Activity-dependent neuroprotective protein (ADNP) is a highly conserved transcription factor comprised of nine-zinc finger domains and a homeobox domain.1 2 It is highly expressed prenatally during critical stages of embryonic brain development.3 Knockout (KO) mouse embryos demonstrate severe neurodevelopmental morphological profiles.4 Although the ADNP KO is lethal, heterozygous embryos demonstrate typical embryogenesis yet display a neurodevelopmental delay phenotype including decreased neuronal survival.3 5
Exome sequencing in the Simons Simplex Collection autism dataset identified ADNP mutations as a putative autism gene candidate.6 7 Helsmoortel et al8 recently reported 10 individuals with autism spectrum disorder (ASD) and mutations in exon 5 of the ADNP gene, nine of which were confirmed de novo. These patients also exhibited intellectual disability (ID) and dysmorphic features such as a prominent forehead. Mutations in the ADNP gene are estimated to be present in at least 0.17% of ASD cases. The current report further expands the ADNP phenotype to include abnormalities in the developing visual system (such as eye movement abnormalities and cortical visual impairment). We advise appropriate screening of eye movement and visual symptoms by clinicians in patients who have mutations in ADNP.
The 6-year-old patient was the first child born to healthy non-consanguineous parents. Pregnancy was notable for placenta previa and early dilation and effacement of the cervix 3 weeks prior to delivery. The patient was born at 40 weeks via C-section secondary to failure to progress and maternal (i.e. maternal hypertension) hypertension weighing 6 pounds 14 ounces. She had a short stay in the neonatal intensive care unit (NICU) for breathe holding and feeding problems. She was also hospitalised at 6 weeks for an acute life-threatening event of multiple cyanotic episodes thought to be due to breath holding. Our patient has been diagnosed with hypotonia and mixed developmental delays, moderate to severe expressive and receptive language delays, fine, gross and oral motor delays, attention deficit hyperactivity disorder (ADHD), and episodic mood disorder not otherwise specified (NOS). She has also been diagnosed with hypermetropia and sleep disturbance NOS. Gastrointestinal problems were gastro-oesophageal reflux disease, feeding problems in infancy and constipation. Notable dysmorphic features were a broad forehead and slightly tented lips. EEG (awake, drowsy and asleep states), brain MRI, magnetic resonance spectroscopy (MRS) and echocardiogram results were normal.
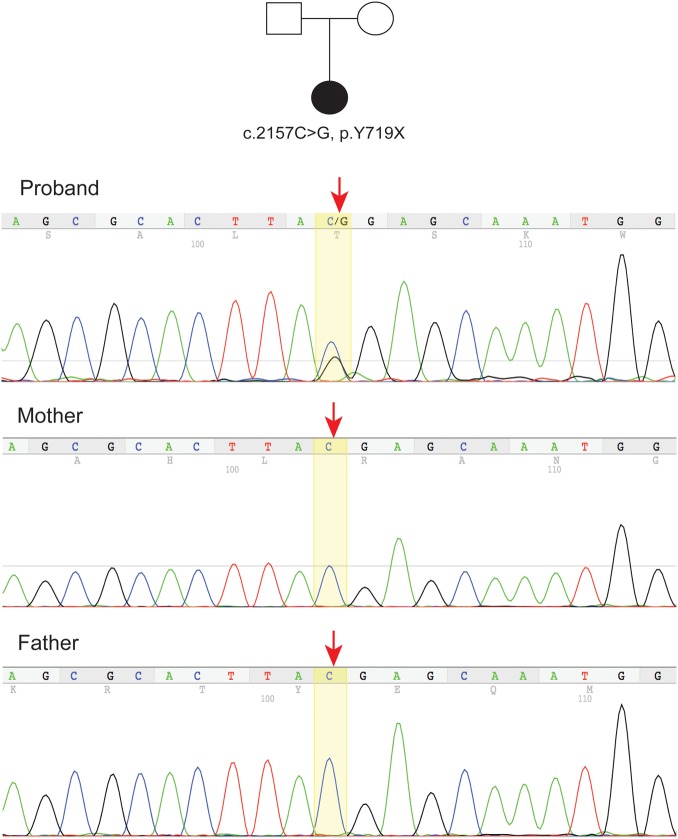
Diagnostic whole exome sequencing in this female patient, performed due to motor/speech delays as well as cyanotic episodes, identified a de novo ADNP mutation (Chr20: 49509094, c.2157C>G, p.Y719X) in exon 5 (figure 1, see online supplementary table 1). The same mutation has been reported in a male patient,8 constituting this mutation as recurrent. The recurrence of this mutation is particularly notable given the large size of the ADNP gene as well as the large size of exon 5 which appears to harbour all mutations discovered to date. This mutation was confirmed by Sanger sequencing and not found in an unaffected brother or biological parents. Prior genome-wide array comparative genomic hybridisation testing did not reveal any pathogenic copy number variants. Her constellation of medical and dysmorphic features is consistent with Helsmoortel et al.8 Notably, our patient is the second ADNP proband diagnosed with a mood disorder.8
Figure 1.
Pedigree and sequence tracing for family with an ADNP mutation. The de novo heterozygous ADNP mutation (Chr20: 49509094, c.2157C>G, p.Y719X) is noted in the proband sequence tracing compared with the same sequence in the parents (highlighted, red arrows). ADNP, activity-dependent neuroprotective protein. Genomic coordinates are reported in the GRC37/hg19 build.
Given the relationship between ADNP mutations and ASD/ID phenotypes, we assessed autistic features and cognitive functioning in our patient at age 6. Interestingly, she did not meet criteria for autism based on the Autism Diagnostic Observation Schedule, Second Edition. Using the ‘some words’ algorithm that requires a score of 12 or greater for an autism diagnosis, she had a total score of 11 (social affect=10, restricted and repetitive behaviour=1) and met criteria for autism spectrum. Her severity score was 5, which is representative of an ASD classification.9 Her full-scale IQ, as measured by the Wechsler Preschool and Primary Scale of Intelligence—Fourth Edition, was 45 indicative of developmental delay. Scores on the Vineland Adaptive Behavior Scale 2nd Edition revealed similar impairment in overall adaptive functioning (standard score=60). Based on these scores, she met criteria for ID.
While hypermetropia has been reported in patients with mutations in ADNP,8 our patient exhibits multiple deficits in the visual system. A novel finding in our patient is cortical visual impairment at 2 years. She was also diagnosed with consecutive exotropia, mild amblyopia and astigmatism at age 2. Although the patient has significant visual impairment, she has made considerable gains particularly in tracking objects. Surgery at 14 months corrected bilateral inferior oblique overaction associated with a marked 25° left head tilt. Visual acuity testing at 5 years showed 20/130 (right eye), 20/170 (left eye) and 20/130 (both eyes). Testing at ages 3 and 4 revealed the patient's uncorrected near visual acuity was 6.8 cycles per degree and 20/190 on a preferential looking test. At her most recent exam, she had normal peripheral vision, normal contrast sensitivity, was able to hold steady fixation on targets and follow moving targets in all directions. Visual fields were full in all quadrants.
In order to understand ADNP function in the developing brain, we examined ADNP mRNA expression in the BrainSpan dataset.10 We downloaded the dataset ‘RNA-Seq Gencode v10 summarized to genes’ from BrainSpan's Developmental Transcriptome (http://www.brainspan.org) and normalised to reads per kilobase per million (RPKM). We applied a log base 2 transformation: log(RPKM+1). Results indicated extensive prenatal expression of ADNP (see online supplementary figure 1). Interestingly, because all mutations described to date are in the last exon, these mutations are unlikely to lead to nonsense-mediated decay. At present, it is unclear if mutations inactivate the protein or alternatively lead to some sort of gain of function. Of note, Helsmoortel et al (2014) demonstrated that the mutant transcript may be upregulated.8 This suggests that the gene expression present here may be perturbed due to mutation.
We also found all the genes positively (Pearson correlation ≥0.9) and negatively (Pearson correlation ≤−0.8) correlated with ADNP (see online supplementary tables S2 and S3). We applied pathway analysis to these correlated genes using the Database for Annotation, Visualization and Integrated Discovery v.6.7.11 Significant pathways for positively correlated genes included transcription (p<0.001), ion binding (p<0.001) and chromatin modification (p<0.001) (see online supplementary table S4). Significant pathways for negatively correlated genes included cellular homeostasis (0.03) and myelination (0.05), especially myelin basic protein (−0.9) (see online supplementary table 5). All p values are reported using Benjamini–Hochberg corrected values.
In summary, we report a patient with a recurrent de novo, protein truncating ADNP mutation with novel clinical features such as visual system impairments. This extends the current phenotypic characteristics and provides a potential role for ADNP in the development of the visual system given its high prenatal expression (see online supplementary figure 1). Our proband is also the second ADNP patient with a mood disorder. Given this additional finding, mood disorders should be investigated as a secondary feature for ADNP mutations. The National Heart, Lung and Blood Institute (NHLBI) Exome Sequencing Project includes exome sequences from 6503 individuals and has been used to clarify X linked ID gene candidates.12 One stop-gain mutation (c.1081C>T, p.Q361*) was found, also in exon 5;8 therefore, while ADNP mutations appear to have a high rate of occurrence in autism, they may not be completely penetrant in all cases. We further investigated ADNP's role in human neurodevelopment, evidenced by its high level of expression in the human fetal brain and association with gene transcriptional regulation. Pathway analysis results of genes highly coexpressed with ADNP show interesting and novel biological processes for future research, particularly in strongly anticorrelated processes like myelination. This suggests a role for ADNP involvement in neuronal/glial cell differentiation.
Supplementary Material
Acknowledgments
This study was approved by the Lifespan Healthcare and Bradley Hospital Institutional Review Boards (IRBs). The authors thank the family for their generous participation in the current study. The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Footnotes
Contributors: MFP and EMM prepared the manuscript. MS performed the bioinformatic analyses and contributed to writing the manuscript. Clinical phenotyping was performed by AJH. MS processed the DNA samples. EDG, WC, JPD, NS, BJ and CP contributed to and commented on the manuscript.
Funding: EMM has received a Career Award in Medical Science from the Burroughs Wellcome Fund and support from NIH NIGMS P20GM103645. This work was supported by grants from the Simons Foundation (SFARI #239834 & #286756 to EMM) and also generous support to EMM from the Nancy Lurie Marks Foundation. Brown Institute for Brain Science (BIBS) and Norman Prince Neuroscience Institute (NPNI).
Competing interests: None.
Ethics approval: Lifespan Healthcare and Bradley Hospital Institutional Review Boards.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Bassan M, Zamostiano R, Davidson A, Pinhasov A, Giladi E, Perl O, Bassan H, Blat C, Gibney G, Glazner G, Brenneman DE, Gozes I. Complete sequence of a novel protein containing a femtomolar-activity-dependent neuroprotective peptide. J Neurochem 1999;72:1283–93 [DOI] [PubMed] [Google Scholar]
- 2.Zamostiano R, Pinhasov A, Gelber E, Steingart RA, Seroussi E, Giladi E, Bassan M, Wollman Y, Eyre HJ, Mulley JC, Brenneman DE, Gozes I. Cloning and characterization of the human activity-dependent neuroprotective protein. J Biol Chem 2001;276:708–14 [DOI] [PubMed] [Google Scholar]
- 3.Pinhasov A, Mandel S, Torchinsky A, Giladi E, Pittel Z, Goldsweig AM, Servoss SJ, Brenneman DE, Gozes I. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res Dev Brain Res 2003;144:83–90 [DOI] [PubMed] [Google Scholar]
- 4.Mandel S, Rechavi G, Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev Biol 2007;303:814–24 [DOI] [PubMed] [Google Scholar]
- 5.Vulih-Shultzman I, Pinhasov A, Mandel S, Grigoriadis N, Touloumi O, Pittel Z, Gozes I. Activity-dependent neuroprotective protein snippet NAP reduces tau hyperphosphorylation and enhances learning in a novel transgenic mouse model. J Pharmacol Exp Ther 2007;323:438–49 [DOI] [PubMed] [Google Scholar]
- 6.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012;485:246–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O'Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012;338:1619–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, van den Ende J, Schuurs-Hoeijmakers JH, Marcelis CL, Willemsen MH, Vissers LE, Yntema HG, Bakshi M, Wilson M, Witherspoon KT, Malmgren H, Nordgren A, Anneren G, Fichera M, Bosco P, Romano C, de Vries BB, Kleefstra T, Kooy RF, Eichler EE, Van der Aa N. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet 2014;46:380–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord 2009;39:693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.BrainSpan: Atlas of the Developing Human Brain [Internet]. Funded by ARRA Awards 1RC2MH089921-01, 1RC2MH090047-01, and 1RC2MH089929-01. © 2011. http://developinghumanbrain.org [Google Scholar]
- 11.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2008;4:44–57 [DOI] [PubMed] [Google Scholar]
- 12.Piton A, Redin C, Mandel JL. XLID-Causing Mutations and Associated Genes Challenged in Light of Data From Large-Scale Human Exome Sequencing. Am J Hum Genet 2013;93:368–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.