Abstract
The HBx protein in Hepatitis B Virus (HBV) is a potential target for anti-liver cancer molecules. Therefore, it is of interest to screen known natural compounds against the HBx protein using molecular docking. However, the structure of HBx is not yet known. Therefore, the predicted structure of HBx using threading in LOMET was used for docking against plant derived natural compounds (curcumin, oleanolic acid, resveratrol, bilobetin, luteoline, ellagic acid, betulinic acid and rutin) by Molegro Virtual Docker. The screening identified rutin with binding energy of -161.65 Kcal/mol. Thus, twenty derivatives of rutin were further designed and screened against HBx. These in silico experiments identified compounds rutin01 (-163.16 Kcal/mol) and rutin08 (- 165.76 Kcal/mol) for further consideration and downstream validation.
Keywords: Hepatitis b virus, HBx, Liver cancer, Natural compounds, Molecular docking
Background
The hepatitis B virus (HBV) is a small enveloped DNA virus that causes acute and chronic hepatitis [1]. Integration of hepatitis B virus into the genome of hepatocellular carcinoma patients will likely increase carcinogenic opportunities in HBVinfected individuals [2]. HBV infection is a worldwide health problem and may lead to lifelong infection, cirrhosis of liver, liver cancer, hepatocellular carcinoma, liver failure and death [3]. Recent epidemiological data have demonstrated that liver cancer incidence has been continuously rising for more than a decade, not only in Asia and Africa but also in America and Europe [4]. Chronic Hepatitis B Virus (HBV) infection is the dominant global cause of HCC, accounting for 55% of cases worldwide and 80% or more in eastern Pacific region and sub- Saharan Africa, the regions with highest rates for incidence of the tumor [5]. Although vaccination programs are implemented in various countries but it is still affecting 350 million to 400 million people worldwide [6]. The mechanisms whereby HBV causes malignant transformation remain uncertain. However, much of the evidence available supports a pathogenetic role for the product of HBV X gene, the HBx protein [7, 8]. HBx gene is the smallest of the four partially overlapping open reading frames of HBV. It comprises 452 nucleotide that encode a 154 amino acid regulatory protein with a molecular mass of 17 kDa. HBx was the name assigned to the gene and protein because the deduced amino acid sequence did not show homology to any known protein [9]. The protein is highly conserved among all eight major genotypes (A to H) of the HBV; it is present in the cytoplasm and, to a lesser extent, the nucleus of hepatocytes. In addition, high levels of HBx lead to an abnormal mitochondrial distribution [10].
The HBx protein is multi-functional and leading source of liver cancer. It functions by protein-protein interaction, and is a promiscuous transactivator of viral and cellular promoters and enhancers [7, 11]. HBx protein does not bind directly to DNA, but causes transcriptional activation by its interaction with nuclear transcription factors and modulation of cytoplasmic signal transduction pathways, including the Ras, Raf, c-jun, MAPK, NFkB, AP-1, Jak-Stat, FAK, and protein kinase C pathways, as well as Src-dependent and phosphatidylinositol-3 kinase signalling cascades [12– 15]. Transcriptional activation is thought to be essential for replication of the virus. Activation of these signalling pathways may also contribute to HBx proteinmediated hepatocellular carcinogenesis through transactivation of cellular signalling cascades and oncogenes that stimulate proliferation of hepatocytes [7].
Some studies demonstrate the correlation between mutation/inactivation of p53 and HBx in hepatocarcinogenesis. In that process, HBx will suppress p53 function which leads to unlimited liver cell division and results in the formation of tumour in liver [15]. p53 is a protein which play essential role for protecting, maintaining and repairing of cells. HBx may cause inactivation of p53 function, therefore if HBxAg-p53 complex is formed, p53 will lose its function, and it is the beginning of cancer development [15].
Historically the majority of new drugs have been generated from natural sources and from compounds derived from natural products [16]. Natural products are becoming an important research area for novel and bioactive molecules for drug discovery [17]. Plant-derived substances have currently become of great interest due to their versatile applications. Medicinal plants are the richest bio-resource of drugs of traditional medicines, modern medicine, nutracetical, pharmaceutical intermediate and chemical entities for synthetic drugs [18]. Several active compounds have been discovered from plants and used directly as patented drugs like Taxol, Artemisinin and Maprouneacin. Due to multitargeting effect, inexpensive and safety of plant-based compounds compared to synthetic agents, there is a necessity for more and more searching and discovery of new novel drugs from plants [19].
Plants have a key source of highly effective conventional drugs for the treatment of many form of cancer; in many instances, the actual compounds isolated from the plants may not serve as the drug, but leads to the development of potential agents. In order to understand the effectiveness of natural compounds as anticancer molecules, there is a need of computational drug designing tools that can identify and analyze active sites and suggest potential drug molecules that can bind to these sites specifically and play vital role in investigation of novel drug molecules. In our study, eight plants derived natural compounds viz., Curcumin, Oleanolic acid, Resveratrol, Bilobetin, Luteoline, Ellagic acid, Betulinic acid and Rutin (Figure 1a) were selected, these natural molecules possess anticancer activity [20– 23]. It may be useful for treatment and prevention of liver cancer and HBV related diseases.
Figure 1.
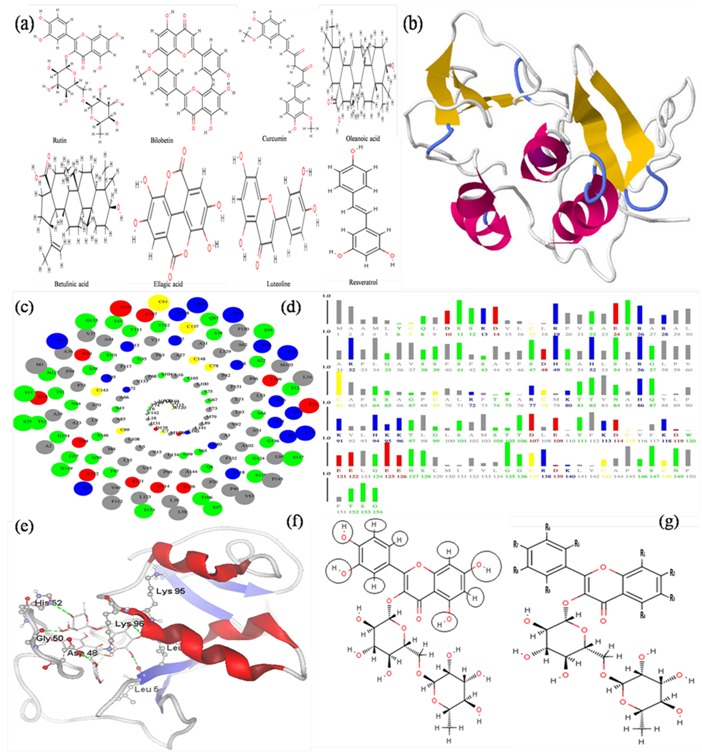
(a) Structure of natural compounds, (b) Structure of HBx protein, (c) Spiral Plot: which shows amino acid residues of HBx, in the order of their solvent accessibility; most accessible residues come on the outermost ring of this spiral. Blue, red, green, gray colors are used for positively charged, negatively charged, polar and non-polar residues respectively. Yellow color is used for Cystein residues. Radius of the solid circles representing these residues corresponds to the relative solvent accessibility, (d) Bar Plots: which shows solvent accessibility of amino acid residues; Length of the bar represents the ASA in units relative to extended state ASA of that residue, (e) Docked structure of HBx with Rutin; shows H bond interactions with LEU5, ASP48, GLY50, HIS52, LEU93, LYS95 & LYS95, (f) Black circle shows modification point in the structure of Rutin (g) and R shows functional groups.
Methodology
Sequence retrieval and analysis:
The FASTA sequence of the HBx protein of HBV (Accession no. ABR68892.1) was retrieved from NCBI ( http:// www.ncbi.nlm.nih. gov /) database; MATLAB Bioinformatics ToolBox (http://www.mathworks.in/) was used for analyzing HBx protein.
Protein structure prediction and validation:
The three dimensional structure of HBx protein from sequence was predicted using threading algorithm by LOMET server (http : //zhanglab.ccmb.med.umich.edu /LOMETS /). For constructing the structure BLAST was performed against PDB database to identify the sequence identitiy, the final structure was done at LOMET server by using amino acid sequence of HBx protein [24]. Structural refinements through energy minimization were performed using SPDB viewer (http://spdbv.vital-it.ch/). The Stability of the predicted model were validated by the Structural Analysis and Verification Server (http:// nihserver .mbi.ucla.edu/SAVES/), which has inbuilt tools such as PROCHECK, WHAT_CHEK, ERRAT, VERIFY_3D, and PROVE; PyMOL (http://www.pymol.org/) was used for the visualization of 3D structure of HBx protein (Figure 1b).
Surface analysis:
Accessible surface area (ASA) analysis of the predicted HBx protein was carried out by ASAView (http://www.abren.net /asaview/), this server provides graphical representation of solvent accessibility of amino acid residues in proteins [25].
Retrieval and preparation of ligand molecules:
The structure of natural compounds derived from plants viz., Curcumin (CID: 969516), Oleanolic acid (CID: 10494), Resveratrol (CID: 445154), Bilobetin (CID: 5315459), Luteoline (CID: 5280445), Ellagic acid (CID: 5281855), Betulinic acid (CID: 64971) and Rutin (CID: 5280805) were retrieved from PubChem database at NCBI (http://pubchem.ncbi.nlm.nih.gov). The three dimensional coordinates of ligand molecules were generated by MarvinSketch (http://www.chemaxon.com /products/ marvin/marvinsketch/) and saved in *.pdb files for docking studies.
Molecular docking approach:
Molegro Virtual Docker (MVD) was used for docking studies. MVD requires a three dimensional structure of both protein and ligand. MVD performs flexible ligand docking, so the optimal geometry of the ligand is determined during the docking [26]. The candidates with the best conformation and energetic results were selected. MVD was used to calculate the interaction energies between ligands and macromolecular systems from the three dimensional structures of the protein and ligand molecule.
Results & Discussion
Binding sites prediction and ASA analysis of HBx:
MVD cavity detection algorithm was used dynamically for predicting the cavities by search algorithm guided differential evolution to focus the search during docking simulation. The volume of cavities present in HBx protein was calculated by MVD; it predicted five cavities by default parameters. Mostly, cavity with the largest size and volumes is associated with the binding site. Therefore cavity with largest volume (Cavity-1 257.024 Å3) has been selected as binding site for HBx protein during molecular docking studies. Accessible surface area (ASA) or solvent accessibility of amino acids in an HBx protein was predicted by ASAView [25]. It display the ASA of amino acid residues in Spiral and bar plots, Spiral plot representing amino acid residues in order of their solvent accessibility and bar plots representing the length of amino acid in terms of solvent accessibility (Figure 1c & 1d).
Molecular docking studies of natural compounds with HBx:
Molecular docking were carried out with each natural compounds viz., Resveratrol, Luteoline, Ellagic acid, Betulinic acid, Oleanoic acid, Curcumin, Bilobatin and Rutin at the specific sites of interaction predicted from MVD. For each compound, out of the many docking poses, only those which possess the highest MolDock score and relatively good hydrogen bond interaction were chosen. Resveratrol was docked with HBx showing binding energy -85.25 Kcal/mol with six hydrogen bonds, Luteoline showing binding energy - 89.42 Kcal/mol with four hydrogen bonds, Ellagic acid showing binding energy -99.88 Kcal/mol with six hydrogen bonds, Betulinic acid showing binding energy -127.47 Kcal/mol with one hydrogen bond, Oleanoic acid showing binding energy -124.15 Kcal/mol with two hydrogen bonds, Curcumin showing binding energy -138.55 Kcal/mol with two hydrogen bonds, Bilobatin showing binding energy -157.37 Kcal/mol with five hydrogen bonds and Rutin showing binding energy - 161.65 Kcal/mol with seven hydrogen bonds Table 1 (see supplementary material).
Prediction and identification of lead molecule:
MVD and its visualizer were used for interaction site analysis. The interaction analysis for binding of natural compounds with HBx protein has been done to find out the residues that are involved in binding. The Rutin shows very high affinity to bind with HBx proteins with energy value -161.65 Kcal/mol (Table 1). It interacted with HBx amino acid residues LYS95, LYS96, LEU93, HIS52, ASP48, GLY50, and LEU5 with seven hydrogen bonds (Figure 1e). The hydrogen bonding is very significant in the interaction of biomolecules. A comparative study with the docking energy values (if the docking energy value are more, the protein-ligand interaction between compounds is low, and vice-versa) reveals that the natural inhibitor Rutin has greater affinity towards the HBx protein of HBV. Hence this information would prove to be important for designing of novel drug molecule against liver cancer.
Designing of Rutin derivatives:
Molecular docking studies of natural compounds with HBx protein of HBV confirmed that the Rutin is the best natural compounds for the treatment of liver cancer. On the basis of this investigation we designed the twenty derivatives of Rutin for improvement of binding affinity towards HBx (Figure 2), Table 2 (see supplementary material). Taking the references from anticancer, antiviral natural and synthetic drug available in market like Indivanir, Tazarotene, Paclitaxol and other commercial drugs, the functional groups -CH3, -OH and -NH2 are commonly found [27, 28]. Protein-ligand interaction studies of Rutin with HBx shows the pharmacophoric part of Rutin that is participating in interaction, other functional group that were not participating in protein ligands interaction were replaced by -CH3, -OH and –NH2 to design the derivative of Rutin (Figure 1f & 1g); the physicochemical properties were calculated by Marvin Sketch.
Figure 2.
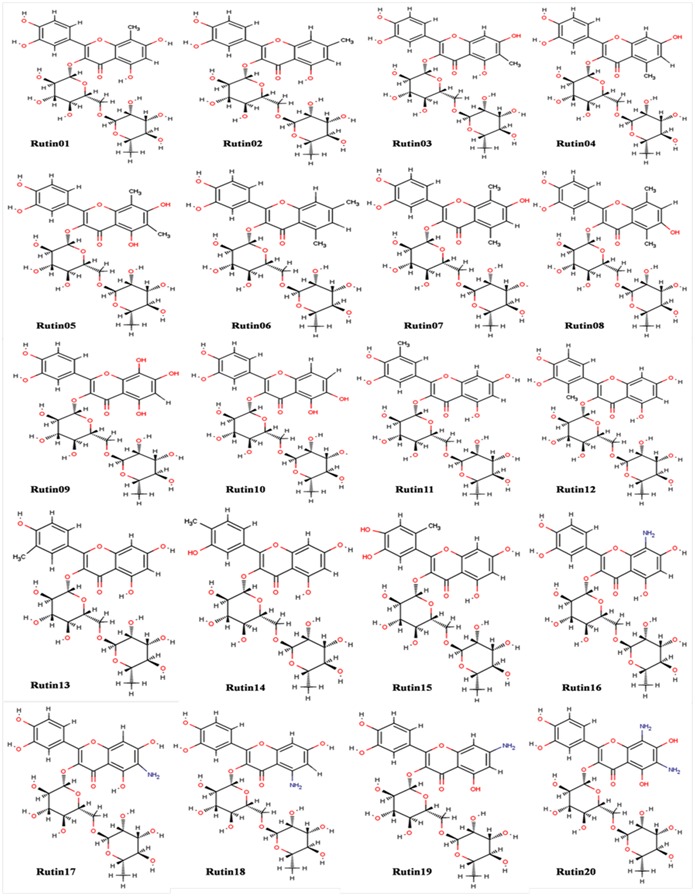
Designed derivative׳s of Rutin (2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3{[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6- ({[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}methyl)oxan-2-yl]oxy}-4H-chromen-4-one).
Molecular docking studies of Rutin derivatives with HBx:
The designed derivatives of Rutin were docked against HBx. Out of the 20 derivatives, Rutin01 and Rutin08 was found to possess best binding affinity towards HBx (Table 2). Rutin01 was found to be binding with HBx by interacting with amino acids residues LEU5, PRO42, ALA70, ARG72, LEU93, LYS96, and SER104 by forming hydrogen bonds. It formed seven hydrogen bonds with high interaction energy of -163.16 Kcal/mol. Similarly Rutin08 was found to bind with HBx by forming five Hydrogen bonds by interacting with amino acids residues LEU5, ARG72, LEU93, LYS96, and LEU98, exhibiting interaction energy of -165.76 Kcal/mol (Figure 3a & 3b).
Figure 3.
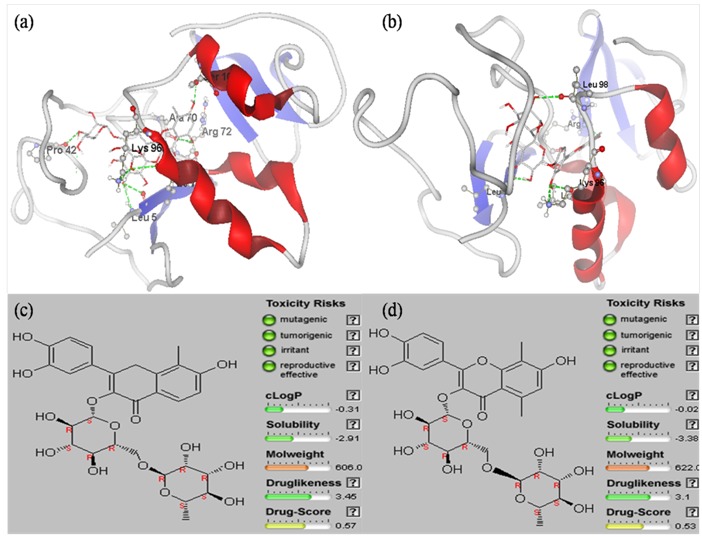
(a) Docked Structure of HBx with Rutin01 shows H-bond interactions with LEU5, PRO42, ALA70, ARG72, LEU93, LYS96 & SER104; (b) Docked Structure of HBx with Rutin08 shows H-bond interactions with LEU5, ARG72, LEU93, LYS96 and LEU98; (c) Drug like properties of Rutin01 and (d) Rutin08: represents drug confirmed behavior.
Drug like properties of Rutin derivatives:
Drug like properties of lead candidates play key roles in drug discovery. Results of molecular docking studies of Rutin derivatives clearly shown that Rutin01 and Rutin08 have greatest affinity towards HBx. The drug like properties of Rutin01 and Rutin08 were predicted by OSIRIS Property Explorer tool (http://www.organic-chemistry.org/prog/ peo/); we have found that Rutin01 and Rutin08 were nonmutagenic, non-toxic and showed drug like properties. In this tool property with high risks of undesired effects like mutagenicity or a poor intestinal absorption are shown in red, whereas a green color indicates drug-confirmed behavior (Figure 3c & 3d).
Investigating the novel molecule for treatment of liver cancer:
From early until now, natural products, especially plants, have been widely used for discovery of new therapeutic agents [23]. The results of present studies clearly reveal that natural compound Rutin is a lead molecule for the prevention of liver cancer. Rutin is found in many medicinal plants; its name comes from the name of Ruta graveolens, a plant that also contain rutin. Rutin is well known as antioxidant, and as natural compound with wide range of medicinal properties [29]. We have found that designed derivative Rutin01 and Rutin08 showed highest affinity towards HBx with drug confirmed behavior, which may be useful for treatment of liver cancer.
Conclusion
The present in silico study provides insights in to the inhibition of HBx protein by natural inhibitors. Rutin was predicted to be the most potent inhibitor amongst all the known selected natural compounds. Twenty derivatives of rutin were further designed and screened to HBx. Of these twenty derivatives, Rutin01 and Rutin08 were observed as the most effective inhibitor to be considered as potential drug for treatment of liver cancer. These results can further be validated through in vitro and in vivo studies.
Supplementary material
Acknowledgments
The authors would like to thank Bioinformatics Lab, Department of Biotechnology, G. B. Pant Engineering College, Pauri Garhwal, Uttarakhand, India for providing all necessary facilities. One of the authors RKP gratefully acknowledges Technical Education Quality Improvement Programme (TEQIP), G. B. Pant Engineering College, Pauri Garhwal, India for the Research fellowship.
Footnotes
Citation:Pathak et al, Bioinformation 10(7): 428-435 (2014)
References
- 1.Riviere L, et al. Recent Results Cancer Res. 2014;193:59. doi: 10.1007/978-3-642-38965-8_4. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Z, et al. Genome Res. 2012;22:593. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia W, et al. Bioorg Med Chem. 2009;17:4569. doi: 10.1016/j.bmc.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Neuveut C, et al. J Hepatol. 2010;52:594. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 5.Kew MC. Pathol Biol (Paris) 2010;58:273. doi: 10.1016/j.patbio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Colombo GL, et al. Clinicoecon Outcomes Res. 2011;3:37. doi: 10.2147/CEOR.S16655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kew MC. J Gastroenterol Hepatol. 2011;1:144. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 8.Kordestani R, et al. Avicenna J Med Biotechnol. 2014;6:3. [PMC free article] [PubMed] [Google Scholar]
- 9.Miller RH, et al. Proc Natl Acad Sci U S A. 1986;83:2531. doi: 10.1073/pnas.83.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huh KW, et al. Mitochondrion. 2002;1:349. doi: 10.1016/s1567-7249(01)00040-x. [DOI] [PubMed] [Google Scholar]
- 11.Feitelson MA, et al. Expert Opin Ther Targets. 2014;18:293. doi: 10.1517/14728222.2014.867947. [DOI] [PubMed] [Google Scholar]
- 12.Benn J, et al. J Virol. 1996;70:4978. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feitelson MA, et al. Am J Pathol. 1997;150:1141. [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami S, et al. J Gastroenterol. 2001;36:651. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- 15.Dewantoro O, et al. Acta Med Indones. 2006;38:154. [PubMed] [Google Scholar]
- 16.Li JW, et al. Science. 2009;325:161. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 17.Pratheeshkumar P, et al. Anticancer Agents Med Chem. 2012;12:1159. doi: 10.2174/187152012803833035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nirmala MJ, et al. Res. Plant Biol. 2011;1:1. http://resplantbiol.com/article /view/243/229. [Google Scholar]
- 19.Abdel-Hameed El-SS, et al. E J of Medicinal Plants. 2012;2:93. [Google Scholar]
- 20. http://www.sciencedomain.org/abstract.php?id=13&aid=378.
- 21.Amin AR, et al. J Clin Oncol. 2009;27:2712. doi: 10.1200/JCO.2008.20.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiyanto E, et al. Asian Pac J Cancer Prev. 2012;13:427. doi: 10.7314/apjcp.2012.13.2.427. [DOI] [PubMed] [Google Scholar]
- 23.Khan RA, et al. BMC Complement Altern Med. 2012;12:178. doi: 10.1186/1472-6882-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Son LH, et al. E J Medicinal Plants. 2014;4:292. http://www.sciencedomain.org/reviewhistory. php?iid=375&id=13&aid=2860. [Google Scholar]
- 25.Wu S, et al. Nucleic Acids Res. 2007;35:3375. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad S, et al. BMC Bioinformatics. 2004;5:51. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomsen R, et al. J Med Chem. 2006;49:3315. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 28.Knox C, et al. Nucleic Acids Res. 2011;39:D1035. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law V, et al. Nucleic Acids Res. 2014;42:D1091. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi JS, et al. J Agric Food Chem. 2009;57:2079. doi: 10.1021/jf803390m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


