Abstract
The 26S proteasome is responsible for most regulated protein turnover and for the degradation of aberrant proteins in eukaryotes. The assembly of this ~2.5 MDa multicatalytic protease requires several dedicated chaperones and, once assembled, substrate selectivity is mediated by ubiquitin conjugation. After modification with ubiquitin, substrates are escorted to the proteasome by myriad factors, including Cdc48 (cell-division cycle 48). Cdc48 also associates with numerous cofactors, but, to date, it is unclear whether each cofactor facilitates proteasome delivery. We discovered that yeast lacking a conserved Cdc48 cofactor, Vms1 [VCP (valosin-containing protein)/Cdc48-associated mitochondrial stress-responsive], accumulate proteasome-targeted ubiquitinated proteins. Vms1 mutant cells also contain elevated levels of unassembled 20S proteasome core particles and select 19S cap subunits. In addition, we found that the ability of Vms1 to support 26S proteasome assembly requires Cdc48 interaction, and that the loss of Vms1 reduced 26S proteasome levels and cell viability after prolonged culture in the stationary phase. The results of the present study highlight an unexpected link between the Cdc48–Vms1 complex and the preservation of proteasome architecture, and indicate how perturbed proteasome assembly affects the turnover of ubiquitinated proteins and maintains viability in aging cells.
Keywords: cell-division cycle 48 (Cdc48), chaperone, p97, proteasome, ubiquitin, yeast
INTRODUCTION
The UPS (ubiquitin–proteasome system) constitutes the major protein degradation machinery in eukaryotic cells and regulates many essential cellular processes, including the cell cycle, signal transduction, transcription and protein quality control [1–6]. As the name implies, UPS substrates are covalently modified by ubiquitin, which is a 76-amino-acid peptide that is most commonly attached via an isopeptide bond on to a lysine residue in a protein substrate [7,8]. The formation of a polyubiquitin chain, containing at least four moieties, is required to target the substrate to a multisubunit catalytic protease, the 26S proteasome [9].
The 26S proteasome is a ~2.5 MDa structure composed of two particles: the core particle (20S or CP), which harbours three distinct proteolytic activities, each in duplicate, and the regulatory particle (19S, PA700 or RP), which recognizes and processes ubiquitinated substrates through ubiquitin receptors and deubiquitinating enzymes respectively [10,11]. When bound to the 20S particle, the 19S particle also opens an obstructed aperture in the 20S particle and unfolds substrates so they can access resident proteases. These events are catalysed by a hexameric AAA-type ATPase ring that neighbours the 20S particle [12,13]. In turn, the central 20S core is flanked on either end by one or two 19S subunits, thus forming the RP-CP and RP2CP proteasome subtypes respectively. Each 20S particle is composed of two central β-type rings and two outer α-rings that exhibit seven-fold symmetry [14–16]. The 19S particle, which contains at least 19 subunits, is more complex, although recent papers have shed light on the placement of the ubiquitin-binding subunits, the deubiquitinating enzyme and the hexameric AAA-ATPases in the particle [17–19].
Assembly pathways have been explored for both the 20S and 19S particles, and, not surprisingly, these are quite complex. The 20S assembly pathway begins with the formation of an α- and β-ring pair (the ‘half-mer’) and is assisted by the proteasome assembly chaperones Pba1 (proteasome biogenesis-associated 1)–Pba4 [11,20–22]. The coalescence of two half-mers, along with another assembly chaperone, Ump1 (ubiquitin-mediated proteolysis 1), precedes the maturation of the β1, 2 and 5 catalytic subunits and the concomitant degradation of Ump1 [23–26]. In contrast, the 19S particle has historically been defined to consist of two subparticles: the base, which directly contacts 20S, and the lid [27]. The assembly of the base, which is composed of the hexameric ATPases Rpt1 (regulatory particle triple-A protein 1)–Rpt6, as well as the Rpn1 (regulatory particle non-ATPase 1), Rpn2 and Rpn13 subunits, requires a cohort of assembly chaperones [Hsm3 (enhanced spontaneous mutability 3), Nas2 (non-ATPase subunit), Nas6 and Rpn14] and proceeds in an established order [28–33]. However, dedicated factors that assist in the assembly of the lid subparticle, which consists of the Rpn3, Rpn5–Rpn9, Rpn11 and Rpn12 subunits, have not been identified [34,35]. How the lid and base subparticles assemble is also not entirely clear, but the incorporation of Rpn10, a ubiquitin-binding receptor, is believed to signify the completion of 19S regulatory particle assembly [27,36]. The final assembly of the 26S proteasome is proposed to occur through one of two possible pathways [37]. The 20S particle may template the assembly of 19S [20,28,29], or 19S may assemble independently before joining 20S [30,32,33]. Regardless, the assembly pathways are most probably redundant since yeast lacking multiple 19S assembly chaperones are viable and, in some cases, the production of 19S or formation of the 26S proteasome is unimpeded [28,30,33].
Once assembled, ubiquitinated substrate delivery to the proteasome requires a cohort of factors, one of which is Cdc48 (cell-division cycle 48) in yeast or its homologue p97 [also known as VCP (valosin-containing protein)] in mammals. Cdc48, which is an AAA-type ATPase, does not act alone and a growing number of Cdc48 partners have been identified, although in most cases the functions of these proteins are mysterious [38,39]. In the present paper we report that the loss of a conserved Cdc48 partner Vms1 (VCP/Cdc48-associated mitochondrial stress-responsive) [40–42] leads to the accumulation of ubiquitinated proteins and is accompanied by increased levels of 20S and 19S proteasome assembly intermediates. Because the observed phenotypes can only be rescued with a copy of Vms1 that interacts with Cdc48, and based on the accumulation of distinct proteasome subunits, we propose that Vms1–Cdc48 maintains proteasome architecture. We have also discovered that Vms1 function maintains the population of capped proteasomes and is required for cell viability in aging yeast cells. The results of the present study explain the mechanism underlying previous observations linking Vms1 function to several UPS-associated phenomena.
EXPERIMENTAL
Yeast strains, growth conditions and plasmids
The yeast strains used in the present study are listed in Supplementary Table S1 (http://www.biochemj.org/bj/458/bj4580459add.htm). Strains were either obtained from Open Biosystems or made by standard genetic procedures [43].Double- and triple-mutant strains were generated by mating yeast mutants of the opposite mating type and sporulating the resulting diploids in 2% potassium acetate. Tetrads were dissected and genotypes were confirmed by screening on selective medium. Strains containing HA (haemagglutinin) epitope-tagged versions of Vms1 were created by amplifying a cassette containing URA3 and Vms1–HA from pRS316-VMS1-HA or pRS316-VMS1ΔVIM-HA [41]. The cassette, which contained homology towards KanMX, was used to replace the vms1ΔKanMX locus. The plasmids used in the present study are listed in Supplementary Table S2 (http://www.biochemj.org/bj/458/bj4580459add.htm).
For all experiments, including stationary-phase analysis, yeast strains were grown at either 26°C or 30°C with shaking at 200 rev./min. The liquid culture medium was either YPD (1% yeast extract, 2% bacto-peptone and 2% dextrose) or SC (synthetic complete) [43]. For spot-growth assays, one D600 of cells was serially diluted 10-fold, spotted on to YPD plates and incubated for 1 day. For the Methylene Blue viability experiments, an aliquot of cells (5–25 µl) was mixed with a 0.1% solution of Methylene Blue (Sigma) and viable and inviable cells were counted with a haemacytometer.
Protein extraction
Total protein extracts for SDS/PAGE were prepared as described previously [44]. When total levels of ubiquitin were assayed, samples were heated at 37°C for 30 min before SDS/PAGE. When protein extracts were prepared to monitor proteasome activity and were analysed by immunoblotting after native PAGE, samples were processed according to published methods, but with minor modifications [45]. In brief, strains were grown to an D600 of 0.6–1.0, harvested and washed with ice-cold water. The cell pellet was then resuspended in 50 mM Tris/HCl (pH 7.4), 5 mM MgCl2, 5mM ATP and 1mM DTT, and the cell slurry was added drop-wise to liquid nitrogen in a mortar. The cells were ground with a pestle for 80 strokes and the resulting powder was snap-frozen in liquid nitrogen and stored at −80°C until used. Where indicated, cell extracts were prepared by glass-bead lysis on a vortex mixer with six 1-min pulses with a 1-min rest on ice in between each pulse. In both cases, lysates were clarified by centrifugation (18000 g for 30 min at 4°C). To immunoprecipitate Cdc48, protein extracts were prepared as follows. Cells were grown to a D600 of 0.6–1.0, treated with 0.1 M sodium azide, and then harvested and washed with ice-cold water. The cells were resuspended in 50 mM Hepes/NaOH (pH 7.4), 150 mM NaCl and 5 mM EDTA, supplemented with a protease inhibitor cocktail (including MG-132) and 10 mMNEM (N-ethylmaleimide), and were lysed with glass beads as described above. Cdc48 and associated proteins were precipitated for 4 h at 4°C with an anti-Myc antibody–agarose conjugate (Santa Cruz Biotechnology) in the same buffer, but supplemented with 0.25% Nonidet P40. The precipitates were washed three times with this buffer and bound proteins were eluted with sample buffer.
Native gel analysis and proteasome activity assays
Native gel analyses were performed as described in [45] using 3.5 or 4.25% non-denaturing PAGE. The gels were prepared in a buffer containing 90 mM Tris base, 90 mM boric acid, 5 mM MgCl2, 0.5mM EDTA, 2.5mM ATP and 1mM DTT, and were polymerized with 0.075% TEMED (N,N,N′,N′-tetramethylethylenediamine) and 0.1% APS (ammonium persulfate). Unless noted otherwise, 50 µg of clarified lysate was mixed with the appropriate amount of 5× sample buffer [250 mM Tris/HCl (pH 7.4), 50%glycerol and 60 ng/ml Xylene Cyanol] and centrifuged for 5 min at 18000 g at 4°C. After the samples were loaded, the gel was run at a 100–110 V constant voltage at 4°C with buffer circulation. The running buffer was the same as the buffer used to prepare the gel except 1 mM ATP was added. To measure the activity of singly and doubly capped proteasomes, the gel was incubated in 50 mM Tris/HCl (pH 7.4), 5 mM MgCl2, 1 mM ATP and 50 µM Suc-LLVY-AMC (N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin; Enzo Life Sciences) for 30 min at 37 °C, and the fluorescent signal was visualized using a Kodak Image Station 440CF. To image the activity of the latent 20S core particle, SDS was added to a final concentration of 0.02% and the gel was incubated for 20 min at 37°C before imaging. For Western blotting, gels were pre-incubated in transfer buffer for 10 min before proteins were transferred on to nitrocellulose membranes and blotted with the indicated antibodies.
Real-time measurements of proteasome activity were performed as reported previously [22].Atotal of 50 µgof clarified yeast extract, prepared by glass-bead lysis as described above, was mixed with an equal volume of 50 mM Tris/HCl (pH 7.4), 5 mM MgCl2, 5mM ATP, 1mM DTT and 20% glycerol. The reaction was brought up to 2 ml with 50 mM Tris/HCl (pH 7.4), 5 mM MgCl2, 5mM ATP, 1mM DTT and 10% glycerol and pre-incubated at 30°C for 30 min. Suc-LLVY-AMC was added to a final concentration of 100 µM and the fluorescent signal was read in an Aminco-Bowman Series 2 Luminescence spectrometer (excitation/emission wavelength, 380/460 nm respectively) at the indicated times.
Antibodies and Western blotting
Western blotting was performed with the indicated antibodies (Supplementary Table S3 at http://www.biochemj.org/bj/458/bj4580459add.htm) and signals were visualized on a Kodak Image Station 440CF using either SuperSignal West Pico or Femto chemiluminescent substrate (Thermo). Image analysis was done with ImageJ v1.46e (NIH). Statistical tests were performed in Microsoft Excel 2007. As determined by Student’s t test, P values less than 0.05 were considered significant.
RESULTS
The Vms1–Cdc48 complex functions in ubiquitinated protein homoeostasis
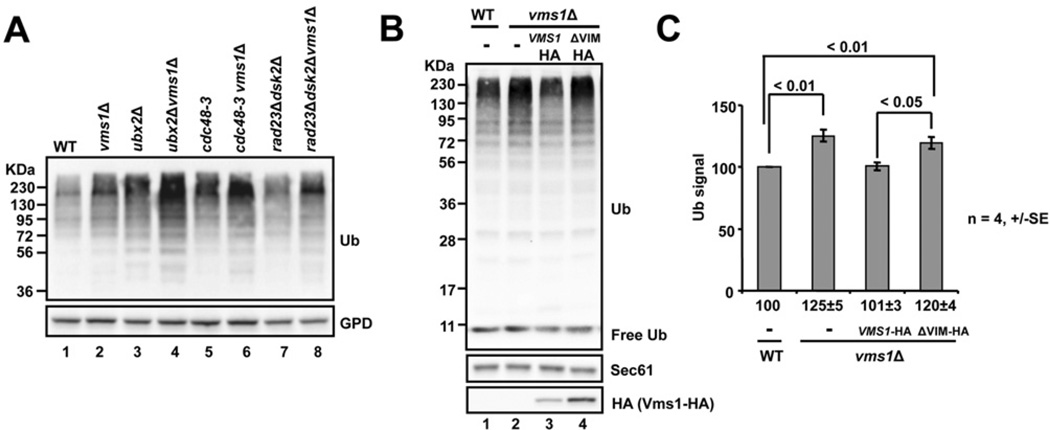
We showed previously that yeast lacking VMS1 accumulate Cdc48-associated ubiquitinated proteins [41]. One explanation for this observation is that ubiquitinated protein levels globally increase; thus, substrate flux to the proteasome rises and Cdc48 captures a portion of these substrates. To test this hypothesis, we probed total precipitated protein extracts from wild-type and vms1Δ yeast for the ubiquitinated protein pool (Figure 1A). As predicted, vms1Δ strains accumulated high-molecular-mass ubiquitinated species to a greater extent when compared with the wild-type strains (Figure 1A, compare lanes 1 and 2). Furthermore, deleting VMS1 in strain backgrounds defective in other UPS-related processes, such as in yeast lacking another Cdc48 partner Ubx2 (ubiquitin regulatory X2) [46], when deleted in the context of the Cdc48 mutant cdc48-3 or when ubiquitinated protein escorts were absent [39] [Rad23 (radiation sensitive 23) and Dsk2], led to an additive increase in the ubiquitinated protein pool (Figure 1A, lanes 3–8). Deleting VMS1 in strains containing mutations in proteasome subunits also gave rise to an additive increase in the ubiquitinated protein pool (Supplementary Figure S1A at http://www.biochemj.org/bj/458/bj4580459add.htm).
Figure 1. Ubiquitinated proteins accumulate in strains lacking VMS1 or expressing a form of Vms1 that is defective for Cdc48 interaction.
(A) The indicated yeast strains were grown to log-phase in rich medium, and total protein was precipitated and resolved by SDS/PAGE. Western blot analysis was used to detect protein-conjugated ubiquitin species (Ub) and GPD (glycerol-3-phosphate dehydrogenase), which served as a loading control. (B) Wild-type (WT) or vms1Δ yeast strains harbouring an empty vector (−), or vectors engineered for the expression of full-length VMS1 (Vms1 HA) or a mutant lacking the VIM domain (ΔVIM HA) were grown in minimal medium to log-phase and processed as in (A) for Western blot analysis. (C) The levels of ubiquitinated proteins > 72 kDa in part (B) were quantified. P values are shown for specific pairwise comparisons.
Because Vms1 associates with Cdc48 though a conserved VIM (VCP-interacting motif) sequence [40,47,48], we next addressed whether the Vms1–Cdc48 interaction was required to restore the ubiquitinated protein pool to wild-type levels. We found that vms1Δ strains harbouring a full-length singly HA-tagged copy of VMS1 reduced the population of ubiquitinated species to those of the wild-type strain, but the expression of the Vms1 ΔVIM mutant was unable to rescue this phenotype (Figures 1B and 1C). The appearance of an increased level of Vms1 ΔVIM is due to the fact that the mutant construct was triply HA tagged, and thus the expression levels of Vms1 ΔVIM and Vms1 are quite similar. In accordance with previous studies, expression of Vms1 lacking VIM also failed to rescue the sensitivity of vms1Δ yeast to cycloheximide and the protein was unable to bind Cdc48 (Supplementary Figures S1B and S1C) [40,47]. These results suggest that the Vms1–Cdc48 complex acts at a late step in the protein degradation pathway.
Yeast lacking Vms1 accumulate free 20S proteasome particles
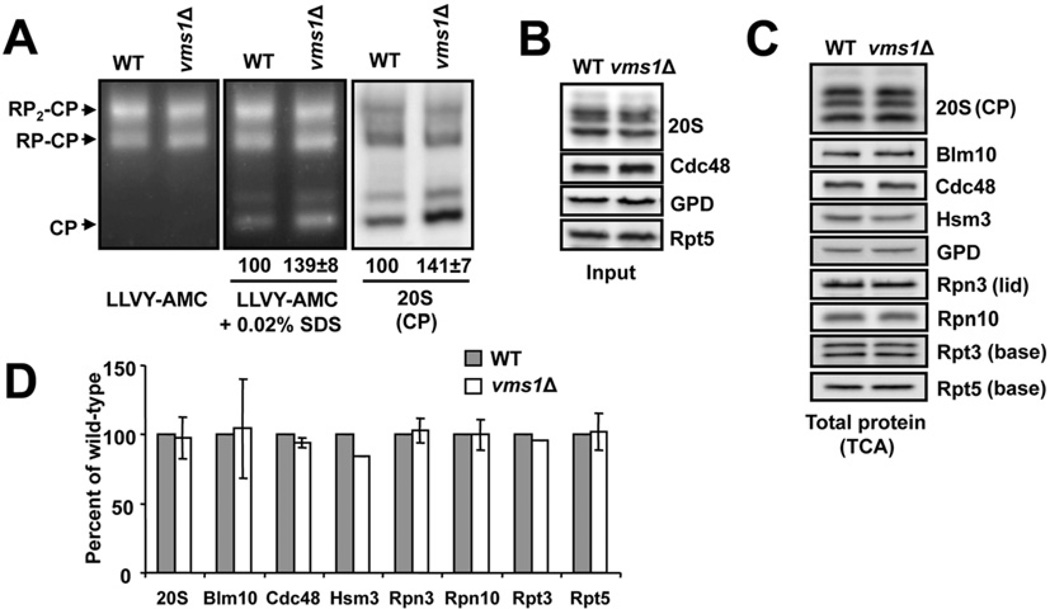
On the basis of the data presented above, one possibility is that the proteasome itself was somehow affected when Vms1 was absent. A search for VMS1 interactors in the DRYGIN synthetic genetic array database [49,50] revealed a number of genes encoding proteasome subunits [e.g. Pre9 (proteinase yscE), Rpn12 and Rpn10]. Therefore we examined whether proteasome function might be impaired in the vms1Δ strain. To this end, protein extracts were resolved on native gels from both wild-type and vms1Δ strains and an in-gel fluorescence reporter assay was used to monitor proteasome activity and stability. In this assay, the gels were incubated with a fluorogenic reporter, Suc-LLVY-AMC (see the Experimental section), and the appearance of AMC-derived fluorescence corresponded to proteasome activity mediated by the resolved RP-CP and RP-CP2 particles [45]. Access to proteases in the 20S particle is restricted in the absence of the 19S, so to visualize 20S (CP) activity the gel can be incubated in a low concentration of SDS, which opens the aperture. When this analysis was performed using lysates from the wild-type and the vms1Δ mutant strain we discovered that loss of Vms1 led to an ~39% increase in the amount of activity associated with the 20S particle, and this effect was accompanied by an identical increase in 20S (CP) protein (Figure 2A). The increase in 20S-mediated proteolytic activity for the fluorescent reporter was also observed in real-time measurements of proteasome activity in solution (Supplementary Figure S2A at http://www.biochemj.org/bj/458/bj4580459add.htm) and was fully recoverable, but only with a copy of Vms1 that interacts with Cdc48 (Supplementary Figure S2B). Because Cdc48 associates with the proteasome [51–54], it was formally possible that the deletion of VMS1 might have grossly affected the integrity of the Cdc48 hexamer. However, the amount of hexamer was unaltered when extracts from a vms1Δ mutant strain were examined by immunoblot analysis after native gel electrophoresis (Supplementary Figure S2C). Because Cdc48 is estimated to be expressed at a 20-fold excess over Vms1 [55], these data suggest that a subpopulation of Cdc48, that which is associated with Vms1, regulates the proteasome.
Figure 2. Yeast lacking VMS1 accumulate free 20S particles.
(A) Lysates from the indicated strains were prepared in liquid nitrogen and total protein was resolved by native PAGE in the presence of ATP. Proteasomes were visualized by incubation with a fluorogenic substrate (Suc-LLVY-AMC) in the absence (left-hand panel) or presence (middle panel) of 0.02% SDS, which activates the proteolytic activity of the 20S particle. An accompanying Western blot was performed for samples resolved by native PAGE (right-hand panel, 20S) and shows an increase in the amount of the core particle (CP). RP2-CP and RP-CP are doubly and singly capped proteasomes respectively. Values below the middle and right-hand panels represent the mean ± S.E.M. of the signal corresponding to the 20S particle (n =14). (B) A total of 2.5 µg of cellular protein used for the analysis in (A) was resolved under denaturing conditions by SDS/PAGE and Western blot analysis was performed to detect the indicated proteins. (C) Total trichloroacetate-precipitated protein was obtained from the indicated strains, resolved under denaturing conditions by SDS/PAGE and the amounts of the indicated species were analysed by Western blotting. (D) Quantification of the Western blots shown in (C). All values are means ± S.D. from three independent samples, except for Hsm3 which was from a single experiment. WT, wild-type.
One explanation for the observations presented in Figure 2(A) is that the increased amount and activity of free 20S core particles was simply due to changes in the cellular concentration of proteasome subunits or proteasome regulators. To address this hypothesis, we performed immunoblot analyses with lysates used for the proteasome activity assay (Figure 2B) as well as protein extracts from the wild-type and vms1Δ strains (Figure 2C). In each case, the levels of the examined proteasome components and proteasome regulators were unchanged (Figures 2C and 2D). To confirm these findings, we created an rpn4Δvms1Δ double mutant and re-examined whether 20S core activity and levels increased. Rpn4 is a transcription factor that triggers the production of proteasome subunits under various stress conditions [56,57]. As shown in Supplementary Figure S2(D), reduced 20S activity and levels were evident in yeast lacking Rpn4, but when Vms1 was also absent an increase in 20S-mediated activity and 20S protein levels were again observed (note the signal corresponding to CP). We also prepared yeast lysates using an alternative method (glass beads) and after proteasome activities were examined using the in-gel assay we measured a 47 ± 7.3% increase in 20S activity in the vms1 mutant. In addition, there was an expected loss of capped proteasomes from 100% to 74 ± 4.9% between lysates from the isogenic wild-type and the vms1Δ strain (results not shown). Together, the loss of Vms1 leads to an altered distribution of capped and uncapped proteasome isoforms without affecting the steady-state levels of proteasome components.
Yeast deleted for VMS1 accumulate 19S assembly intermediates
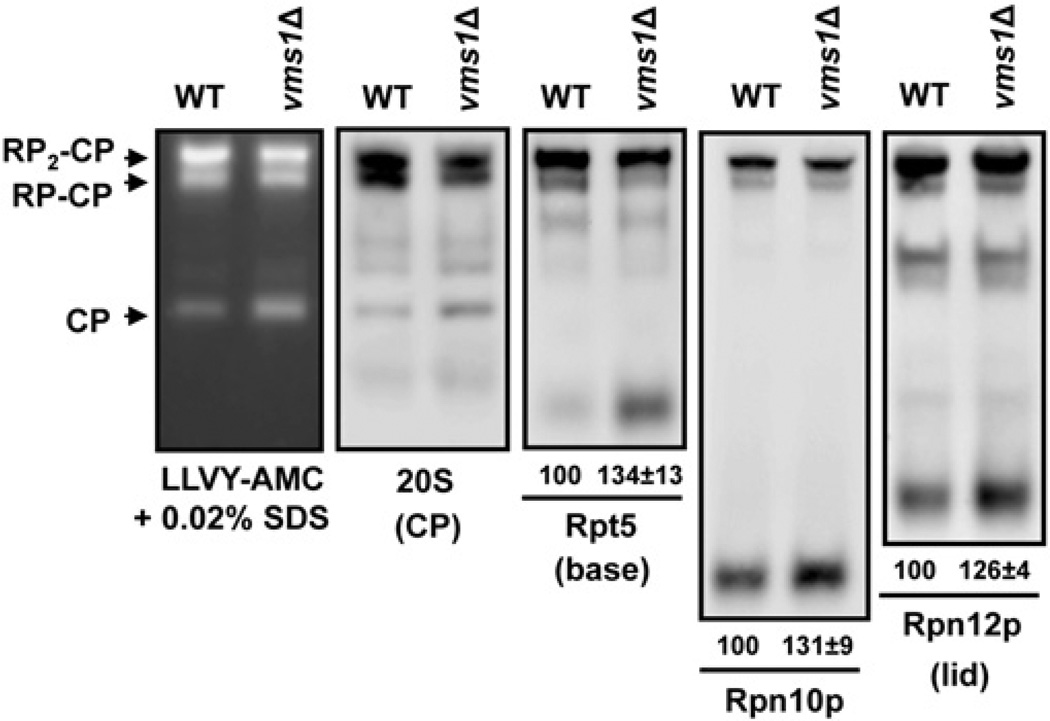
The accumulation of 20S core particles without an accompanying increase in the amount of resident proteins in vms1Δmutant yeast suggests that there is a defect in 19S assembly or in the association of the 19S and 20S subunits. If there is a defect in 19S assembly, one might expect 19S assembly intermediates to accumulate in the vms1Δ strain. To test this hypothesis, we again resolved lysates from wild-type and vms1Δ mutant yeast, but in this case we immunoblotted for distinct proteins associated with either the 19S base (Rpt5), lid (Rpn12) or both subassemblies (Rpn10). As shown in Figure 3, an increase in the amount of a rapidly migratingRpt5-immunoreactive species was evident in the vms1Δ strain. On the basis of its migration, this species appears to be a 19S base assembly intermediate described previously, the Rpt5–Rpt4 complex [30]. In addition, we observed increased levels of the described free Rpn10 and Rpn12 subunits, of which the latter is referred to as the ‘LP1’ subunit [35,58,59]. These results indicate that 19S components accumulate in vms1Δ yeast and are consistent with the observed accumulation of 20S core particles.
Figure 3. Strains lacking VMS1 accumulate 19S assembly intermediates.
Lysates from the indicated strains were resolved by 4.25% native PAGE and the gels were incubated with the fluorogenic proteasome substrate Suc-LLVY-AMC in the presence of 0.02% SDS and were imaged (left-hand panel). A Western blot analysis with lysates from the indicated strains was performed in parallel after resolution by native PAGE. Antibodies used include one that detects the 20S core particle (CP) and others that are directed against proteins in different regions of the 19S cap: Rpt5, base; Rpn10, the connecting subunit; and Rpn12, lid. Where indicated, the faster-migrating species in the vms1Δ strain was quantified relative to the wild-type (WT) strain in at least nine independent experiments. Values under the blots are means ± S.E.M.
Vms1 maintains proteasome levels in the stationary phase
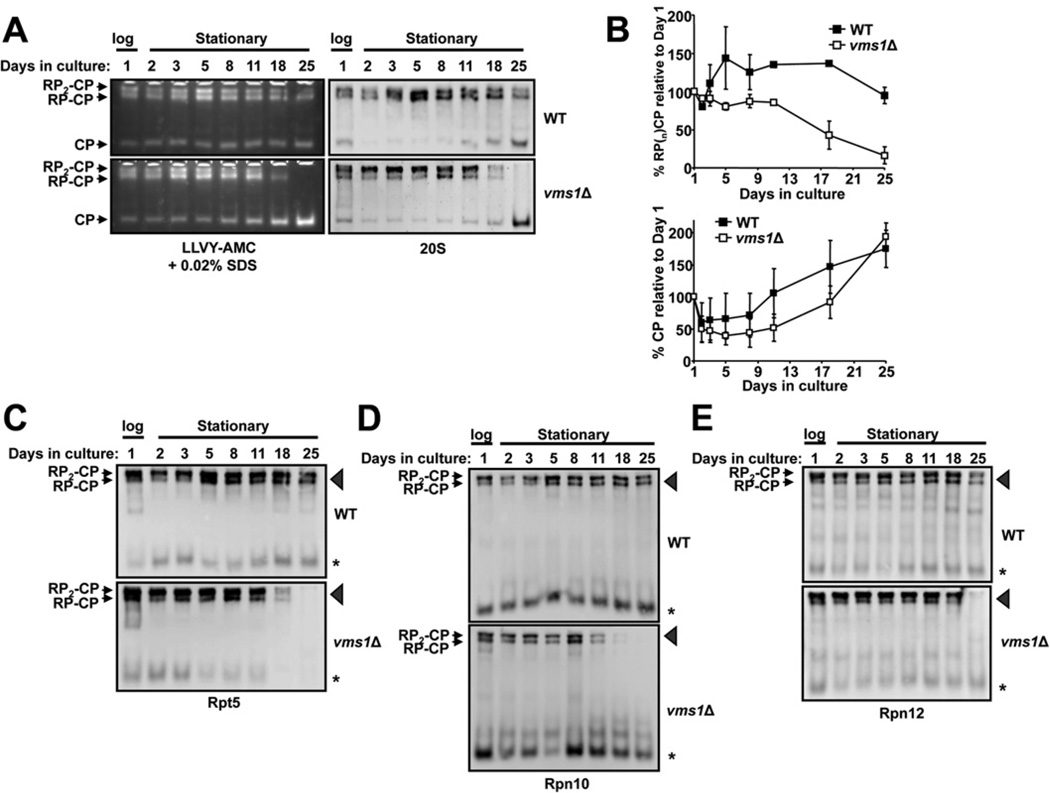
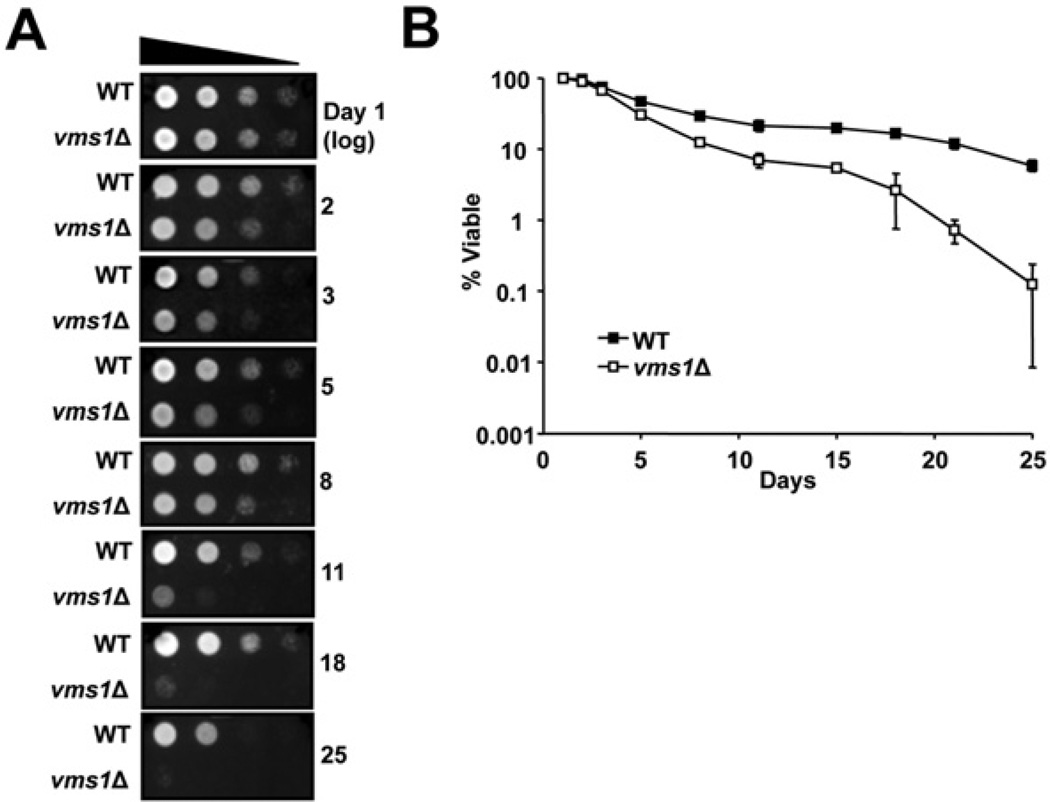
Cells lacking VMS1 exhibit several phenotypes, including sensitivity to cycloheximide and rapamycin, defects in the degradation of a misfolded membrane protein in the ER (endoplasmic reticulum) and in the turnover of a mitochondrial membrane protein, and a loss of viability during prolonged culture in the stationary phase [40–42,60,61]. Of these phenotypes, prolonged growth in stationary phase is also known to down-regulate the activity of capped proteasomes [62]. To examine whether Vms1 function contributes to proteasome homoeostasis in the stationary phase, and whether the deletion of VMS1 enhances the decline in capped proteasome activity in aging cells, we cultured wild-type and vms1Δ cells for 25 days with constant aeration. Aliquots were taken at various times, and cell lysates were prepared. We found that vms1Δ cells displayed reduced RP2-CP and RP-CP proteasome activity and levels after 18 and 25 days in the stationary phase (Figures 4A and 4B). In parallel, and consistent with data indicating a proteasome defect in vms1Δ yeast (Figure 3), significantly lower levels of Rpt5, Rpn10 and Rpn12 were present in the gels at positions corresponding to the RP2-CP and RP-CP proteasomes (Figures 4C–4E, arrowheads). We note that both wild-type and vms1Δcells accumulate free 20S core particles during the stationary phase (Figure 4A, CP). This suggests that Vms1 plays a positive role in maintaining the RP2-CP and RP-CP population. In contrast with what was observed in logarithmically growing cells, we did note variable levels of unassembled 19S components in each case (Figures 4C–4E, *). This may arise from complex changes in metabolic activity and protein synthesis in the stationary phase, along with changes in protein degradation pathways (e.g. autophagy) under these conditions [63,64]. Nevertheless, the loss of 19S components in the RP2-CP and RP-CP proteasomes and decreased activity after 18 and 25 days were mirrored by a dramatic decrease in viability when vms1Δ cells in the stationary phase were transferred to rich medium (Figure 5A). The enhanced decrease in viability of vms1Δ cells was also observed when cells were exposed to a vital dye, Methylene Blue, and viable cells were counted (Figure 5B). These collective data indicate thatVms1 maintains the integrity of singly and doubly capped proteasomes during prolonged culture at the stationary phase. We propose that this phenomenon explains the loss of viability in vms1Δ in aging yeast and the decreased lifespan of nematodes lacking the Vms1 homologue [40] (see the Discussion section).
Figure 4. VMS1 maintains 26S proteasome stability after prolonged incubation at the stationary phase.
(A) The indicated yeast strains were cultured with constant aeration for 25 days at 25°C. Samples were taken over time, lysates were prepared and equal amounts of protein were resolved by native PAGE. Proteasomes were detected with the fluorogenic substrate Suc-LLVY-AMC in the presence of 0.02% SDS (left-hand panels) and by Western blot analysis (right-hand panels) using antibodies directed against proteins in the 20S core particle (CP). Note the more rapid disappearance of the singly (RP-CP) and doubly (RP2-CP) capped core particles in the vms1Δ strain. Data are representative of three independent experiments. (B) Quantification of singly and doubly capped proteasomes (upper panel) and free core particle (lower panel) from (A). Results are means ± S.D. from three independent experiments. (C–E) A Western blot analysis was performed using antibodies directed against the indicated components in the 19S particle. As in (A), note the more rapid disappearance of components in the RP-CP and RP2-CP particles (denoted by arrowheads), especially at days 18 and 25, in the vms1Δ strain. The asterisk denotes the faster-migrating species observed in Figure 3. WT, wild-type.
Figure 5. Loss of Vms1 reduces cell viability after prolonged incubation at the stationary phase.
(A) The indicated strains were cultured with constant aeration for 25 days at 25°C. Serial dilutions of the cultures, taken at different times, were plated on rich medium and incubated for 2 days at 30°C. (B) Methylene Blue staining to detect dead cells was performed by visual inspection with a haemacytometer. The plot shows the log percentage of viable cells (i.e. the unstained cells) relative to total cell numbers during the indicated time course. Data at each time point represent the mean ± S.E.M. for four separate cultures. A minimum of 100 cells was analysed in each culture. WT, wild-type.
DISCUSSION
The Cdc48 cofactor Vms1 has been linked to ubiquitin-mediated degradation of select protein substrates in the ER, mitochondria and nucleus [40–42,61]. In the present study, we show that vms1 mutant yeast accumulate ubiquitinated proteins and that this effect correlates with an altered distribution of proteasome particles and an increase in 19S assembly intermediates. The increased concentration of total ubiquitinated proteins provides an explanation for our observation that more ubiquitinated proteins are bound to Cdc48 in vms1Δ strains [41]. Our combined data also provide a clear view of how a defect in proteasome assembly translates into a defect in ubiquitinated protein turnover. Moreover, these collective phenomena may explain the source of the degradation defects observed previously in vms1Δ strains [40–42].
We found that both free 20S core particles and components of the 19S cap accumulate when Vms1 is absent. On the basis of the accumulation of these species, we posit a role for Vms1 in 19S assembly since other mutants that affect 19S base (e.g. hsm3Δ, rpt3Δ-1, rpt4Δ-1) and lid [e.g. rpn12(211–274)] assembly also accumulate free 20S core and 19S intermediates [28–30,33,35]. Some chaperones that affect 19S base assembly, such as Nas2 and Hsm3, interact directly with the ATPase subunits [28–30,33]. Vms1, which binds to Cdc48, could similarly interact with 19S ATPase subunits to facilitate their assembly. However, we were unable to detect a stable interaction between Vms1 with the proteasome (J.R. Tran and J.L. Brodsky, unpublished work). It is possible that Vms1 associates transiently with proteasome assembly intermediates, and/or with proteasome chaperones. On the basis of the accumulation of polyubiquitinated proteins in the vms1Δ strain, Vms1 may alternatively function as an adaptor for any one of a number of ubiquitin-binding proteins that deliver substrates to the proteasome, and as a result proteasome integrity and function are altered when VMS1 is deleted. Like Vms1, most of the adaptors and the assembly chaperones are not essential. Yet another possibility is that Vms1 regulates Cdc48–proteasome interaction. Cdc48 was found recently to associate with 20S and form a proteasome subtype [54], but no difference in the Cdc48–proteasome interaction was observed when we isolated a functional tagged version of Cdc48 from wild-type and vms1Δ yeast (J.R. Tran and J.L. Brodsky, unpublished work). Furthermore, proteasome assembly was robust in strains containing a temperature-sensitive allele in CDC48, cdc48-3, and the deletion of VMS1 in this genetic background yielded only a modest increase in free 20S particles (J.R. Tran and J.L. Brodsky, unpublished work).
Yeast deleted for VMS1 exhibit reduced fitness in the stationary phase, but the mechanism underlying this phenomenon was mysterious [40]. We propose that reduced fitness arises as a consequence of the observed decline in the amount of capped proteasomes and in capped proteasome activity (Figures 4 and 5). The profile of proteasome subcomplexes is also altered in the stationary phase when another gene, SPG5 (stationary phase gene 5), is deleted [65]. However, the proteasome profiles of stationary phase cultures of vms1Δ and spg5Δ cells differ. Cells lacking VMS1 lose capped proteasomes and maintain free 20S core particles (Figure 4), whereas spg5Δ cells lose free 20S core particles, but maintain doubly capped proteasomes. Paradoxically, both vms1Δ and spg5Δ show decreased viability during the stationary phase, albeit at different time scales, indicating that a balance of proteasome subtypes is necessary for viability. A simple testable hypothesis based on work from our laboratory and the Finley group is that Vms1 and Spg5 function antagonistically to maintain a balance of proteasome subtypes. The prediction is that loss of SPG5 in a background lacking VMS1 restores the balance of proteasome subtypes. To begin to address this possibility, we examined the activities of proteasome particles in extracts from logarithmically growing wild-type, vms1Δ, spg5Δ and spg5Δvms1Δ yeast. We show in Supplementary Figure S3 that the loss of SPG5 in the vms1Δ strain led to no recovery of the phenotype. The simplest conclusion is that Spg5 and Vms1 may simply function independently of one another, and only Vms1 (which unlike Spg5 is conserved in higher cell types) facilitates the maintenance of proteasome architecture in rapidly growing cells. Further work is needed to clarify the respective roles of Vms1 and Spg5 in the ubiquitin–proteasome pathway.
An intriguing previous observation is that Caenorhabditis elegans lifespan is reduced when VMS-1 expression is attenuated by RNAi [40]. We suggest that the decreased lifespan of nematodes lacking VMS-1 arises from compromised proteasome homoeostasis, which is associated with a variety of age-related disorders in model organisms and in mammals [66,67]. An analysis of the effects of Vms1 and its homologues on proteasome homoeostasis, aging and physiology in higher organisms is clearly warranted.
Vms1was identified as a protein quality control component that affects the turnover of select substrates in the ER, mitochondria and nucleus [40–42]. Why are so few protein substrates affected even though ubiquitinated proteins accumulate in vms1Δ cells [41,42]? One explanation is that functionally redundant factors contribute to degradation in the absence of Vms1 function, and that the degree of ubiquitination required to degrade a given protein differs among proteasome-targeted substrates. In other words, substrates that are overtly affected may be exceptionally sensitive to proteasome modulation. This idea is supported by our previous finding that additive degradation defects are evident when vms1 and ufd2 or ubx4 mutants are combined [41], and that defects in proteasome assembly, brought about by deleting the ADD66 (α1-proteinase inhibitor-degradation deficient 66) gene, affect the degradation of only select substrates [22]. Thus the increased ubiquitin signal in the vms1Δ strain may result from the accumulation of a relatively minor substrate pool. The source of this pool is unknown, but Vms1 is found along with Cdc48 as a component of the RQC (ribosomal quality control) complex [68]. The purification of the RQC complex captured Vms1, as well as Shp1 (suppressor of high-copy PP1 1), Ubx2 and Ubx4 [69], which we showed previously exhibit negative genetic interactions with VMS1 [41]. Because 1–15% of nascent translated proteins are ubiquitinated [70,71], it is possible that these substrates, which are subjected to the RQC pathway, might be overrepresented in the vms1Δ mutant. Finally, Vms1 could promote transfer between select Cdc48-bound clients to downstream components of the UPS, such as ubiquitin receptors, deubiquitination enzymes or the polyubiquitin-extending enzyme Ufd2 (ubiquitin fusion degradation 2) [41,42]. Why then might the loss of Vms1 affect proteasome assembly? It is possible that Rpt5 decline in the vms1Δ mutant (Figure 4B) triggers proteasome disassembly. Alternatively, Vms1 may couple proteasome assembly with the degradation of specific proteins or Vms1 may help assemble a particular proteasome variant that handles a distinct substrate ensemble. Several proteasome variants exist, and the functional distinctions and substrate specificities between these are poorly understood [72,73]. Future work is clearly needed to distinguish between these models.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Alexander Buchberger (Universität Würzburg, Würzburg, Germany), Professor Daniel Finley (HarvardMedical School, Boston, MA, U.S.A.), Professor Rasmus Hartmann-Petersen (University of Copenhagen, Copenhagen, Denmark), Professor Mark Hochstrasser (Yale University, New Haven, CT, U.S.A.), Professor Susan Michaelis (Johns Hopkins University School of Medicine, Baltimore, MD, U.S.A.), Professor Randy Schekman (University of California, Berkeley, Berkeley, CA, U.S.A.) and Professor Dorota Skowyra (Saint Louis University School of Medicine, St. Louis, MO, U.S.A.) for reagents; Dr Suzanne Elsasser (Harvard Medical School, Boston, MA, U.S.A.), Professor Daniel Finley, Dr Derek Joyce (The Salk Institute for Biological Studies, La Jolla, CA, U.S.A.) and Dr Robert Tomko Jr (Yale University, New Haven, CT, U.S.A.) for valuable discussions during the course of the present study; and Dr Teresa Buck, Ms Jennifer Goeckler, Dr Christopher Guerrerio, Dr Allyson O’Donnell and Dr Eric Spear for their assistance during the course of the present study.
FUNDING
This work was supported by the National Institutes of Health [grant number GM75061 (to J.L.B.)].
Abbreviations
- Cdc48
cell-division cycle 48
- ER
endoplasmic reticulum
- HA
haemagglutinin
- Hsm3
enhanced spontaneous mutability 3
- Nas2
non-ATPase subunit
- Rpn
regulatory particle non-ATPase
- Rpt
regulatory particle triple-A protein
- RQC
ribosomal quality control
- SPG5
stationary phase gene 5
- Suc-LLVY-AMC
N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin
- Ubx
ubiquitin regulatory X
- Ump1
ubiquitin-mediated proteolysis 1
- UPS
ubiquitin–proteasome system
- VCP
valosin-containing protein
- VIM
VCP-interactingmotif
- Vms1
VCP /Cdc48-associated mitochondrial stress-responsive.
Footnotes
AUTHOR CONTRIBUTION
Joseph Tran planned the study, performed the experiments, analysed the data and composed the paper. Jeffrey Brodsky helped plan the experiments, analysed the data and edited the paper before submission.
REFERENCES
- 1.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr. Opin. Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso V, Friedman PA. Minireview: Ubiquitination-regulated G protein-coupled receptor signaling and trafficking. Mol. Endocrinol. 2013;27:558–572. doi: 10.1210/me.2012-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finley D. Recognition and processing of ubiquitin–protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varshavsky A. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 7.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 12.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 13.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 14.Kish-Trier E, Hill CP. Structural biology of the proteasome. Annu. Rev. Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem. Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 16.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 17.Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 21.Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Scott CM, Kruse KB, Schmidt BZ, Perlmutter DH, McCracken AA, Brodsky JL. ADD66, a gene involved in the endoplasmic reticulum-associated degradation of α-1-antitrypsin-Z in yeast, facilitates proteasome activity and assembly. Mol. Biol. Cell. 2007;18:3776–3787. doi: 10.1091/mbc.E07-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the β7 subunit and activator complexes stabilize nascent 20S proteasomes and promote their maturation. J. Biol. Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 24.Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 25.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. β-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 28.Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C-termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 35.Tomko RJ, Jr, Hochstrasser M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol. Cell. 2011;44:907–917. doi: 10.1016/j.molcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Forster F, Baumeister W. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1479–1484. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Besche HC, Peth A, Goldberg AL. Getting to first base in proteasome assembly. Cell. 2009;138:25–28. doi: 10.1016/j.cell.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 2011;36:515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran JR, Tomsic LR, Brodsky JL. A Cdc48p-associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J. Biol. Chem. 2011;286:5744–5755. doi: 10.1074/jbc.M110.179259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek GH, Cheng H, Kim I, Rao H. The Cdc48 protein and its cofactor Vms1 are involved in Cdc13 protein degradation. J. Biol. Chem. 2012;287:26788–26795. doi: 10.1074/jbc.M112.351825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 44.Tran JR, Brodsky JL. Assays to measure ER-associated degradation in yeast. Methods Mol. Biol. 2012;832:505–518. doi: 10.1007/978-1-61779-474-2_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- 46.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stapf C, Cartwright E, Bycroft M, Hofmann K, Buchberger A. The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. J. Biol. Chem. 2011;286:38670–38678. doi: 10.1074/jbc.M111.274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballar P, Shen Y, Yang H, Fang S. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J. Biol. Chem. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- 49.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koh JL, Ding H, Costanzo M, Baryshnikova A, Toufighi K, Bader GD, Myers CL, Andrews BJ, Boone C. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 2010;38:D502–D507. doi: 10.1093/nar/gkp820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 53.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 56.Metzger MB, Michaelis S. Analysis of quality control substrates in distinct cellular compartments reveals a unique role for Rpn4p in tolerating misfolded membrane proteins. Mol. Biol. Cell. 2009;20:1006–1019. doi: 10.1091/mbc.E08-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, Erdjument-Bromage H, Tempst P, Buranda T, Sklar LA, Baumler J, et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 60.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 61.Esaki M, Ogura T. Cdc48p/p97-mediated regulation of mitochondrial morphology is Vms1p-independent. J. Struct. Biol. 2012;179:112–120. doi: 10.1016/j.jsb.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 62.Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 63.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 64.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanna J, Waterman D, Boselli M, Finley D. Spg5 protein regulates the proteasome in quiescence. J. Biol. Chem. 2012;287:34400–34409. doi: 10.1074/jbc.M112.390294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011;3:a004440. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaiswal M, Sandoval H, Zhang K, Bayat V, Bellen HJ. Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu. Rev. Genet. 2012;46:371–396. doi: 10.1146/annurev-genet-110711-155456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lopez AD, Tar K, Krugel U, Dange T, Ros IG, Schmidt M. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol. Biol. Cell. 2011;22:528–540. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, Van den Eynde BJ. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.