Abstract
Muscle ankyrin-repeat proteins (MARPs) have been shown to serve diverse functions within cardiac and skeletal muscle cells. Apart from their interactions with sarcomeric proteins like titin or myopalladin that locate them along myofilaments, MARPs are able to shuttle to the nucleus where they act as modulators for a variety of transcription factors. The deregulation of MARPs in many cardiac and skeletal myopathies contributes to their use as biomarkers for these diseases.
Many of their functions are attributed to their domain composition. MARPs consist of an N-terminal coiled-coil domain responsible for their dimerization. The C-terminus contains a series of ankyrin-repeats, whose best-characterized function is to bind to the N2A-region of the giant sarcomeric protein titin.
Here we investigate the nature of their dimerization and their interaction with titin more closely. We demonstrate that the coiled-coil domain in all MARPs enables their homo- and hetero-dimerization in antiparallel fashion. Protein complementation experiments indicate further antiparallel binding of the ankyrin-repeats to titin’s N2A-region. Binding of MARP to titin also affects its PKA mediated phosphorylation. We demonstrate further that MARPs themselves are phosphorylated by PKA and PKC, potentially altering their structure or function. These studies elucidate structural relationships within the stretch-responsive MARP/titin complex in cross-striated muscle cells, and may relate to disease relevant posttranslational modifications of MARPs and titin that alter muscle compliance.
Keywords: muscle, MARP, titin, phosphorylation, kinase
Introduction
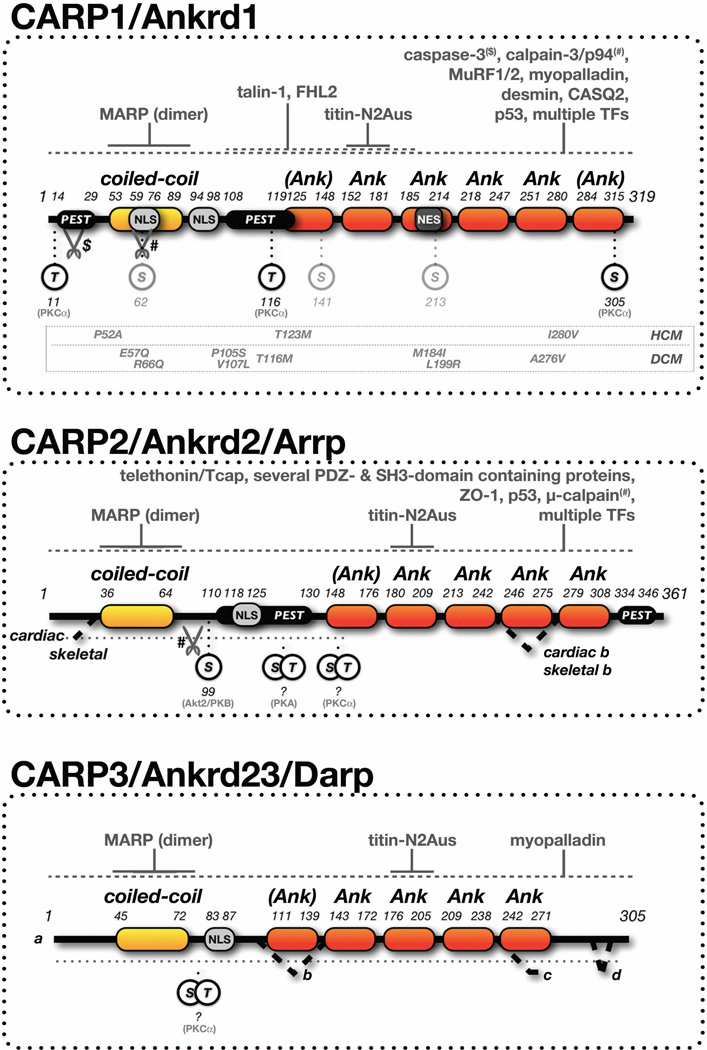
CARP1 (Ankrd1), CARP2 (Ankrd2/Arrp) and CARP3 (Ankrd23/DARP) are members of the muscle ankyrin-repeat protein family (MARPs) that have been shown to play important roles for muscle gene expression and myofilament organization (Kojic et al., 2010; Laure et al., 2010; Witt et al., 2004; Zou et al., 1997). Proteins of the MARP family contain a single coiled-coil domain at the N-terminus, followed by several conserved ankyrin-repeats towards the C-terminus (Figure 1). Both domains have been demonstrated to serve interactions with a multitude of proteins.
Figure 1. MARP structure, interaction partners and posttranslational modifications.
Isoforms, domain structure, signaling and peptide motifs, as well as known and novel posttranslational modification sites of the three members of the MARP family: CARP1/Ankrd1, CARP2/Ankrd2 and CARP3/Ankrd3. Minimal binding sites for MARPs and titin (solid lines), and other interaction partners (dashed lines, see references in the introduction), protease sites, and phosphorylation sites including their modifying protein kinases are shown. Domain labels in brackets indicate newly predicted domains. Characterized disease linked mutations for human CARP1 are shown.
Abbreviations and symbols: Ank - ankyrin-repeat domain; PEST - proline (P), glutamic acid (E), serine (S), threonine (T)-rich sequence; NLS - nuclear localization sequence; NES - nuclear export sequence; TFs - transcription factors; $ - caspase cleavage site; # - calpain-3/p94 or µ-calpain cleavage site; HCM - hypertrophic cardiomyopathy; DCM - dilated cardiomyopathy. Residue numbers correspond to human versions of CARP1, CARP2 and CARP3 (see Materials and Methods).
While the main function of the N-terminal coiled-coil domain in all MARPs has been assigned to the formation of homo-dimers (Witt et al., 2005), the biological role for the C-terminal ankyrin-repeats are more diverse and partially specific for each of the MARP family members. The best characterized role for the MARP C-terminus is its interaction with the N2A-region of titin/connectin (Miller et al., 2003), which may also be responsible for the striking I-band localization of CARP1 (Bang et al., 2001).
Apart from titin, CARP1 has been shown to interact with other sarcomeric and ‘extra’-sarcomeric proteins, like myopalladin (Arimura et al., 2009; Bang et al., 2001), the intermediate filament desmin (Witt et al., 2005), talin-1 and FHL2 (Moulik et al., 2009). In addition, CARP1 was found to bind to calsequestrin-2 (CASQ2), implicating a role for the protein in the modulation of calcium signaling and excitation-contraction coupling (Torrado et al., 2005). The role as “modulator” of signaling pathways is also reflected in the finding that CARP1 acts as co-regulator for many transcription factors, among them NFkB (Laure et al., 2010), YB1 (Zou et al., 1997), p53, KLF12, and LDB1 (Kojic et al., 2010). Bioinformatical investigation of the CARP1 protein sequence revealed that the protein may harbor many sites for posttranslational modifications, as well as signal peptide sequences for degradation and localization (Jeyaseelan et al., 1997; Zou et al., 1997). Indeed, several nuclear import and export signals may be responsible for the various subcellular localizations reported for the protein and its dual role at the myofibrils and in the nucleus. Reports indicate also that CARP1 may be interaction partner and substrate for p94/calpain-3 (Laure et al., 2010) and caspase-3 (Ju et al., 2007) proteases during times of cellular stress. Particularly its interaction and cleavage by calpain-3, another interaction partner for titin-N2A in skeletal muscles, was demonstrated to influence NFkB signaling (Laure et al., 2010).
CARP1 protein levels are regulated in cardiomyocytes in a spatial and temporal manner. Developmentally, CARP1 expression (mRNA) is tightly regulated under control of the Nkx2.5 transcription factor in atria and ventricles, as well as the Sp3 and Hey2 transcription factors in ventricles only, and can be detected as early as embryonic stage E8.5 within the developing myocardium (van Loo et al., 2007; Zou et al., 1997). CARP1 remains expressed in hearts until adulthood, with higher CARP1 expression reported for atrial compared to ventricular tissue, and some reports suggesting an overall left vs. right myocardial asymmetry in the levels of CARP1 protein (Mikhailov and Torrado, 2008; Torrado et al., 2004; Torrado et al., 2006). Experiments investigating the cardiotoxic effect of doxorubicin revealed further that CARP protein levels are extremely sensitive to this chemotherapeutic agent (Jeyaseelan et al., 1997). CARP1 protein levels are also regulated through its rapid degradation by the ubiquitin-proteasome (UPS) pathway (Badi et al., 2009). CARP1 half-life in transfected HeLa cells was determined to be approximately 30 minutes, and to be dependent on the presence of PEST sequences (Rechsteiner and Rogers, 1996; Rogers et al., 1986). The degradation through the UPS pathway is further supported by experiments with animals deficient in the muscle specific E3-ubiquitin ligases of the MuRF family (muscle-specific RING finger). Knockouts of either MuRF1 or MuRF2, or double knockouts have slightly, or significantly increased CARP1 protein levels, respectively. In addition it was reported that CARP1 directly interacts with MuRF1 and MuRF2 (Witt et al., 2008).
The second best characterized member of the MARP family is CARP2, which shares many features with CARP1. Apart from its homo-dimerization and interaction with titin-N2A, CARP2 was also found to bind many of the same transcription factors identified for CARP1: among them YB-1, p53 (Belgrano et al., 2011; Kojic et al., 2004). CARP2 also interacts with several PDZ-domain containing proteins, among them ZO-1 and ZO-2, several SH3-domain containing proteins, as well as telethonin/Tcap (Belgrano et al., 2011). Although the biological relevance for the interaction of CARP2 with telethonin remains to be explored, it is noteworthy that telethonin was characterized as a substrate for the mechano-sensitive kinase domain in titin (Mayans et al., 1998), thereby putatively merging the mechano-responsive properties of MARP complex with the titin-kinase pathway.
While many of the posttranslational modifications for CARP1 have been hypothesized, CARP2 was demonstrated to be phosphorylated by Akt2/PKB (Cenni et al., 2011). In addition, CARP2 can be proteolytically cleaved by µ-calpain (Hayashi et al., 2008). However, in contrast to CARP1, which demonstrated increased binding to titin after calpain-mediated proteolysis (Laure et al., 2010), proteolytically cleaved CARP2 displays unchanged N2A-binding affinity.
Although most CARP2 expression can be found in skeletal muscles, presumably under control of the MyoD and NFkB transcription factors (Pallavicini et al., 2001), a cardiac splice variant is reported to be expressed at lower levels in cardiomyocytes (Miller et al., 2003). Like MyoD, CARP2 expression is developmentally upregulated during myoblast differentiation, coinciding with a change in subcellular localization from the nucleus to the cytoplasm (Pallavicini et al., 2001).
Comparatively little is known about the third member of the MARP family, CARP3. Like the other MARPs, CARP3 forms homo-dimers (Witt et al., 2005) and interacts with the sarcomeric proteins titin and myopalladin (Miller et al., 2003). Similar to CARP1, CARP3’s subcellular localization and expression was found to be responsive to mechanic stimuli. While CARP3 expression was found primarily in skeletal muscles, its cardiac expression is induced in the hearts of a type-2 diabetes insulin-resistant mouse model (Ikeda et al., 2003), a characteristic that garnered it its alias DARP (diabetes-related ankyrin-repeat protein). Bioinformatic analysis also indicated the occurrence of several CARP3 splice isoforms that primarily influence the number and composition of its C-terminal ankyrin-repeats (Figure 1).
A common characteristic of all MARP family members is their induction upon cellular stress, a quality that designated them as “stress response molecules” (Miller et al., 2003) and suggested their use as diagnostic biomarkers (Kojic et al., 2011). Particularly CARP1 and CARP2 were shown to be highly elevated for many cardiac and skeletal myopathies, including hypertrophic and dilated cardiomyopathy (HCM and DCM, respectively), as well as several sub-types of muscular dystrophy and congenital myopathies (for a review see (Kojic et al., 2011)). CARP1 mRNA analysis also indicated differential atrial vs. ventricular regulation (Torrado et al., 2006) and aberrant splice variants characterized by retention of 3‘ introns in the failing myocardium (Torrado et al., 2009). Several mutations in CARP1 have been implicated to be causative of HCM (Arimura et al., 2009; Crocini et al., 2013) or DCM (Duboscq-Bidot et al., 2009; Moulik et al., 2009). However, these mutations represent most likely gain-of-function rather than loss-of-function mutants, as single, double and triple knockouts for MARP family members are viable and were only shown to be more affected by acute eccentric contraction injury (Barash et al., 2007).
The best characterized interaction partner for all members of the MARP family is the giant sarcomeric protein titin (connectin). Within cross-striated muscle cells, titin acts as a “master regulator” for myofibrillogenesis and structural integrity. It offers binding sites for the actin and myosin filament systems, and for a multitude of other sarcomeric proteins important for the structure of the sarcomere, like α-actinin and myosin-binding protein C (Kontrogianni-Konstantopoulos et al., 2009; Lange et al., 2006). In addition, titin harbors interaction sites for many ‘extra’-sarcomeric proteins that may be dispensable for myofibrillogenesis, but have been shown to serve important roles for muscle specific signaling pathways involved in the ‘fine-tuning’ of muscle contraction and for muscle maintenance (Kruger and Linke, 2011; Lange et al., 2006). Some of these ‘extra’-sarcomeric and sarcomere-associated proteins like the four-and a half lim-domain protein family (FHL proteins) or the MARP family bind to unstructured regions within the titin molecule (Lange et al., 2002; Miller et al., 2003; Sheikh et al., 2008). In addition to their role as docking sites for proteins, the titin PEVK, N2B and N2A unstructured regions have also been demonstrated to serve as elastic elements during contraction, whose passive mechanic spring-like properties can be tuned through phosphorylation by various protein kinases (Kruger and Linke, 2011). This duality of function led to the notion that the elastic unstructured regions within the titin molecule, in combination with their interaction partners may be one mechanism for muscle cells to sense and respond to mechanic stimuli. Indeed, it was recently demonstrated that FHL proteins that bind to the N2B region of cardiac titin are able to recruit elements of the MAPK signaling pathway to the myofilaments (LeWinter and Granzier, 2010; Sheikh et al., 2008), and may directly modulate titin-N2B phosphorylation levels (Raskin et al., 2012).
Similar to the titin-N2B/FHL complex, the titin-N2A/MARP complex counts among the stretch sensitive elements with mechano-responsive potential. This is evidenced by the ability of cAMP-dependent protein kinase (PKA) to phosphorylate the N2A-region of titin and lower its passive stiffness in cardiac muscles (Kruger and Linke, 2006). Moreover, MARPs that bind to titin-N2A have been demonstrated to show stretch-dependent subcellular localization: CARP1 shuttles between its sarcomeric localization within the I-band to the nucleus, whereas CARP3 is found increasingly at intercalated disks in stretched neonatal cardiomyocytes (Miller et al., 2003).
We set out to investigate the two molecular functions associated with all MARP family members in closer detail: their dimerization and interaction with the N2Aus-region of titin. We reasoned that further characterization of these interactions may reveal as of yet unrecognized and under-appreciated structural features and functions of MARP family proteins. We also elucidate the influence that MARPs play on the PKA mediated phosphorylation of titin, and study their own posttranslational modification potential by protein kinases. Additionally, effects of MARP proteins on titin phosphorylation are discussed as they may result in significant changes to muscle compliance. This feature may be of particular pathophysiological importance as MARPs were found to be upregulated for many cardiac and skeletal muscle myopathies.
Materials and Methods
Generation of constructs
The human versions of CARP1/Ankrd1 (NM_014391), CARP2/Ankrd2 (NM_020349), CARP3/Ankrd23 (NM_144994) and the N2A unique region of human titin (NM_133378) were all cloned via PCR from cardiac cDNA libraries (Clontech, Mountain View, CA).
Generation of protein complementation constructs was done through in-frame ligation of either full-length CARP1, coiled-coil region of CARP1 (amino acids 1–122), CARP1 ankyrin-repeats (amino acids 105–319), the coiled-coil region of CARP2 (amino acids 1–146), the coiled-coil region of CARP3 (amino acids 1–109), or the unique sequence within the N2A-region of titin (N2Aus; amino acids 8528–8661) into the protein complementation vectors (split-YFP) YN-C1, YC-C1, YC-N1 or YN-C1/N1-YC (Zou et al., 2006). This effectively generates either N-terminally tagged fusion proteins with the N-terminal (YN) or C-terminal (YC) half of YFP, or C-terminally tagged fusion proteins with the C-terminal half of YFP, or a fusion protein tagged at the N-terminus with YN and at the C-terminus with YC. In addition, a full-length HA-tagged versions of CARP2 was done by in-frame insertion of CARP2 coding sequence into the pHA-N1 vector, a modified version of the pEGFP-N1 vector, where GFP has been replaced by the HA-tag (Lange et al., 2012).
Generation of the GST-fusion proteins was done by in-frame cloning of the full-length, coiled-coil domain or ankyrin-repeats of CARP1, full-length of CARP2 or full-length of CARP3, as well as the titin-N2Aus region into the GST-C1 vector (Lange et al., 2012).
Site directed mutagenesis was done as previously described (Lange et al., 2012) using the following oligonucleotides (with mutations in lower case): T11A.mut GAAAGTAGAGGAACTGGTCgCTGGAAAGAAGAATGGC; S62A.mut GGGAGCAACAGTGGAAAgcCGAGAAACAACGAGAGGC; T116A.mut CCAGAACCTGAAATCATTgCGGAACCTGTGGATGTGCC; S213A.mut GAATAAAGGAGCAAAAATTgcCGCCCGAGATAAGTTGCTCAG and S305A.mut CCAAAGCAATATTCGACgcCCTCAGAGAGAACTCCTAC.
All constructs and mutations have been verified for correct and in frame integration through sequencing.
Cell culture
Cos-1 cells (Gluzman, 1981) were cultured as described previously (Lange et al., 2002). For transfection of DNA, a mixture of 1µg DNA, 200µl DMEM and 3µl Lipofectamine-2000 (Life Technologies, Carlsbad, CA) was added to medium of Cos-1 cells growing at 60–80% confluency. Cells were grown for another three days before fixation or lysis.
For GST pulldown assays, transfected cells from 60mm dishes were washed once in PBS and lysed into 200µl ice-cold IP buffer (150mM NaCl, 10mM Tris-HCl pH8, 1mM DTT, 0.2% NP-40, 0.2% SDS) supplemented with protease and phosphatase inhibitor cocktails (Complete EDTA-free and PhosSTOP; Roche, Indianapolis, IN). Insoluble proteins were removed through centrifugation in a tabletop centrifuge at maximum speed at 4°C for 10 minutes, and the soluble supernatant was subsequently used for the biochemical interaction assays.
Immunofluorescence
Transfected Cos-1 cells were washed once with phosphate-buffered saline (PBS) and fixed for 5 minutes with 4% paraformaldehyde in PBS, briefly washed in PBS and permeabilized with 0.2% Triton-X100 in PBS for another 5 minutes. Cells were then incubated with primary antibody diluted into gold buffer (GB; 20 mM Tris-HCl, pH 7.5, 155 mM NaCl, 2 mM ethylene glycol tetraacetic acid, 2 mM MgCl2, 5% bovine serum albumin [BSA]) for 1 hour. After washing three times with PBS for 5 minutes each, cells were then incubated with secondary antibody diluted in GB for one hour, followed by three washes of PBS. Cells were mounted in fluorescent mounting medium (DAKO, Carpinteria, CA) and imaged on an Olympus Fluoview confocal microscope equipped with a 40× oil immersion objective in sequential scanning mode, and zoom rates between 1 and 3. DAPI (4′,6-diamidino-2-phenylindole; 2 mg/ml; Sigma-Aldrich) was incubated with the secondary antibody mixture.
Antibodies
To identify cells transfected with the protein complementation constructs (split-YFP), and for the detection of HA-tagged fusion proteins in immunoblots we utilized antibodies raised against the green fluorescent protein (GFP) or the HA-tag from Roche (Indianapolis, IN). Secondary antibodies for immunofluorescence were from Jackson ImmunoResearch (West Grove,PA), while secondary antibodies for immunoblot analysis were from DAKO (Carpinteria, CA).
Protein expression, GST-pulldown assay
Biochemical protein-protein interaction assay (GST-pulldown) was done as previously described (Raskin et al., 2012). Expression of GST-fusion proteins was done using BL21 cells (Invitrogen; Life Technologies, Carlsbad, CA). In short, BL21 cells transformed with the GST-expression vector were grown in 400ml LB-medium supplemented with the appropriate antibiotic on an orbital shaker at 37°C to an OD600 of >1. After incubation on ice for 15 minutes, protein expression was induced by addition of IPTG (Isopropyl β-D-1-thiogalactopyranoside) to a final concentration of 0.2mM. Cells were transferred into a temperature controlled orbital shaker and protein expression was allowed to proceed over night at 18°C. Cells were collected by centrifugation, lysed into 40ml ice cold lysis buffer (150mM NaCl, 10mM Tris-HCl pH 8, 1% Triton X100) and sonicated for 1 minute at 4°C at 70% output (Vibracell, Sonics & Materials Inc., Newtown, CT). Insoluble cell debris was removed by ultracentrifugation (45 minutes at 11000rpm, 4°C) and the supernatant was incubated with 400µl glutathione-sepharose beads (Amersham Biosciences) for 2 hours at 4°C on a shaker. After washing of beads with ice cold PBS, bound protein was eluted with GST-elution buffer (150 mM NaCl, 50mM Tris-HCl pH 7.4, 150 mM reduced glutathione) and dialyzed over night at 4°C against dialysis buffer (20mM HEPES KOH pH7.4, 10mM MgSO4, 0.1mM CaCl2). Protein concentration was determined by bradford assay (Biorad) or through densitometry of coomassie stained SDS-page gels.
For GST pulldown assays, soluble proteins from transfected Cos-1 cells were incubated with approximately 2µg of GST or GST-fusion proteins for 2 hours on ice. After addition of 15µl glutathione-sepharose beads and continuing incubation on a shaker at 4°C for 1 hour, beads were pelleted using a tabletop centrifuge at 8000rpm for 30 seconds. Pellets were washed three times with ice-cold PBS, resuspended in SDS-sample buffer and bound proteins were analyzed through SDS-page followed by immunoblot analysis as described before (Lange et al., 2002).
In-vitro kinase assay
For in-vitro kinase assays, 2µg of GST fusion proteins were combined with either purified PKCα (Sigma-Aldrich) or PKA (NEB) in kinase buffer (20mM HEPES KOH pH7.4, 10mM MgSO4, 0.1mM CaCl2) supplemented with radioactively labeled [γ-32P]ATP (Amersham Biosciences). For PKCα in-vitro kinase assays, the reaction mix was additionally supplied with a combination of 1µg/µl phosphatidylserine (1,2-Diacyl-sn-glycero-3-phospho-L-serine) and 2µg/µl diacylglycerol (DAG; 1,2-Dioleoyl-sn-glycerol) dissolved in resuspension buffer (10 mM HEPES KOH pH7.4, 0.3% Triton X-100). To measure the effect of MARPs on N2A phosphorylation, increasing amounts of either GST-CARP1 (or GST-CARP3) or GST alone were added to the reactions. The mixture was subsequently incubated at 30°C for 30 minutes. After incubation, samples were mixed with SDS-sample buffer and separated using SDS-page. Gels were coomassie-stained, dried on a Biorad gel dryer and radioactively labeled proteins were analyzed using X-ray films and autoradiography. For densitometric quantification of phosphorylation levels, X-ray films were scanned and band intensity was measured using ImageJ (NIH).
Statistical analysis, image processing and bioinformatic analysis
Statistical analysis was done using Excel (Microsoft). Data presented are mean values ± standard errors. Significance was evaluated by the two-tailed student’s t-test. Sample size (n-values) and p-values are indicated in the figures and/or figure legends.
Images were processed using ImageJ (NIH) equipped with the LOCI bio-formats plugin and Photoshop (Adobe).
For isoform and sequence analysis the following programs have been used: EPESTFIND http://bips.u-strasbg.fr/EMBOSS/, SMART http://smart.embl-heidelberg.de/, secondary structure prediction http://www.compbio.dundee.ac.uk/www-jpred/, LOGICOIL oligomeric state probability prediction of a coiled-coil sequence http://coiledcoils.chm.bris.ac.uk/LOGICOIL/, helical wheel projections http://rzlab.ucr.edu/scripts/wheel/wheel.cgi, the NIH nucleotide database http://www.ncbi.nlm.nih.gov/nucleotide/, and the ENSEMBL database http://www.ensembl.org/.
Results
MARPs form antiparallel dimers through their coiled-coil domains
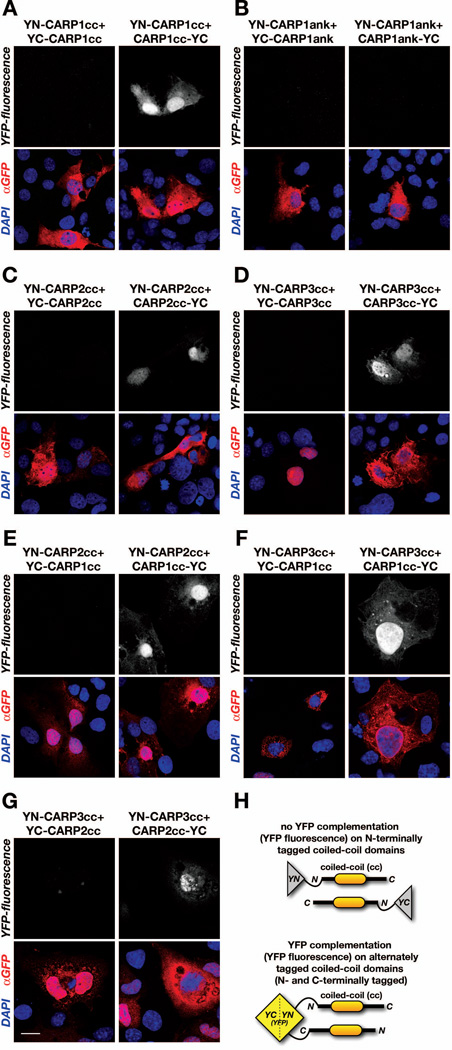
CARP1 (Ankrd1) has been demonstrated to dimerize through its N-terminal coiled-coil domain (Witt et al., 2005). Experiments utilizing yeast two-hybrid assays indicated further that all members of the CARP protein family: CARP1, CARP2 (Arpp/Ankrd2) and CARP3 (Ankrd23/DARP) are capable of forming homo-dimers. We set out to investigate the nature of this interaction more closely. Using protein complementation assays (split-YFP) we confirmed that the coiled-coil domain of CARP1 mediates the dimerization of the protein. Alternate tagging of the YFP halves to either the N-terminus or C-terminus of the coiled-coil domain revealed further that the dimerization occurs in anti-parallel fashion (Figure 2A). We also tested the C-terminal ankyrin-repeat domains and found no association in parallel or anti-parallel fashion in our protein complementation (Figure 2B). These results agree with previously published results (Witt et al., 2005), whereby ankyrin-repeat domains were not involved in the formation of CARP1 dimers in yeast-two hybrid assays.
Figure 2. MARPs form antiparallel homo- and heterodimers.
A. Protein complementation (split-fluorescent protein) assay probing for the nature of the CARP1 homo-dimerization. Cos-1 cells were transfected with CARP1 coiled-coil domains (CARP1cc) tagged at the N-terminus with YFP N-terminal half (YN-CARP1cc) in combination with either CARP1cc tagged at the N-terminus (left panels) or at the C-terminus (right panels) with YFP C-terminal half (YC-CARP1cc or CARP1cc-YC, respectively). Reconstitution of functionally active YFP demonstrated antiparallel dimerization (signal in YFP-fluorescence channel, right panel). B. Protein complementation assay (as in A) testing dimerization of CARP1 ankyrin-repeats (CARP1ank). Cos-1 cells were transfected with YN-CARP1ank in combination with either YC-CARP1ank (left panels) or CARP1ank-YC (right panels). No YFP-fluorescence was observed in transfected cells (top panels), indicating no close association between the two tagged CARP1ank domains. C-D. Protein complementation assay (as in A) probing for homo-dimerization of CARP2 (C) or CARP3 (D) coiled-coil domains (CARP2cc, CARP3cc, respectively). Cos-1 cells were transfected with YN- or YC-tagged CARP2cc (C) or YN- or YC-tagged CARP3cc (D). YFP fluorescence indicated antiparallel association of CARP2cc (C, right panels), as well as antiparallel association of CARP3cc (D, right panel). No YFP signal was found when coiled-coil domains were tagged at the N-terminus with YN or YC (C & D, left panels). E-G. Protein complementation assay (as in A) investigating hetero-dimerization potential between CARP1cc and CARP2cc (E), CARP1cc and CARP3cc (F), or CARP2cc and CARP3cc (G). Similar to observed homo-dimerizations for CARP1cc, CARP2cc or CARP3cc, all investigated coiled-coil domains form antiparallel hetero-dimers, as indicated by a positive signal in the YFP-fluorescence channel (E-G, top-right panels). A-G. Transfection efficiency was validated with a GFP antibody (red in the overlay), and DAPI (blue in overlay) was used as counterstain. Scale bar = 20µm. H. Model for MARP coiled-coil domain interaction, and interpretation of absence (top) or presence of YFP-signal (bottom) in the protein complementation assay.
Next we wondered if the antiparallel association through the coiled-coil domains demonstrated for CARP1 is preserved for the other two members of the MARP family. Using protein complementation, we probed for the dimerization potential and nature of the interaction of the coiled-coil domains of CARP2 and CARP3. Indeed, the coiled-coil domains of both CARP2 and CARP3 are able to form homo-dimers. Similar to the coiled-coil domain for CARP1, the dimer formation for CARP2 and CARP3 occurs in antiparallel fashion (Figures 2C, 2D).
Previous reports demonstrated that MARP hetero-dimerization does not occur in forced yeast-two hybrid assays (Miller et al., 2003; Witt et al., 2005). Nevertheless, we employed protein complementation assays to test for the formation of CARP1-CARP2, CARP1-CARP3 and CARP2-CARP3 hetero-dimers through interaction of the respective coiled-coil domains. We found that all members of the MARP family were indeed capable to form hetero-dimers that were, as shown for the homo-dimers, formed in anti-parallel fashion (Figures 2E–G).
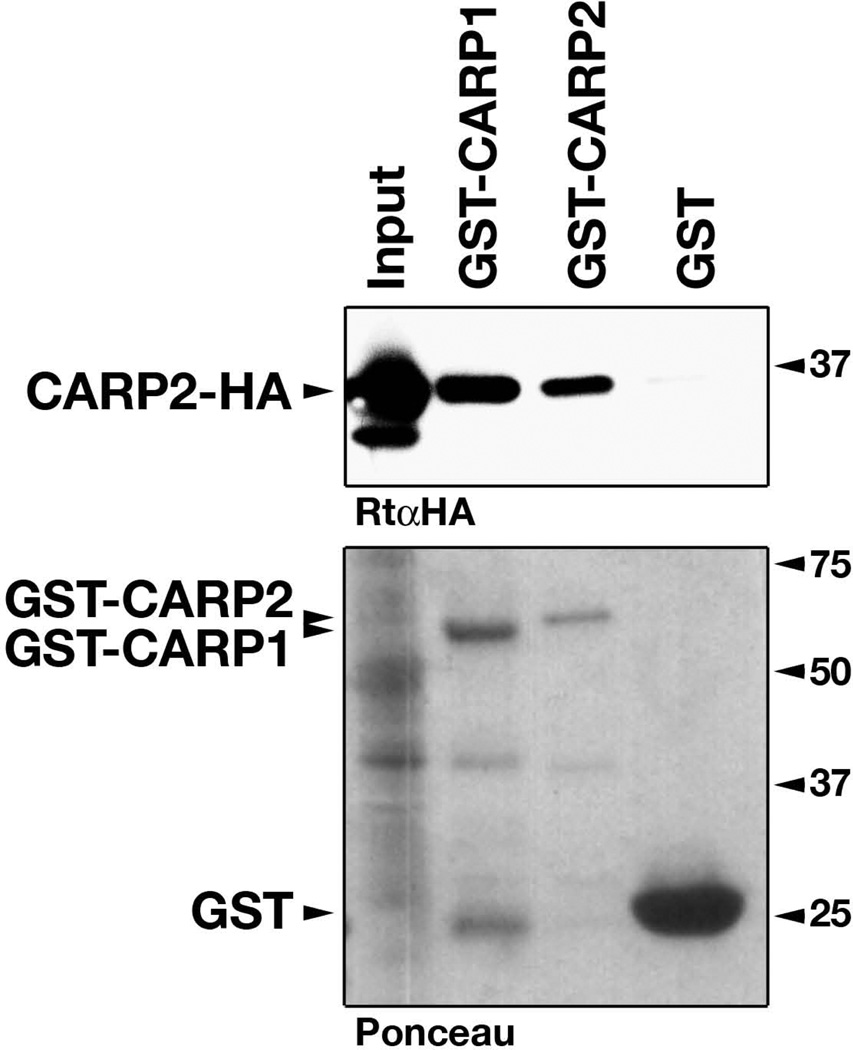
To validate this result, we tested for interaction of CARP2 with either CARP1 or CARP2 in a biochemical interaction assay (GST-pulldown) and observed strong binding of CARP2 with either CARP1 (hetero-dimer) or CARP2 (homo-dimer; Figure 3). In addition, there was no noticeable difference in the intensity of the CARP2 binding to CARP2 or CARP1, which may serve as an indicator of similar binding affinities.
Figure 3. Biochemical evaluation of MARP hetero-dimer formation.
HA-tagged CARP2 (CARP2-HA, upper panel) from transfected Cos-1 cells (Input) was used to probe for interaction with either GST-tagged CARP1, CARP2 or GST itself (visible in the ponceau staining, lower panel). Presence of CARP2-HA in the bound fractions of GST-CARP1 indicates heterodimerization between CARP1 and CARP2. CARP2 homo-dimerization (band in GST-CARP2 lane), and GST alone was used as positive and negative controls, respectively. Molecular weights in kDa.
In summary, results from our protein complementation assay validate the association of MARPs through their N-terminal coiled-coil domains. In contrast to previous studies we further demonstrated the capability of all MARP family members to form hetero-dimers, again through their respective coiled-coil domains. We could also show that the association for all MARPs occurs in antiparallel fashion (Figure 2H), as opposed to a parallel interaction.
CARP1 ankyrin-repeats interact in antiparallel fashion with the N2A-region of titin
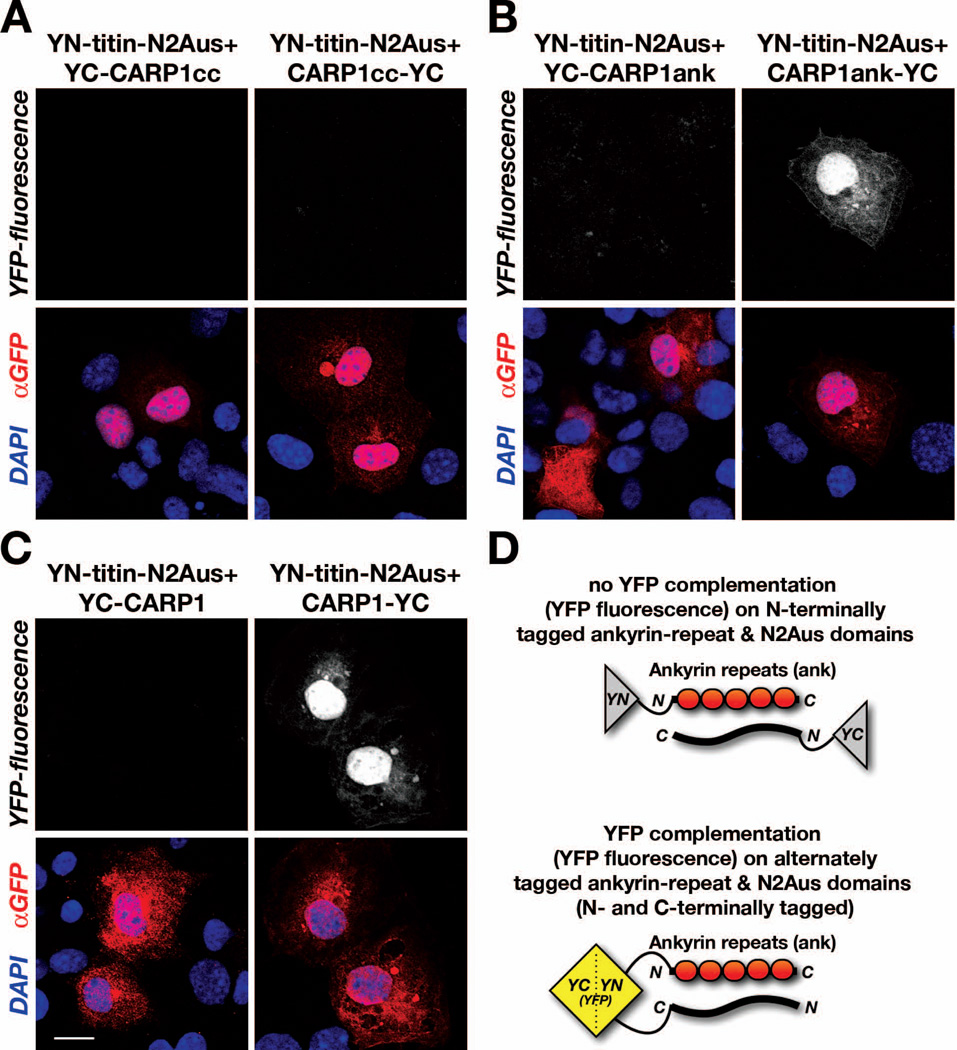
One of the most prominent features of the MARP family is their myofibrillar localization and interaction with the N2A-region of titin in cross-striated muscle cells (Laure et al., 2010; Miller et al., 2003; Witt et al., 2004). Two binding sites within CARP1 have been identified for the interaction with the unique unstructured region in titin-N2A (N2Aus). A highly conserved predominant binding site within CARP1 was mapped to ankyrin-repeat number two of the CARP1 C-terminus. A weaker secondary site was mapped to the unstructured N-terminus of CARP1 (amino acids 21–33) (Miller et al., 2003).
Using protein complementation assays (split-YFP) we tested for the association of either the N-terminal coiled-coil domain or the C-terminal ankyrin-repeats in CARP1 with the minimal binding site in the N2Aus-region of titin. We found no association of the coiled-coil domains with titin-N2Aus in parallel or antiparallel fashion (Figure 4A). However, we were able to confirm the association of CARP1 ankyrin-repeats with titin-N2Aus (Figure 4B). In addition, we were able to show that the interaction of either full-length CARP1 or CARP1 ankyrin-repeat domains with titin-N2Aus occurs in antiparallel fashion (Figures 4B, 4C, right panels; Figure 4D).
Figure 4. CARP1 ankyrin-repeats interact with titin-N2Aus in antiparallel fashion.
A–C. Protein complementation assay (split-fluorescent protein) probing for interaction of titin-N2A unique sequence (N2Aus) with either (A) CARP1 coiled-coil domains (CARP1cc), (B) CARP1 ankyrin-repeats (CARP1ank), or (C) full-length CARP1. A. No association (absence of YFP-fluorescence, top panels) was observed upon cotransfection of YN-titin-N2Aus with either YC-CARP1cc (left panels) or CARP1cc-YC (right panels). B. Reconstitution of YFP (YFP-fluorescence, top-right panel) in Cos-1 cells transfected with YN-titin-N2Aus and CARP1ank-YC (right panel), but not in cells transfected with YN-titin-N2Aus and YC-CARP1ank (left panel), indicated antiparallel association between titin and ankyrin-repeats in CARP1. C. Utilization of full-length CARP1 tagged at the N-terminus (YC-CARP1, left panels) or C-terminus (CARP1-YC) in combination with YN-titin-N2Aus also indicated presence of antiparallel association between titin and CARP1, as judged by YFP-fluorescence (top-right panel).
A–C. Scalebar = 20µm. Transfection efficiency was validated with a GFP antibody (red in the overlay), and DAPI (blue in overlay) was used as counterstain. Scale bar = 20µm.
D. Model for CARP1ank interaction with titin, and interpretation of absence (top) or presence of YFP-signal (bottom) in the protein complementation assay.
MARPs affect titin phosphorylation by PKA
Like all unstructured regions within titin, the N2A-region has been shown to regulate muscle compliance in a phosphorylation dependent manner (Kruger and Linke, 2006). We recently characterized that binding partners for another unstructured region within titin, the N2B–us3 region, can exert changes to the phosphorylation level of this giant sarcomeric protein (Raskin et al., 2012). Using in-vitro kinase assays we set out to investigate, whether binding of MARP to titin-N2A may influence its phosphorylation status.
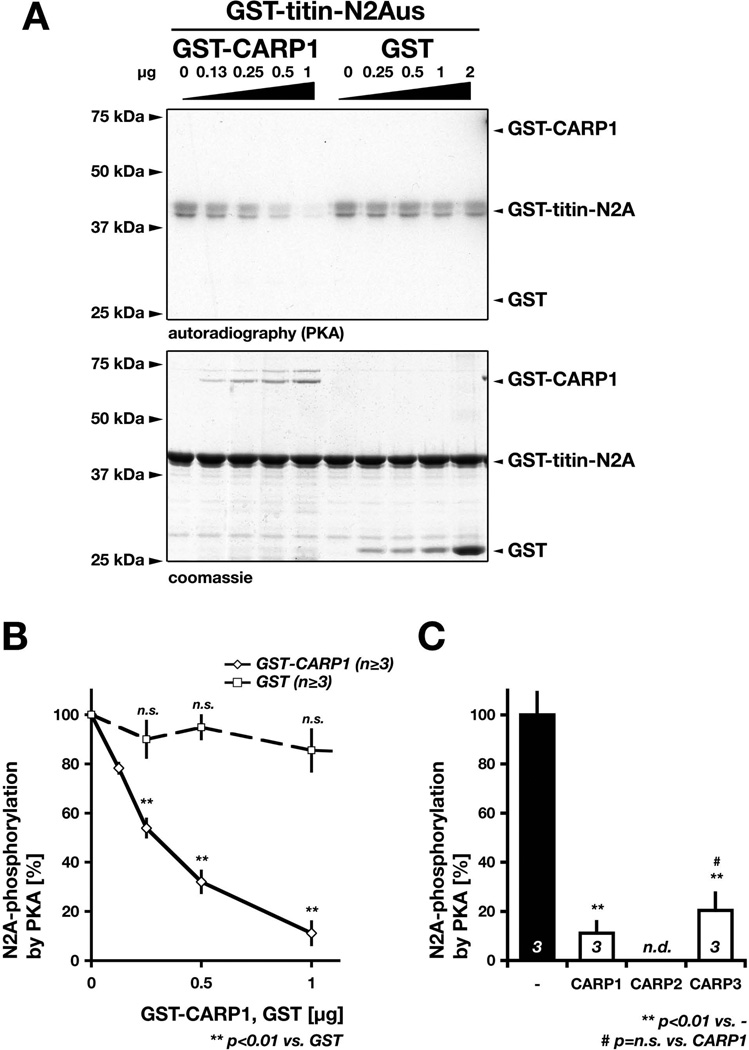
We found that increasing amounts of CARP1 in our kinase assays lead to a marked reduction in titin-N2Aus phosphorylation efficiency by PKA (Figure 5A), while increasing levels of GST had no effect. Remarkably, CARP1 reduced N2Aus-phosphorylation levels by approximately 50% at a ratio of 1/8 (w/w) between CARP1 and titin (Figure 5B). Because binding to titin’s N2A-region is conserved between all members of the MARP family, we also tested the capability of CARP3 to affect titin-N2Aus phosphorylation levels. Intriguingly, CARP3 is capable to reduce titin-N2Aus phosphorylation by PKA to a similar extent as CARP1 (Figure 5C), indicating that all members of the MARP family may exert this effect. This result may also point to a sterical hindrance of the kinase as molecular mechanism of action. MARP binding to titin-N2Aus may mask or potentially engage the phosphorylation site in the interaction, as to affecting the protein kinase directly. A similar mechanism has been recently proposed for FHL action on the ERK2-mediated phosphorylation of titin’s N2B–region (Raskin et al., 2012). In addition, the data indicate that binding of MARP to titin may not be dependent on phosphorylation of titin’s N2Aus-region.
Figure 5. MARPs modulate titin-N2Aus phosphorylation by PKA.
A. In-vitro kinase assay using radioactively labeled [γ-32P]ATP, PKA and GST-titin-N2Aus (2µg) as substrate in combination with increasing amounts of either GST-CARP1 (left side; 0, 0.13, 0.25, 0.5 or 1µg GST-CARP1) or GST (right side; 0, 0.25, 0.5, 1 or 2µg GST). Phosphorylation of titin-N2Aus was assessed through autoradiography (top panel), while presence of proteins was validated through coomassie staining (bottom image). Shown are typical gel-images obtained for the assay. B. Quantification of N2Aus phosphorylation by PKA and effect of increasing amounts of GST-CARP1 (diamond) or GST (square) on N2Aus phosphorylation efficiency by PKA (as in A). Results are expressed as the % of N2A phosphorylation in absence of either GST-CARP1 or GST, which was arbitrarily set to 100%. Sample size (n) and p-values are indicated. Changes in N2Aus phosphorylation in presence of up to 1µg GST are not significant (n.s.) compared to absence of GST. C. Quantification of the GST-N2Aus (2µg) phosphorylation by PKA in absence (-; arbitrarily set to 100% as in B) or presence of either 2µg GST-CARP1 or 2µg GST-CARP3. Influence of GST-CARP2 on N2Aus phosphorylation was not determined (n.d.). Shown are mean values with standard errors. Sample size (n, bottom of each bar graph) and p-values are indicated.
Posttranslational modification of MARPs by PKA and PKC
Bioinformatical analysis of the CARP1 protein sequence revealed several putative protein motifs and sites for posttranslational modifications through protein kinases. Most notably, the sequence analysis revealed five putative protein kinase C (PKC) phosphorylation sites, one presumed cAMP-dependent protein kinase (PKA) and four putative casein kinase II (CKII) phosphorylation sites (Jeyaseelan et al., 1997). While investigating the effect of MARP family members on titin-N2A phosphorylation, we noticed that CARP1 was not identified as a substrate for PKA (Figure 5A). However, upon testing of all members of the MARP family through in-vitro kinase assays, we readily detected phosphorylation of CARP2 by PKA (Figure 6A). Indeed, a contaminating truncated form of GST-CARP2 that presumably consists of an N-terminal protein fragment (asterisk, Figure 6A) located the phosphorylation site within the first 20kDa of CARP2. Similar to CARP1, CARP3 was not phosphorylated by PKA.
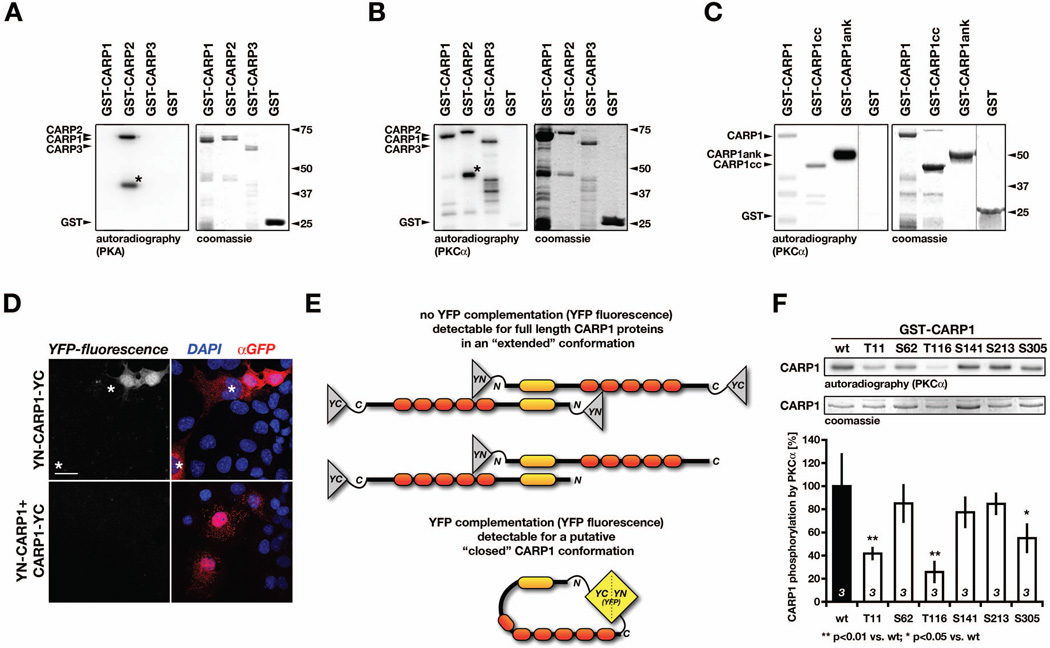
Figure 6. MARP phosphorylation by PKA and PKC.
A–B. Phosphorylation of either 2µg GST-CARP1, GST-CARP2, GST-CARP3 or GST by (A) PKA or (B) PKCα in an in-vitro kinase assay using [γ-32P]ATP. Shown are representative images for the autoradiography (left panel) identifying phosphorylated proteins, as well as the coomassie stained gels (right panel). A. PKA only phosphorylated full-length or a truncated (asterisk) GST-CARP2, while GST-CARP1, GST-CARP3 and GST were not recognized as PKA substrates. B. PKCα phosphorylated GST-CARP1, full-length or a truncated (asterisk) GST-CARP2 and GST-CARP3, while GST alone was not phosphorylated. C. In vitro-kinase assay using PKCα with [γ-32P]ATP in combination with either 2µg full-length GST-CARP1, GST-CARP1 coiled-coil domain (CARP1cc), GST-CARP1 ankyrin-repeats (CARP1ank) or GST alone. Shown are representative images for the autoradiography (left panel) identifying phosphorylated proteins, as well as the coomassie stained gel (right panel). Please note that the dotted lines between lanes denotes that samples were run on the same gel but were non-contiguous. D. Protein complementation assay (split-YFP) in Cos-1 cells transfected with full-length CARP1 tagged either at both ends with YN and YC (YN-CARP1-YC; top panels) or transfected with a combination of N-terminally and C-terminally tagged CARP1 (YN-CARP1 + CARP1-YC). Occasionally observed YFP-fluorescence for YN-CARP1-YC transfected cells (top-left panel) may indicate conformational change within CARP1. Note transfected, but YFP negative cells (asterisks). Cos-1 cells co-transfected with YN-CARP1 and CARP1-YC displayed no YFP-fluorescence and served as negative controls (bottom panels). E. Schematic interpretation of (D), showing no YFP complementation for CARP1 tagged at both ends in “extended” conformation (YN-CARP1-YC) or in case of the negative control (YN-CARP1 + CARP1-YC; top panel), while the putative “closed” CARP1 conformation leads to close vicinity of CARP1 N- and C-terminus, and reconstitution of functional YFP (bottom panel). F. Identification of major PKCα phosphorylation sites within CARP1. In-vitro kinase assay using PKCα with [γ-32P]ATP in combination with either 2µg wildtype (wt), Thr11Ala (T11), Ser62Ala (S62), Thr116Ala (T116), Ser141Ala (S141), Ser213Ala (S213) or Ser305Ala (S305) GST-CARP1. Shown are representative images for the autoradiography (top panel) identifying phosphorylated proteins, as well as the coomassie stained gel (middle panel). Quantification of mutant CARP1 phosphorylation compared to wildtype CARP1, which was arbitrarily set to 100% (bottom panel). Shown are mean values, with sample sizes (n-values, bottom of the bar graph) as well as p-values indicated.
Although we were unable to confirm the putative cAMP-dependent protein kinase site in CARP1 (Jeyaseelan et al., 1997), we tested if CARP1, CARP2 or CARP3 may be subjected to phosphorylation by protein kinase C (PKCα), as previously predicted. Using in-vitro kinase assays with PKCα, we were able to show phosphorylation of all three MARP family members (Figure 6B). CARP2 displayed again the characteristic phosphorylation of its truncated form (asterisk Figure 6B), indicating presence of the PKC phosphorylation site within its N-terminal 20kDa. To narrow down PKC phosphorylation sites within CARP1, we investigated full-length CARP1, N-terminal CARP1 coiled-coil domains (CARP1cc) and C-terminal ankyrin-repeats (CARP1ank) by in-vitro kinase assays (Figure 6C). Full-length CARP1 and all investigated CARP1 fragments were phosphorylated, suggesting presence of multiple PKC sites throughout CARP1. The phosphorylation intensity of CARP1ank however, was more intense in the assay compared to full-length CARP1 or CARP1cc. This result may indicate conformational/sterical hindrance of the phosphorylation site(s) located in the C-terminal ankyrin-repeats by the CARP1 N-terminus and exposure of a cryptic phosphorylation site within CARP1 upon loss/truncation of the coiled-coil domains or conformational change of the protein. We further investigated the possibility that CARP1 may exist in different conformations within cells by using the protein complementation assay. We employed full-length CARP1 that has been tagged on both ends with YFP halves (YN-CARP1-YC) and observed occasionally functionally active YFP (Figure 6D, upper panels), indicating close vicinity of the CARP1 N- and C-terminus (Figure 6E, lower panel). Intriguingly, neighboring cells that also expressed the construct displayed no complementation (asterisks in Figure 6D), suggesting the presence of a hinge region that may be susceptible for putative conformational change (Figure 6E lower panel). A negative control employing alternately tagged full-length CARP1 never displayed functional complementation of YFP (Figure 6D, lower panels). This excludes the possibility of complementation through two independently tagged CARP1 proteins (Figure 6E, upper panel).
Next, we investigated what sites within CARP1 may serve as substrates for PKCα. We expressed full-length CARP1 serine-alanine or threonine-alanine mutants of the putative PKC sites previously identified through sequence analysis: Thr11, Ser62, Ser141, Ser213 and Ser305 (Jeyaseelan et al., 1997). In addition, we constructed a threonine-alanine mutant of Thr116, a residue recently implicated in the development of familial dilated cardiomyopathy (DCM) in humans (Badi et al., 2009; Duboscq-Bidot et al., 2009). Analysis of wildtype and mutant CARP1 proteins in PKCα in-vitro kinase assays revealed decreased phosphorylation for all investigated mutants, with Thr11Ala (T11), Thr116Ala (T116) and Ser305Ala (S305) displaying significantly reduced phosphorylation efficiencies compared to wildtype CARP1 (Figure 6F). No investigated mutant showed complete absence of phosphorylation by PKC, again indicating the presence of several minor and major phosphorylation sites for PKCα within CARP1.
In summary, we identified CARP2 as a substrate for PKA and all members of the MARP family as substrates for PKCα through in-vitro kinase assays. An N-terminally truncated CARP1 consisting only of the ankyrin-repeats displayed markedly increased CARP1 phosphorylation compared to either the full-length protein, or CARP1 consisting of the coiled-coil domain only. This indicated the presence of a cryptic phosphorylation site that may be only exposed upon a conformational change of CARP1. Protein complementation of N- and C-terminally tagged full-length CARP1 supported the hypothesis that CARP1 may indeed undergo conformational changes. In addition, we identified several PKC sites within CARP1, as CARP1 phosphorylation by PKCα was significantly reduced upon mutagenesis of Thr11, Thr116 or Ser305 to alanine.
Discussion
We investigated several major functional aspects of MARP action: their dimerization and interaction with the N2Aus-region within titin, and their potential for posttranslational modifications by protein kinases.
Dimer formation
The dimer formation promoted through the N-terminal coiled-coil domains is intrinsic to all members of the MARP family. Up to now, only the formation of homo-dimers has been validated through experimental results from forced yeast-two hybrid assays (Miller et al., 2003; Witt et al., 2005). Although formation of hetero-dimers was tested, no positive interaction as measured by yeast growth on selective plates was found (Witt et al., 2005). While investigating the nature of the dimer through protein complementation, we also tested for the putative formation of hetero-dimers. Our results indicated that CARP1-CARP2, CARP1-CARP3 and CARP1-CARP3 interactions are indeed possible through binding of the respective coiled-coil domains. This result was validated for the CARP1-CARP2 hetero-dimer through a pulldown assay. Indeed, overlapping myofibrillar localization of MARPs and their interaction with the titin N2A-region, their co-expression in cardiac and skeletal muscle, as well as nearly identical domain patterns and high degree of sequence conservation may further substantiate hetero-dimer formation. Well-known limitations of the yeast-two hybrid system in testing for protein-protein interactions may be one of the reasons these interactions were not previously detected (Van Criekinge and Beyaert, 1999).
Structurally, our protein complementation assays indicated antiparallel interaction between two coiled-coil domains of proteins of the MARP family. Both, parallel and antiparallel coiled-coil domain interactions have been described, and can be found in crystal structures of proteins containing this domain type. Secondary structure predictions of coiled-coil domains in MARPs indicate the presence of relatively short (approximately 10–24 residues), uninterrupted alpha-helices. Computation of the oligomeric state probability of MARP coiled-coil sequences using LOGICOIL (Vincent et al., 2012) correctly predicted occurrence of antiparallel dimers only for CARP2. Raw scores for CARP1 antiparallel dimers (score=1) were lower compared to a predicted tetrameric state (score=1.2) or parallel dimer (score=1.07). CARP3 antiparallel dimer formation was also predicted with lower probability (score=1.01) compared to formation of tetramers (score=1.12). Helical wheel projections (not shown) indicate the involvement of hydrophobic residues in the formation of putative binding interfaces, namely Leu70, Leu81 and Leu84 in CARP1; Val38, Ile45 and Leu49 in CARP2; as well as Leu69 and Leu72 in CARP3. Dimeric coiled-coils often display a characteristic heptad repeat, in which hydrophobic residues are typically 3–4 residues apart (Grigoryan and Keating, 2008; Lupas, 1996). However, this regular pattern may be disrupted upon distortion of the alpha-helical turns, resulting in patterns that position the sidechains after two turns (or seven residues; (Lupas, 1996)). Moreover, the pattern of hydrophobic residues may be interrupted with charged side-chains, as reported for myosin coiled-coil interactions (Straussman et al., 2005; Straussman et al., 2007). Mutational analysis of MARP coiled-coil domain key-residues and the analysis of crystal structures obtained for homo- or hetero-dimers may solve the issue of how these domains interact and which residues are involved in the formation of the binding interface.
MARPs as titin-filament cross-linker
Protein complementation also demonstrated that the interaction of CARP1 ankyrin-repeats with the unstructured region of titin-N2A occurs in antiparallel fashion. This result in combination with the overall antiparallel shape of the MARP dimer reveals a novel concept for the biological functions of MARPs: as titin-filament cross-linkers in the sarcomeric I-band (Figure 7). Filament cross-linkers have been well established as important for the structural integrity of the sarcomere at the Z-disc (α-actinin (Young et al., 1998), telethonin (Zou et al., 2006)), A-band (myosin binding protein-C (Welikson and Fischman, 2002)) and M-band (myomesin protein family, (Agarkova and Perriard, 2005; Lange et al., 2005)). However, the idea of a filament cross-linker at the I-band may be controversial, particularly with regards to the flexible nature of the unstructured region that binds to MARPs.
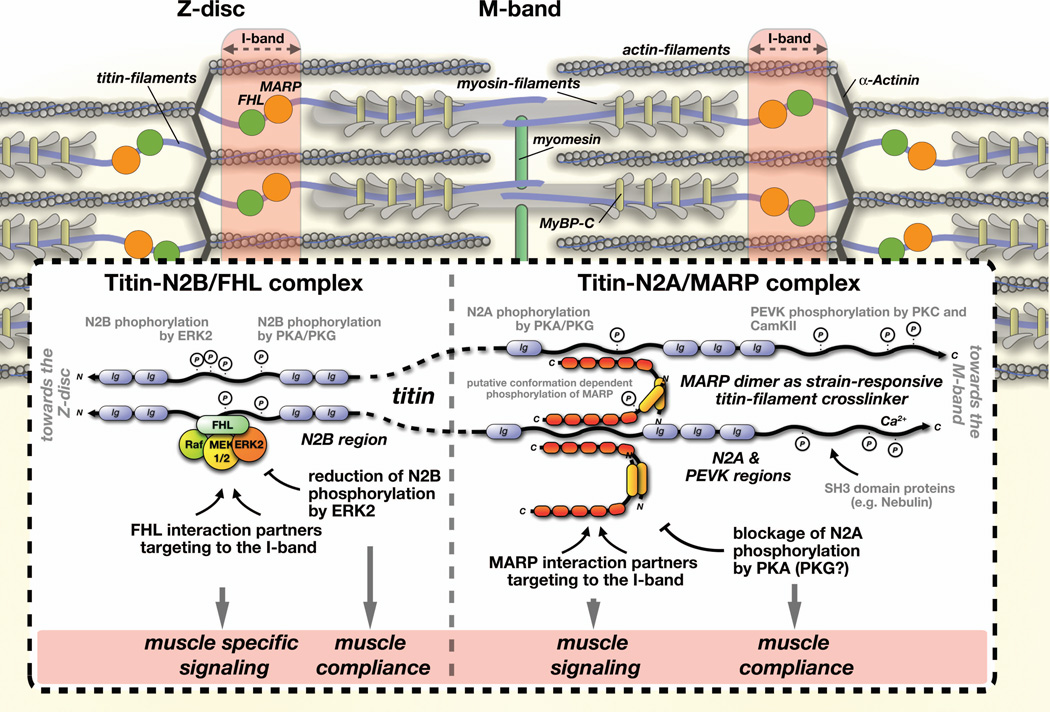
Figure 7. Titin-linked mechano-responsive complexes at the I-band.
Schematic representation of mechano-responsive complexes linked to the N2B- and N2A-regions of titin and their dual roles in modulating muscle specific signaling and altering muscle compliance. Note the similarities between functions of FHL and muscle ankyrin-repeat proteins. Both have been shown to bind to a large number of interaction partners, potentially targeting these proteins to the sarcomeric I-band. In addition, MARPs and FHL proteins have been demonstrated to shuttle to the nucleus where they may act as transcriptional modifiers.
Structurally, a MARP antiparallel dimer may connect two adjacent titin molecules, thereby serving as titin-filament cross-linker with strain-sensing potential (through exposure of cryptic phosphorylation sites). Binding of FHL to titin-N2B, and interaction of CARP1 with titin-N2A have been demonstrated to either reduce, or block phosphorylation of titin, potentially resulting in changes to muscle compliance. Because MARPs and FHL proteins are dysregulated in many forms of skeletal and cardiac myopathies, their effect on muscle specific signaling and physiology may have ramifications for the disease etiology.
It would be interesting to investigate whether MARPs interact differentially to stretched vs. relaxed titin-N2A. The observation that all members of this protein family alter their subcellular localization or cellular signaling pathways upon muscle strain (Miller et al., 2003; Mohamed et al., 2010) may support this hypothesis.
Another intriguing theory would be that MARP binding may directly affect compliance of the N2A-region (besides their modulation of titin phosphorylation). Elasticity measurement of recombinant titin constructs in presence or absence of MARP by atomic-force microscopy (AFM) may reveal intriguing insights into both hypotheses.
MARPs as modulators of muscle compliance
The I-band portion of titin contains two more elastic elements in addition to the N2A-region: the N2B- and PEVK-regions (Kruger and Linke, 2011). All three regions have been demonstrated to influence muscle compliance by acting as spring-like elements that extend upon stretch and exert passive retractive force (Linke and Grutzner, 2008). Hence, alternate splice variants of titin that insert different combinations of these regions into the protein have been demonstrated as adaptive response for cardiac muscle cells to meet cardiac development and counteract physiological strain (Kruger et al., 2008; Voelkel and Linke, 2011). A second mechanism that influences compliance of titin is the phosphorylation of serine and threonine residues within titin’s elastic regions by protein kinases, such as cAMP- and cGMP-dependent protein kinases (PKA and PKG, respectively; (Kruger and Linke, 2006; Kruger et al., 2009)), protein kinase-C (PKC; (Hidalgo et al., 2009)), Ca2+/calmodulin-dependent protein kinase II (CamKII; (Hamdani et al., 2013)) and mitogen-activated protein kinase (MAPK, ERK1/2; (Raskin et al., 2012)). We recently demonstrated that binding of FHL proteins to the N2B–region is able to influence titin phosphorylation by ERK2 (Raskin et al., 2012). Our results on the effect that MARPs exert on the phosphorylation of titin-N2Aus by PKA display surprising similarities between protein complexes at the N2B- and N2A-regions (Figure 7). Both MARP and FHL inhibit phosphorylation of titin by PKA or ERK2, respectively.
Physiologically it has been demonstrated for titin-N2A and -N2B that phosphorylation by PKA or PKG leads to an increase in muscle compliance (decrease in titin-based passive stiffness;(Kruger and Linke, 2006; Kruger et al., 2009)). Complete absence of MARP from cross-striated muscles should therefore be reflected by increased muscle compliance (decreased titin-based muscle-stiffness), caused by putative hyper-phosphorylation of titin at the N2A-region. Indeed, triple MARP knockout fibers were found to be “less stiff, tended to have longer resting sarcomere lengths, and expressed a longer isoform of titin” (Barash et al., 2007). The mechanistic insight and hypothesis for MARP functions grant a reevaluation of the MARP knockout mice, particularly with respect to the surprisingly mild cardiac and skeletal muscle phenotype. In addition, the biological implications on muscle physiology are particularly interesting on the pathophysiological level, as MARP (Arber et al., 1997), activity of protein kinases (like PKCα (Simonis et al., 2007)), and titin splice pattern (Nagueh et al., 2004) or phosphorylation (Kruger et al., 2010) are often altered in many forms of cardiac and skeletal muscle myopathies.
MARP phosphorylation
Posttranslational modifications are generally thought to alter protein functions. Computational analysis of the CARP1 protein sequence revealed a surprising abundance of putative phosphorylation sites (Jeyaseelan et al., 1997). While phosphorylation of CARP1 has not been validated experimentally, CARP2 was characterized to be phosphorylated at Ser99 by Akt2/PKB (Cenni et al., 2011). The biological role for protein phosphorylation at this site was found in the repression of myoblast differentiation in response to cellular stress. In contrast, nothing is known about posttranslational modifications for the third member of the MARP family.
Here we identified CARP2 as a novel substrate for PKA, while CARP1 and CARP3 were not phosphorylated in vitro by this protein kinase. Results of a truncated form of CARP2 indicated presence of the PKA phosphorylation site within the proteins N-terminus. In addition, we could demonstrate that all members of the MARP family were phosphorylated by PKCα, likely at multiple sites within each protein. Similar to its phosphorylation by PKA, the truncated version of CARP2 indicated presence of a major PKCα phosphorylation site within the CARP2 N-terminus. Mutational analysis of predicted PKC phosphorylation sites within CARP1 identified Thr11, Thr116 and Ser305 as major targets for this protein kinase. Particularly Thr116, whose mutation to methionine was found to be linked to the development of dilated cardiomyopathy in humans may require further analysis. Curiously, truncated versions of CARP1 suggested presence of a cryptic PKCα phosphorylation site within the CARP1 C-terminus, which became only exposed upon loss of the N-terminal coiled-coil domain. This lead us to the hypothesis that CARP1 may undergo a conformational change, a theory that gained some support by our protein complementation assay. It is intriguing to speculate if a conformational change of CARP1 caused by mechanical strain may result in its increased phosphorylation due to exposed cryptic phosphorylation sites. Phosphorylation of MARPs may then serve as a trigger for their differential subcellular localization and cellular activity in stretched vs. static cross-striated muscle cells. Although this hypothesis needs further experimental validation and cell-biological analysis, the proposed cross-linker function of MARP dimers at the N2A-region locate it ideally to serve as mechano-sensitive/responsive element. Future studies that investigate changes to MARP phosphorylation by PKC and PKA in vivo and analyses that attempt to correlate MARP phosphorylation states with healthy and diseased muscles may shed further light on the physiological relevance for their posttranslational modification.
Acknowledgements
We would like to express our gratitude to Meagan Wu for help with the sample preparation and imaging, Jennifer Santini and the UCSD Microscopy Core Facility (grant number: P30 NS047101) and Enrico Girardi (La Jolla Institute for Allergy and Immunology) for technical assistance, as well as Elisabeth Ehler for critical reading of the manuscript. S.L. received funding from the NIH/NHLBI (HL107744). JC was supported by funds received from the NIH/NHLBI (HL066100).
Literature cited
- Agarkova I, Perriard JC. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J, Jr., Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Arimura T, Bos JM, Sato A, Kubo T, Okamoto H, Nishi H, Harada H, Koga Y, Moulik M, Doi YL, Towbin JA, Ackerman MJ, Kimura A. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- Badi I, Cinquetti R, Frascoli M, Parolini C, Chiesa G, Taramelli R, Acquati F. Intracellular ANKRD1 protein levels are regulated by 26S proteasome-mediated degradation. FEBS Lett. 2009;583:2486–2492. doi: 10.1016/j.febslet.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Bang ML, Mudry RE, McElhinny AS, Trombitas K, Geach AJ, Yamasaki R, Sorimachi H, Granzier H, Gregorio CC, Labeit S. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol. 2001;153:413–427. doi: 10.1083/jcb.153.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash IA, Bang ML, Mathew L, Greaser ML, Chen J, Lieber RL. Structural and regulatory roles of muscle ankyrin repeat protein family in skeletal muscle. Am J Physiol Cell Physiol. 2007;293:C218–C227. doi: 10.1152/ajpcell.00055.2007. [DOI] [PubMed] [Google Scholar]
- Belgrano A, Rakicevic L, Mittempergher L, Campanaro S, Martinelli VC, Mouly V, Valle G, Kojic S, Faulkner G. Multi-tasking role of the mechanosensing protein Ankrd2 in the signaling network of striated muscle. PLoS One. 2011;6:e25519. doi: 10.1371/journal.pone.0025519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenni V, Bavelloni A, Beretti F, Tagliavini F, Manzoli L, Lattanzi G, Maraldi NM, Cocco L, Marmiroli S. Ankrd2/ARPP is a novel Akt2 specific substrate and regulates myogenic differentiation upon cellular exposure to H(2)O(2) Mol Biol Cell. 2011;22:2946–2956. doi: 10.1091/mbc.E10-11-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocini C, Arimura T, Reischmann S, Eder A, Braren I, Hansen A, Eschenhagen T, Kimura A, Carrier L. Impact of ANKRD1 mutations associated with hypertrophic cardiomyopathy on contraction parameters of engineered heart tissue. Basic Res Cardiol. 2013;108:349. doi: 10.1007/s00395-013-0349-x. [DOI] [PubMed] [Google Scholar]
- Duboscq-Bidot L, Charron P, Ruppert V, Fauchier L, Richter A, Tavazzi L, Arbustini E, Wichter T, Maisch B, Komajda M, Isnard R, Villard E. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J. 2009;30:2128–2136. doi: 10.1093/eurheartj/ehp225. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdani N, Krysiak J, Kreusser MM, Neef S, Dos Remedios CG, Maier LS, Kruger M, Backs J, Linke WA. Crucial role for Ca2(+)/calmodulin-dependent protein kinase-II in regulating diastolic stress of normal and failing hearts via titin phosphorylation. Circ Res. 2013;112:664–674. doi: 10.1161/CIRCRESAHA.111.300105. [DOI] [PubMed] [Google Scholar]
- Hayashi C, Ono Y, Doi N, Kitamura F, Tagami M, Mineki R, Arai T, Taguchi H, Yanagida M, Hirner S, Labeit D, Labeit S, Sorimachi H. Multiple molecular interactions implicate the connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J Biol Chem. 2008;283:14801–14814. doi: 10.1074/jbc.M708262200. [DOI] [PubMed] [Google Scholar]
- Hidalgo C, Hudson B, Bogomolovas J, Zhu Y, Anderson B, Greaser M, Labeit S, Granzier H. PKC phosphorylation of titin’s PEVK element: a novel and conserved pathway for modulating myocardial stiffness. Circ Res. 2009;105:631–638. doi: 10.1161/CIRCRESAHA.109.198465. 617 p following 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Emoto N, Matsuo M, Yokoyama M. Molecular identification and characterization of a novel nuclear protein whose expression is up-regulated in insulin-resistant animals. J Biol Chem. 2003;278:3514–3520. doi: 10.1074/jbc.M204563200. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan R, Poizat C, Baker RK, Abdishoo S, Isterabadi LB, Lyons GE, Kedes L. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem. 1997;272:22800–22808. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- Ju W, Valencia CA, Pang H, Ke Y, Gao W, Dong B, Liu R. Proteome-wide identification of family member-specific natural substrate repertoire of caspases. Proc Natl Acad Sci U S A. 2007;104:14294–14299. doi: 10.1073/pnas.0702251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic S, Medeot E, Guccione E, Krmac H, Zara I, Martinelli V, Valle G, Faulkner G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J Mol Biol. 2004;339:313–325. doi: 10.1016/j.jmb.2004.03.071. [DOI] [PubMed] [Google Scholar]
- Kojic S, Nestorovic A, Rakicevic L, Belgrano A, Stankovic M, Divac A, Faulkner G. A novel role for cardiac ankyrin repeat protein Ankrd1/CARP as a co-activator of the p53 tumor suppressor protein. Arch Biochem Biophys. 2010;502:60–67. doi: 10.1016/j.abb.2010.06.029. [DOI] [PubMed] [Google Scholar]
- Kojic S, Radojkovic D, Faulkner G. Muscle ankyrin repeat proteins: their role in striated muscle function in health and disease. Crit Rev Clin Lab Sci. 2011;48:269–294. doi: 10.3109/10408363.2011.643857. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Linke WA. Protein kinase-A phosphorylates titin in human heart muscle and reduces myofibrillar passive tension. J Muscle Res Cell Motil. 2006;27:435–444. doi: 10.1007/s10974-006-9090-5. [DOI] [PubMed] [Google Scholar]
- Kruger M, Sachse C, Zimmermann WH, Eschenhagen T, Klede S, Linke WA. Thyroid hormone regulates developmental titin isoform transitions via the phosphatidylinositol-3-kinase/ AKT pathway. Circ Res. 2008;102:439–447. doi: 10.1161/CIRCRESAHA.107.162719. [DOI] [PubMed] [Google Scholar]
- Kruger M, Kotter S, Grutzner A, Lang P, Andresen C, Redfield MM, Butt E, dos Remedios CG, Linke WA. Protein kinase G modulates human myocardial passive stiffness by phosphorylation of the titin springs. Circ Res. 2009;104:87–94. doi: 10.1161/CIRCRESAHA.108.184408. [DOI] [PubMed] [Google Scholar]
- Kruger M, Babicz K, von Frieling-Salewsky M, Linke WA. Insulin signaling regulates cardiac titin properties in heart development and diabetic cardiomyopathy. J Mol Cell Cardiol. 2010;48:910–916. doi: 10.1016/j.yjmcc.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Kruger M, Linke WA. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J Biol Chem. 2011;286:9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- Lange S, Himmel M, Auerbach D, Agarkova I, Hayess K, Furst DO, Perriard JC, Ehler E. Dimerisation of myomesin: implications for the structure of the sarcomeric M-band. J Mol Biol. 2005;345:289–298. doi: 10.1016/j.jmb.2004.10.040. [DOI] [PubMed] [Google Scholar]
- Lange S, Ehler E, Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Lange S, Perera S, Teh P, Chen J. Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover. Mol Biol Cell. 2012;23:2490–2504. doi: 10.1091/mbc.E12-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laure L, Daniele N, Suel L, Marchand S, Aubert S, Bourg N, Roudaut C, Duguez S, Bartoli M, Richard I. A new pathway encompassing calpain 3 and its newly identified substrate cardiac ankyrin repeat protein is involved in the regulation of the nuclear factor-kappaB pathway in skeletal muscle. FEBS J. 2010;277:4322–4337. doi: 10.1111/j.1742-4658.2010.07820.x. [DOI] [PubMed] [Google Scholar]
- LeWinter MM, Granzier H. Cardiac titin: a multifunctional giant. Circulation. 2010;121:2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA, Grutzner A. Pulling single molecules of titin by AFM--recent advances and physiological implications. Pflugers Arch. 2008;456:101–115. doi: 10.1007/s00424-007-0389-x. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Mayans O, van der Ven PF, Wilm M, Mues A, Young P, Furst DO, Wilmanns M, Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Mikhailov AT, Torrado M. The enigmatic role of the ankyrin repeat domain 1 gene in heart development and disease. Int J Dev Biol. 2008;52:811–821. doi: 10.1387/ijdb.082655am. [DOI] [PubMed] [Google Scholar]
- Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, Granzier H, McElhinny AS, Gregorio CC, Labeit S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003;333:951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Mohamed JS, Lopez MA, Cox GA, Boriek AM. Anisotropic regulation of Ankrd2 gene expression in skeletal muscle by mechanical stretch. FASEB J. 2010;24:3330–3340. doi: 10.1096/fj.10-158386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulik M, Vatta M, Witt SH, Arola AM, Murphy RT, McKenna WJ, Boriek AM, Oka K, Labeit S, Bowles NE, Arimura T, Kimura A, Towbin JA. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol. 2009;54:325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- Pallavicini A, Kojic S, Bean C, Vainzof M, Salamon M, Ievolella C, Bortoletto G, Pacchioni B, Zatz M, Lanfranchi G, Faulkner G, Valle G. Characterization of human skeletal muscle Ankrd2. Biochem Biophys Res Commun. 2001;285:378–386. doi: 10.1006/bbrc.2001.5131. [DOI] [PubMed] [Google Scholar]
- Raskin A, Lange S, Banares K, Lyon RC, Zieseniss A, Lee LK, Yamazaki KG, Granzier HL, Gregorio CC, McCulloch AD, Omens JH, Sheikh F. A novel mechanism involving four-and-a-half LIM domain protein-1 and extracellular signal-regulated kinase-2 regulates titin phosphorylation and mechanics. J Biol Chem. 2012;287:29273–29284. doi: 10.1074/jbc.M112.372839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Raskin A, Chu PH, Lange S, Domenighetti AA, Zheng M, Liang X, Zhang T, Yajima T, Gu Y, Dalton ND, Mahata SK, Dorn GW, 2nd, Brown JH, Peterson KL, Omens JH, McCulloch AD, Chen J. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118:3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis G, Briem SK, Schoen SP, Bock M, Marquetant R, Strasser RH. Protein kinase C in the human heart: differential regulation of the isoforms in aortic stenosis or dilated cardiomyopathy. Mol Cell Biochem. 2007;305:103–111. doi: 10.1007/s11010-007-9533-3. [DOI] [PubMed] [Google Scholar]
- Straussman R, Squire JM, Ben-Ya’acov A, Ravid S. Skip residues and charge interactions in myosin II coiled-coils: implications for molecular packing. J Mol Biol. 2005;353:613–628. doi: 10.1016/j.jmb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Straussman R, Ben-Ya’acov A, Woolfson DN, Ravid S. Kinking the coiled coil--negatively charged residues at the coiled-coil interface. J Mol Biol. 2007;366:1232–1242. doi: 10.1016/j.jmb.2006.11.083. [DOI] [PubMed] [Google Scholar]
- Torrado M, Lopez E, Centeno A, Castro-Beiras A, Mikhailov AT. Left-right asymmetric ventricular expression of CARP in the piglet heart: regional response to experimental heart failure. Eur J Heart Fail. 2004;6:161–172. doi: 10.1016/j.ejheart.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Torrado M, Nespereira B, Lopez E, Centeno A, Castro-Beiras A, Mikhailov AT. ANKRD1 specifically binds CASQ2 in heart extracts and both proteins are co-enriched in piglet cardiac Purkinje cells. J Mol Cell Cardiol. 2005;38:353–365. doi: 10.1016/j.yjmcc.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Torrado M, Nespereira B, Bouzamayor Y, Centeno A, Lopez E, Mikhailov AT. Differential atrial versus ventricular ANKRD1 gene expression is oppositely regulated at diastolic heart failure. FEBS Lett. 2006;580:4182–4187. doi: 10.1016/j.febslet.2006.06.073. [DOI] [PubMed] [Google Scholar]
- Torrado M, Iglesias R, Nespereira B, Centeno A, Lopez E, Mikhailov AT. Intron retention generates ANKRD1 splice variants that are co-regulated with the main transcript in normal and failing myocardium. Gene. 2009;440:28–41. doi: 10.1016/j.gene.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Van Criekinge W, Beyaert R. Yeast Two-Hybrid: State of the Art. Biol Proced Online. 1999;2:1–38. doi: 10.1251/bpo16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo PF, Mahtab EA, Wisse LJ, Hou J, Grosveld F, Suske G, Philipsen S, Gittenberger-de Groot AC. Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol Cell Biol. 2007;27:8571–8582. doi: 10.1128/MCB.01350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent TL, Green PJ, Woolfson DN. LOGICOIL--multi-state prediction of coiled-coil oligomeric state. Bioinformatics. 2012;29:69–76. doi: 10.1093/bioinformatics/bts648. [DOI] [PubMed] [Google Scholar]
- Voelkel T, Linke WA. Conformation-regulated mechanosensory control via titin domains in cardiac muscle. Pflugers Arch. 2011;462:143–154. doi: 10.1007/s00424-011-0938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welikson RE, Fischman DA. The C-terminal IgI domains of myosin-binding proteins C and H (MyBP-C and MyBP-H) are both necessary and sufficient for the intracellular crosslinking of sarcomeric myosin in transfected non-muscle cells. J Cell Sci. 2002;115:3517–3526. doi: 10.1242/jcs.115.17.3517. [DOI] [PubMed] [Google Scholar]
- Witt CC, Ono Y, Puschmann E, McNabb M, Wu Y, Gotthardt M, Witt SH, Haak M, Labeit D, Gregorio CC, Sorimachi H, Granzier H, Labeit S. Induction and myofibrillar targeting of CARP, and suppression of the Nkx2.5 pathway in the MDM mouse with impaired titin-based signaling. J Mol Biol. 2004;336:145–154. doi: 10.1016/j.jmb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt SH, Labeit D, Granzier H, Labeit S, Witt CC. Dimerization of the cardiac ankyrin protein CARP: implications for MARP titin-based signaling. J Muscle Res Cell Motil. 2005;26:401–408. doi: 10.1007/s10974-005-9022-9. [DOI] [PubMed] [Google Scholar]
- Young P, Ferguson C, Banuelos S, Gautel M. Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of alpha-actinin. EMBO J. 1998;17:1614–1624. doi: 10.1093/emboj/17.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Pinotsis N, Lange S, Song YH, Popov A, Mavridis I, Mayans OM, Gautel M, Wilmanns M. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
- Zou Y, Evans S, Chen J, Kuo HC, Harvey RP, Chien KR. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2–5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]