Abstract
Rodents have long been recognized as the principal reservoirs of hantaviruses. However, with the discovery of genetically distinct and phylogenetically divergent lineages of hantaviruses in multiple species of shrews, moles, and insectivorous bats from widely separated geographic regions, a far more complex landscape of hantavirus host distribution, evolution, and phylogeography is emerging. Detailed phylogenetic analyses, based on partial and full-length genomes of previously described rodent-borne hantaviruses and newly detected non-rodent-borne hantaviruses, indicate an Asian origin and support the emerging concept that ancestral non-rodent mammals may have served as the hosts of primordial hantaviruses.
Keywords: hantavirus, Eulipotyphla, Chiroptera, evolution, host-switching
A new frontier
A new frontier Guided by decades-old historical accounts associating hantaviruses (family Bunyaviridae, genus Hantavirus) with shrews [1–4] and moles [5], and empowered by molecular technology and the generosity of museum curators and field mammalogists who willingly granted access to their archival tissue collections, opportunistic investigations have resulted in the identification of genetically distinct and phylogenetically divergent lineages of hantaviruses in multiple species of shrews and moles (order Eulipotyphla, families Soricidae and Talpidae) [6–27] and insectivorous bats (order Chiroptera; see Glossary) [28–32] from widely separated geographic regions. These newfound hantaviruses broaden our knowledge about their reservoir host distribution significantly beyond that of rodents (order Rodentia, families Muridae and Cricetidae). In addition, the discovery of genetically distinct eulipotyphla- and chiroptera-associated hantaviruses enriches our understanding about their evolutionary origins and indicates that their phylogeography is far more complex and ancient than originally contemplated. As such, an emerging new frontier in hantavirus research is now focused on filling major gaps in our understanding about the ecology, host diversity, transmission dynamics, and pathogenic potential of these previously unrecognized, still-orphan hantaviruses, before the next new disease outbreak is documented.
Early history of hantavirus discovery and epidemic activity
The seminal discovery of Hantaan virus (HTNV), as the etiologic agent and prototype virus of hemorrhagic fever with renal syndrome (HFRS), in a Korean striped field mouse (Apodemus agrarius corea) trapped in 1976 [33], serves as a milestone in modern-day hantavirology. The isolation of HTNV has made possible the identification of multiple other HFRS-causing hantaviruses, including Puumala virus (PUUV) in the bank vole (Myodes glareolus) [34], Seoul virus (SEOV) in the Norway rat (Rattus norvegicus) [35], and Dobrava virus (DOBV) in the yellow-necked mouse (Apodemus flavicollis) [36]. Although HFRS has been recognized for more than 1000 years [37], and hantaviruses themselves may have existed for thousands [38] to tens of millions [39–43] of years, renewed attention to these once exotic rodent-borne viruses was prompted by a terrifying outbreak of a rapidly progressive, frequently fatal respiratory disease, now known as hantavirus cardiopulmonary syndrome (HCPS), caused by Sin Nombre virus (SNV) transmitted by the deer mouse (Peromyscus maniculatus) in southwestern USA in 1993 [44–47]. HFRS- and HCPS-causing hantaviruses are harbored by different lineages of rodent subfamilies in the Old and New Worlds, respectively. However, the two clinical syndromes represent a spectrum of disease such that HFRS cases commonly have cardiopulmonary features and HCPS cases frequently exhibit renal insufficiency or dysfunction [48,49].
Evolutionary lessons from rodent reservoirs
To date, rodents have been the only reservoir hosts associated with pathogenic hantaviruses. Therefore, historically, rodents have been an important focus of hantavirus surveillance and research. The breadth of rodent taxa harboring hantaviruses spans two families (Muridae and Cricetidae) and four subfamilies within the suborder Myomorpha of the order Rodentia, and includes mice, rats, lemmings and voles (Table S1 in the supplementary material online; representative human cases are included). Generally, each rodent host species has its own hantavirus species [43], although in some cases various hantavirus genotypes have been described from multiple closely related host species (e.g., [50]). Overall, the rodent-borne hantaviruses can be divided into two major lineages: a highly diverse lineage distributed in New World mice and rats, lemmings, and voles (Cricetidae subfamilies Arvicolinae, Neotominae, and Sigmodontinae), and another distinct, less diverse lineage distributed in Old World mice and rats (Muridae subfamily Murinae) that is sister to a highly diverse and globally distributed lineage of soricid and talpid-borne hantaviruses (Figure 1). Thus, within the rodent-borne lineages, hantaviruses are geographically structured in distribution similar to their host subfamilies, the Old World Murinae, New World Neotominae and Sigmodontinae, and northern hemisphere Arvicolinae. The average sequence divergences at the amino acid level of 26% for the small genomic segment (S), 29% for the medium segment (M), and 23% for the large segment (L) among the rodent-borne hantaviruses represented in Table S1 are substantial and may reflect a high degree of host specialization.
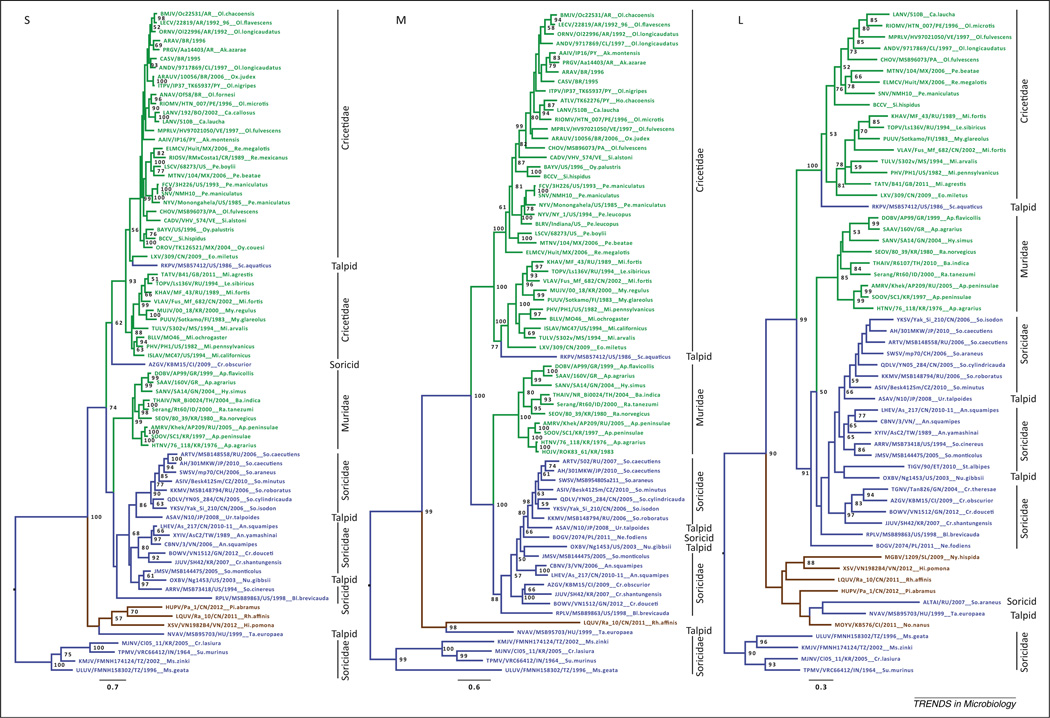
Figure 1.
Hantavirus phylogenies, based on sequences of the S (small), M (medium), and L (large) genomic segments, derived using maximum likelihood methods. The full-length coding region was used for each segment when available. Bayesian methods produced similar topologies. Bootstrap support values for nodes of interest supported >50% of the time (1000 replicates, implemented in RAxML under the GTR+gamma model of evolution [81]) are shown at nodes. Virus lineages are color-coded according to host order (green, Rodentia; blue, Eulipotyphla; brown, Chiroptera), with rodent and eulipotyphlan families indicated to right. Scales bars indicate substitutions per site. Abbreviations: GTR, general time reversible; RAxML, randomized axelerated maximum likelihood. Figure created using RAxML Blackbox (http://embnet.vital-it.ch/raxml-bb/).
In many instances the relatedness among host taxa and the relatedness among their hantaviruses correspond [43], with the most notable exception being the lack of support for monophyly of the rodent-borne hantaviruses in light of their relationships with those found in shrews, moles, and bats [12,31,51]. That each species of rodent hosts a specific hantavirus, or in some cases a few closely related hantaviruses, and the phylogenetic congruency between host and virus within many rodent lineages, has led to the perception that hantaviruses are a model of host–parasite coevolution in the strictest meaning, that of co-divergence. The timing of hantavirus divergence remains a still-unanswered question (Box 1). However, mounting phylogenetic evidence from the recently described hantaviruses across a range of other mammalian hosts confirms that rodent-borne hantaviruses are polyphyletic. That is, they do not form a single lineage, but one that probably involved one or more exchanges of ancestral viruses with other mammals, such as shrews, moles, and/or bats [12,31], or their common ancestor. Evolutionary forces have continued to shape hantavirus diversification in the different host groups and geographic settings.
Box 1. Outstanding questions.
What is the true host range of hantaviruses: are there other host taxonomic orders yet to be discovered that will shed more light on their origins and phylogeography?
Do reservoir hosts share particular properties that would indicate mechanisms of selection operating on hantaviruses?
Do hantaviruses exert selection pressures on their hosts, and could changing traits in hosts be identified that are suggestive of a truly trait-for-trait coevolutionary relationship?
Multi-locus molecular data indicate ongoing uncertainty about some of the mammalian host evolutionary relationships – how might this resolution affect our understanding of the patterns and mechanisms of evolution in hantaviruses?
What drives hantavirus evolutionary diversification, and how can we best test adaptive phenotypes for viruses in the absence of experimental systems for significant host groups such as shrews, moles, and bats? Although Thottapalayam virus and Imjin virus have been isolated from the Asian house shrew and the Ussuri white-toothed shrew, respectively, none of the other newfound hantaviruses from shrews, moles, and bats has been successfully isolated in cell culture: which of these newfound hantaviruses are of highest priority to isolate to better understand their biology and evolution?
What alternative independent methods would allow us to date hantavirus divergences more definitively?
Given the increasing affordability and power of next-generation sequencing methods, what are the possibilities of primer-independent surveillance tools in hantavirus discovery? This review describes how recent discoveries of several divergent hantaviruses have changed our views of the group’s origin and evolution: what is the potential for these new sequencing methods to reveal more divergent viruses that exist beyond the limits of primer-based detection and whose discovery could have even larger impact on our current understanding of their evolutionary history?
New hosts discovered, redefining hantavirus evolutionary trajectories and origins
Although not recognized at the time, Thottapalayam virus (TPMV), a once unclassified virus isolated from an Asian house shrew (Suncus murinus) captured in southern India in 1964 [1], was technically the first hantavirus. However, even after TPMV was shown to be a hantavirus [3] it was assumed to represent a spillover event from a rodent host. Shrews and moles were generally ignored in the ecology and evolution of hantaviruses despite the finding of HFRS antigens or antibodies in the Eurasian common shrew (Sorex araneus), Eurasian pygmy shrew (Sorex minutus), Eurasian water shrew (Neomys fodiens), European mole (Talpa europaea), Chinese mole shrew (Anourosorex squamipes), and northern short-tailed shrew (Blarina brevicauda) [2,4,5,52,53]. Each of these species, and many other species of shrews and moles, representing five subfamilies and two families of Eulipotyphla, have been shown to host hantaviruses (Table S1) whose genetic diversity far surpasses that of hantaviruses carried by rodents [6–27]. In addition, highly divergent lineages of hantaviruses have been identified in seven species of insectivorous bats, including the banana pipistrelle (Neoromicia nanus) in Côte d’Ivoire [28,32], hairy slit-faced bat (Nycteris hispida) in Sierra Leone [29], Pomona roundleaf bat (Hipposideros pomona) in Vietnam [30,32], and Japanese house bat (Pipistrellus abramus), Chinese horseshoe bat (Rhinolophus sinicus), Formosan lesser horseshoe bat (Rhinolophus monoceros), and intermediate horseshoe bat (Rhinolophus affinis) in China [31] (Table S1).
Based on the genetic data available from these and other studies, the shrew- and mole-borne hantaviruses, similarly to those harbored by rodents, do not form a single comprehensive lineage (Figure 1). These molecular data represent near-complete gene sequences for up to all three segments, analyzed in a phylogenetic framework using maximum-likelihood based methods as the most robust and least biased approach to reveal evolutionary relationships. From the resulting phylogenies, one ancient lineage emerges that includes hosts from the soricid subfamilies Crocidurinae and Myosoricinae. Another major lineage shares an ancestor with the Old World rodent-borne lineage, but is much more diverse in sequence divergence, host range, and geographic distribution (Figures 1–3). This lineage includes hosts from two Eulipotyphla families (Talpidae and Soricidae) and three subfamilies (Scalopinae and Talpinae, and Soricinae). This lineage is further subdivided into two Old World lineages and one North American lineage (Figure 3). These overall patterns concur across several phylogenetic presentations [20,22,26,31,43].
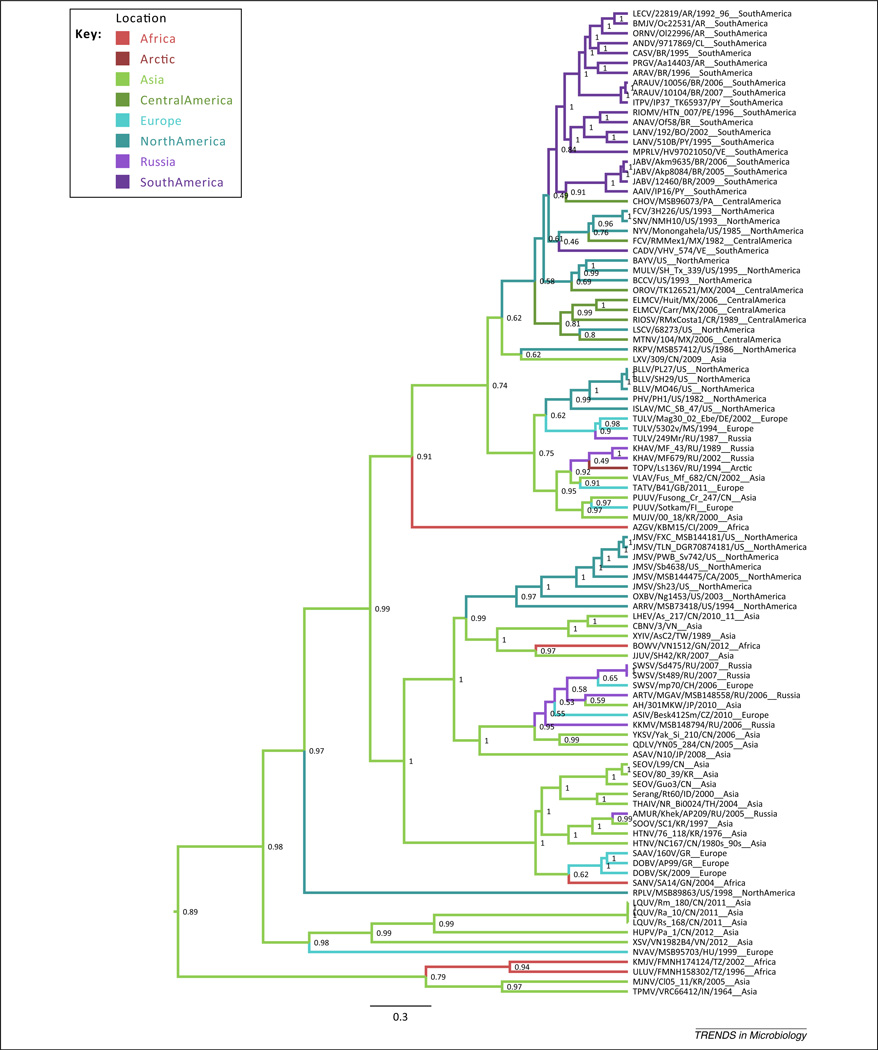
Figure 3.
Phylogeography of hantaviruses. Maximum clade credibility tree based on the S segment from BEAST v1.8 (MCMC run for 50 million generations [82]), with estimates of node geographic state indicated by color of the descending branch and probability at node. The scale bar indicates substitutions per site. Abbreviations: see legends to Figures 1,2. Figure created using BEAST v1.8 (http://beast.bio.ed.ac.uk/Main_Page) and the included TreeAnnotator v.1.8.
Hantaviruses detected in insectivorous bat species, all reported from the Old World (Africa and Asia), form a distinct lineage ancestral to the rodent-borne and the derived shrew-borne lineages, but are probably descended from a shrew-borne ancestral hantavirus according to phylogenetic analysis presented here. The basal position of a Eulipotyphla lineage represented by the prototype TPMV suggests that the origins of hantaviruses were in shrews and moles (Figure 1). Guo and colleagues [31] also recognized that shrews or moles could represent the ancestral host, although their data suggest bats with equal probability. Two independent methods support the basal position of TPMV and its sister taxa: Imjin virus (MJNV) in the Ussuri white-toothed shrew (Crocidura lasiura) [15], Uluguru virus (ULUV) in the Geata mouse shrew (Myosorex geata), and Kilimanjaro virus (KMJV) in the Kilimanjaro mouse shrew (Myosorex zinki) [54]. First, hantavirus sequences were aligned and rooted to another member of the family Bunyaviridae, Bunyamwera virus (type species of the Orthobunyavirus genus) for the glycoprotein- coding region, or M segment (the only segment reliably alignable with other genera in the family) [55], and maximum-likelihood phylogenetic estimation placed this shrew-borne lineage basal to the other hantaviruses relative to the Bunyamwera outgroup (Figure S1). Second, estimation of the tree root for the hantavirus ingroup for all segments was executed in BEAST (Bayesian evolutionary analysis by sampling trees) v2.0.2 [56], using a Bayesian MCMC (Markov Chain Monte Carlo) framework, and stably converged on the TPMV/MJNV lineage in the topology shown in Figure 1.
Given that a lineage of shrew-borne hantaviruses forms the root of the hantavirus diversification, it is likely that the primordial host of hantaviruses was a shrew or mole (within the order Eulipotyphla). Ancestral state reconstruction based on Bayesian methods (BayesTraits v2.0, [57]) identified the probability of the root host state being a rodent as 0.011). Guo and coworkers [31] reported similar findings based on their extensive multi-year survey for hantaviruses in bats and shrews in China. Together, these data suggest that rodents, as we know them today, were not the original hosts of hantaviruses. Others have suggested that the ancestral host may have been an early placental mammal from which shrews, moles, bats, and rodents diverged, along with their viruses, and that this ancestral mammal may have acquired its hantaviruses from insects, where other bunyaviruses occur [12,43].
Coevolution, co-divergence, and host-switching
As has been noted, hantaviruses show an astonishing degree of phylogenetic correspondence with their hosts. Specifically, closely related hantaviruses are generally found in closely related hosts as opposed to more distant hosts. Topological congruence in divergence patterns between hantaviruses and their hosts is widespread throughout hantavirus evolutionary history, and in particular hantavirus lineages is significantly supported over other patterns, such as host-switching [13,31,58]. Overall, hantavirus diversification is highly structured by host identity at the host subfamily, family, and order levels {Bayesian tip-association significance testing (BaTS) program statistics, association index (AI), and Fitch parsimony statistic (PS) P = 0 indicate that the probability of the observed degree of phylogenetic correlation or structure in the data occurring by chance is zero [59]}. Clearly these groups are coupled in evolution but, because hantaviruses and their hosts presumably evolve at vastly different rates, strict coevolution between hantaviruses and their hosts, defined as reciprocal change over the same timescales, remains a question (Box 1).
In some of its earliest uses, coevolution has variously been described as gene-for-gene changes in the parasite and host due to the selective pressures they exerted on each other [60], or more generally the evolutionary influences that plants and herbivorous insects exert on each other, without the restriction of direct gene-for-gene reciprocity or temporal congruence (occurring on the same timescale). Furthermore, coevolution has been used to describe not only specific changes between reciprocating partners diverging simultaneously (strictest use) but also the diffuse indirect evolutionary interactions between groups of taxa, such as the evolution of immune defense and pathogen avoidance in a general sense. These various scales of evolutionary interaction can all lead to congruence in the diversification patterns of interacting taxa. However, the process of co-divergence, or parallel cladogenesis, requires that speciation in both partners occurs in concert, resulting in topological and temporal congruence (reviewed in [60]).
Dating hantavirus divergence and estimating rates of evolution
Although hantaviruses and their mammalian hosts show significant topological congruity throughout their evolutionary histories (Figure 2), it is not known whether their divergence occurred on similar timescales. The mammalian host taxonomic order Eutheria (infraclass Placentalia), which includes all mammals indigenous to North America, Europe, Africa, and Asia (except the opossum), arose on the order of 160 million years before present [61]. This is much earlier than viral origins projected under the slowest evolutionary rates observed or estimated in hantaviruses (which leave no fossil traces): the slowest rate estimate based on a subset of hantaviruses sampled over different time periods and scales is 4.245 × 10−4 (0.000299–0.00055) substitutions per site per year [62]. Extrapolated back through time, this rate would place the origin of hantaviruses at ~3500 years before present. Similar methods have placed the origin of the rodent-borne hantaviruses at 2000 years [38]. However, extrapolation assumes that the estimated rates are representative of and have been similar throughout the evolutionary history of hantaviruses and across lineages, which is probably not the case given that these rates are estimated on a small subset of the diversity and that there is evidence of lineage-specific selection.
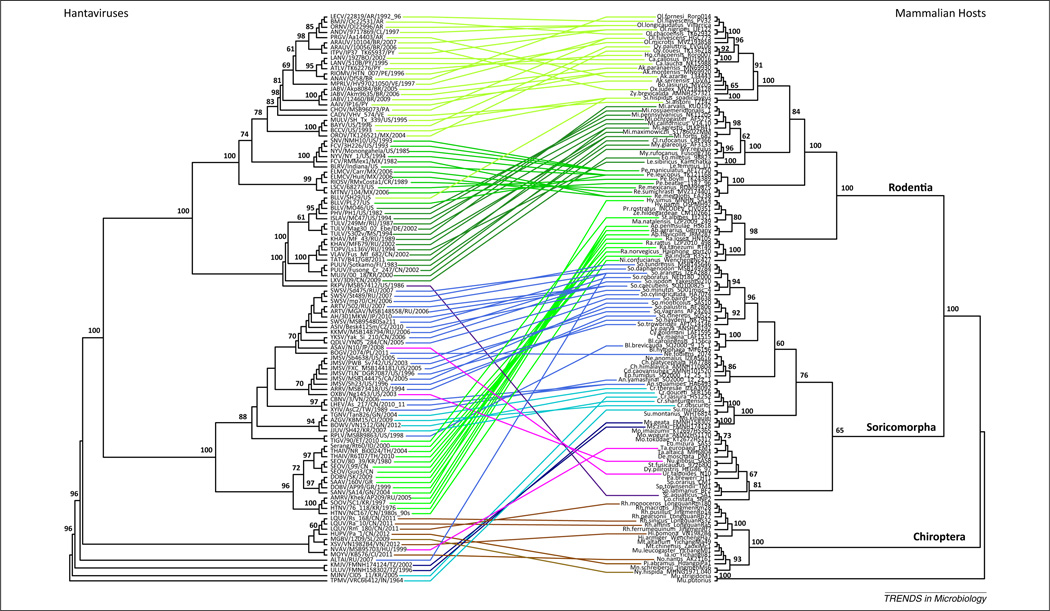
Figure 2.
Hantavirus and mammalian host co-phylogenies. Maximum likelihood (ML) phylogenetic trees of hantaviruses based on concatenated S, M, and L coding sequences (left) and their hosts based on cytochrome b sequences (right). Hantavirus tree is rooted on TPMV and relatives as indicated by BEAST v1.8 MCMC analyses to 50 million generations [69]. Host tree is rooted on Carnivora representatives. Horizontal lines represent host associations, colored by host class (green, Rodentia; blue/purple/violet, Eulipotyphla; brown, Chiroptera) and further shaded within these classes by host subfamily. Bootstrap support, based on 1000 ML replicates implemented in RAxML Blackbox under the GTR+gamma model of evolution [81], is shown at key nodes. Abbreviations: BEAST, Bayesian evolutionary analysis by sampling trees (http://beast.bio.ed.ac.uk/); MCMC, Markov chain Monte Carlo; TMPV, Thottapalayam virus; other abbreviations are given in Figure 1 legend. Figure created using RAxML Blackbox and R statistical program, ape package (http://cran.r-project.org/web/packages/ape/).
Determining the date of hantavirus origin and the rates of their diversification into major lineages correlated with host lineages is challenging in the absence of an independent method of calibration. In most cases, the evolutionary history of a group is calibrated against a fossil record or using specimens drawn from different times. Several virus evolutionary histories have been calibrated the latter way, from viruses sampled over time [38,58,62–65], with best results when the span of sampling is of a scale with the evolutionary history and when rates are relatively consistent through time and across lineages. Tip-dated methods have given variable estimates of the rate of evolution of hantaviruses depending on the lineage, on the order of 10−2 to 10−4 substitutions per site per year [38,58,62,65]. Direct measurements of evolutionary rates in specific hantaviruses currently circulating in nature have been reported on the order of 10−3 substitutions per site per year [66,67].
The alternative method uses the fossil record of the host groups to calibrate hantavirus evolutionary rates [40,42]. The justification for this is based on parsimony: repeated closely matched (in pattern) bifurcations in the evolutionary histories of parasites and hosts, along with the strict dependencies of parasite life histories on their hosts, could be consistent with co-divergence. If this co-divergence began as early as the diversification of the placental mammals into the current hantavirus host groups, then hantavirus origins in mammals could date back to 90–100 million years before present [43]. Host-calibrated estimates of hantavirus evolutionary rates range from 10−6 to 10−7 substitutions per site per year [40,42] based on virus synonymous substitutions over timescales of the host fossil record and major geological events such as the separation of New and Old World Microtus 1.8–2.0 million years ago by the Bering Sea [68].
Mechanisms of hantavirus divergence
The topological congruence of evolutionary divergence patterns between hantaviruses and their hosts may alternatively arise due to host tracking, sometimes described as phylogenetic conservatism or preferential host-switching [69,70]. Hantaviruses show a high degree of host specificity, in which a given mammalian species is usually infected with a single hantavirus species, and distinct hantaviruses in turn are associated with only one host species or occasionally multiple but closely related species (Figure 2). This suggests a requirement for high host fidelity to be successful, and the force of this selection would be particularly strong based on strict dependence of the virus on a single host for its life cycle. Strong selection for specific compatibility with the host would lead to patterns of congruency in the phylogenetic trees of the interacting taxa, a pattern that, uncalibrated, would appear consistent with co-divergence or co-speciation.
Natural selection has been an important driver of hantavirus divergence. Natural selection consists of both negative selective forces curtailing the propagation of less fit forms as well as positive selection leading to the increase and eventual fixation of more fit forms. Hantavirus diversity has been limited and shaped by negative selection, as suggested by proportionally fewer substitutions affecting phenotype (as in the encoded amino acids) than expected [66,67,71]. At the same time, positive selection has presumably played a role in shaping hantavirus diversity, although the data are limited and the precise mechanisms are still unknown. Lin and colleagues [58] report non-significant phylogenetic signals for positive selection in S and M segments for particular internal branches at clade-defining sites within the Murinae-borne hantavirus lineage. However, Razzauti and coworkers [71] suggest that particular ‘preferred’ genotypes of PUUV may persist over time, based on a 5 year study of bank voles from Finland. It is possible that positive selection plays an episodic role in shaping hantavirus diversity, for example following an opportunity for a host switch. Positive-selective sweeps may have operated only early in the origination of the major lineages, as suggested by Sironen and Plyusnin [72]. Episodic, rare positive selection is difficult to detect using current phylogenetic methods owing to a lack of statistical power. The form and mechanisms of positive selection remain unresolved mysteries in understanding the processes of diversification of hantaviruses (Box 1).
Genetic drift is the alternative force of evolutionary change that, unlike natural selection, does not act on fitness differences but instead is neutral with respect to the phenotype of a variant. Under genetic drift, variation can become randomly fixed or eliminated, resulting in change over generations. Genetic drift has presumably been the underlying force of hantavirus diversification in the absence of selection whenever viruses have become isolated in new hosts.
The diversification of hantaviruses across disparate host groups has involved host switches at specific points in their evolutionary history. One possible scenario supported by phylogenies presented here and by Guo and coworkers [31] would have involved a host switch from a shrew ancestor into bats, and subsequently into rodents, followed by additional cross-species transmission events within some of these orders [13,20,58,73], that may have happened twice (first into New World and globally distributed rodents, and then into Old World rodents). If the ancestral state for hantaviruses was a chiropteran host, then again a switch or switches into shrews as well as two switches into rodents may have occurred. If instead a rodent representative were the ancestral host of hantaviruses, a host switch or switches would have occurred into shrews, moles, and bats. Finally, if an ancestral placental mammal to all these host groups was the original host, at least one host switch has still been proposed between Muridae rodents and shrew hosts [43]. The recent discoveries of novel hantaviruses and host records has exposed these and numerous other host-switching events within several hantavirus lineages involving different host families, subfamilies and genera, disrupting the concordant virus–host pattern of coevolution and underlying the further diversification of hantaviruses [8,13,17,22,28,31, 58,70,74]. Some patterns have begun to emerge that bear further investigation. For example, all of the hantavirus-infected moles have been involved in host-switching events (Figure 2, purple lines). That is, their hantaviruses each come from very different lineages of hantaviruses that do not otherwise occur in talpids and, furthermore, in many cases these viruses appear less derived than their relatives: Rockport virus (RKPV) [20] is basal to the globally distributed rodent-borne hantavirus lineage, Asama virus (ASAV) [10] is basal to a globally distributed shrew-borne clade, and Nova virus (NVAV) [12] falls out as distantly related to other shrew-borne and bat-borne lineages. This might mean that talpids played an important role in the origin and diversification of hantaviruses. By contrast, strains of Bloodland Lake virus (BLLV) are found in disparate hosts, spilling over from a Microtis host origin into Peromyscus and Sigmodon species (Table S1), suggesting that certain hantaviruses may have an enhanced propensity for host capture.
As enveloped viruses with tripartite negative-sense, single-stranded RNA genomes, another important mechanism of hantavirus diversification may be reassortment of gene segments or recombination within the same segments of different individuals. Incongruent evolutionary histories among the different gene segments, sometimes affecting the ancestral identity of major lineages [31], highlight the importance of reassortment in evolutionary innovation. For example, RKPV and Azagny virus (AZGV) [19] are talpid- and soricid-borne hantaviruses, respectively, that both show evidence of being reassortants because their gene segments differ from each other in their evolutionary histories (Figure 1). Some or all segments of both viruses also show a phylogenetic association with rodent-borne lineages instead of other shrew-borne viruses, suggesting the possibility that a reassortment event may underlie a host-switching event, leading to the origination of particular rodent-borne lineages. Instances of reassortment have been reported in various hantaviruses [65,66,71,75–77], attributed in one case to host spill-over [75]. Recombination is reported less often [78], but phylogenetic evidence reflected in weak bootstrap support at relevant nodes (Figure 1) suggests that the S segments of both RKPV and AZGV could have been subject to recombination. Thus, reassortment and possibly recombination may be important sources of diversity in hantaviruses, specifically affecting host capture.
The emerging picture of hantavirus evolution, in light of the growing data on host distribution, remains marked by a high degree of congruence in the diversification patterns of virus and host over different periods and lineages. The timing and mechanisms behind virus divergence are areas for further discussion, as is the importance of reassortment or other factors such as geographic or ecological opportunities in host-switching (Box 1).
Phylogeographic patterns
Phylogeography has played an important role in the evolution of hantaviruses and accounts for significant phylogenetic structure in hantavirus diversification. Geographic structure has been reported at multiple spatial scales [31,67,79] and is greatly affected by host distribution [65,67]. Within both shrew-borne and rodent-borne lineages there are groups restricted to New World versus Old World distributions, as well as lineages that are globally distributed (Figure 3). In some cases, virus exchange among distant hosts has been facilitated by geographic proximity, as in Oxbow virus (OXBV) [13], Jemez Springs virus (JMSV) [11], ASAV [10], and relatives [31]. Estimates of the geographic origins of the major lineages of hantaviruses place Asia at the origin of the entire group [19,31], with a posterior probability of 0.89 based on the data reviewed here (Figure 3; executed in BEAST v1.8, under models by [80]). Figure 3 shows that, from a probable Asian origin, hantaviruses independently spread to the Americas separately in shrews and rodents, and that the rodent niche of the Americas was a particularly good one for hantaviruses, given their subsequent radiation into a variety of rodent hosts across this region.
Concluding remarks
Until recently hantaviruses were viewed as being hosted exclusively by rodents. Today the picture of hantaviruses is that of a highly diverse genus distributed globally across diverse taxonomic orders and ecological niches, whose biology, evolution, and propensity to cause disease and/or spill over into humans remains largely unknown. That shrews, moles, and insectivorous bats harbor hantaviruses which exhibit far greater genetic diversity than those carried by rodents heralds a compelling conceptual framework for reconstructing the evolutionary origins of hantaviruses. In addition, because all other members of the Bunyaviridae family involve insect or arthropod vectors, the evolutionary history of hantaviruses may have originated with the emergence of a primordial virus through species jumping from insect or arthropod hosts into an early eulipotyphlan or chiropteran ancestor, with multiple subsequent host-switching events. New knowledge from these still-early investigations will continue to redefine our understanding and bring new insights into the mechanisms of hantavirus evolution and emergence.
Supplementary Material
Acknowledgments
Accelerated acquisition of new knowledge about the spatial and temporal distribution, host range, genetic diversity, and evolution of hantaviruses was made possible through the collaborative contributions of the following individuals: Sergey A. Abramov, Luck Ju Baek, Chris Conroy, Joseph A. Cook, Christiane Denys, Laurie Dizney, Paul E. Doetsch, Sylvain Dubey, Jacob A. Esselstyn, Janusz Hejduk, Monika Hilbe, Andrew G. Hope, Jean-Pierre Hugot, Francois Jacquet, Blaise Kadjo, Michael Y. Kosoy, Takeshi Kurata, Eileen Lacey, Aude Lalis, Pawel P. Liberski, Burton K. Lim, Janusz Markowski, Shigeru Morikawa, Vivek R. Nerurkar, Violaine Nicolas, Satoshi D. Ohdachi, Nobuhiko Okabe, Maria Puorger, Luis A. Ruedas, Nuankanya Sathirapongsasuti, Beata Sikorska, Jin-Won Song, Ki-Joon Song, William T. Stanley, Laarni Sumibcay, Ninh U. Truong, Thang T. Truong, Liudmila N. Yashina, and Hon-Tsen Yu. We also thank Brian Simison, of the California Academy of Sciences Center for Comparative Genomics, for computational resources, and Durrell Kapan for conceptual comments. This work was supported in part by the National Institutes of Health (grants R01AI075057, U54AI065359 and P20GM103516), the Japan Society for the Promotion of Science (grant 24405045), and grant H25-Shinko-Ippan-008 for Research on Emerging and Re-emerging Infectious Diseases.
Glossary
- Chiroptera
an order of placental mammals, the bats. They are grouped in the Laurasiatheria lineage together with Eulipotyphla and some other mammalian orders that do not include Rodentia.
- Co-divergence or co-speciation
terms used to refer to the reciprocal differentiation or speciation of interacting taxa, presumably in response to evolutionary pressures exerted by one on the other and vice versa. In the strictest sense, this would involve ‘one-for-one’ changes in taxon pairs and occur on similar timescales.
- Eulipotyphla
an order of placental mammals that includes most taxa formerly in Insectivora (dissolved when it was shown to be polyphyletic), such as the families Soricidae (true shrews) and Talpidae (moles and shrew moles). Used here in place of Soricomorpha to refer to soricids and talpids because Soricomorpha is paraphyletic: Soricidae are more closely related to the family Erinaceidae (which includes hedgehogs) than Talpidae.
- Genetic drift
the fixation of genotypes through random processes, tending to build-up over time and differentiate populations isolated from gene flow. Noted to be an important mechanism of evolution in hantaviruses that are largely isolated in specific hosts.
- Gene flow
the movement of genetic forms into new populations, such as those formerly isolated by geography, or even by host type.
- Host-switching or host capture
the stable establishment of a parasite into a new and distinct host species.
- Monophyletic
a evolutionary term used in phylogenetics to refer to a group of taxa that includes an ancestral species and all its descendants. See also paraphyletic and polyphyletic.
- Natural selection
the differential representation of types in future generations caused by fitness differences (where fitness is the ability to reproduce, or survive to reproduce). Negative selection results in proportionally fewer individuals of a given type in subsequent generations and ultimately leads to the disappearance of that type, whereas positive selection results in an increase in the proportion of individuals of a given type in future generations and can ultimately lead to the fixation of that type to 100%.
- Paraphyletic
a evolutionary term used in phylogenetics to refer to a group of taxa that includes an ancestral species and most of its descendants minus a monophyletic group. See also monophyletic and polyphyletic.
- Polyphyletic
a evolutionary term used in phylogenetics to refer to groups of taxa that do not exclusively share a common ancestor. See also monophyletic and paraphyletic.
- Reassortment
an exchange of segment(s) between parental viruses. A potentially important source of innovation for segmented viruses, particular hantaviruses show evidence of reassortment. For example, AZGV, RKPV, and LXV all show incongruities in terms of their phylogenetic positions across S, M, and L segments, suggesting that these segments have different evolutionary histories.
- Recombination
the recombining of genetic material from two ‘parental’ viruses. Because the hantavirus genome is separated into three segments, we use ‘recombination’ here to refer specifically to rearrangement of the genomic material between parental viruses within a given segment. AZGV, RKPV, and RPLV represent recombinants for the S segment: weak bootstrap support values reflect uncertainty in their evolutionary histories.
- Rodentia
an order of placental mammals, the rodents, that includes several families discussed here, such as Muridae (Old World mice, rats, and gerbils) and Cricetidae (New World mice and rats, hamsters, voles, lemmings). Rodentia falls into the lineage Euarchontoglires, as opposed to the group Laurasiatheria that includes shrews, moles (Eulipotyphla), and bats (Chiroptera).
Footnotes
Publisher's Disclaimer: Disclaimer statement
The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.tim.2014.04.008.
References
- 1.Carey DE, et al. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India. Indian J. Med. Res. 1971;59:1758–1760. [PubMed] [Google Scholar]
- 2.Gavrilovskaya IN, et al. Features of circulation of hemorrhagic fever with renal syndrome (HFRS) virus among small mammals in the European U.S.S.R. Arch. Virol. 1983;75:313–316. doi: 10.1007/BF01314898. [DOI] [PubMed] [Google Scholar]
- 3.Zeller HG, et al. Electron microscopic and antigenic studies of uncharacterized viruses. II. Evidence suggesting the placement of viruses in the family Bunyaviridae. Arch. Virol. 1989;108:211–227. doi: 10.1007/BF01310935. [DOI] [PubMed] [Google Scholar]
- 4.Gligic A, et al. Hemorrhagic fever with renal syndrome in Yugoslavia: epidemiologic and epizootiologic features of a nationwide outbreak in 1989. Eur. J. Epidemiol. 1992;8:816–825. doi: 10.1007/BF00145326. [DOI] [PubMed] [Google Scholar]
- 5.Tkachenko EA, et al. Potential reservoir and vectors of haemorrhagic fever with renal syndrome (HFRS) in the U.S.S.R. Ann. Soc. Belg. Med. Trop. 1983;63:267–269. [PubMed] [Google Scholar]
- 6.Arai S, et al. Hantavirus in northern short-tailed shrew, United States. Emerg. Infect. Dis. 2007;13:1420–1423. doi: 10.3201/eid1309.070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klempa B, et al. Novel hantavirus sequences in shrew, Guinea. Emerg. Infect. Dis. 2007;13:520–522. doi: 10.3201/eid1303.061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J-W, et al. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus) Virol. J. 2007;4:114. doi: 10.1186/1743-422X-4-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J-W, et al. Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg. Infect. Dis. 2007;13:1784–1787. doi: 10.3201/eid1311.070492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arai S, et al. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides) Proc. Natl. Acad. Sci. U.S.A. 2008;105:16296–16301. doi: 10.1073/pnas.0808942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arai S, et al. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am. J. Trop. Med. Hyg. 2008;78:348–351. [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HJ, et al. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea) PLoS ONE. 2009;4:e6149. doi: 10.1371/journal.pone.0006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HJ, et al. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii) Virology. 2009;388:8–14. doi: 10.1016/j.virol.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang HJ, et al. Genetic diversity and phylogeography of Seewis virus in the Eurasian common shrew in Finland and Hungary. Virol. J. 2009;6:208. doi: 10.1186/1743-422X-6-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J-W, et al. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura) J. Virol. 2009;83:6184–6191. doi: 10.1128/JVI.00371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HJ, et al. Novel hantavirus in the flat-skulled shrew (Sorex roboratus) Vector Borne Zoonotic Dis. 2010;10:593–597. doi: 10.1089/vbz.2009.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yashina LN, et al. Seewis virus: phylogeography of a shrew-borne hantavirus in Siberia, Russia. Vector Borne Zoonotic Dis. 2010;10:585–591. doi: 10.1089/vbz.2009.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu SH, et al. Genetic diversity of Imjin virus in the Ussuri white-toothed shrew (Crocidura lasiura) in the Republic of Korea, 2004–2010. Virol. J. 2011;8:56. doi: 10.1186/1743-422X-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang HJ, et al. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d’Ivoire. Virol. J. 2011;8:373. doi: 10.1186/1743-422X-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang HJ, et al. Shared ancestry between a newfound moleborne hantavirus and hantaviruses harbored by cricetid rodents. J. Virol. 2011;85:7496–7503. doi: 10.1128/JVI.02450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HJ, et al. Genetic diversity of Thottapalayam virus, a Hantavirus harbored by the Asian house shrew (Suncus murinus) in Nepal. Am. J. Trop. Med. Hyg. 2011;85:540–545. doi: 10.4269/ajtmh.2011.11-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arai S, et al. Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology. 2012;424:99–105. doi: 10.1016/j.virol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu SH, et al. Complete genome sequence and molecular phylogeny of a newfound hantavirus harbored by the Doucet’s musk shrew (Crocidura douceti) in Guinea. Infect. Genet. Evol. 2013;20:118–123. doi: 10.1016/j.meegid.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu SH, et al. High prevalence of Nova hantavirus infection in the European mole (Talpa europaea) in France. Epidemiol. Infect. 2014;142:1167–1171. doi: 10.1017/S0950268813002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu SH, et al. Boginia virus, a newfound hantavirus harbored by the Eurasian water shrew (Neomys fodiens) in Poland. Virol. J. 2013;10:160. doi: 10.1186/1743-422X-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radosa L, et al. Detection of shrew-borne hantavirus in Eurasian pygmy shrew (Sorex minutus) in Central Europe. Infect. Genet. Evol. 2013;19:403–410. doi: 10.1016/j.meegid.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Resman K, et al. Molecular evidence and high genetic diversity of shrew-borne Seewis virus in Slovenia. Virus Res. 2013;177:113–117. doi: 10.1016/j.virusres.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 28.Sumibcay L, et al. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d’Ivoire. Virol. J. 2012;9:34. doi: 10.1186/1743-422X-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss S, et al. Hantavirus in bat, Sierra Leone. Emerg. Infect. Dis. 2012;18:159–161. doi: 10.3201/eid1801.111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arai S, et al. Novel bat-borne hantavirus, Vietnam. Emerg. Infect. Dis. 2013;19:1159–1161. doi: 10.3201/eid1907.121549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo W-P, et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu SH, et al. Molecular phylogeny of hantaviruses harbored by insectivorous bats in Côte d’Ivoire and Vietnam. Viruses. 2014;6:1897–1910. doi: 10.3390/v6051897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HW, et al. Isolation of the etiologic agent of Korean hemorrhagic fever. J. Infect. Dis. 1978;137:298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- 34.Brummer-Korvenkontio M, et al. Nephropathia epidemica: detection of antigen in bank voles and serologic diagnosis of human infection. J. Infect. Dis. 1980;141:131–134. doi: 10.1093/infdis/141.2.131. [DOI] [PubMed] [Google Scholar]
- 35.Lee HW, et al. Isolation of Hantaan virus, the etiologic agent of Korean hemorrhagic fever, from wild urban rats. J. Infect. Dis. 1982;146:638–644. doi: 10.1093/infdis/146.5.638. [DOI] [PubMed] [Google Scholar]
- 36.Avsic-Zupanc T, et al. Characterization of Dobrava virus: a hantavirus from Slovenia, Yugoslavia. J. Med. Virol. 1992;38:132–137. doi: 10.1002/jmv.1890380211. [DOI] [PubMed] [Google Scholar]
- 37.Klein SL, Calisher CH. Emergence and persistence of hantaviruses. Curr. Top. Microbiol. Immunol. 2007;315:217–252. doi: 10.1007/978-3-540-70962-6_10. [DOI] [PubMed] [Google Scholar]
- 38.Souza WM, et al. Phylogeography and evolutionary history of rodent-borne hantaviruses. Infect. Genet. Evol. 2014;21:198–204. doi: 10.1016/j.meegid.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Plyusnin A, et al. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 40.Hughes AL, Friedman R. Evolutionary diversification of protein-coding genes of hantaviruses. Mol. Biol. Evol. 2000;17:1558–1568. doi: 10.1093/oxfordjournals.molbev.a026254. [DOI] [PubMed] [Google Scholar]
- 41.Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 2001;256:47–75. doi: 10.1007/978-3-642-56753-7_4. [DOI] [PubMed] [Google Scholar]
- 42.Sironen T, et al. Molecular evolution of Puumala hantavirus. J. Virol. 2001;75:11803–11810. doi: 10.1128/JVI.75.23.11803-11810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plyusnin A, Sironen T. Evolution of hantaviruses: co-speciation with reservoir hosts for more than 100 MYR. Virus Res. 2014 doi: 10.1016/j.virusres.2014.01.008. http://dx.doi.org/10.1016/j.virusres.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 44.Nerurkar VR, et al. Genetically distinct hantavirus in deer mice. Lancet. 1993;342:1058–1059. doi: 10.1016/0140-6736(93)92917-i. [DOI] [PubMed] [Google Scholar]
- 45.Nichol ST, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 46.Childs JE, et al. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 1994;169:1271–1280. doi: 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- 47.Nerurkar VR, et al. Genetic evidence for a hantavirus enzootic in deer mice (Peromyscus maniculatus) captured a decade before the recognition of hantavirus pulmonary syndrome. Virology. 1994;204:563–568. doi: 10.1006/viro.1994.1570. [DOI] [PubMed] [Google Scholar]
- 48.Jonsson CB, et al. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clement J, et al. Hemorrhagic fever with renal syndrome in the new, and hantavirus pulmonary syndrome in the old world: paradi(se)gm lost or regained? Virus Res. 2014 doi: 10.1016/j.virusres.2013.12.036. http://dx.doi.org/10.1016/j.virusres.2013.12.036. [DOI] [PubMed] [Google Scholar]
- 50.Chu Y-K, et al. Phylogenetic and geographical relationships of hantavirus strains in eastern and western Paraguay. Am. J. Trop. Med. Hyg. 2006;75:1127–1134. [PMC free article] [PubMed] [Google Scholar]
- 51.Yanagihara R, et al. Hantaviruses: rediscovery and new beginnings. Virus Res. 2014 doi: 10.1016/j.virusres.2013.12.038. http://dx.doi.org/10.1016/j.virusres.2013.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee PW, et al. Partial characterization of Prospect Hill virus isolated from meadow voles in the United States. J. Infect. Dis. 1985;152:826–829. doi: 10.1093/infdis/152.4.826. [DOI] [PubMed] [Google Scholar]
- 53.Chen SZ. Strains of epidemic hemorrhagic fever virus isolated from the lungs of C. russula and A. squamipes. Zhonghua Yu Fang Yi Xue Za Zhi. 1986;20:261–263. (in Chinese) [PubMed] [Google Scholar]
- 54.Kang HJ, et al. Expanded host diversity and geographic distribution of hantaviruses in sub-Saharan Africa. J. Virol. 2014 doi: 10.1128/JVI.00285-14. http://dx.doi.org/10.1128/JVI.00285-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King AM, et al. Virus Taxonomy: Classification and Nomenclature of Viruses. 9th edn. Elsevier Academic Press; 2012. [Google Scholar]
- 56.Bouckaert R, et al. BEAST2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pagel M, et al. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 58.Lin X-D, et al. Cross-species transmission in the speciation of the currently known murinae-associated hantaviruses. J. Virol. 2012;86:11171–11182. doi: 10.1128/JVI.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker J, et al. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 2008;8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Futuyma DJ, Slatkin M. Coevolution. Sinauer Associates; 1983. [Google Scholar]
- 61.Luo Z-X, et al. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 62.Ramsden C, et al. High rates of molecular evolution in hantaviruses. Mol. Biol. Evol. 2008;25:1488–1492. doi: 10.1093/molbev/msn093. [DOI] [PubMed] [Google Scholar]
- 63.Lemey P, et al. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Twiddy SS, et al. Inferring the rate and time-scale of dengue virus evolution. Mol. Biol. Evol. 2003;20:122–129. doi: 10.1093/molbev/msg010. [DOI] [PubMed] [Google Scholar]
- 65.Lin X-D, et al. Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today. J. Virol. 2012;86:972–981. doi: 10.1128/JVI.00725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Black WC, et al. Temporal and geographic evidence for evolution of Sin Nombre virus using molecular analyses of viral RNA from Colorado, New Mexico and Montana. Virol. J. 2009;6:102. doi: 10.1186/1743-422X-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres-Perez F, et al. Spatial but not temporal co-divergence of a virus and its mammalian host. Mol. Ecol. 2011;20:4109–4122. doi: 10.1111/j.1365-294X.2011.05241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffmann RS, Koebbl JW. Zoogeography. In: Tamarin RH, editor. Biology of New World Microtus. American Society of Mammalogists Special Publication No. 8, American Society of Mammalogists; 1985. pp. 84–115. [Google Scholar]
- 69.Charleston MA, Robertson DL. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 2002;51:528–535. doi: 10.1080/10635150290069940. [DOI] [PubMed] [Google Scholar]
- 70.Ramsden C, et al. Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence. Mol. Biol. Evol. 2009;26:143–153. doi: 10.1093/molbev/msn234. [DOI] [PubMed] [Google Scholar]
- 71.Razzauti M, et al. Microevolution of Puumala hantavirus during a complete population cycle of its host, the bank vole (Myodes glareolus) PLoS ONE. 2013;8:e64447. doi: 10.1371/journal.pone.0064447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sironen T, Plyusnin A. Genetics and evolution of hantaviruses. In: Plyusnin A, Elliott RM, editors. Bunyaviridae: Molecular and Cellular Biology. Caister Academic Press; 2011. pp. 61–94. [Google Scholar]
- 73.Vapalahti O, et al. Isolation and characterization of a hantavirus from Lemmus sibiricus: evidence for host switch during hantavirus evolution. J. Virol. 1999;73:5586–5592. doi: 10.1128/jvi.73.7.5586-5592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nemirov K, et al. Phylogenetic evidence for host switching in the evolution of hantaviruses carried by Apodemus mice. Virus Res. 2002;90:207–215. doi: 10.1016/s0168-1702(02)00179-x. [DOI] [PubMed] [Google Scholar]
- 75.Chu Y-K, et al. Phylogenetic exploration of hantaviruses in Paraguay reveals reassortment and host switching in South America. Virol. J. 2011;8:399. doi: 10.1186/1743-422X-8-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han G-Z, Worobey M. Homologous recombination in negative sense RNA viruses. Viruses. 2011;3:1358–1373. doi: 10.3390/v3081358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J, et al. Genetic analysis of hantaviruses and their rodent hosts in central-south China. Virus Res. 2012;163:439–447. doi: 10.1016/j.virusres.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Nikolic V, et al. Evidence of recombination in Tula virus strains from Serbia. Infect. Genet. Evol. 2014;21:472–478. doi: 10.1016/j.meegid.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 79.Medina RA, et al. Ecology, genetic diversity, and phylogeographic structure of Andes virus in humans and rodents in Chile. J. Virol. 2009;83:2446–2459. doi: 10.1128/JVI.01057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemey P, et al. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stamatakis A, et al. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 82.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.