Abstract
Wheat, the major source of vegetable protein in human diet, provides staple food globally for a large proportion of the human population. With higher protein content than other major cereals, wheat has great socio- economic importance. Nonetheless for wheat, three important fungal pathogens i.e. rust, smut and bunt are major cause of significant yield losses throughout the world. Researchers are putting up a strong fight against devastating wheat pathogens, and have made progress in tracking and controlling disease outbreaks from East Africa to South Asia. The aim of the present work hence was to develop a fungal pathogens database dedicated to wheat, gathering information about different pathogen species and linking them to their biological classification, distribution and control. Towards this end, we developed an open access database Tripath: A biological, genetic and genomic database of economically important wheat fungal pathogens – rust: smut: bunt. Data collected from peer-reviewed publications and fungal pathogens were added to the customizable database through an extended relational design. The strength of this resource is in providing rapid retrieval of information from large volumes of text at a high degree of accuracy. Database TRIPATH is freely accessible.
Availability
http://www.gbpuat-cbsh.ac.in/departments/bi/database/tripath/
Keywords: Wheat, Rust, Smut, Bunt, Tripath
Background
Wheat is one of the most important cereal crop grown worldwide. Occupying 17% (one-sixth) of crop acreage of the world [1], wheat is a staple food for 35% of the world’s population and provides more calories and protein in the global diet than any other cereal. Apart from constituting the bulk of staple diet, it also contributes essential amino acids, minerals, vitamins, and beneficial phytochemicals and dietary fiber components. Wheat can be consumed variously as flour for breads, cookies, pasta, noodles etc. [2] and also for fermentation to make beer [3] and other alcoholic beverages. In the Indian sub-continent the major wheat producing states are placed in the northern regions of the country with the states of UP, Uttrakhand, Punjab and Haryana contributing to nearly 80% of the total wheat production. While wheat is of great socio-economic importance, it gets infected with many fungal diseases among rust, smut and bunt are the most common ones. These diseases are borned by fungal pathogens like rust disease is caused by Puccinia sp., smut disease by Urocystis sp. and Ustilago sp., and bunt disease from Tilletia sp respectively. These diseases cause harvest losses, affect the quality of the harvested grains, and cause degradation during storage. Nonetheless, yield losses can be linked to genetically determined resistance and tolerance of the wheat cultivars to specific diseases, diversity and level of the pathogen inoculums present, and the local climate conditions [4].
Rusts are the most important foliar diseases of wheat in Australia. Additionally, rust causes serious epidemics in other parts of the world such as North America, Mexico, South America and is a devastating seasonal disease in India. Of the three types of rust, stripe, stem and leaf, rust the latter is potentially the least damaging in susceptible varieties, though conditions are conducive for this disease in most seasons. Stem rust occurs worldwide wherever wheat is grown. Over a large area, losses from stem rust can be severe, ranging from 50-70% [5]. Stripe rust has been reported particularly in Central and West Asia, North Africa, Australia, Europe, China and United States. The smuts and bunts diseases are a group of related fungi that infect developing grain. Cool, humid weather accompanied by light showers or heavy dews is most favorable for these infections. A special mention is Karnal bunt or partial bunt which was first reported from a city named karnal and also from Punjab area in 1930. This integrated database aims to provide a compendium of browsable information for the three economically important fungal pathogen of wheat. Primarily, collation of genomic information of the wheat species with the pathogen diversity greatly facilitates researchers who wish to use this information to understand disease aetiology and crop vigour, pathogenesis and resistance mechanisms.
Methodology
Database Construction and Content:
Data were collected from various literature sources from the web. Tripath database is containing information about wheat fungal diseases viz. Rust, Smut and Bunt. The database can be accessed using fungus name for information on specific rust, bunt and smut. Database architecture is designed by Adobe dream weaver CS6 using hypertext mark-up language (html). Web interfaces were written in PHP, html and JavaScript. XAMPP software was used for whole compilation of data. Data was compiled by the Apache with MySQL relational database storage. Tripath database can be accessed at http://www.gbpuat-cbsh.ac.in/ departments/bi/database/ tripath/. All database information is compiled sequentially including systematic position, geographical distribution, morphology and symptoms, weather conditions, hosts, significance and control of the diseases. The database includes all information about the rust, smut and bunt diseases and also the genetic and genomic information related to the species.
Database Description:
Database is a collection of whole information of wheat fungal diseases viz. Rust, Smut and Bunt. In these diseases, rust contains 197 species, smut contains 105 species and bunt contains total 81 species. Database including the information of these diseases is called TRIPATH, i.e., information of three fungal pathogens of wheat. The database entry side contain ‘Home’, ‘Rust’, ‘Smut’, ‘Bunt’, ‘Genetic/Genomic information’, ‘About us’ and ‘Contact us’ sections. ‘Home’ section contains general information about wheat and its importance. ‘Rust’ section contains three important types of Rusts i.e. Leaf rust (caused by P. recondita), Stem rust (caused by P. graminis), Stripe rust (caused by P. striiformis), and its distribution, significance and control. In ‘Smut’ section, it is devastating disease of cereals contains mainly two important types i.e. Loose smut of wheat (caused by Ustilago tritici), Flag smut of wheat (caused by Urocystis agropyri), and its distribution, significance and control. In ‘Bunt’ section, it is also devastating disease of cereals contains mainly three common types i.e. Common bunt (caused by T. tritici & T. laevis), Dwarf bunt (caused by T. controversa), Karnal bunt (caused by T. indica), and its distribution, significance and control. Genetic marker, genetic map, and chromosome, EST, protein, genomic, mRNA and WGS information were also added in Genetic/Genomic information section. For retrieving entries of interest, Tripath provides a search tool using keywords of rust, smut & bunt name. The search fields can be individually or simultaneously utilized to filter entries. Figure 1 is showing a screen shot of the different pages of TRIPATH database.
Figure 1.
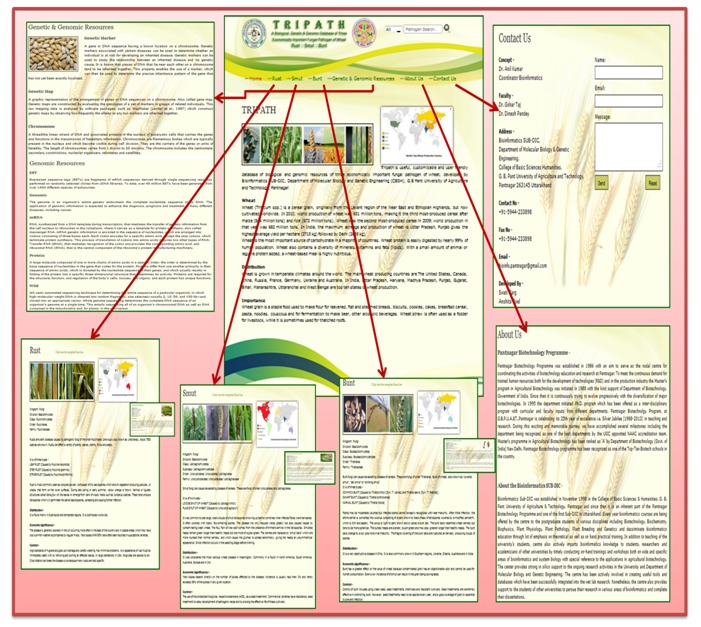
The Tripath database web resource
Utility
TRIPATH database provides authentication of fungal species of three important pathogens of wheat i.e. rust, smut and bunt. With the help of this database users can identify and characterize of fungal diversity amongst three pathogens. TRIPATH also develops our understanding for the etiology of disease and central measures. It generates molecular and genomic data for understanding the mechanism of pathogenesis and defense and molecular detection.
Future development
Continuous updates shall be released to include other pathogens of cereals plant. The present access method shall be upgraded for faster access to accommodate the growing number of data to browse records on the studied fungal pathogens.
Acknowledgments
Authors are grateful to Sub-DIC, Bioinformatics at G.B. Pant University of Agriculture and Technology, Pantnagar, India for providing computational facility.
Footnotes
Citation:Garg et al, Bioinformation 10(7): 466-468 (2014)
References
- 1.Gupta PK, et al. Int J Plant Genomics. 2008;2008:896451. doi: 10.1155/2008/896451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang HM, et al. APCBEE Procedia. 2012;12:16. [Google Scholar]
- 3.Palmer, John J. Defenestrative Pub Co. 2001;233 [Google Scholar]
- 4.Maria RS, et al. International J Agronomy. 2012;76:9048. [Google Scholar]
- 5.Mumtaz S, et al. African J Biotechnology. 2009;8:1188. [Google Scholar]


