteins, a larger panel of RBPs would need to be generated and tested. It would have special interest to test MSI closely related RBPs containing RRMs with some sequence similarity (e.g. HuR).
Most importantly, we did not identify any of the aminoglycoside antibiotics present in the screen as putative MSI inhibitors (Table 4), suggesting that the binding pocket of our proteins are unique and are not affected by these classical RNA-protein disruptive molecules. This data bolsters our approach and provides additional evidence that selective MSI inhibitors can be identified. Inhibitors for RNA-protein interactions have been targeted in influenza [26], Rift Valley fever virus [32] or Hepatitis C virus [33] but have not been explored in RNA-binding proteins involved in cancer yet. Our study provides a strategy for the development of additional assays for identifying inhibitors for other RNA-binding proteins.
In summary, our assay represents a novel high-throughput drug screen to identify inhibitors for the MSI family of RNA binding proteins. We provide a platform for the discovery of small molecules that can target these clinically relevant cancer targets. Moreover, we plan to expand our studies in the context of a larger screen and to functionally validate our identified MSI inhibitors in both cell based and animal models.
Acknowledgments
The authors would like to thank members of the HTS Core Facility for their help during the course of the study. We would also like to acknowledge De-Ming Chau, Danica Chau and Glorymar Ibañez for their help and advice with the GST-tagged protein purification and optimization of the proteins used in this study and James Taggart for his suggestions during the preparation of the manuscript. The HTS Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and by a NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44. MGK was supported by the US National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases Career Development Award, V-foundation and Sidney Kimmel Award. This work was supported by the Louis V Gerstner Young Investigator Award and by the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center.
ABREVIATIONS
- RBP
RNA-binding protein
- MSI
Musashi
- GST
Glutathione S-Transferase
- SYBR
N′,N′-dimethyl-N-[4-[(E)-(3-methyl-1,3-benzothiazol-2-ylidene)methyl]-1-phenylquinolin-1-ium-2-yl]-N-propylpropane-1,3-diamine
- EMSA
Electrophoresis Mobility Shift Assays
- HTS
High Throughput Screening
- BSA
Bovine Serum Albumin
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- DMSO
Dimethyl Sulfoxide
- EDTA
EthylenediaminetetraaceticAcid
- TCEP
Tris(2-carboxyethyl)phosphine
- FI
fluorescence intensity
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interests.
References
- 1.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–25. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, Ferrando AA, Grutz G, Turner M. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–24. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, Gozo M, Einhorn W, Lane SW, Scholl C, Frohling S, Fleming M, Ebert BL, Gilliland DG, Jaenisch R, Daley GQ. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–8. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–58. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 7.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–74. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, Ra YS, Zilberberg K, McLeod J, Scherer SW, Sunil Rao J, Eberhart CG, Grajkowska W, Gillespie Y, Lach B, Grundy R, Pollack IF, Hamilton RL, Van Meter T, Carlotti CG, Boop F, Bigner D, Gilbertson RJ, Rutka JT, Taylor MD. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–72. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbouti A, Hoglund M, Johansson B, Lassen C, Nilsson PG, Hagemeijer A, Mitelman F, Fioretos T. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23) Cancer Res. 2003;63:1202–6. [PubMed] [Google Scholar]
- 11.Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, Goldman J, Goh H, Kim SH, Kim DW, Chuah C, Oehler VG, Radich JP, Jordan CT, Reya T. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–8. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mu Q, Wang Y, Chen B, Qian W, Meng H, Tong H, Chen F, Ma Q, Ni W, Chen S, Jin J. High expression of Musashi-2 indicates poor prognosis in adult B-cell acute lymphoblastic leukemia. Leuk Res. 2013;37:922–7. doi: 10.1016/j.leukres.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–56. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Park SM, Deering RP, Lu Y, Tivnan P, Lianoglou S, Al-Shahrour F, Ebert BL, Hacohen N, Leslie C, Daley GQ, Lengner CJ, Kharas MG. Musashi-2 controls cell fate, lineage bias, and TGF-beta signaling in HSCs. J Exp Med. 2014;211:71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le SY, Shapiro BA, Penalva LO. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem. 2009;284:12125–35. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–53. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antczak C, Wee B, Radu C, Bhinder B, Holland EC, Djaballah H. A High-Content Assay Strategy for the Identification and Profiling of ABCG2 Modulators in Live Cells. Assay Drug Dev Technol. 2014;12:28–42. doi: 10.1089/adt.2013.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antczak C, Mahida JP, Bhinder B, Calder PA, Djaballah H. A high-content biosensor-based screen identifies cell-permeable activators and inhibitors of EGFR function: implications in drug discovery. J Biomol Screen. 2012;17:885–99. doi: 10.1177/1087057112446174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antczak C, Shum D, Radu C, Seshan VE, Djaballah H. Development and validation of a high-density fluorescence polarization-based assay for the trypanosoma RNA triphosphatase TbCet1. Comb Chem High Throughput Screen. 2009;12:258–68. doi: 10.2174/138620709787581729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turek-Etienne TC, Small EC, Soh SC, Xin TA, Gaitonde PV, Barrabee EB, Hart RF, Bryant RW. Evaluation of fluorescent compound interference in 4 fluorescence polarization assays: 2 kinases, 1 protease, and 1 phosphatase. J Biomol Screen. 2003;8:176–84. doi: 10.1177/1087057103252304. [DOI] [PubMed] [Google Scholar]
- 22.Harvey BJ, Levitus M. Nucleobase-specific enhancement of Cy3 fluorescence. J Fluoresc. 2009;19:443–8. doi: 10.1007/s10895-008-0431-1. [DOI] [PubMed] [Google Scholar]
- 23.Li D, Peng X, Yan D, Tang H, Huang F, Yang Y, Peng Z. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann Surg Oncol. 2011;18:2074–83. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 24.Nakano A, Kanemura Y, Mori K, Kodama E, Yamamoto A, Sakamoto H, Nakamura Y, Okano H, Yamasaki M, Arita N. Expression of the Neural RNA-binding protein Musashi1 in pediatric brain tumors. Pediatr Neurosurg. 2007;43:279–84. doi: 10.1159/000103307. [DOI] [PubMed] [Google Scholar]
- 25.Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH, Anderson S, Haegert A, Brahmbhatt S, Shukin R, Mo F, Li E, Fazli L, Hurtado-Coll A, Jones EC, Butterfield YS, Hach F, Hormozdiari F, Hajirasouliha I, Boutros PC, Bristow RG, Jones SJ, Hirst M, Marra MA, Maher CA, Chinnaiyan AM, Sahinalp SC, Gleave ME, Volik SV, Collins CC. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J Pathol. 2012;227:53–61. doi: 10.1002/path.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho EJ, Xia S, Ma LC, Robertus J, Krug RM, Anslyn EV, Montelione GT, Ellington AD. Identification of influenza virus inhibitors targeting NS1A utilizing fluorescence polarization-based high-throughput assay. J Biomol Screen. 2012;17:448–59. doi: 10.1177/1087057111431488. [DOI] [PubMed] [Google Scholar]
- 27.Katsamba PS, Myszka DG, Laird-Offringa IA. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J Biol Chem. 2001;276:21476–81. doi: 10.1074/jbc.M101624200. [DOI] [PubMed] [Google Scholar]
- 28.Visco C, Perrera C, Thieffine S, Sirtori FR, D’Alessio R, Magnaghi P. Development of biochemical assays for the identification of eIF4E-specific inhibitors. J Biomol Screen. 2012;17:581–92. doi: 10.1177/1087057112438554. [DOI] [PubMed] [Google Scholar]
- 29.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol. 2002;9:912–7. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 30.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–59. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Zeng F, Liu Q, Liu H, Liu Z, Niu L, Teng M, Li X. The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr D Biol Crystallogr. 2013;69:373–80. doi: 10.1107/S0907444912047828. [DOI] [PubMed] [Google Scholar]
- 32.Ellenbecker M, Lanchy JM, Lodmell JS. Identification of Rift Valley fever virus nucleocapsid protein-RNA binding inhibitors using a high-throughput screening assay. J Biomol Screen. 2012;17:1062–70. doi: 10.1177/1087057112448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26:1019–27. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]