Abstract
Dengue infection has turned into a serious health concern globally due to its high morbidity rate and a high possibility of increase in its mortality rate on the account of unavailability of any proper treatment for severe dengue infection. The situation demands an urgent development of efficient and practicable treatment to deal with Dengue virus (DENV). Flavonoids, a class of phytochemicals present in medicinal plants, possess anti-viral activity and can be strong drug candidates against viruses. NS1 glycoprotein of Dengue virus is involved in its RNA replication and can be a strong target for screening of drugs against this virus. Current study focuses on the identification of flavonoids which can block Asn-130 glycosylation site of Dengue virus NS1 to inhibit viral replication as glycosylation of NS1 is required for its biological functioning. Molecular docking approach was used in this study and the results revealed that flavonoids have strong potential interactions with active site of NS1. Six flavonoids (Deoxycalyxin A; 3,5,7,3',4'-pentahydroxyflavonol-3-O-beta-D-galactopyranoside; (3R)-3',8-Dihydroxyvestitol; Sanggenon O; Epigallocatechin gallate; Chamaejasmin) blocked the Asn-130 glycosylation site of NS1 and could be able to inhibit the viral replication. It can be concluded from this study that these flavonoids could serve as antiviral drugs for dengue infections. Further in-vitro analyses are required to confirm their efficacy and to evaluate their drug potency.
Keywords: Dengue, NS1, Glycosylation site, Medicinal Plants, Flavoniods, Molecular Docking
Background
Flavivirus is a viral genus of the family flaviridae comprising of viruses with enveloped RNA genome and human pathogenic nature. Flaviviruses, most likely found in the tropical and temperate parts of the world are a root cause of dengue fever, West Nile fever, tick-borne encephalitis, and yellow fever in human [1]. One of the major infectious flavivirus is the dengue virus (DENV-1, DENV-2, DENV-3, and DENV-4) having infected Aedes aegypti mosquito bite as its route of transmission. According to statics, Dengue virus claims approximately 25,000 lives and infects over 50 million human beings annually [2, 3]. Dengue viruses are etiologic agents of dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS) [2]. DENV infection has become a serious problem and dreadful threat for life in major tropical and subtropical areas of the world including Asia, Central and South America and Africa [4, 5]. It is unfortunate that no vaccine has passed the clinical trial till date, inspite of the fact that massive research is conducted for the development of a DENV vaccine [6]. The 11kb genome of DENV encodes a single polyprotein which is then proteolyticaly cleaved into three structural and seven nonstructural proteins. These ten viral proteins are capsid (C), premembrane (prM/M), envelope protein (E), NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5. The structural proteins are involved in structural formation of virus and non-structural proteins have their role in viral replication [7]. The flavivirus NS1 protein bears certain interesting and peculiar properties as compared to nonstructural proteins encoded by other RNA viruses. NS1 of Dengue virus, a highly conserved glycoprotein of 46 kDa size, is found both intracellularly and extracellularly and sometimes also attached to cell surfaces. As NS1 is synthesized, it moves towards endoplasmic reticulum where it modifies into a homodimer of partial hydrophobic nature. NS1 remains confined to intracellular membranes that are presumed sites of viral RNA synthesis [8]. The analyses of yellow fever virus NS1 suggest a high possibility of NS1 involvement in viral RNA replication process [9, 10]. For the replication of NS1 its glycosylation sites are significant. Three glycosylation sites are positioned at Asn130, Asn175 and Asn207 residues in NS1 [11, 12]. Asn-207 is involved in secretion and cell surface expression of protein while Asn-130 has imperative role in NS1 structural stability and interaction with complement proteins [13]. The results of previous studies revealed the role of Asn-130 in viral growth, NS1 secretion, cytopathic effect in cells [8, 10, 14, 15, 16]. Different in-vivo experiments results proved that loss of ASN-130 in NS1 results in attenuated dengue virus infection in mice [17, 18, 19].
Asn-130 is thus considered an essential target to screen and calculate effects of various drug candidates on NS1. Phytochemicals are found abundantly in Medicinal Plants [20]. These phytochemicals act as a strong defense mechanism for plants and also safeguard human bodies and animals against various contagious viruses and epidemics [21]. A broad range of phytochemicals can be traced in medicinal plants including organosulfur compounds, limonoids, furyl compounds, alkaloids, polyines, coumarins, thiophenes, peptides, flavonoids, terpenoids, polyphenolics and saponins. These phytochemicals serves their remedial function by scavenging and hindering viral entry and DNA\RNA replication against a wide range of viruses [22]. According to previous studies, Flavonoids can play an active role in the cure of Dengue virus infections due to their efficient anti-viral activity [23]. Treatment of Dengue Virus with medicinal plant costs less as compared to good old traditional methods [24]. It may also be preferred because of the multiple target activities, little probable to cause resistance and nominal side-effects [25].
Recent advancements in computational biology techniques have broadened the horizons of research in the field of drug development. These days, in drug designing and screening of newly discovered antiviral compounds against threatening diseases, Molecular docking for prediction of predominant binding mode of a ligand with a protein of known threedimensional structure is considered a key technique [26]. Hence, the aim of the present study is to screen 2200 flavonoids of 405 antiviral medicinal plants against Dengue virus NS1 using in-silico techniques. The focal idea of this study is to target the N-glycosylation sites (Asn-130) of Dengue virus NS1 to screen novel flavonoids that could help to restraint the DENV infection. The result of this study will provide worthwhile information regarding drug development and would also prove beneficial in computer aided screening of the drugs against DENV infection.
Methodology
This study involves the docking of 2200 flavonoids of 405 antiviral medicinal plants against Dengue virus NS1 protein. Molecular Operating Environment (MOE) software package was used to carry out docking [27].
Ligand׳s searching and database preparation:
An extensive literature survey was performed to hunt for flavonoids from antiviral medicinal plants, which have shown their potential as drugs against viral diseases specially against Dengue Virus. Chemical structures of Flavonoids were downloaded from MAPS Database [28], PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Pubmed Central (http://www.ncbi.nlm.nih.gov/pmc/), Pubchem (http://www.ncbi.nlm.nih.gov/pccompound), Zinc Database [29] and ChEMBL ( http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245175/). Structures were stored in .mol format. Structural optimization was done by adding hydrogen atoms using MOE software. Energy minimization of selected molecules was performed with parameters (gradient: 0.05, Force Field: MMFF94X, Chiral Constraint and Current Geometry). All these refined flavonoid structures were then saved in .mdb database to be further analyzed through molecular docking.
Refinement of receptor protein:
Three-dimensional (3D) structure of the Dengue virus NS1 was retrieved from the Protein Data Bank (PDB) using PDB ID: 4O6B [1] and was optimized by removing water molecules, 3D protonation and Energy minimization using Molecular Operating Environment (MOE). Moreover, energy minimization was done using parameters (Force Field: AMBER99+Solvation, gradient: 0.05, and Chiral Constraint: Current Geometry). This refined structure was then used as receptor for docking analyses.
Molecular docking:
Active site Finder tool of MOE was used to identify and calculate active sites in the receptor molecule from the 3D atomic coordinates of the receptor. By default, all calculated sites were appeared as selected. Active site containing ASN 130 was selected for future docking and that particular site was isolated from rest of the structure. Secondary structures were removed from the receptor and docking of ligand database against Asn-130 of Dengue virus NS1 was then executed. The parameters were set (Re-scoring function: London dG , placement: triangle matcher, Retain: 10, Refinement: Force field, and Re-scoring 2: London dG). Docking program of MOE provides correct conformation of the ligand so as to obtain minimum energy structure. After docking, S score was considered the criteria to select best conformation for flavonoids and these were then further studied to analyze the hydrogen bonding/π-π interactions through LigX tool of MOE.
Drug Scan:
Drug scan of flavonoids was performed using the ligand properties checking tool of MOE to ensure that the compounds possess suitable molecular properties to be a drug candidate.
Results
The 3D X-Ray diffraction structure of Dengue virus NS1 with resolution of 3.00 Å was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb) using PDB ID: 4O6B. This section presents the results obtained by docking of 2200 flavonoids from all medicinal plants against Asn-130 glycosylation site of NS1.
Molecular docking:
Ten different conformations for each flavonoid had been provided by MOE. All these conformations were sorted according to S score with minimum S score being the criteria for the best conformation. Out of all the flavonoids used for docking top 100 flavonoids with minimum S score were selected for further Hydrogen bonding/π-π interactions.
Interaction analysis:
Out of 100 selected flavonoids used for interaction analysis only 6 flavoniods ( Deoxycalyxin A, 3,5,7,3',4'- pentahydroxyflavonol-3-O-beta-D-galactopyranoside, (3R)- 3',8-Dihydroxyvestitol, Sanggenon O, Epigallocatechin gallate and Chamaejasmine ) have shown significant interactions with Asn130 and bound deeply inside the binding pocket. Chemical structures of selected flavonoids are shown in (Figure 1). Deoxycalyxin A ranked at top had potential interactions with Asn130, His129, Lys85 and Asn76. Moreover, 3,5,7,3',4'- pentahydroxyflavonol-3-O-beta-D-galactopyranoside which was ranked next to Deoxycalyxin A had interacted strongly with Asn130, His129 and Lys85 and (3R)-3',8- Dihydroxyvestitol had also shown strong interactions with Asn130, Ser80 of NS1. Sanggenon O, Epigallocatechin gallate and Chamaejasmine had also shown to have potential interactions with N-glycosylation site Asn-130 of NS1. Selected flavonoids S score and Interacting residues of the NS1 with flavonoids are shown in Table 1 (see supplementary material). Interactions between receptor and ligands in 3D and 2D form are shown in (Figure 2). Binding mode of ligands with receptor protein is shown in (Figure 3). Flavonoids have shown strong interactions with Asn-130 glycosylation site of DENV NS1 along with having minimum S Score. These results elucidated that flavonoids could be strong drug candidates against NS1.
Figure 1.
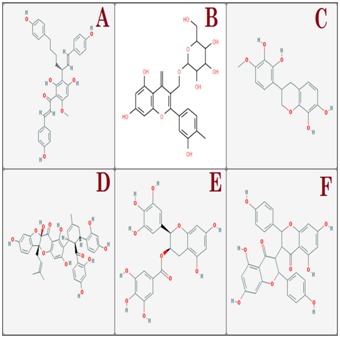
Chemical structure of A) Deoxycalyxin A; B) 3,5,7,3',4'-pentahydroxyflavonol-3-O-beta-Dgalactopyranoside; C) (3R)-3',8-Dihydroxyvestitol; D) Sanggenon O; E) Epigallocatechin gallate; F) Chamaejasmin.
Figure 2.
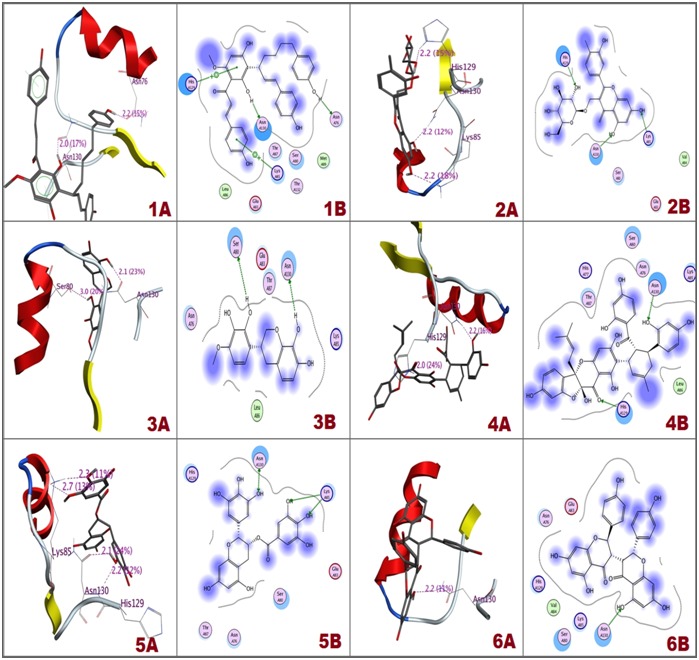
3D and 2D interaction images respectively 1A, 1B) Deoxycalyxin A interaction with DENV NS1 ; 2A, 2B) 3,5,7,3',4'- pentahydroxyflavonol-3-O-beta-D-galactopyranoside interaction with DENV NS1; 3A, 3B) (3R)-3',8-Dihydroxyvestitol interaction with DENV NS1 ; 4A, 4B) Sanggenon O interaction with DENV NS1 ; 5A, 5B) Epigallocatechin gallate interaction with DENV NS1 ; 6A, 6B) Chamaejasmin interaction with DENV NS1.
Figure 3.

A) Docked Deoxycalyxin A complex with DENV NS1 pocket; B) Docked 3,5,7,3',4'-pentahydroxyflavonol-3-O-beta-Dgalactopyranoside complex with DENV NS1 pocket; C) Docked (3R)-3',8-Dihydroxyvestitol complex with DENV NS1 pocket; D) Docked Sanggenon O complex with DENV NS1 pocket ; E) Docked Epigallocatechin gallate complex with DENV NS1 pocket; F) Docked Chamaejasmin complex with DENV NS1 pocket.
Drug Scan:
Final selected flavonoid compounds were analyzed using the Ligand properties checking tool of MOE which assessed the molecular properties and practicability of these compounds of being drug candidates on the basis of “Lipinski׳s Rule of Five” [30]. The rule describes molecular properties important for a drug׳s pharmacokinetics in the human body, including their absorption, distribution, metabolism and excretion. These compounds were examined for their drug-suitableness and the results are shown in Table 2 (see supplementary material). Our results showed that all the flavonoid compounds used in this study fulfill the criteria of being drug candidates.
Discussion
Dengue infection has become a major medical concern for the world now. Every year over 50 million people become a victim of dengue and about 25000 lose the battle of life against this fatal infection [2, 3].The causative agent of this life claiming infection is a mosquito-borne flavivirus, DENV. The DENV has a highly conserved nonstructural glycoprotein NS1. Previous studies reflect that dengue victims who developed DHF rather than DF have high concentrations of NS1 in their sera, which indicates that the severity of DENV-infected patients depends on the concentration of NS1 [31, 32]. NS1 contains three Nlinked glycosylation sites located at ASN-130, Asn-175 and Asn-207. NS1 is involved in replication and pathogenicity of DENV [7, 8]. Asn-130 site plays a vital role in biological activity of NS1. Different studies have shown that if we mutate Asn-130 or block this site, DENV replication attenuated [10, 16, 17, 18, 19]. Extensive researches have been performed to develop vaccines against this infection but these efforts have not proven successful till today. There is a need to design new approaches to combat this viral infection [6]. Use of natural pharmaceuticals like medicinal plants may prove an efficient treatment for this unresolved medical problem [24]. Flavonoids can play an important role in the cure of Dengue virus infections [28]. For past few years, binding analyses of compounds through in-silico techniques prior to their lab manufacturing and examination has become a common and useful practice among researchers. One of the popular computational techniques used in medical researches is molecular docking which is used to find the binding behaviors of small molecules against their target proteins. This technique proves helpful in providing a clear molecular vision of viral genes and identification of novel inhibitory compounds against fatal viral infections [26]. Therefore current study exploits the fact that blocking of Asn-130 from NS1 inhibits replication of DENV and anti-viral activity of flavonoids from medicinal plants. 2200 flavonoids from 405 medicinal plants have been docked against Asn-130 glycosylation site of NS1. Flavonoids have strong interactions with Asn-130 of NS1. Deoxycalyxin A not only interacts well with Asn130 but also with His129, Lys85 and Asn76. Some other flavonoids also show stable interactions at multiple residues of selected active site of NS1. Strong binding of NS1 and flavonoids suggests a strong possibility of flavonoids being used as drugs against dengue virus.
Conclusion
Dengue, for more than a half century now, has been a major health concern world widely with one third of the world׳s population at the risk of this infection. Dengue has proven to claim the lives of a million people yearly but no vaccine has been successfully developed for this dreadful disease up till now. It has now become inevitable to design new and novel strategies for developing drug candidates that can target DENV. Current study focuses on the assessment of flavonoids from medicinal plants as drug candidates against NS1. Molecular docking of flavonoids against NS1 has revealed strong interactions between flavonoids (Deoxycalyxin A, 3,5,7,3',4'-pentahydroxyflavonol-3-O-beta-D- alactopyranoside, (3R)-3',8-Dihydroxyvestitol, Sanggenon O, Epigallocatechin gallate and Chamaejasmine ) and the Asn-130 glycosylation site of NS1. The information acquired through this study on the binding mode of flavonoids and NS1 will highly facilitate the synthesis and testing of flavonoids as drugs against DENV. On a concluding note, this study has suggested that flavonoids will be strong future drug candidates against DENV.
Supplementary material
Footnotes
Citation:Qamar et al, Bioinformation 10(7): 460-465 (2014)
References
- 1.Akey DL, et al. Science. 2014;343:881. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. Clin Microbiol Rev. 1998;11:480. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halstead SB. Lancet. 2007;370:1644. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 4.Das S, et al. J Clin Microbiol. 2008;48:3276. doi: 10.1128/JCM.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatima Z, et al. BMC Microbiol. 2011;11:200. doi: 10.1186/1471-2180-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson SE, et al. Vaccine. 2002;4:31. [Google Scholar]
- 7.Chambers TJ, et al. Annu Rev Microbiol. 1990;44:649. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 8.Mason PW. Virology. 1989;169:354. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackenzie JM, et al. Virology. 1996;220:232. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 10.Muylaert IR, et al. J Virol. 1997;71:291. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avirutnan P, et al. J Infect Dis. 2006;193:1078. doi: 10.1086/500949. [DOI] [PubMed] [Google Scholar]
- 12.Rodenhuis-Zybert IA, et al. Cell Mol Life Sci. 2010;67:2773. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Somnuke P, et al. Virology. 2011;413:253. doi: 10.1016/j.virol.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pryor MJ, et al. Traffic. 2007;8:795. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 15.Crabtree MB, et al. Arch Virol. 2005;150:771. doi: 10.1007/s00705-004-0430-8. [DOI] [PubMed] [Google Scholar]
- 16.Pletnev AG, et al. J Virol. 1993;67:4956. doi: 10.1128/jvi.67.8.4956-4963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muylaert IR, et al. Virology. 1996;222:159. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- 18.Pryor MJ, et al. J Gen Virol. 1998;79:2631. doi: 10.1099/0022-1317-79-11-2631. [DOI] [PubMed] [Google Scholar]
- 19.Whiteman MC, et al. Vaccine. 2010;28:1075. doi: 10.1016/j.vaccine.2009.10.112. [DOI] [PubMed] [Google Scholar]
- 20.Calixto JB. Braz J Med Biol Res. 2000;33:179. doi: 10.1590/s0100-879x2000000200004. [DOI] [PubMed] [Google Scholar]
- 21.Kubmarawa D, et al. J Med Plants Res. 2008;2:352. [Google Scholar]
- 22.Idrees S, et al. Asian Pac J Trop Biomed. 2013;3:232. doi: 10.1016/S2221-1691(13)60057-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senthilvel P, et al. Bioinformation. 2013;9:889. doi: 10.6026/97320630009889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qamar TU, et al. Bioinformation. 2014;19:115. doi: 10.6026/97320630010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jassim SA, Naji MA. J Appl Microbiol. 2003;95:412. doi: 10.1046/j.1365-2672.2003.02026.x. [DOI] [PubMed] [Google Scholar]
- 26.Lengauer T, Rarey M. Curr Opin Struc Biol. 1996;6:402. doi: 10.1016/s0959-440x(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 27.1010 Sherbooke St West, Suite #910, Montreal, QC, Canada: Chemical Computing Group Inc; 2012. MOE: Molecular Operating Environment (MOE) H3A 2R7. [Google Scholar]
- 28.Ashfaq UA, et al. Bioinformation. 2013;9:993. doi: 10.6026/97320630009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irwin JJ, et al. J Chem Inf Model. 2012;52:1757. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipinski CA, et al. Adv Drug Deliv Rev. 2001;46:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 31.Wong SJ, et al. J Clin Microbiol. 2003;41:4217. doi: 10.1128/JCM.41.9.4217-4223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libraty DH, et al. J Infect Dis. 2002;186:1165. doi: 10.1086/343813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


