Abstract
To answer the question as to which commercial high-density SNP chip covers most of the human genome given a fixed budget, we compared the performance of 12 chips of different sizes released by Affymetrix and Illumina for the European, Asian, and African populations. These include Affymetrix' relatively new population-optimized arrays, whose SNP sets are each tailored toward a specific ethnicity. Our evaluation of the chips included the use of two measures, efficiency and cost–benefit ratio, which we developed as supplements to genetic coverage. Unlike coverage, these measures factor in the price of a chip or its substitute size (number of SNPs on chip), allowing comparisons to be drawn between differently priced chips. In this fashion, we identified the Affymetrix population-optimized arrays as offering the most cost-effective coverage for the Asian and African population. For the European population, we established the Illumina Human Omni 2.5-8 as the preferred choice. Interestingly, the Affymetrix chip tailored toward an Eastern Asian subpopulation performed well for all three populations investigated. However, our coverage estimates calculated for all chips proved much lower than those advertised by the producers. All our analyses were based on the 1000 Genome Project as reference population.
Keywords: coverage, efficiency, SNP chip, DNA array, linkage disequilibrium
INTRODUCTION
During the design stage of a genome-wide association study (GWAS), researchers typically need to decide which commercial SNP chip to use. Given approximately equivalent chip qualities, the chip to be selected should be the one that provides the greatest genomic coverage for the largest number of individuals as possible within the budgetary constraints.1 Of course, other considerations, such as experience with protocols and the extent to which a chip assays other genetic variations, may influence the final decision.
However, comparisons of different SNP chips are becoming ever more challenging, owing to the increasing number of chips on the market, each with its own specific properties. Illumina (San Diego, CA, USA), one of the two market leaders in the mass production of SNP chips, increased the number of SNPs from 500K (HumanHap550) to around 2.5 million (HumanOmni2.5-8), whereas Affymetrix (Santa Clara, CA,USA) reduced the number of SNPs from around 1 million (Genome-Wide SNP Array 6.0) to approximately 600K (eg, Axiom Genome-Wide Human EU and Axiom Genome-Wide ASI). This reduction allows Affymetrix to market their new chips at a relatively low price. Nevertheless, including fewer SNPs on a chip usually results in a loss of coverage, which Affymetrix aimed to prevent by designing chips separately for each of the three main ethnicities (European, Asian, and African). Hence, any direct comparison of these chips is not appropriate using conventional means, such as coverage2, 3 or simulated power,4, 5 as neither measure factors financial cost into its calculation.
To this end, we introduced two new measures to assess the performance of different SNP chips: efficiency and cost–benefit ratio (CBR). Both measures factor in either cost ratio of two chips or the number of SNPs on a chip as well as the extent to which the genetic variation in the human genome is captured. For efficiency measures, we used the chip size measured by the number of SNPs on the chip as a substitute for price, after demonstrating that there is a strong positive linear correlation between the two variables. We avoided using the absolute values of the quoted prices, because these vary considerably over time and depend on individual negotiations. Furthermore, chip efficiencies were calculated with regard to a range of linkage disequilibrium (LD) thresholds, therefore allowing a more complete picture of the genomic coverage each chip provides.
We used our new measures to compare eight more recently released chips by Affymetrix and Illumina. Additionally, we determined conventional coverage, as defined by Li et al,3 with regard to the whole genome, consecutive regions, and genes. Conventional coverage is indeed informative for comparison purposes when chips have roughly the same price (or same chip size). In this case, local and gene coverage enable investigators to make comparisons between chips on a finer scale.
Coverage, efficiencies, and CBRs were determined separately for each of the three main ethnicities (European, Asian, and African). Investigating ethnicities separately is common practice in modern GWASs, because discretely admixed samples, which include individuals with different genetic backgrounds, have been shown to inflate levels of false positives.6 We also calculated all measures for older chips as a benchmark, as we are the first study to use the 1000 Genome Project (1000GP) as a reference set for extensive chip comparisons across different ethnicities.
MATERIALS AND METHODS
Data
We investigated the performance of recently released Illumina and Affymetrix chips. The Affymetrix chips we investigated are all tailored towards specific human populations or subpopulations, while the Illumina chips are generally optimized with respect to Europeans. Table 1 summarizes their approximate sizes, design properties, and targeted minor allele frequency (MAF). Table 1 also lists the abbreviations for each chip used throughout the article. As benchmark, we additionally analyzed the following older chips (though some of these are no longer commercially available):
Illumina: HumanHap550 (∼550K SNPs), HumanHap650 (∼650K SNPs)
Affymetrix: Axiom Genome-Wide SNP Human Array 5.0 (∼500K SNPs), Axiom Genome-Wide SNP Human Array 6.0 (∼1 million SNPs)
Table 1. Chip size, the lowest MAF covered by the chip, the number of non-synonymous SNPs, and design notes of recent Illumina and Affymetrix chips according to their datasheets provided by the companies.
| Chip size in number (SNPs) | Lowest MAF captured | Number (non-synonymous SNPs) | Based on | Note | |
|---|---|---|---|---|---|
| Affymetrix | |||||
| Axiom Genome-Wide Human EU (Axiom GW EU) | ∼600 000 | 1% | 10 648 | HapMap, Single Nucleotide Polymorphism database (dbSNP), 1000 GP | Targeting European population |
| Axiom Genome-Wide Human ASI (Axiom GW ASI) | ∼600 000 | 1% | 10 346 | HapMap, dbSNP, 1000 GP | Targeting Asian population |
| Axiom Genome-Wide Human CHB (Axiom GW CHB) | ∼1 200 000 | 2% | 10 560 | HapMap, dbSNP, 1000 GP | Targeting CHB subpopulation |
| Axiom Genome-Wide Human PanAFR (Axiom GW PanAFR) | ∼2 200 000 | 2% | 12 250 | HapMap, dbSNP, 1000 GP, Southern African Genomes Project | Targeting African population |
| Illumina | |||||
| Human OmniExpress | ∼700 000 | 5% | 15 062 | HapMap | Optimized tag SNP |
| Human Omni1S-8 | ∼1 000 000 | 5% | 5641 | 1000GP | Optimized tag SNP |
| Human Omni2.5-8 | ∼2 500 000 | 2.5% | 41 900 | 1000GP | Targeting common and rare variants |
| Human Omni2.5S-8 | ∼2 500 000 | 1% | 57 360 | 1000GP | Targeting rare variants |
http://www.affymetrix.com/support/technical/datasheets/axiom_ceu_arrayplate_datasheet.pdf, http://www.affymetrix.com/support/technical/datasheets/axiom_asi_arrayplate_datasheet.pdf, http://www.affymetrix.com/support/technical/datasheets/axiom_chb_1_2_array_plate_set_datasheet.pdf, http://www.affymetrix.com/support/technical/datasheets/axiom_panafr_arrayplate_datasheet.pdf, http://www.illumina.com/documents/products/datasheets/datasheet_human_omni_express.pdf, http://res.illumina.com/documents/products/datasheets/datasheet_human_omni1s.pdf, http://res.illumina.com/documents/products/datasheets/datasheet_human_omni2.5.pdf, http://res.illumina.com/documents/products/datasheets/datasheet_omni25s.pdf.
In order to evaluate common and low-frequency variants, we use Version 3 (April 2012, NCBI Build 37) of the 1000GP. It contains genotypes of 1092 individuals of different ancestries for 38.2 million variants on 23 chromosomes (1–22, X). For our analysis, 286 individuals from the GBR, IBS, CEU, and TSI 1000GP samples formed the European reference population; 286 individuals from the CHB, CHS, and JPT 1000GP samples formed the Asian reference population; and 246 individuals from the ASW, LKW, and YRI 1000GP samples formed the African reference population. Unfortunately, analysis for individual subpopulations was not possible owing to the relatively small sizes of each sample. We filtered all reference population data only to contain SNPs with MAF≥1%, as none of the studied arrays was designed to cover variants with MAF<1%. After filtration, the final reference sets included 8 846 061, 7 855 506, and 15 827 437 SNPs for the European, Asian, and African populations, respectively. Note that the use of the 1000GP is likely to result in systematic overestimation of all measures due to overfitting.5 Overfitting refers to potential bias incurred by using the same data for both training and evaluation. Here, the data from the 1000GP were used to design some of the chips (compare Table 1) as well as to calculate the measures used in their comparison.
To determine gene coverage, we additionally required an appropriate gene annotation, which we obtained from the Ensembl Genes database (Version 72, NCBI Build 37).7 Each gene was assigned the maximum-sized region using the transcription start and end position (with 50 Kbp upstream and downstream), unless it contained fewer than five SNPs. Such genes were excluded to prevent unreliable results in accordance with Li et al.3 Following exclusion, we were left with 56 473, 56 458, and 56 528 genes for the European, Asian, and African reference sets, respectively. These numbers include protein-coding as well as non-coding genes according to the Ensembl Genes database.7
Efficiency
The success of a chip design for GWAS use depends crucially on two abilities:
exploitation of LD structures, because this ensures the use of a minimal number of SNPs and, in turn, keeps costs low; and
coverage of SNPs that are not strongly correlated with any other SNPs, as these have a high tendency to be non-synonymous.8
Thus we define two efficiency measures: LD efficiency and non-taggable SNPs' efficiency. On the one hand, LD efficiency, EffLD, evaluates the fraction of all taggable SNPs that the considered chip is able to tag. Thereby, a taggable SNP is defined as a SNP in the reference population that reaches a pre-defined simple pairwise correlation (r2) threshold with at least one other SNP in the same reference. On the other hand, non-taggable SNPs' efficiency, EffNT, reflects how well a chip covers SNPs that are non-taggable given a predefined LD threshold. Both measures are adjusted with the help of the chip size to allow comparison of chips with different prices. Chip size, which correlates strongly with chip price linearly (compare Supplementary Figure S1), was used instead of absolute prices, as it proved more robust. Chip prices vary considerably over time and depend on numerous factors, including negotiation skills and the number of chips ordered. Hence, we suggest defining the efficiencies as
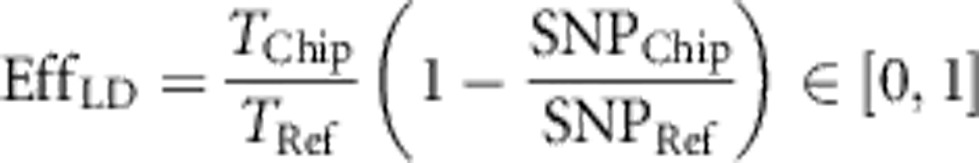 |
and
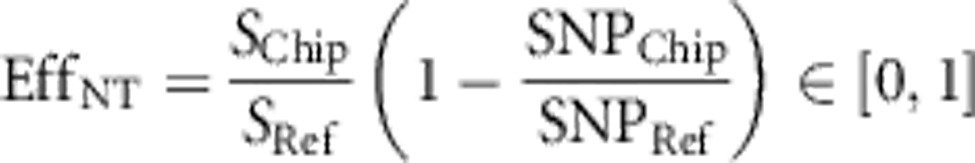 |
where SNPChip and SNPRef denote the total number of SNPs on the chip and in the reference set, respectively. TChip is the number of SNPs in the reference tagged by SNPs on the chip, while TRef denotes the total number of taggable SNPs in the reference. SChip and SRef refer to the number of non-taggable SNPs on the chip and in the reference, respectively. We calculated both efficiencies for several values (from 0.1 to 1; step size=0.1) of the predefined pairwise LD threshold.
CBR
The CBR facilitates direct comparison of two chips either in terms of LD exploitation or coverage of non-taggable SNPs. Its construction requires the absolute price:
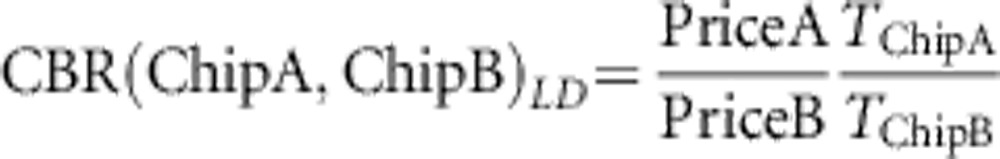 |
for LD and
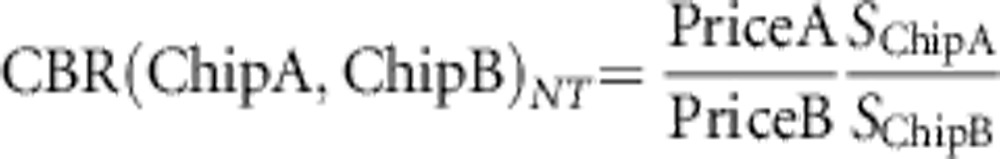 |
for non-taggable SNPs. If CBR(Chip A, Chip B)>1, chip A has the better cost–benefit compared with chip B and vice versa. The results presented in this paper are based on informal price quotes, so we would strongly encourage investigators to recalculate the cost–benefit using their own price quotes. Furthermore, we only evaluated CBR at the typical LD threshold of r2≥0.8.
Coverage
Coverage rate was calculated using the equation defined by Li et al.3 For a certain region in the genome, which could be a local window or a gene with start and end position on a chromosome, we denote G, R, T, and L as the number of all SNPs in the region, in the reference set, on the chip, and tagged. According to Li et al,3 coverage rate is defined by
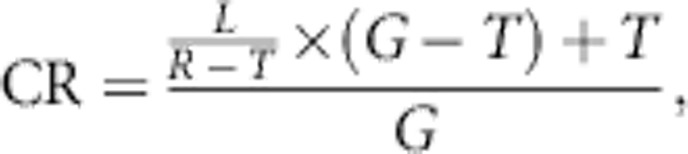 |
which represents the fraction of all SNPs in the region that can be captured by the chip. However, the definition of CR ignores the SNPs that are on the chip but not in the reference set. So, in correspondence with Li et al,3 CR may be regarded as a lower bound of the coverage rate. Li et al3 suggests to update the coverage by defining m as the number of SNPs on the chip but not in the reference set and R1+m, T1=T+m, and L1=T1/T × L. The updated equation is given by
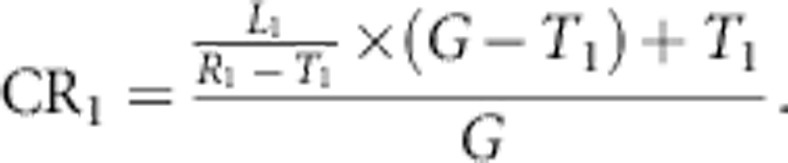 |
CR1 assumes that the number of tagged SNPs increases linearly with the number of SNPs on the chip. This assumption is overly optimistic, and we therefore believe that it tends to overestimate coverage. As CR tends to underestimate and CR1 to overestimate coverage rate, Li et al3 compensate by taking the average of both equations.
To obtain G, we needed an estimate of the total number of SNPs in the human genome. As of July 2013, validated SNPs with a MAF≥1% in the NCBI database counted 19 million. Thus, there are approximately 6809 SNPs in each 1 Mbp region. The calculation of LD was based on the LD threshold of r2≥0.8.
Coverage estimates were calculated for the whole genome, each gene separately, and consecutive local regions. Local coverage was determined in regions of 1 Mbp, which were moved 200 Kbp at each turn across each chromosome.
Results
Coverage
All our calculated whole-genome coverage estimates were <50% (see Table 2) and thus are much lower than the coverages advertised by the respective manufacturers. A similar result was also found by Jiang et al9 for the East Asian HapMap3 reference population. They observed 18% lower coverage rate for the Human OmniExpress than the coverage rate reported by Illumina. These differences in coverage are probably due to the existence of multiple definitions of coverage. They may also stem from the use of the more comprehensive 1000GP as a reference. As we expected, coverage increased with the number of SNPs genotyped. When comparing chips with <1 million SNPs, Illumina chips were found to have greater coverage than Affymetrix chips for all reference populations. Interestingly, the Axiom GW EU, which specifically targets the European population, was outperformed in terms of coverage of the European genome by the Axiom GW ASI, which was actually designed to target the Asian population. For chips including one million SNPs to two million SNPs, Axiom GW CHB offered the greatest coverage for all three populations. Coverage for Axiom GW PanAFR and Human Omni2.5-8, both with >2 million SNPs, proved about equal. Independent of the chip size, coverage was considerably lower for the African reference population than for both other reference populations, probably owing to shorter length of haplotypes in the African population.10 Older chips were generally unable to reach the coverage estimates of newer chips with an approximately equal number of SNPs. (Note that our coverage estimates reported for the old chips were much lower than those found by Li et al,3 because we used the more comprehensive 1000GP as a reference set and updated estimates of the total number of SNPs in the human genome to current knowledge).
Table 2. Whole-genome coverage of different chips evaluated for European, Asian, and African reference populations.
| European | Asian | African | |
|---|---|---|---|
| <1 million SNPs | |||
| Axiom GW EU | 0.16 | 0.21 | 0.06 |
| Axiom GW ASI | 0.17 | 0.21 | 0.06 |
| Axiom GW SNP Array 5.0 | 0.15 | 0.18 | 0.05 |
| Human OmniExpress | 0.21 | 0.26 | 0.08 |
| Human Hap550 | 0.18 | 0.22 | 0.06 |
| Human Hap650Y | 0.20 | 0.25 | 0.07 |
| 1–2 million SNPs | |||
| Axiom GW CHB | 0.30 | 0.37 | 0.12 |
| Axiom GW SNP Array 6.0 | 0.25 | 0.31 | 0.10 |
| Human Omni1S-8 | 0.22 | 0.29 | 0.10 |
| >2 million SNPs | |||
| Axiom GW PanAFR | 0.43 | 0.52 | 0.20 |
| Human Omni2.5-8 | 0.44 | 0.53 | 0.20 |
| Human Omni2.5S-8 | 0.30 | 0.48 | 0.15 |
Chips are separated in three categories according to their size as measured in the number of SNPs. Note that all coverage calculations are based on r2 threshold of 0.8.
The same observations held largely in terms of gene coverage (compare Supplementary Figure S2) as well as local coverage (compare Supplementary Figure S3). However, local coverage strongly varies across the genome. In particular, the Human OmniS2.5-8 had strictly lower local coverage rates than chips of comparable size with the exception of the chromosome 6 region between 25 and 35 Mbp, which locates the MHC region with an extensive LD structure and a high number of genes.11 The Human Omni1S-8 poorly covers the sex chromosome X compared with the rest of the genome. On average, the majority of chips did not exhibit significantly elevated coverage for genes compared with the rest of the genome. However, the coverage of different genes exhibited extreme variations. Note that general patterns in local and gene coverage did not vary considerably between the different reference populations.
Efficiency
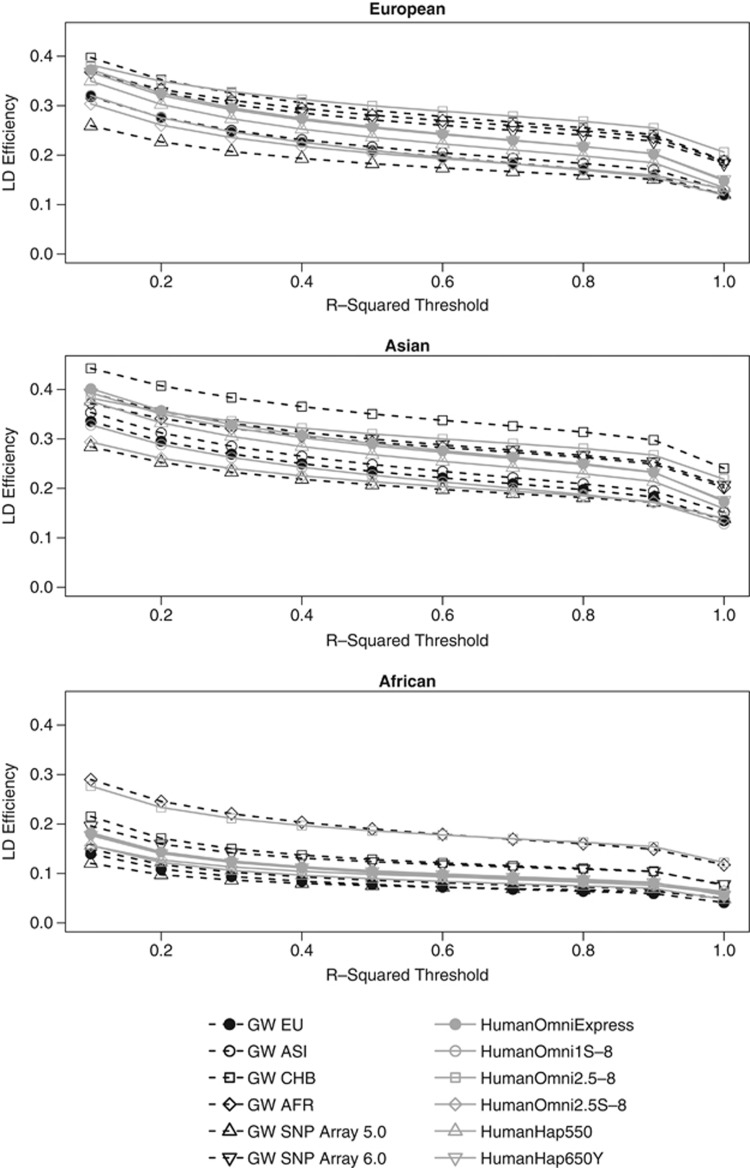
The LD efficiency of all chips in all reference populations decreased monotonically on increasing r2 threshold, as illustrated in Figure 1. Generally, chips with a greater overall efficiency were affected more strongly by an increase in the r2 threshold as manifested by steeper curves. For the Asian reference population, the Axiom GW CHB had the greatest LD efficiency independent of r2 threshold. For Africans, the Axiom GW PanAFR and the HumanOmni2.5-8 displayed considerably greater LD efficiency than all other investigated chips. Unlike for Asians and Africans, there were several highly performing chips in terms of LD efficiency for the European reference population. Ultimately, the greatest LD efficiency was achieved by the Human Omni2.5-8. in a similar fashion to coverage; we observed that the Axiom GW ASI was at least as good as or better in terms of LD efficiency than the Axiom GW CEU for Europeans. The other chip designed for an Asian population, the Axiom GW CHB, also performed more efficiently than the Axiom GW CEU despite size adjustment.
Figure 1.
LD efficiencies of different chips evaluated for European, Asian, and African reference populations. In the efficiency calculations, the r2 threshold varied from 0.1 to 1 in steps of size 0.1.
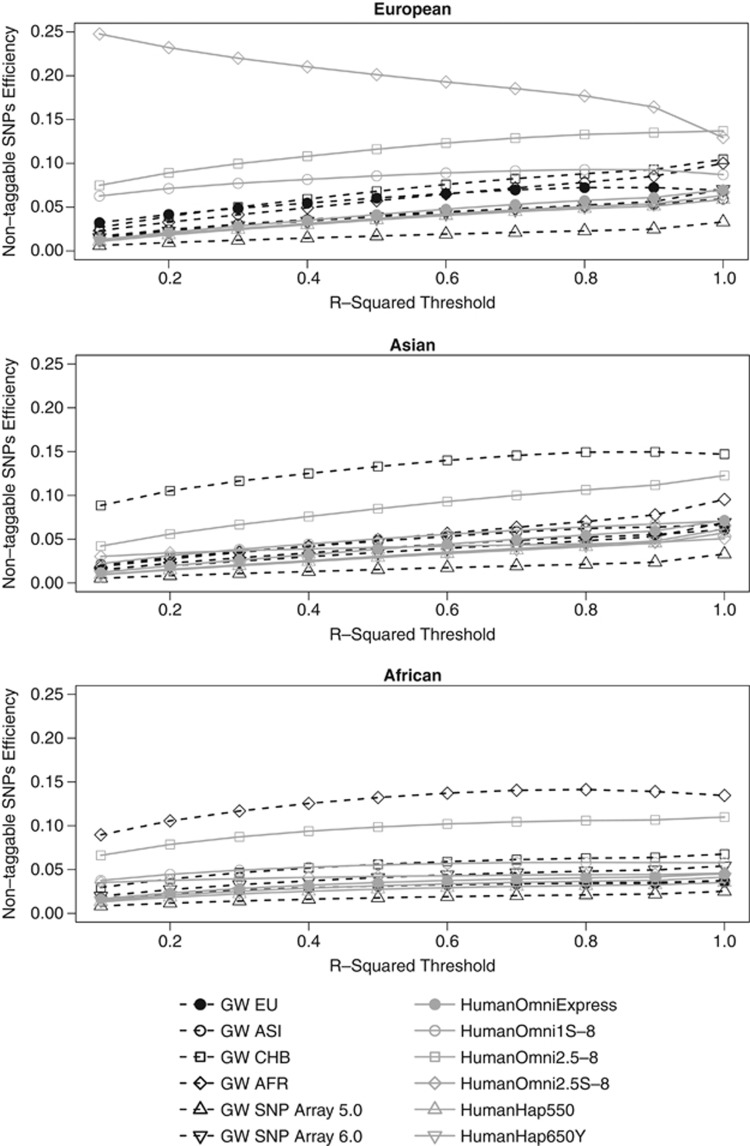
For most chips investigated, non-taggable SNPs' efficiency increased steadily with increasing r2 threshold up to r2=0.9 (refer to Figure 2). The exception was the Human Omni2.5S-8 with a monotonically decreasing curve for the European reference population. A possible explanation for this is the focus of the Human Omni2.5S-8 on low-frequency variation within the European population. If the r2 threshold is small, there are fewer non-taggable SNPs. These non-taggable SNPs are, in turn, likely to be low-frequency SNPs with lower levels of LD. The focus of this chip therefore implies that it covers a relatively high percentage of non-taggable SNPs at low r2 thresholds. At extremely high r2 thresholds, the Human Omni2.5S-8 is no more efficient in terms of non-taggable SNPs than the Human Omni2.5-8 for Europeans. For the Asian reference population, the Axiom GW CHB had the greatest non-taggable SNPs efficiency, despite being designed specifically for an Eastern Asian subpopulation. We believe that the overall good performance of Axiom GW CHB for the entire Asian reference population may be explained by the fact that Asian subpopulations, such as CHB, are genetically very similar. This is also evident from principle component plots of the 1000GP, in which all Eastern Asian subpopulations appear to cluster together. For the African reference population, the Axiom GW PanAFR outperformed all other chips in terms of covering non-taggable SNPs efficiently. For all populations, the Axiom Genome-Wide SNP Human Array 5.0 consistently had the lowest LD and non-taggable SNPs' efficiencies.
Figure 2.
Non-taggable SNPs' efficiencies of different chips evaluated for European, Asian, and African reference populations. In the efficiency calculations, the r2 threshold was varied from 0.1 to 1 in steps of size 0.1.
CBR
Comparing the different chips using our newly developed CBR measures yielded interesting results. However, readers are advised that these should not be understood as endorsements or recommendations, as they are based solely on our price quotes. For the Asian reference population, CBR measures suggested, unsurprisingly, that the Axiom GW CHB was more cost-effective than any other investigated chip with regard to covering the genome as well as capturing non-taggable SNPs (refer to Supplementary Table S1). For the African reference population, LD as well as non-taggable SNP CBRs favored the Axiom GW PanAFR and Human Omni2.5-8 (see Supplementary Table S2). Given the lack of strong LD patterns for the African population, we did indeed expect these larger chips to dominate cost–benefit comparisons for Africans.
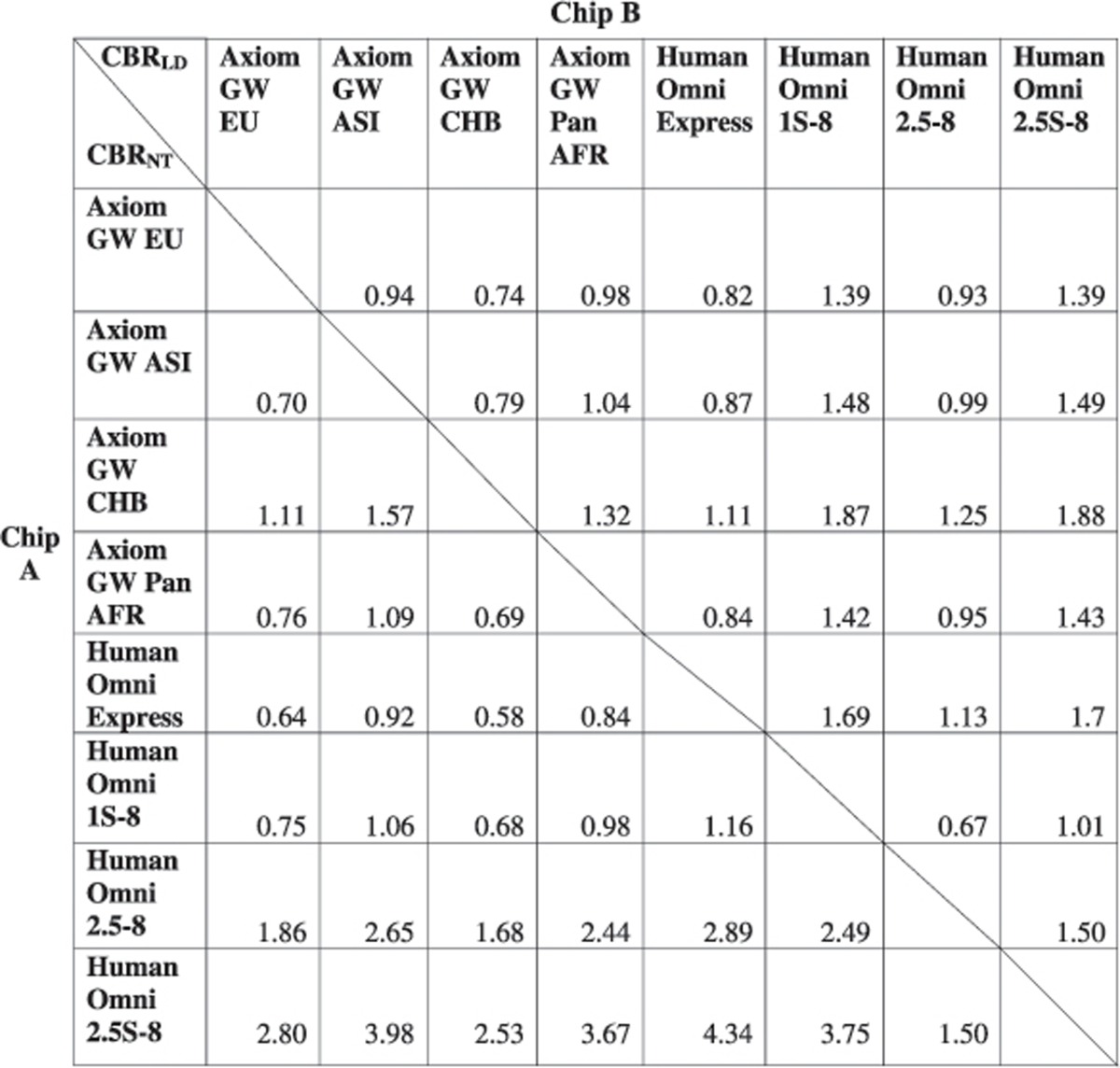
Results for the European reference population were more challenging to interpret. Table 3 illustrates the LD and non-taggable SNPs' CBR measures for the European population. In similarity with the Asian reference population, the Axiom GW CHB and the Human Omni2.5-8 yield good CBRs for both LD and non-taggable SNPs. The Human OmniExpress targeting SNPs with MAF≥5% demonstrates high CBR values for LD but not for non-taggable SNPs. We observed the opposite for the Human Omni2.5S-8. This chip, which targets SNPs with an MAF as small as 1%, offers the most cost-effective capture of non-taggable SNPs but seems to struggle in terms of cost-effective coverage. This was also evident for the LD efficiency measure, for which the Human Omni2.5S-8 only attained slightly improved LD efficiencies than the older Affymetrix Axiom Genome-Wide SNP Human Array 5.0. However, LD efficiencies for the Human Omni2.5-8 might be underestimated, as <50% of the SNPs included on this chip were genotyped as part of the 1000GP reference. As observed previously for coverage and efficiencies, the Axiom GW CEU is not as cost-effective with respect to LD or non-taggable SNPs as the Axiom GW CHB.
Table 3. Cost–benefit ratio CBR(Chip A, Chip B) of different chips with respect to LD and non-taggable SNPs' (NT) efficiency evaluated for the European population.
Discussion
Even though the final choice for a particular SNP chip is often subjective, efficiency measures and CBRs serve as useful comparative tools. Unlike coverage, they adjust for price or its substitute chip size. Using informal prices quoted to us directly by Affymetrix and Illumina, both efficiency and CBR measures suggested that Axiom GW CHB and Axiom GW PanAFR were the most cost-effective and efficient chips for the Asian and African population, respectively. For the European population, we established Human Omni2.5-8 and again the Axiom GW CHB as the preferred choices. As the Axiom GW CHB also demonstrated reasonable cost–benefit and efficiencies for the African population, we would recommend using this chip for discretely admixed samples as well as for the Asian and European samples. It should be noted that the Human Omni2.5-8 was nearly as efficient and offered more genomic coverage. However, according to the price quotes we received this chip was not as competitively priced as the Axiom GW CHB.
Our results also shed new light on the strategy of designing population-optimized SNP chips. Indeed, such a strategy seems compelling for the African population as corroborated by the good performance of the Axiom GW PanAFR for Africans. This population exhibits smaller LD blocks than, for instance, the European population12 as well as greater genetic variation.13 For the African population, real gains are therefore achievable by population optimization in the design of the chips. Furthermore, we were able to confirm the conclusion reached by Jiang et al9 that the use of ‘genotyping platforms tailored to East Asian populations could improve coverage in future studies'. However, designing chips specifically for the European population failed to bring such gains.
An accepted tool to improve coverage is the imputation of SNPs by exploiting LD structures obtained using haplotype maps and/or reference populations. Even though imputation performance was beyond the scope of our work, we believe that a high degree of LD efficiency is an indication of imputation success. Yu and Schaid14 demonstrated that the accuracy of imputation was greatly improved when SNPs were in strong LD. Furthermore, results by Jiang et al9 as well as by Spencer et al4 reveal that imputation is most advantageous to chips that also had a high degree of LD efficiency in our investigations. Another way to enhance the coverage of particular genomic regions of interest is to use customizable chips, now offered by both Illumina and Affymetrix. A well-known example is the MetaboChip with around 200 000 SNPs designed to study metabolic traits.15 In designing such a chip, investigators select SNPs of particular interest to their research question. However, typically SNPs typed as part of conventional chips are added to provide a backbone. Thus, the need for suitable and cost-effective conventional SNP chips remains.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) research training group ‘Scaling Problems in Statistics' (RTG 1644). We also wish to thank Andrew Entwistle for proofreading the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Anderson CA, Pettersson FH, Barrett JC, et al. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet. 2008;83:112–119. doi: 10.1016/j.ajhg.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16:635–643. doi: 10.1038/sj.ejhg.5202007. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K, Schadt EE, Storey JD. Calibrating the performance of SNP arrays for whole-genome association studies. PLoS Genet. 2008;4:e1000109. doi: 10.1371/journal.pgen.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, König I. Weinheim: Jon Wiley & Sons; 2010. A statistical approach to genetic epidemiology. [Google Scholar]
- Flicek P, Ahmed I, Amode MR, et al. Ensembl 2013. Nucleic Acids Res. 2012;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Taylor MS, Cardon LR. Singleton SNPs in the human genome and implications for genome-wide association studies. Eur J Hum Genet. 2008;16:506–515. doi: 10.1038/sj.ejhg.5201987. [DOI] [PubMed] [Google Scholar]
- Jiang L, Willner D, Danoy P, Xu H, Brown MA. Comparison of the performance of two commercial genome-wide association study genotyping platforms in Han Chinese samples. G3. 2013;3:23–29. doi: 10.1534/g3.112.004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Yu HX, Chia JM, Bourque G, et al. A population-based LD map of the human chromosome 6p. Immunogenetics. 2005;57:559–565. doi: 10.1007/s00251-005-0002-4. [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3:1–12. doi: 10.1186/gb-2002-3-7-comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Schaid DJ. Methods to impute missing genotypes for population data. Hum Genet. 2007;122:495–504. doi: 10.1007/s00439-007-0427-y. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kang HM, Ding J, et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e100279. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.