Abstract
There is substantial interest in the role of rare genetic variants in the etiology of complex human diseases. Several gene-based tests have been developed to simultaneously analyze multiple rare variants for association with phenotypic traits. The tests can largely be partitioned into two classes – ‘burden' tests and ‘joint' tests – based on how they accumulate evidence of association across sites. We used the empirical joint site frequency spectra of rare, nonsynonymous variation from a large multi-population sequencing study to explore the effect of realistic rare variant population structure on gene-based tests. We observed an important difference between the two test classes: their susceptibility to population stratification. Focusing on European samples, we found that joint tests, which allow variants to have opposite directions of effect, consistently showed higher levels of P-value inflation than burden tests. We determined that the differential stratification was caused by two specific patterns in the interpopulation distribution of rare variants, each correlating with inflation in one of the test classes. The pattern that inflates joint tests is more prevalent in real data, explaining the higher levels of inflation in these tests. Furthermore, we show that the different sources of inflation between tests lead to heterogeneous responses to genomic control correction and the number of variants analyzed. Our results indicate that care must be taken when interpreting joint and burden analyses of the same set of rare variants, in particular, to avoid mistaking inflated P-values in joint tests for stronger signals of true associations.
Keywords: rare variants, site frequency spectra, burden tests, population stratification, type I error
Introduction
Recent large-scale sequencing studies have identified an abundance of rare variation in the human genome, likely resulting from recent population expansion and purifying selection against deleterious variants.1, 2 Coding regions of the genome are enriched for rare, putatively functional variants,3, 4 attractive candidates for explaining missing heritability in complex diseases.5, 6, 7 A variety of gene-based tests that simultaneously analyze multiple rare variants have been proposed; the majority can be partitioned into two categories based on the assumptions of their genotype–phenotype model.8 The first category, based on the concept of rare variant ‘burden', tests for a significant correlation between a disease phenotype and an aggregate rare variant summary statistic computed for each individual in a data set. For example, burden test summary statistics include an indicator for presence of at least one rare allele,9 the total count of rare alleles,10, 11 and a weighted count of rare alleles.12 In contrast, ‘joint' or ‘dispersion' tests model the marginal effects of individual rare alleles and combine this information across multiple sites to test for association, specifically modeling variants with opposite directions of risk effect. Two popular examples of joint tests include the Sequence Kernel Association Test13 (SKAT) and C-Alpha.14
Comparative analyses have shown that performance varies among rare variant tests, particularly with respect to the underlying phenotype model and the inclusion of noncausal variants.8, 15 For example, joint tests have more power to identify regions containing a mix of risk and protective rare variants, whereas burden tests can have more power when all rare variants either increase or decrease risk. A common strategy for increasing power to detect associations is analyzing the same set of rare variants with multiple tests. Understanding the behaviors of each test is critical for correctly interpreting results. Here, we report that the two classes of gene-based tests respond differently to forms of rare variant population structure, leading to unique patterns of population stratification.
Population stratification arises when cases and controls are sampled at differential rates from genetically divergent populations.16, 17 Frequencies of individual rare alleles differ between populations due to geographic localization and limited sharing of rare variation.3, 18 Also, populations can differ in the total quantity of rare alleles they harbor due to differences in effective population sizes, demographic events, bottlenecks, or selective pressures.3, 18, 19 For example, African populations contain a larger number of rare variant sites than European populations, and within Europe, there is an increasing gradient of cumulative rare variation moving from north to south.3, 19 Stratification in single marker tests depends only on differences in population allele frequencies at individual sites.17, 20 In contrast, gene-based tests, which aggregate information across multiple sites, must contend with population differences in both individual allele frequencies and the total quantities of rare variants.
Several recent papers address stratification in gene-based tests. Mathieson and McVean21 and Kiezun et al22 initially demonstrated that burden-style tests are prone to inflation due to underlying population structure, and that the degree of inflation can differ from single-marker tests. Liu et al23 reported differential levels of stratification between C-Alpha and a collapsing test in data simulated using a specific coalescent model. In Addition, burden tests had lower levels of inflation relative to C-Alpha in a recent analysis of rare variation in autism spectrum disorders.24 In this paper, we investigated the specific patterns of rare variant population structure that affect the type I error of gene-based tests. In particular, we find that frequency differences of individual rare variants have a much stronger effect on joint tests than burden tests. In contrast, population differences in overall abundance of rare alleles inflate only burden tests. This difference leads to differential inflation between gene-based rare variant tests. We quantified the rare variant patterns in European populations and conclude that the pattern responsible for inflating joint tests is likely more prevalent in real data.
We designed an analysis around the joint site frequency spectra (JSFS) of rare, nonsynonymous variants identified as part of a previously published sequencing study initially designed to identify and characterize variation in 202 drug-target genes in 14 002 worldwide individuals.3 The JSFS is a common tool in population genetics to summarize the configuration of observed allele counts between two groups of samples, typically from different populations.18 Here, we used the JSFS as probabilistic models from which we generated examples of case-control data sets containing realistic patterns of population structure, but without any true genotype–phenotype association. We focused on the JSFS from four geographically-defined European populations: Central, Western, Northwestern, and Northern Europeans (see map in Figure 1). The genetic diversity in our JSFS reflects population structure that could reasonably be present in an association study of European samples, and provides an ideal method to study realistic gene-based test inflation.
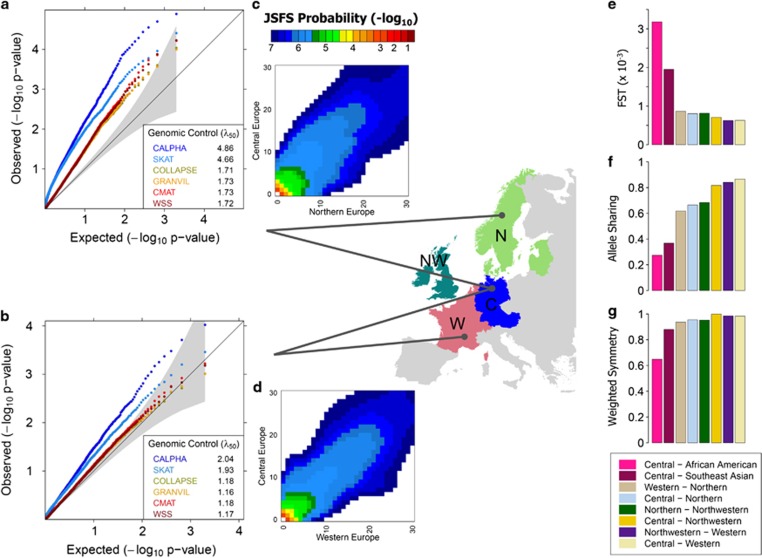
Figure 1.
Rare variant diversity statistics and P-value distributions for gene-based tests in structured European populations. We focused on the empirical JSFS of rare, nonsynonymous variants in 202 drug target genes identified by sequencing of Northern, Northwestern, Western, and Central European population samples (labeled N, NW, W, and C, respectively, in map insert). Heatmaps of the JSFS, pictured for (c) Central and Northern European and (d) Central and Western European population comparisons, provide a graphical representation of the distribution of rare alleles between populations. We quantified aspects of between-population rare variant structure using: (e) the FST statistic of overall rare variant population divergence, (f) the allele sharing statistic to measure variation in population-specific frequencies of individual rare alleles, and (g) the weighted symmetry statistic to measure the evenness of cumulative rare variant load between the populations. To study the effect of these population structures on inflation in gene-based tests, we analyzed data sets simulated from each JSFS that contained population structure but no genotype-phenotype association. QQ plots provide the distribution of P-values for several gene-based rare variant tests in data sets containing a mix of (a) Central and Northern Europeans and (b) Central and Western Europeans. Genomic control values (λ50) quantify the inflation of the P-value distributions relative to a uniform null distribution. For illustrative purposes, we display the QQ plots for an extreme sampling scenario in which all cases are sampled from one population and all controls are sampled from the other population. Results for less extreme scenarios are shown in Figure 2. We find that data sets from more divergent populations (Central and Northern European) produce higher levels of P-value inflation for each gene-based test than data sets from more closely related populations (Central and Western Europeans). Furthermore, the joint tests SKAT and C-Alpha (blue dots in QQ plots) consistently show much higher inflation than the burden tests Collapsing, GRANVIL, CMAT, and WSS tests (red dots in QQ plots) across all population comparisons.
Our JSFS-based simulation strategy was motivated by the fact that although the Nelson et al data set contains sequence data from many populations, the number of samples within individual populations does not allow for standard ‘resampling' techniques. The joint distribution of rare alleles between pairs of populations, summarized in the JSFS, provided a means for unlimited sampling of population allele counts from their empirical distributions. As gene-based tests operate directly on the JSFS of cases and controls, our approach retained the critical population-level information that confounds gene-based tests without requiring individual-level sequence data.
Materials and Methods
Joint site frequency spectra (JSFS)
Consider a sample containing sequence data for N haplotypes from population 1 and N haplotypes from population 2. For a given polymorphic site in the data set, let φ(i, j) denote the probability that i copies of the non-reference allele are observed among the N population 1 haplotypes and j copies are observed among the N population 2 haplotypes. Then, we define Φ={φ(i, j)∣i,j∈(0, N)} to be the JSFS of populations 1 and 2.
The empirical JSFS for multiple worldwide populations were previously computed and reported as part of Nelson et al.3 Briefly, 202 drug-target genes were deep sequenced in a total of 14 002 samples, including European (N=12 514), African-American (N=594) and Southern Asian (N=566, mostly from India) individuals. The sequenced samples were derived from several case-control data sets. Within each disease study, individuals with pairwise relatedness of 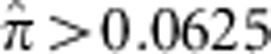 were removed to eliminate closely related individuals. Previous rare variant analyses of these disease studies discovered no significant associations.3 We focused our analysis on four European subpopulations that were geographically classified according to the UN geoscheme for Europe: Northwestern European (Great Britain and Ireland), Northern Europeans (excluding Finnish), Western European (Belgium, France, Luxembourg, and The Netherlands) and Central Europe (Austria, Germany, and Switzerland). To account for differences in population sample sizes, the JSFS were computed by averaging over downsampled realizations of 474 individuals per population. We focused on rare, putatively functional variants likely to be included in gene-based tests by restricting attention to the JSFS of nonsynonymous variants with sample minor allele frequency <1%.
were removed to eliminate closely related individuals. Previous rare variant analyses of these disease studies discovered no significant associations.3 We focused our analysis on four European subpopulations that were geographically classified according to the UN geoscheme for Europe: Northwestern European (Great Britain and Ireland), Northern Europeans (excluding Finnish), Western European (Belgium, France, Luxembourg, and The Netherlands) and Central Europe (Austria, Germany, and Switzerland). To account for differences in population sample sizes, the JSFS were computed by averaging over downsampled realizations of 474 individuals per population. We focused on rare, putatively functional variants likely to be included in gene-based tests by restricting attention to the JSFS of nonsynonymous variants with sample minor allele frequency <1%.
JSFS summary statistics
We quantified rare variant population structure within a JSFS using three summary statistics. To focus on rare variants, we computed each summary statistic over allele counts i, j for which the pooled sample allele frequency 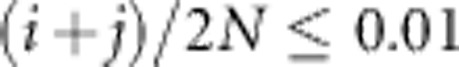 . We calculated an overall measure of genetic diversity using a variation on the standard FST statistic:
. We calculated an overall measure of genetic diversity using a variation on the standard FST statistic:
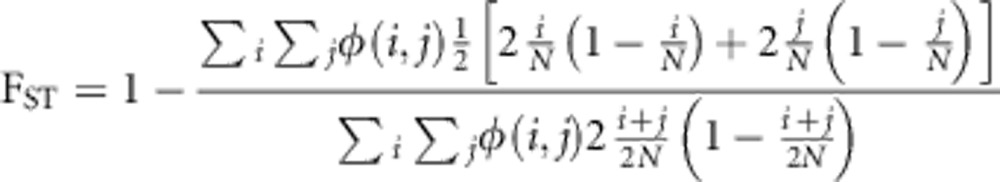 |
Allele sharing18 is the probability that two individuals carrying an allele of count n in the sample come from different populations, normalized by the expected probability in a panmictic population:
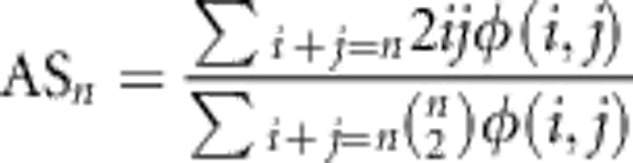 |
The allele sharing statistic (AS) for an entire JSFS of rare alleles is defined as the weighted average of ASn
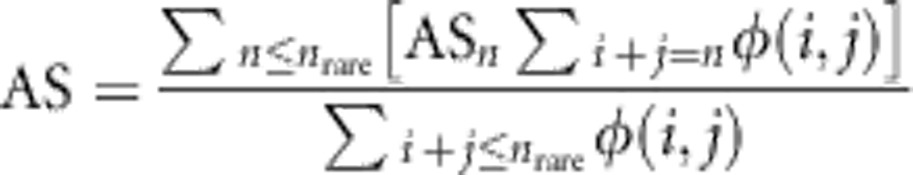 |
where nrare=2N × 0.01 denotes alleles below 1% frequency in the sample.
Weighted symmetry (WS) measures how evenly rare alleles are distributed between the two populations,
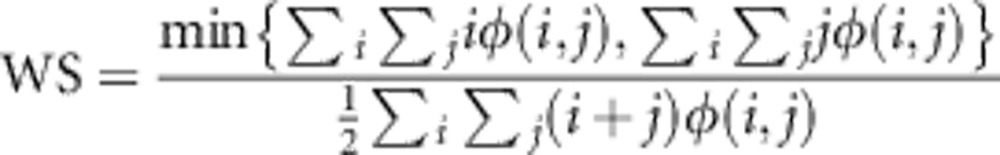 |
A graphical interpretation of the allele sharing and weighted symmetry statistics is provided in Supplementary Figure 2.
JSFS transformations
To isolate the effects of allele sharing and weighted symmetry on test statistic inflation, we designed two transformations that redistribute probability within a JSFS. For each transformation we began with a panmictic JSFS18 with weighted symmetry WS=1 and allele sharing AS=1.
The first transformation created a sequence of JSFS with fixed weighted symmetry and decreasing allele sharing by iteratively applying the following function:
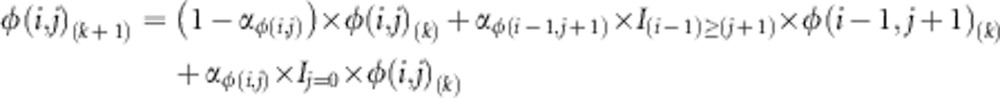 |
for i>j and
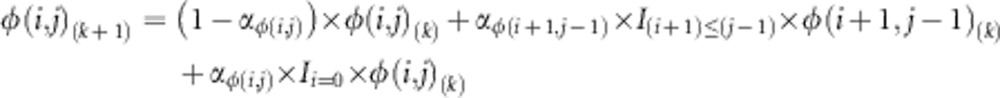 |
for i<j, where φ(i, j)(k) is the (i, j)th element of the kth iteration in the sequence of JSFS and αφ(i, j) is a weight, which decreases as the transformation moves away from the y=x line. This transformation moves probability away from the x=y line, increasing the probability of observing larger differences between population allele counts i and j.
Second, we created a sequence of JSFS with a fixed value of allele sharing and decreasing weighted symmetry by iteratively moving probability across the x=y line from one half of the spectrum to the other using the following transformation:
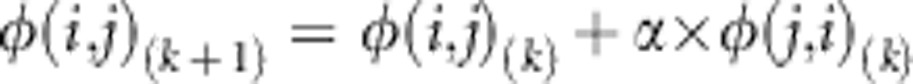 |
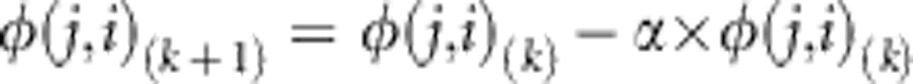 |
Where, φ(i, j)(k) is (i, j)th element of the kth iteration in the sequence of JSFS. As in the previous transformation, the probability of observing a variant with n total minor alleles in the 2N haplotypes does not change. The probability of observing i>j where i and j are the number of minor alleles observed in populations 1 and 2, respectively, increases.
Simulation of data sets
For each of the six Nelson et al inter-European comparisons, we treated the respective JSFS as a joint probability distribution from which we simulated sequence data. As the JSFS depends on sample size and our empirical JSFS were computed using 474 individuals from each population, we simulated genotypes within a single gene for 948 total individuals, 474 individuals (N=948 haplotypes) from each of the two populations. For each genic realization, we sampled pairs of allele counts (is, js∣1≤s≤S) for S different rare variant sites, each with probability according to the JSFS. At the sth site, we randomly distributed the is copies of the minor allele among N=948 population 1 haplotypes and js copies among N=948 population 2 haplotypes. Allele counts for the S different sites were independently drawn from the JSFS, implicitly assuming a lack of correlation between rare variants in a gene. Although this may not reflect the true relationship between all rare variants, it does not affect test performance as each test is designed to account for correlation between sites.
To induce varying degrees of population structure, we first created diploid samples by randomly pairing together haplotypes within each population group. We then assigned a phenotype status to each diploid sample based solely on population affiliation. Treating r as a mixing parameter (0.5≤r≤1.0), we randomly selected r × N/2 samples from the first population to be cases and the remaining (1−r) × N/2 to be controls. We then assigned (1−r) × N/2 and r × N/2 haplotypes from the second population to be cases and controls, respectively. Data sets constructed with r=0.5 contained equal numbers of cases and controls from each population. Alternatively, r=1.0 indicated an extreme sampling scenario where all cases were from one population and all controls were from the other population.
Measures of test statistic inflation
We analyzed each data set with four burden tests: Collapsing,9 CMAT,10 GRANVIL,25 Weighted Sum Statistic (WSS)12, and two joint tests: SKAT13 and C-Alpha.14 We quantified inflation in the distribution of P-values of each test relative to the expected uniform null distribution using a variation on the genomic control statistic of Devlin and Roeder.17 For p(50) and p(90), the median and 90th percentile values for a test statistic's observed P-value distribution, we define
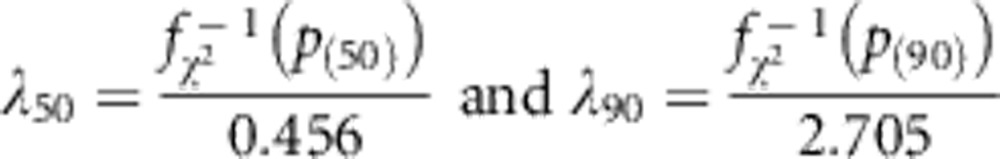 |
as the median and 90th percentile genomic control values, where 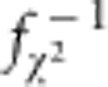 (p(q)) is the quantile function for a 1-degree of freedom χ2 random variable. The denominators for λ50 and λ90 are the median and 90th percentile values for a 1-degree of freedom χ2 random variable, respectively.
(p(q)) is the quantile function for a 1-degree of freedom χ2 random variable. The denominators for λ50 and λ90 are the median and 90th percentile values for a 1-degree of freedom χ2 random variable, respectively.
Results
We simulated data sets containing various degrees of rare variant population differentiation using the empirical JSFS of rare, nonsynonymous variation in four geographically-defined European populations: Central, Western, Northwestern and Northern Europeans.3 Pairwise rare variant Fst computed on the JSFS ranged from 6.26 × 10−4 to 8.66 × 10−4 (Figure 1e), indicating low overall genetic divergence.
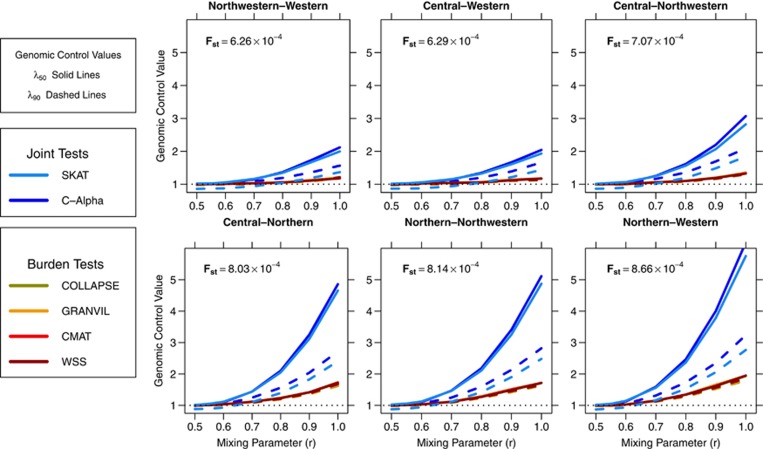
We analyzed the data sets with four burden tests: Collapsing,9 CMAT,10 GRANVIL,25 Weighted Sum Statistic12, and two joint tests: SKAT13 and C-Alpha.14 For each test, we reported the P-value distribution for 1000 genes (averaged across 10 replicate runs) over a range of mixing parameters r, assuming a fixed sample size of 474 cases and 474 controls, and a fixed number of variants (S=30). In Figure 2, we summarized P-value inflation of each test as genomic control values.17 The standard genomic control value (λ50) quantifies inflation at the median of the P-value distribution. As we observed different levels of inflation in the tails of the P-value distributions, we also report λ90, the genomic control value computed for the 90th percentile of the P-value distributions.
Figure 2.
Genomic control (GC) values for gene-based rare variant tests in structured European data sets. Median GC values (λ50, solid lines) and 90th percentile GC values (λ90, dashed lines) are shown at a range of mixing parameters (r) for each inter-European population comparison. For scenarios containing population structure (r>0.5), the joint tests (blue lines) consistently have higher λ50 values than the burden tests (red lines) in all population scenarios. In addition, λ90<<λ50 in many scenarios for the joint tests, indicating that inflation in the joint tests is not consistent across the P-value distribution.
Data sets simulated with balanced population sampling (r=0.5) yielded median genomic control values of λ50≈1.00 for all tests in each population comparison, indicating no inflation. Joint tests were deflated in their tails (λ90<1), consistent with the conservative nature of these tests for smaller samples sizes and at stringent alpha levels.13 Genomic control values increased for each test and population comparison as the mixing ratio increased from r=0.5 to r=1.0, indicating P-value inflation due to population structure. More divergent populations, as quantified by Fst, showed higher levels of inflation for each test. For example, at mixing ratio r=0.8, the genomic control of the Collapsing test was λ50=1.05 in the Central European and Western European comparison (Fst=6.29 × 10−4) but λ50=1.23 in the more divergent Central European and Northwestern European comparison (Fst=7.07 × 10−4). In many cases, inflation in the medians of the P-value distributions was larger than in the tails (ie λ50>λ90) as evidenced by the difference between dashed (λ90) and solid lines (λ50) in each panel of Figure 2. The inconsistent inflation was more pronounced in joint tests, and increased in magnitude with both increasing r and increasing population diversity. As a result, standard genomic control severely overcorrected inflated P-values more often for joint tests than for burden tests (Supplementary Figure 1).
Comparing inflation statistics between tests, we observed two consistent patterns across all scenarios. First, the level of P-value inflation for the different tests clustered into two distinct groups, one consisting of the joint tests: SKAT and C-Alpha, and the other containing the burden tests: CMAT, Collapsing, WSS, and GRANVIL. Within each group, the level of inflation was similar between tests. For example, in data sets of Central and Western Europeans with r=0.7, each burden test had λ50≈1.04, whereas SKAT and C-Alpha had λ50 values near 1.15. The distinct patterns of inflation for the two classes of tests can be seen in Figures 1a and b where burden tests (red dots) clustered together tightly, and are clearly separated from joint tests (blue dots). The second consistent pattern in the analysis was higher inflation for joint test statistics relative to burden test statistics; the difference increasing with both the divergence of the underlying populations and the mixing parameter r. For example, the difference in inflation between CMAT and SKAT rose from λ50=1.04 and 1.13, respectively, in Central and Western data sets to λ50=1.15 and 1.56 for the JSFS of the more divergent Northern and Western Europeans at r=0.7.
We hypothesized that the observed patterns of P-value inflation for burden and joint tests could be explained by underlying rare variant population structures. To test this, we quantified specific patterns of population structure within the JSFS using two statistics: allele sharing and weighted symmetry (see Materials and Methods, Supplementary Figure 2). The allele sharing (AS) statistic18 quantifies interpopulation differences in individual allele frequencies for a JSFS. AS=1 indicates allele frequency differences consistent with panmictic population sampling and the statistic decreases towards zero as differences in population allele frequencies increase. We developed the weighted symmetry (WS) statistic to summarize the difference in overall rare allele abundance between populations. Weighted symmetry of WS=1 indicates an equal quantity of rare alleles in each population and decreases towards zero with increasing inequality in rare allele abundance.
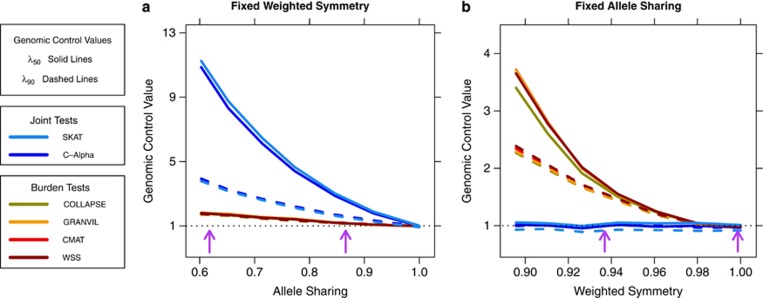
We isolated the effects of weighted symmetry and allele sharing on test statistic inflation by analyzing data sets simulated from JSFS where one statistic was fixed and the other decreased in value (see Materials and Methods). We first analyzed JSFS with weighted symmetry fixed at WS=1 (Figure 3a). When allele sharing AS also equaled one, the JSFS is equivalent to panmictic sampling and there is no inflation for any test. As allele sharing decreased, genomic control values quickly increased for the joint tests, indicating P-value inflation. In comparison, there was only a slight increase in inflation for the burden tests. Next, we considered JSFS with allele sharing fixed at AS=1 and allowed weighted symmetry to decrease. P-value inflation for every burden test increased with decreasing WS, but both SKAT and C-Alpha were unaffected (Figure 3b). Taken together, these results imply that the two classes of tests have opposite responses to decreasing weighted symmetry and decreasing allele sharing. Inflation in burden tests is primarily due to unequal contributions of rare alleles between the two populations, whereas joint test inflation is driven solely by differences in population-specific frequencies of individual rare alleles.
Figure 3.
The isolated effects of weighted symmetry and allele sharing on P-value inflation in gene-based rare variant tests. (a) For data set simulated with weighted symmetry fixed at WS=1 and decreasing allele sharing, inflation grows much larger for the joint tests than for the burden tests. (b) In contrast, for data sets simulated with allele sharing fixed at AS=1 and decreasing values of weighted symmetry, inflation in each burden test increases, whereas the joint tests remain well-controlled. Thus, the two classes of gene-based tests have differing responses to these patterns of rare variant population structure. The purple arrows in each plot indicate the minimum and maximum values of that statistic observed in the European JSFS. The range of empirical values explains why we observed higher levels of inflation in the joint tests.
Having established that burden test inflation correlates strongly with weighted symmetry and joint test inflation with allele sharing, we computed these quantities for our European JSFS (Figures 1f and g). Allele sharing ranged between 0.86 and 0.62, with the lowest values observed for JSFS containing the Northern Europeans. We observed weighted symmetry values as high as 0.99 for the JSFS of Central and Northwestern populations and as low as 0.94 for Northern and Western Europeans. The lower weighted symmetry values for JSFS containing Northern Europeans are indicative of fewer rare alleles in that population, consistent with the hypothesis that a historical bottleneck event decreased the population's effective size. Our simulations with fixed weighted symmetry and allele sharing provide context for the differential inflation observed in the inter-European data sets. Allele sharing between European populations was sufficiently low to produce large inflation in the joint tests (purple arrows in Figure 3a). Alternatively, weighted symmetry between European populations did not decrease to levels that produced substantial inflation in burden tests (Figure 3b).
For comparison, we also computed allele sharing and weighted symmetry for the JSFS between our European samples and both African-American and South Asian samples from the same Nelson et al data set.3 As expected we saw smaller values of both statistics for these intercontinental population comparisons (Figures 1f and g). Allele sharing between Europeans and African-Americans ranged from 0.22 to 0.28, and from 0.27 to 0.37 between Europeans and the South Asians. Weighted symmetry between the European populations and South Asian took values of ∼0.90, slightly less than the inter-European comparisons. Weighted symmetry between the African-Americans and Europeans however was much lower, between 0.62 and 0.66, highlighting the larger difference in the total number of rare alleles between these populations. Extrapolating on the theoretical results in Figure 3, the values of weighted symmetry between Europeans and African-Americans or Europeans and South Asians are capable of significantly inflating burden tests. However, for these comparisons, allele sharing is even lower and inflation would still be larger for the joint tests.
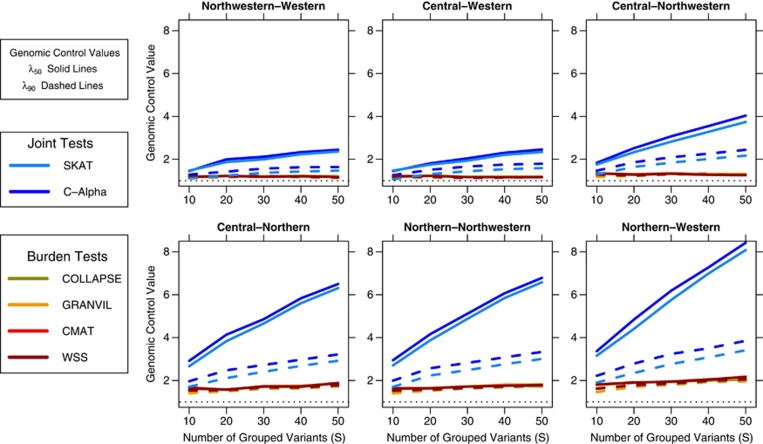
Till now, we have assumed a fixed number of rare variants within each gene (S=30). In reality, the number of rare variants combined into a gene-based test varies depending on several factors, including gene length, sample size, population genetic diversity, annotation, and frequency thresholds. To understand the impact that the number of variants per gene has on stratification we repeated our simulations over a range of values for the number of pooled variants S (Figure 4). The two classes of tests responded quite differently to a varying number of pooled sites: joint tests showed a clear increase in inflation as S increased, whereas inflation in burden tests remained effectively constant. The differential sources of stratification explain this result. In these closely related European populations, the cumulative quantity of rare alleles is quite similar (WS≈1) but most individual allele frequencies vary slightly between populations. Additional variants do not alter the cumulative allele balance tested for by burden statistics. However, each additional variant provides further evidence of differing allele frequencies between cases and controls, leading to the increasing inflation for joint tests. We expect that in a scenario of populations with smaller WS, inflation in burden tests would also increase with the number of variants.
Figure 4.
The effect of number of rare variant sites (S) pooled together in a gene-based tests on P-value inflation (shown for mixing parameter r=1.0). There is a clear increase in inflation for the joint tests (blue lines) as the number of rare variant sites pooled into a gene-based test increases. Inflation in the burden tests (red lines) remains relatively consistent as the number of sites increases.
Discussion
We used the JSFS as a model to study the structure of rare variants within European populations and its effect on gene-based tests. By quantifying specific patterns in the JSFS, we established that different aspects of population differentiation are responsible for inflating the type I error rates in the two classes of gene-based tests. Our results build on those of previous studies examining rare variant population stratification. We independently demonstrated different levels of inflation in C-Alpha and burden tests previously reported in both coalescent simulations23 and real sequencing data.24 We found that the pattern of differential inflation held more broadly for burden and joint tests over a range of population sampling scenarios. Modeling our data sets using the empirical JSFS from several European populations illustrated the magnitude of stratification in realistic samples. Moreover, we identified the precise underlying characteristics of rare variant population structure responsible for the differential stratification, namely, imbalance in rare allele load and overdispersion of individual rare allele frequencies. By looking at the empirical weighted symmetry and allele sharing values observed between multiple European populations we explained the patterns of population stratification observed in gene-based rare variant tests.
The primary advantage of joint tests over burden tests is greater power to detect association in genes containing rare variants with opposite directions of effect. Interestingly, it is precisely this ability to accommodate a mix of risk and protective variants that makes joint tests more vulnerable to stratification in real population scenarios. Joint tests view the population-specific differences in allele frequency at each variant site, regardless of direction, as signal for association. Alternatively, burden tests require the population-specific differences to be predominantly in the same direction, a more stringent criterion. Intuitively, differences in allele frequency (low allele sharing) are more pronounced between populations than differences in the number of rare variants (low weighted symmetry) because all forms of population differentiation resulting in genetic drift lead to allele frequency differences. Creating a significant imbalance in the total quantity of rare variants requires more specific models, for example, a recent bottleneck, unequal migration or differential growth rates. Thus, the forms of population structure that produce inflation in joint tests are more prevalent in real data and we predict that, although joint tests provide more power to detect many true rare variant associations, they also require more caution to avoid spurious results.
We anticipate that differential inflation will be particularly problematic for interpreting burden and joint test results of sequencing studies that target only a handful of candidate genes. It is straightforward to determine if differential inflation exists when many genes are sequenced (ie exome sequencing) by comparing the distributions of P-values for the joint and burden tests. This may not be possible in a targeted sequencing data set, and if population structure exists, smaller P-values in joint tests could easily be interpreted as stronger signals of true associations rather than increased susceptibility to inflation.
The effect of the number of variants per gene on joint statistic inflation provides a practical approach for recognizing stratification. Typically, only rare variants predicted to be deleterious (eg nonsynonymous) are included in a gene-level analysis. The remaining ‘excluded' rare variants, which likely outnumber the predicted deleterious variants, are presumably null with respect to phenotype status, yet still contain signal for population structure (Supplementary Table 1). Thus, a joint test analysis of the excluded variants is more powerful for detecting population stratification than the analysis of the fewer predicted deleterious variants. We therefore recommend performing the same joint test analysis planned for the predicted deleterious variants on the excluded variants as a method to test for population stratification. This method could be particularly helpful for interpreting joint test P-values in targeted sequencing studies.
Previous studies have emphasized the challenge of correcting for rare variant population structure in multi-marker gene-based tests. Kiezun et al22 corrected the stratification using a modified permutation algorithm requiring that population labels be both discrete and either known or accurately estimated, neither of which may be satisfied in real data sets. Mathieson and McVean21 and Liu et al23 each showed the standard application of principle components could not correct for all scenarios in either single marker or gene-based analyses of rare variants. In light of our finding that inflation differs according to the type of gene-based test, the appropriate correction strategy may be context specific depending on the test and populations. We illustrated this point using genomic control. Even in a set of homogenous genes with a uniform number of variants and identical underlying JSFS, we often observed that the median of the P-value distribution was more highly inflated than the tail of the distribution (λ50>λ90). Under these conditions applying a standard genomic control correction based on λ50 overcorrects the most significant genes in the analysis (Supplementary Figure 1), which reduces power for real associations. The overcorrection was more severe for joint tests, implying that genomic control may be more appropriate for burden tests.
Presently, attempts to identify rare risk variants using the pooling approach of gene-based tests have had limited success. Despite the potential for stratification seen here, real data sets have often identified no statistically significant genes rather than too many. This lack of significant findings, even false positives, is likely the result of current studies being underpowered due to insufficient sample sizes. Larger sample sizes in future sequencing studies will increase power to find true signals, but will also increase the likelihood of subtle population structure and the number of variants pooled within genes, both of which increase the potential for rare variant population stratification.
Acknowledgments
This research was supported by NIH/NHGRI grants HG005855 (MZ, SZ), HG000040 (MR), HG000376 (MR), HG007089 (JN), a Humboldt Fellowship to SZ and the Searle Scholars Program to JN (DW). We thank Hyun Min Kang for providing assistance with the EPACTS software and Michael Boehnke and Rebecca Rothwell for valuable feedback.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppell M, Boehnke M, Zollner S. FTEC: a coalescent simulator for modeling faster than exponential growth. Bioinformatics. 2012;28:1282–1283. doi: 10.1093/bioinformatics/bts135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases. Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Wegmann D, Trynka G, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyghe JR, Jackson AU, Fogarty MP, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Fast S, Zawistowski M, Tintle NL. A geometric framework for evaluating rare variant tests of association. Genet Epidemiol. 2013;37:345–357. doi: 10.1002/gepi.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawistowski M, Gopalakrishnan S, Ding J, et al. Extending rare-variant testing strategies: analysis of noncoding sequence and imputed genotypes. Am J Hum Genet. 2010;87:604–617. doi: 10.1016/j.ajhg.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, et al. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladouceur M, Dastani Z, Aulchenko YS, et al. The empirical power of rare variant association methods: results from sanger sequencing in 1,998 individuals. PLoS Genet. 2012;8:e1002496. doi: 10.1371/journal.pgen.1002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC. Population subdivision with respect to multiple alleles. Ann Hum Genet. 1969;33:23–29. doi: 10.1111/j.1469-1809.1969.tb01625.x. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Nicolae DL, Chen LS. Marbled inflation from population structure in gene-based association studies with rare variants. Genet Epidemiol. 2013;37:286–292. doi: 10.1002/gepi.21714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sabo A, Neale BM, et al. Analysis of rare, exonic variation amongst subjects with autism spectrum disorders and population controls. PLoS Genet. 2013;9:e1003443. doi: 10.1371/journal.pgen.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi R, Kumar A, Morris AP. Assessing the impact of missing genotype data in rare variant association analysis. BMC Proc. 2011;5:S107. doi: 10.1186/1753-6561-5-S9-S107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.