Abstract
The importance of miRNA in cellular regulation is gaining momentum. Therefore, it is of interest to study miRNA in human genes. Hence, the humanmRNA sequences (12,175) were searched for miRNA binding sites and 2,563predicted sites were found. We observed that the miR-3960 has more than 1000mRNA binding sites with high affinity (with ΔG/ΔGm values greater than or equal to 90%) for 375genes. The miR-3960 has 565 binding sites in the 5'UTRs and 515 sites in theCDS of mRNAs. Nucleotide sequences of the binding sites in CDS encode for polyalanine orpolyproline. It is observed that miR-3960 has binding sites in 73 mRNAs of target genesencoded transcription factors. Thus, we document predictedproperties (polysites, sites in CDS) of uncharacterized miR-3960 binding sites. The studying of the miRNA properties is important for creation of diagnostic methods of cancer.
Keywords: MicroRNA, Genes, mRNA, miR-3960
Background
The importance of miRNA studies is steadily increasing over two decades [1]. It is known that many miRNAs regulate the expression of genes involved intumourigenesis [2–4]. For example, changes in miRNA concentrations have beenobserved during the development of lung cancer [2], breast cancer [3] andother cancers [4]. It has been proposed that miRNA binding to mRNAs occurs mainly in the 3'UTRs of mRNAs [5]. However, it has been shown, that miRNAbinding sites can be located in 5'UTR and CDS of several mRNAs [6, 7]. Therefore, it is importance to document a comprehensive study on miRNAusing available data in the public domain. Hence, we studied miR-3960 withpreferential binding to 5'UTR and CDS of several genes. Most of these genesare involved in the development of cancer; therefore, it is necessary toidentify the features for miRNA binding sites. Identification of newinteractions with miR-3960 in mRNAs will increase our understanding of thepost-transcriptional regulation of gene expression.
Methodology
Sequence dataset:
The complete nucleotide sequences of precursor mRNAs (premRNAs) of human genes were downloaded from NCBI GenBank (http://www.ncbi.nlm.nih.gov) in FASTA format. The nucleotide sequences of human mature miR-3960 were downloaded from the miRBase database (http://mirbase.org) [8].
Sequence Analysis:
Nucleotide sequences of pre-mRNAs were shorten into mature mRNA sequences by Lextractor002 script (http:// sites. google.com/site/malaheenee/software). This script create file with mRNA sequence and indicate its bounds between 5'UTR, CDS, and 3'UTR according Genbank information about each gene.
Target prediction using MirTarget program:
The target genes for the tested miRNAs were identified using the MirTarget program, which was developed in our laboratory. The workflows to scanning mirna are shown in (Figure 1 & Figure 2)
Figure 1.
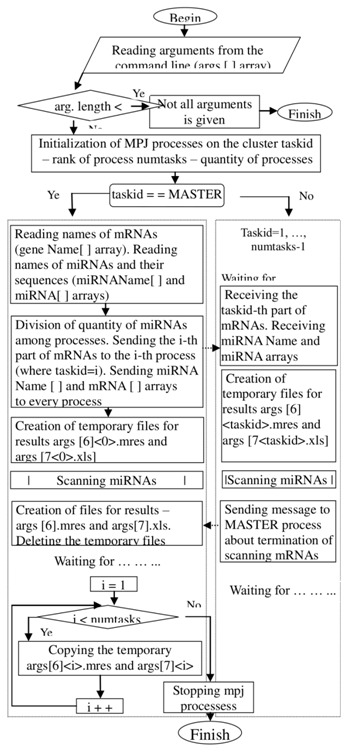
Workflow to MirTarget program (parallel algorithm).
Figure 2.
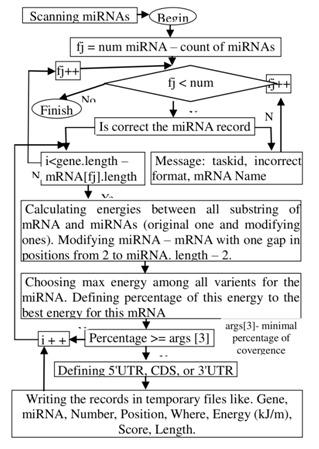
Workflow to scanning mRNAs.
This program defines the following features of binding sites: a) the start point of an miRNA binding site in mRNA; b) the localization of miRNA binding sites in the 5'-untranslated region (5'UTR), coding domain sequence (CDS) and 3'UTR of the mRNAs; c) the free energy of hybridization (ΔG, kJ/mole); and d) the schemes of nucleotide interactions between the miRNAs and the mRNAs. The ΔG/ΔGm (%) ratio was counted for each site, where ΔGm equalled the free energy of a miRNA binding with its perfect complementary nucleotide sequence. The miRNA binding sites located on the mRNAs had ΔG/ΔGm ratios of 90% or more. We also noted the position of the binding sites on the mRNA, beginning from the first nucleotide of the mRNA's 5' UTR. The MirTarget program computed the interactions between the nucleotides of the miRNA and those of the target gene mRNAs. It was found that there are hydrogen bonds between adenine (A) and uracil (U), guanine (G) and cytosine (C), and G and U, as well as between A and C [9]. The distance between A and C was equal to the GC, A-U, and G-U distances [10]. The numbers of hydrogen bonds in the G-C, A-U, G-U and A-C interactions were found to be 3, 2, 1 and 1, respectively. The free binding energies of these nucleotide pairs were accepted as the same values (3:2:1:1).
Results
Features of miR-3960 binding with mRNAs target genes:
miR-3960 is in an exon of the cyclin-dependent kinase 9 gene, located on chromosome 9. miR-3960 has a length of 20 nt and a GC-content of 96%. The maximum free energy of miR-3960 binding to mRNA is -125.3 kJ/mole. We found that miR-3960 has 1100 binding sites on 375 target mRNAs with ΔG/ΔGm values of 90% or more. Of those, 20 miR-3960 binding sites are located in 3'UTRs, 565 sites are located in 5'UTRs and 515 sites are located in CDSs. miR-3960 has 19 binding sites in the mRNA of the RHOT1 gene, 18 binding sites in the mRNAs of the AFF2 and KIF3B genes, 16 binding sites in the 5'UTRs of the mRNAs of the DAZAP1 and BCL2L11 genes and 14 binding sites in the CDSs of the mRNA of the BTBD2 gene. Approximately half of miR-3960 binding sites are located in the protein-coding regions of mRNAs. The ΔG/ΔGm value for all miR-3960 binding sites in the mRNA of target genes is more than 90% and is 98.2% for the mRNAs of the ADRB1, CEBPB, RANBP9 and SKOR2 genes. The nucleotide sequences of the miR-3960 binding sites in the CDSs of 47 genes are shown in Figure 3. The binding sites are clearly conserved as indicated by comparing them with the adjacent nucleotide sequences of the CDSs.
Figure 3.
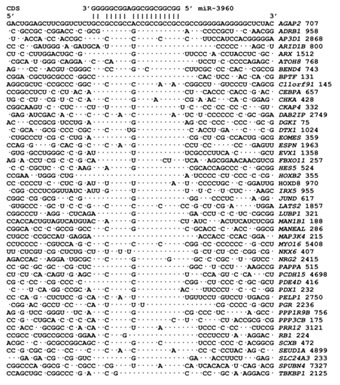
Aligned nucleotide sequences in the CDS of mRNAs containing miR-3960 binding sites. We selected 47 of 375 mRNA of target genes which have single, not regular sites. Тhe number after gene name is start point of the miRNA binding site.
The characteristics of the mRNA binding sites of miR-3960 in several genes are shown in Table 1 (see supplementary material). The presence of several miR-3960 binding sites within a single mRNA increases the probability that miR-3960 will bind to the mRNA and increases its ability to inhibit protein synthesis. The oligonucleotide sequences of the binding sites located in CDSs encode for oligopeptides containing mainly alanine or proline depending on the reading frame Table 1. These data indicate the importance of preserving the nucleotide sequences of the miR-3960 binding sites but not the amino acid sequences of the oligopeptides in the encoded protein. The mRNAs of some target genes have miR-3960 binding sites located with an interval of one, two or three nucleotides. 73 out of 2109 genes of transcription factors are targets for miR-3960. For example, the ZIC5 gene encodes a transcription factor, and its mRNA has 12 miR-3960 binding sites located through one or two nucleotides. Consequently, the probability of miR-3960 binding with the ZIC5 mRNA is high. The miR-3960 binding sites in the ZIC5 mRNA encode an oligopeptide predominantly composed of proline. The mRNA of several genes also have conserved nucleotide sequences at their miR-3960 binding sites in comparison with the amino acid composition of the corresponding oligopeptides. Thus, the oligonucleotides of the miR-3960 binding sites encode for oligopeptides with different amino acid sequences, as shown in Table 1 (see supplementary material).
Many target genes contain miR-3960 binding sites in the 5'UTRs of their mRNAs; this includes 565 binding sites. This localisation of miR-3960 binding sites allows miR-3960 to decrease the protein synthesis of those genes. The nucleotide sequences of miR-3960 binding sites in the 5'UTRs of mRNAs of target genes are highly conserved. mRNAs of many genes have miR-3960 binding sites in their 5'UTRs with displacement in one, two or three nucleotides. The number of the miR-3960 binding sites varied from five to 18. The ΔG/ΔGm value of the miR-3960 binding sites varied from 91.5% to 96.6% and corresponded to a high degree of interaction between the miRNAs with the mRNAs. miR-3960 binding sites are located predominantly in the 5'UTRs and CDSs of the studied mRNAs, and only 1.8% of the binding sites are in 3'UTRs. The miR-3960 binding sites of the mRNA of ten genes were spaced two or three nucleotides apart. miR-3960 binding sites are located in the 5'UTRs, CDSs and 3'UTRs of mRNAs. The 3'UTRs of the mRNAs of several genes have high homology to miR-3960 binding sites.
Discussion
The average GC-content value in 5'UTRs and CDSs is known to be higher than that in 3'UTRs. Retaining the nucleotide sequence of miRNA binding sites within the high GC-content in the 3'UTR is difficult because of the involvement of the 3'UTR in the formation of the 2 and 3 dimensional structures of mRNAs. The presence of a complementary binding site of the same length is necessary for the generation of stable secondary and tertiary mRNA structures. A high GC-content in the miRNA binding site promotes its presence on the surface of the 3D-structure of the mRNA because G and C nucleotides are more hydrophilic than A and U nucleotides. Thus, the probability of miRNAs binding to regions of 5'UTRs and CDSs with high GC-content is higher than the probability of miRNAs binding to regions of 3'UTRs [11].
Another way to increase the probability of miRNA binding to mRNA is increasing the number of sites in each mRNA. For example, miR-3960 has multiple binding sites in the 5'UTRs and CDSs of many genes. It is difficult to conserve the multiple independent miRNA binding sites in these regions; therefore, it is easier to increase the probability of miRNA binding by creating multiple binding sites located side by side. This was the case for the miR-3960 binding sites in the 5'UTRs and CDSs of many studied mRNAs Table 1 (see supplementary material).
We propose that the localisation of miRNA binding sites to 5'UTRs and CDSs will aid in the termination of protein synthesis. Such localisation saves the cell time and energy compared to translation inhibition in the case of localisation of miRNA binding sites to 3'UTRs. The results of the present study showed that in most cases miRNA binding sites located in CDSs are near the start codon.
Unfortunately, there is not sufficient experimental verification of the formation of miR-3960 binding sites. However, available data confirm the high predictive power of our developed program. For example, the overexpression of miR-3960 promotes BMP2-induced osteoblastogenesis. The miR- 2861~miR-3960 cluster was found to be associated with bone homeostasis induced by BMP2-Runx2 [12]. Chatterjee et al. showed that miR-3960 has a binding site in the mRNA of the Hoxa2 gene, which is a repressor of Runx2 gene expression [13]. Some target genes of miR-3960 encode transcription factors: SKOR2, PRDM12, FOXF2, FOXE1, BTBD2, AFF2, ZSWIM6, ZNF219, ZNF703, TRIM67 and RANBP9. Inhibition of the translation of the corresponding proteins causes many pathologies, including tumourigenesis. For example, miR-3960 can supress BCL11A gene expression resulting in the development of lymphoma [14]. Alterations in the gene expression of CDH3 and NDRG3 have been observed in colorectal and prostate cancer [15– 16]. The observed changes in expression of these genes were most likely due to the action of miR-3960 on their mRNAs.
Conclusion
In this study, we have shown that miR-3960 has 1100 binding sites on 375 target mRNAs with ΔG/ΔGm values of 90% or more. miR-3960 binding sites are located predominantly in the 5'UTRs and CDSs of the studied mRNAs. Another feature of this miRNA is the presence of multiple binding sites in the mRNA of its target genes. Dysregulation of miR-3960 expression can lead to cancer, because of presence great number of target genes with high affinity of the binding sites. Therefore the study is important for creation of diagnostic methods of cancer.
Supplementary material
Footnotes
Citation:Ivashchenko et al, Bioinformation 10(7): 423-427 (2014)
References
- 1.Tétreault N, De Guire V. Clin Biochem. 2013;46:842. doi: 10.1016/j.clinbiochem.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, et al. PLoS One. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong S, et al. Gene. 2013;531:8. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, et al. Zhonghua Bing Li Xue Za Zhi. 2013;42:20. doi: 10.3760/cma.j.issn.0529-5807.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Zhou P, et al. PLoS One. 2013;8:e68204. doi: 10.1371/journal.pone.0068204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berillo O, et al. Comput Biol Med. 2013;43:1374. doi: 10.1016/j.compbiomed.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Hausser J, et al. Genome Res. 2013;23:604. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S, et al. Nucleic Acids Res. 2006;34:D140. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kool ET. Annu Rev Biophys Biomol Struct. 2001;30:1. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Leontis N, et al. Nucleic Acids Res. 2002;30:3497. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issabekova A, et al. Bioinformation. 2012;8:513. doi: 10.6026/97320630008513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu R, et al. J Biol Chem. 2011;286:12328. doi: 10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, et al. Trends Dev Biol. 2012;6:45. [PMC free article] [PubMed] [Google Scholar]
- 14.Satterwhite E, et al. Blood. 2001;98:3413. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- 15.Broussard EK, et al. Cancer Prev Res (Phila) 2013;6:666. doi: 10.1158/1940-6207.CAPR-12-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. Int J Cancer. 2009;124:521. doi: 10.1002/ijc.23961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


