Abstract
Androgen deprivation is the standard systemic treatment for advanced prostate cancer (PCa), but most patients ultimately develop castration-resistance. We show here that MYB is transcriptionally activated by androgen deprivation or impairment of androgen receptor (AR) signaling. MYB gene silencing significantly inhibited PCa growth in vitro and in vivo. Microarray data revealed that c-Myb shares a substantial subset of DNA damage response (DDR) target genes with AR, suggesting that c-Myb may replace AR for the dominant role in the regulation of their common DDR target genes in AR inhibition-resistant or AR-negative PCa. Gene signatures comprising AR, MYB, and their common DDR target genes are significantly correlated with metastasis, castration-resistance, recurrence, and shorter overall survival in PCa patients. We demonstrated in vitro that silencing of MYB, BRCA1 or TOPBP1 synergized with poly (ADP-ribose) polymerase (PARP) inhibitor olaparib (OLA) to increase cytotoxicity to PCa cells. We further demonstrated that targeting the c-Myb-TopBP1-ATR-Chk1 pathway by using the Chk1 inhibitor AZD7762 synergizes with OLA to increase PCa cytotoxicity. Our results reveal new mechanism-based therapeutic approaches for PCa by targeting PARP and the c-Myb-TopBP1-ATR-Chk1 pathway.
INTRODUCTION
Androgen deprivation is the standard systemic treatment for advanced prostate cancer (PCa), but most patients ultimately develop castration-resistance. The role of the androgen receptor (AR) in the development of castrate resistant prostate cancer (CRPC) is complex and, despite decades of research, remains poorly understood (1, 2). Increased AR expression and stimulation of specific AR target genes have been shown to contribute through various mechanisms, (3, 4). Castration-resistance is also associated with de-repression of a specific set of AR target genes (3). A subset of aggressive tumors that display clinical features characteristic of small-cell prostate carcinoma show complete loss of AR expression (5). CRPC also exhibits resistance to other therapeutic agents (6, 7). Understanding the selection mechanisms and genetic pathways associated with drug resistance remains one of the most important problems in developing potentially curative therapies for advanced PCa.
In an effort to identify molecular-pathologic events associated with the development of CPRC, we initially performed candidate gene expression profiling of bone metastases derived from mCRPC patients and compared them to matched primary tumors or benign prostate tissue. Our analysis demonstrated significantly higher c-Myb expression in bone metastases, suggesting that c-Myb may play a role in the development of CRPC. MYB is a transcription factor with diverse cellular functions including hematopoiesis, cell proliferation, differentiation, survival and tumorigenesis (8). Gene fusion and copy-number alterations of MYB suggest an oncogenic role in the progression of some breast, prostate and head and neck cancers (9, 10). c-Myb increased abundance of multiple genes involved in the progression of various malignant cells, including proliferation genes MYC, CCNA1, CCNB1 and CCNE1, survival genes BCL2, HSPA5, HSP70, AURKA and TPX2, and invasion and metastasis genes SNAI2, MMP1, MMP9 and CSTD (8, 11–13). While c-Myb expression has recently been associated with PCa cell proliferation, survival, and invasion in vitro (14), the regulation, function, and mechanistic associations of c-Myb in PCa remain largely unknown.
Abundant DNA translocations and deletions arise in a highly interdependent manner in the PCa genome (15). For example, a large percentage of PCa’s harbors fusions of TMPRSS2 to ERG or ETV1, which are considered drivers for advanced PCa (16). Maintenance of genomic integrity by DDR is critical to preventing tumorigenesis, and DDR gene defects are common in multiple malignancies, including PCa (17). Importantly, the loss of components of one DNA repair pathway may be compensated for by the increased activity of other pathways, which can provide necessary “stress support” for genetically unstable cancer cells (18). However, DDR gene defects or targeting specific DDR genes can present opportunities for cancer treatment. PARP1 is a key protein in the regulation of multiple forms of DNA repair processes (19, 20). Numerous studies have demonstrated “synthetic lethality” of PARP1 inhibitors with BRCA1/2 deficiency in cancer treatment (21, 22). It was also reported that pharmacological inhibition of PARP1 inhibits ETS-positive, but not ETS-negative, prostate cancer xenograft growth (23). Chk1 has also been shown to be a central player in DDR network. Upon sensing DNA damage or replication stress, the ataxia telangiectasia mutated- and Rad3-related (ATR) kinase is activated, which in turn phosphorylates Chk1, leading to the activation of G2/M checkpoint and DNA repair (24, 25). In this study we tested (i) whether c-Myb upregulation in advanced PCa is related to ADT or impairment of AR signaling; (ii) whether AR and c-Myb contribute to DDR and correlate with PCa progression; (iii) whether targeting PARP and c-Myb-regulated specific DDR signaling pathways can generate synergistic cytotoxicity to PCa. Our results reveal a new mechanism-based, therapeutic strategy for PCa based on targeting PARP and the c-Myb-TopBP1-ATR-Chk1 signaling pathway.
RESULTS
c-Myb is transcriptionally activated by ADT or impairment of AR signaling
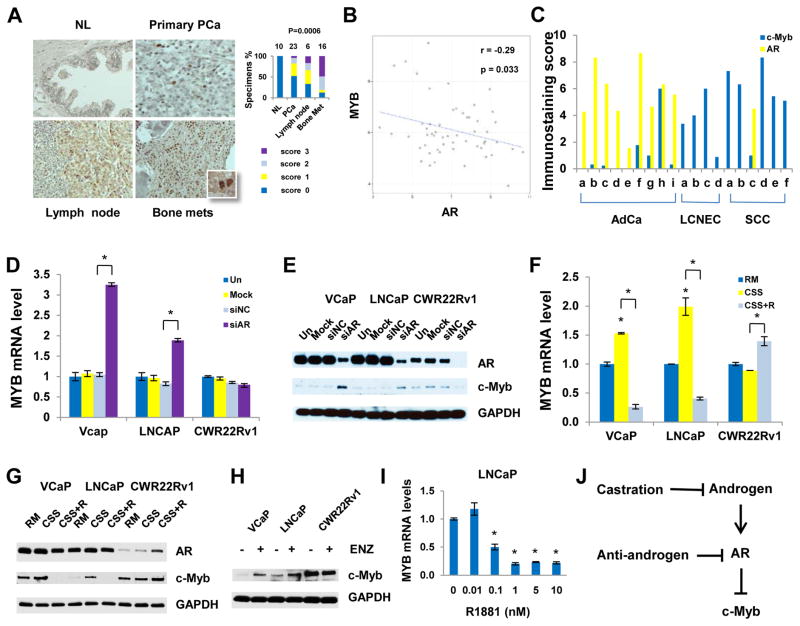
Our immunohistochemical (IHC) analyses demonstrated significantly higher c-Myb expression in bone metastases of CRPC patients than their primary tumors or benign prostate tissue (Fig. 1A). Analysis of independently published gene expression microarray data that include 4 normal bone marrow specimens, 22 local primary PCa specimens, and 29 metastatic PCa specimens (GSE32269) (26) revealed that MYB gene expression was negatively correlated with AR (r= −0.29, P= 0.033) (Fig. 1B). This negative correlation was markedly increased in bone metastatic prostate tumors (r = −0.32) compared with local primary tumors (r= −0.11) (Supplementary Fig. S1). IHC analysis of PCa xenograft tissue microarrays revealed c-Myb overexpression in AR-negative human PCa variants, including large neuroendocrine prostate carcinoma and small-cell prostate carcinoma, whereas both AR-positive PCa xenografts and AR-positive human primary PCa tissues had lower levels of c-Myb expression (Fig. 1C). Together, our data and published data suggest that androgen-AR signaling may negatively regulate c-Myb.
Fig. 1. Activation of c-Myb in CRPC and under experimental conditions that impair AR signaling.
(A) IHC analysis for expression of c-Myb in human PCa tissues. The numbers on the top of bar graphs denotes the number of specimens. P values were derived using Kruskal-Wallis rank testing. (B) Correlation analysis of MYB and AR mRNA expression in a published data set (GSE32269). (C) Human PCa xenograft tissue microarray. AdCa, adenocarcinoma; LCNEC, large cell neuroendocrine carcinoma; SCC, small cell prostate carcinoma. X axis labeling represent individual xenograft specimens. The information regarding patient derived xenografts is summarized in Table S2. (D and E) PCa cells were untreated, mock transfected or transfected with 20 nM siAR or negative control siRNA (siNC) for 48 hours. (D) qRT-PCR analysis (E) Western blotting analysis. (F and G) PCa cells were grown in regular medium (RM), charcoal-stripped serum medium (CSS) or CSS plus 10 nM R1881 (CSS+R) for 48 hours. (F) qRT-PCR analysis. (G) Western blotting analysis. (H) Western blotting analysis showing c-Myb abundances after the treatment of 1 μM enzalutamide (ENZ) for 48 hours. (I) qRT-PCR analysis 48 hours after R1818 treatment. (J) Summary of AR regulation of c-Myb. *P < 0.05.
To test this concept, we examined MYB mRNA and c-Myb protein expression under a series of experimental conditions that included androgen withdrawal and impairment of AR signaling. Gene expression microarray (Supplementary Fig. S2), quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1D) and Western blotting (Fig. 1E) analyses showed that AR silencing activated MYB mRNA and c-Myb protein expression in AR-positive, androgen-dependent VCaP and LNCaP, but not in AR-positive, castration-resistant CWR22Rv1 PCa cells. AR suppression of c-Myb expression in VCaP and LNCaP cells was shown to be androgen-dependent: charcoal-stripped serum (CSS) and the 2nd-generation AR inhibitor enzalutamide increased c-Myb expression, whereas R1881, a synthetic androgen, strongly suppressed c-Myb expression (Fig. 1, F–H). Androgen titration experiments revealed that physiologic concentrations of androgen (≥1 nM of R1881), but not castrate concentrations (≤10 pM), could efficiently suppress MYB expression (Fig. 1I). Together, our data demonstrated an AR inhibition-mediated mechanism for the activation of c-Myb in PCa cells (Fig. 1J).
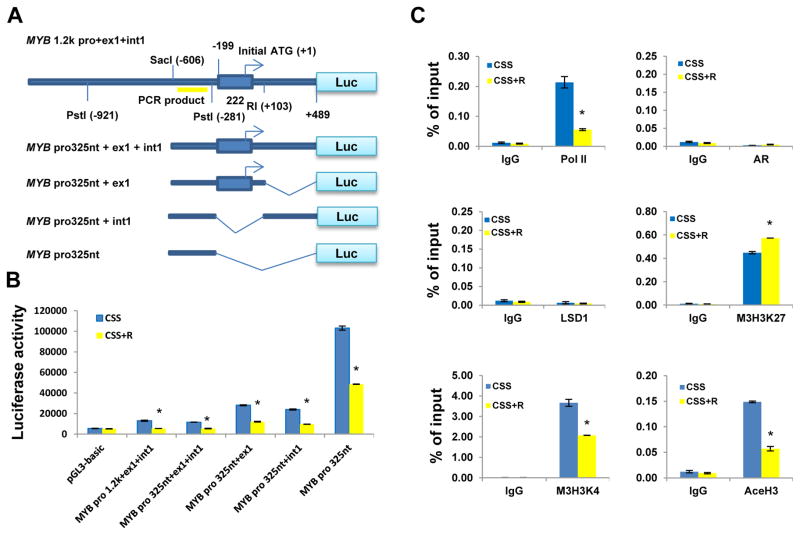
To identify the promoter region responsible for the downregulation of MYB by androgen-AR signaling, we constructed a series of MYB–luciferase reporters composed of different lengths of MYB promoter DNA with or without exon 1 and/or intron 1 (Fig. 2A). Luciferase reporter assays using LNCaP cells treated with CSS and CSS plus R1881 (CSS+R) showed that the minimum DNA fragment for the androgen-AR suppression of c-Myb is the proximal 325 nucleotides of MYB promoter (−734 to −281) (Fig. 2B). Our data also revealed androgen-independent negative regulatory elements in exon 1 and intron 1, as MYB expression was lower in all reporters with exon 1 or intron 1 than in the MYB 325-nucleotide promoter fragment (pro325nt) (Fig. 2B). To determine whether androgen-AR signaling suppresses MYB transcription through AR direct binding to or modification of the chromatin architecture of this 325-nucleotide promoter fragment, we performed chromatin immunoprecipitation (ChIP) assays to assess the binding activity of AR; RNA polymerase II; lysine-specific demethylase 1 (LSD1), which is reportedly involved in AR auto-suppression (3); and some widely used transcription marks. The results showed increased amounts of the transcription inactivation mark trimethyl-histone 3 lysine 27, and decreased amounts of RNA polymerase II and the transcription activation marks trimethyl-histone 3 lysine 4 and acetyl-histone 3 lysine 27 in the 325-nucleotide c-Myb promoter region in response to R1881 stimulation. In contrast, the amounts of AR and LSD1 binding to this promoter region were very low and relatively unchanged (Fig. 2C). Overall, our data indicate that AR may transcriptionally suppresses MYB, in part, through chromatin modification of the MYB promoter.
Fig. 2. AR transcriptionally suppresses MYB through modification of chromatin architecture on MYB promoter.
(A) The structural illustration of different MYB promoter luciferase reporters. (B) Luciferase reporter assays. LNCaP cells were transfected with MYB promoter reporters or control pGL3-basic empty vector and β-gal–expressing vector (internal control) for 24 h, followed by treatment with CSS or CSS plus 10 nM R1881 (CSS+R) for 24 h. (C) ChIP assays. LNCaP cells were treated with or without 10 nM R1881 in CSS medium for 24 h prior to ChIP assays. The data are expressed as percentage of input. Pol II, RNA polymerase II; LSD1, lysine-specific demethylase 1; M3H3K27, trimethyl-histone 3 lysine 27; M3H3K4, trimethyl-histone 3 lysine 4; AceH3, acetyl-histone 3 lysine 27. *P < 0.05.
MYB gene silencing significantly suppresses PCa growth in vitro and in vivo
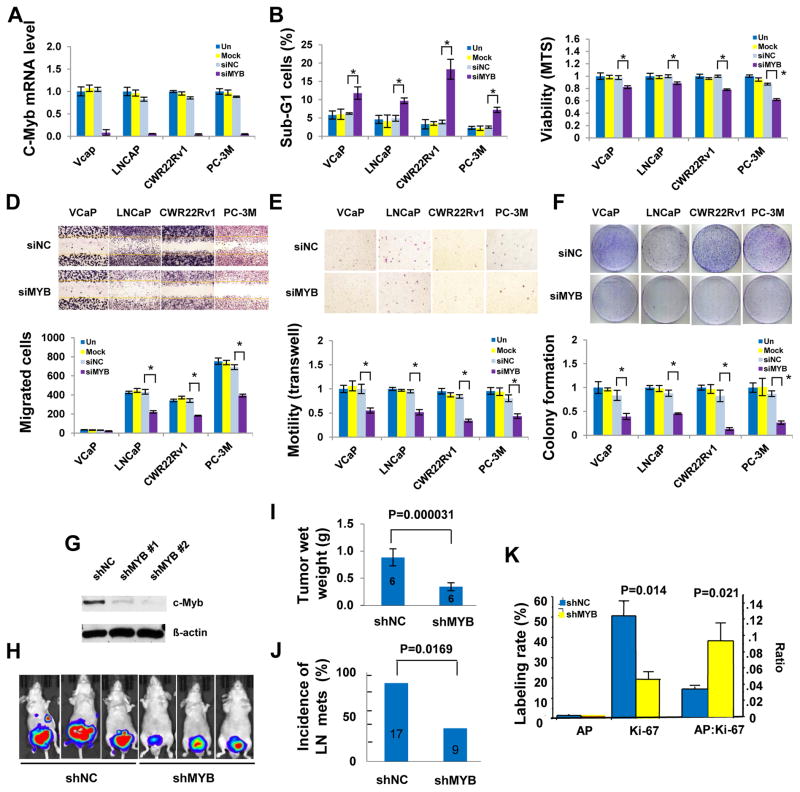
To gain insight into the functions of c-Myb in PCa progression, we tested the effects of c-Myb on cell survival, cell motility, and clonogenic growth in cultured PCa cells using small interference (si)RNA. Suppression of MYB (Fig. 3A) significantly increased sub-G1 cells (Fig. 3B) and significantly reduced viability and cell proliferation (Fig. 3C), migrated cells in wound healing assay (Fig. 3D) and Boyden chamber assay (Fig. 3E), and colony formation (Fig 3F). To demonstrate the in vivo function of c-Myb, in the absence of AR, i.e., conditions in which c-Myb is derepressed and fully functional, we determined the effect of c-Myb on tumor growth and metastasis using a MYB small hairpin (sh)RNA stably transfected PC-3M orthotopic xenograft mouse model. Our results showed that silencing of MYB (Fig. 3G) significantly reduced xenograph tumor growth and spontaneous metastasis as indicated by bioluminescence (Fig. 3H), tumor wet weight (Fig 3I) and lymph node metastasis (Fig. 3J). IHC analysis of Day 28 (end point) tumors did not show differences in apoptosis between the shMYB and shNC groups, however, the apoptotic activity relative to proliferative activity, as expressed as AP: Ki-67 ratio, was significantly higher in the shMYB tumors than in the shNC control as a result of significantly reduced proliferation (Fig. 3K). We speculate that low apoptosis rate in shMYB tumor may be the consequence of selection of apoptosis-resistant cells in this model.
Fig. 3. c-Myb silencing inhibits PCa Growth in vitro and in vivo.
(A) qRT-PCR analysisfor validation of MYB siRNA efficiency. MYB silencing led to increased sub-G-1 cells in flow cytometry analysis (B), reduced cell proliferation and survival in MTS assay (C), fewer migrated cells in wound healing assay (D) and in Boyden chamber assay (E), and decreased colony growth (F) of PCa cells. Top panels of D–F arerepresentative images and bottom panels are average of data from triplicate experiments. (G to K) PC-3M orthotopic xenograft model. (G) Western blotting analysis of c-Myb expression in input PC-3M cells. (H) Representative bioluminescent images (day 21). (I) The tumor wet weights in shMYB mice were significantly reduced compared with those in the shNC 28 days after cancer cell injection. (J) shMYB significantly reduced incidence of nymph node metastasis. (K) Apoptosis (TUNEL), cell proliferation (Ki67 staining) and ratio of apoptosis to cell proliferation (AP: Ki67) were compared between the shNC (n=6) and the shMYB tumors (n=5). The data are presented as mean ± standard errors. The numbers in the bars indicate total number of mice. *P < 0.05 unless indicated otherwise.
AR- and c-Myb-regulated DDR gene signature is strongly correlated with metastasis, castration resistance, and recurrence
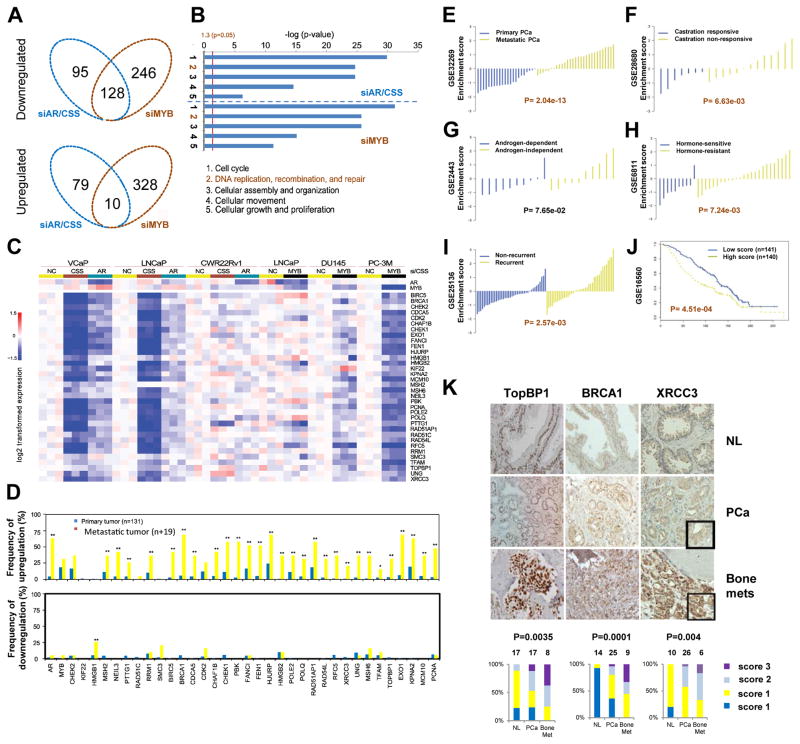
Through microarray analysis of both siRNA- and CSS-treated PCa cell samples we identified specific gene sets that are regulated by AR and c-Myb (Fig. 4A). Gene ontology enrichment analysis using Ingenuity Pathway Analysis (Ingenuity Systems) revealed a striking similarity in the top-regulated biologic processes between AR and c-Myb (Fig. 4B) and a subset of DDR genes regulated by both AR and c-Myb (Fig. 4C). Using published microarray data (27), we found that the DDR genes listed in Fig. 4C have high frequencies of overexpression in human PCa patient samples, especially in metastatic PCa (Fig. 4D). These data suggested that c-Myb and/or AR activities lead to upregulation of DDR genes in PCa and that c-Myb may replace AR for the dominant role in the regulation of their common DDR target genes in castration-resistant or AR-negative PCa. In addition, AR- and/or c-Myb-regulated DDR genes were selected during PCa progression.
Fig. 4. Integrated analysis of AR siRNA/CSS and MYB siRNA responsive genes and Correlation analysis of the DDR gene signature.
(A) Venn diagram showing genes targeted by siAR or CSS, siMYB, or both. (B) Ingenuity Pathway Analysis gene ontology enrichment analysis for siAR or CSS and MYB siRNA negatively regulated top five biologic processes. (C) Heat map for DDR genes negatively regulated by siAR or CSS and siMYB. (D) mRNA expression of AR, MYB, and their DDR target genes in primary and metastatic PCa by the analysis of online data (www.cbioportal.org/public-portal) by Taylor et al. (27). Statistical analysis was performed using chi-square analysis. (E–J) Correlation analysis of the DDR gene signature. (E) Primary PCa versus metastatic PCa. (F–H) Androgen-dependent versus androgen-independent. (I) Non-recurrence versus recurrence. (J) Kaplan-Meier curves that compare the survival of patients that have a higher-than-median signature score (red) with those that have a lower-than-median score (blue). (K) Upregulation of DDR genes TOPBP1, BRCA1, and XRCC3 in PCa bone metastases. Normal human prostate (NL), primary PCa, or bone metastasis (bone mets) tissues were evaluated by IHC. The percentages of each type of specimen with a given immunostaining score were compared. P values were derived using Kruskal-Wallis rank testing.
To test the potential clinical significance of AR- and c-Myb–regulated DDR genes, we generated a DDR gene signature (AR, MYB, and 34 AR and c-Myb coregulated DDR genes). Using six independent prostate-specific gene expression microarray datasets from the Gene Expression Omnibus (GEO) database, we calculated enrichment scores for our DDR gene signature for each sample in every dataset. The results showed that the enrichment scores of our DDR gene signature were positively correlated with PCa metastasis, castration resistance, recurrence and reduced overall survival of PCa patients (Fig. 4, E–J). IHC analysis of three selected DDR genes—BRCA1, TOPBP1, and XRCC3 confirmed that these three DDR genes were overexpressed at the protein concentrations in human PCa specimens, especially in bone metastases from CRPC patients (Fig. 4K). Together, the data from these analyses suggest that our DDR gene–derived signature is significantly, positively correlated with major features of aggressive PCa and demonstrate promising predictive potential for clinical application.
Silencing of MYB, BRCA1 or TOPBP1 synergizes with PARP inhibitor to increase PCa cytotoxicity
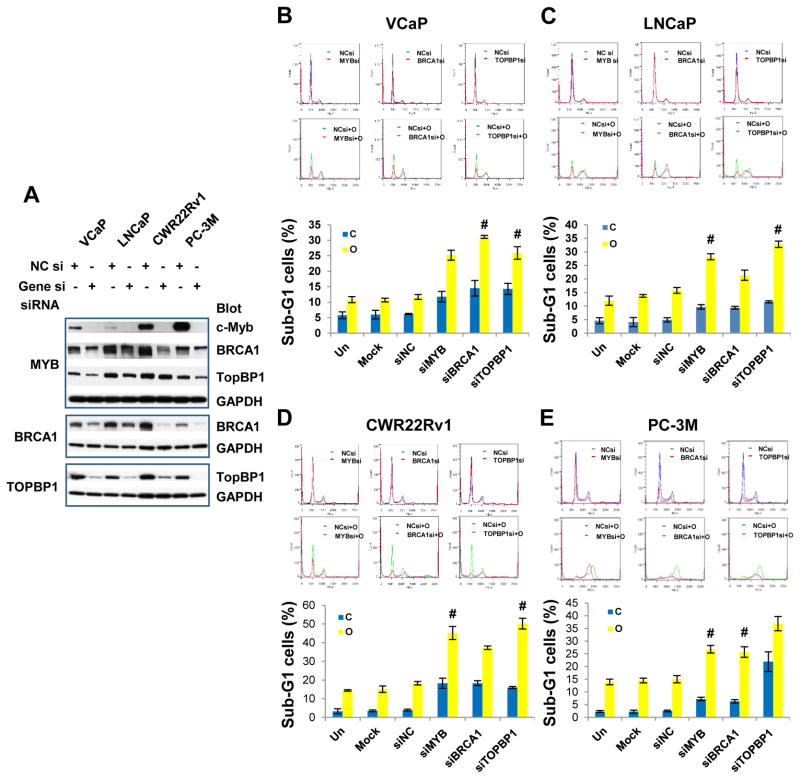
To address whether silencing of MYB or selected DDR signature genes could synergize with PARP inhibition to increase cytotoxicity to PCa, we performed cell cycle analysis for combinations of DDR gene silencing and PARP inhibition. Among DDR signature genes we selected BRCA1 and TOPBP1 because “synthetic lethality” from PARP1 inhibitor and deficiency of BRCA1/2, and the crucial role of TOPBP1 in the activation of ATR-Chk1 pathway. Transfection of siRNAs targeting MYB, BRCA1, or TOPBP1 into PCa cells effectively suppressed expression of their corresponding target genes (Fig. 5A). Consistent with our microarray and qRT-PCR data (Fig. 4C and Supplemental Fig. S3), MYB gene silencing led to reduced protein abundance of BRCA1 and TopBP1 (Fig. 5A). Cell cycle analysis showed that silencing of c-Myb, BRCA1 or TOPBP1 typically increased the sub-G1 cell population, with an accompanying decrease in the G1 cell population (Fig 5, B–E and Table S3); OLA, a PARP inhibitor, significantly reduced G1 cells and increased sub-G1 cells in all PCa cell lines, and markedly increased G2/M cells in LNCaP and PC-3M; The combination of DDR gene silencing and OLA further reduced G1 and G2 cells and further increased sub-G1 cells (Fig 5B–E top panels and Table S3). Synergistic cytotoxic effects were achieved in most cases by the combination of siMYB, siBRCA1, or siTOPBP1 and OLA (Fig. 5, B–E and Supplementary Table S4).
Fig. 5. DDR gene silencing synergizes with PARP inhibition to increase cytotoxicity to PCa cells.
(A) Western blotting analysis showing efficiency of MYB, BRCA1 and TOPBP1 siRNAs. Note MYB gene silencing led to downregulation of BRCA1 and TopBP1. (B–E), Cell cycle analysis. PCa cells were untreated, mock transfected or transfected with specific siRNAs (20 nM) and grown for 24 h, and then followed by the treatment of PARP inhibitor olaparib (OLA; 10 μM) for 48 h. Top panels are representative cell cycle profiles. Bottom bar graphs are average of triplicates. C, vehicle control; O, olaparib; Data are presented as mean ± standard deviation. # indicates synergistic effect. Synergy quantification was summarized in Supplementary Table S4.
Targeting c-Myb-TopBP1-ATR-Chk1 pathway using the Chk1 inhibitor AZD7762 synergizes with OLA in the treatment of PCa
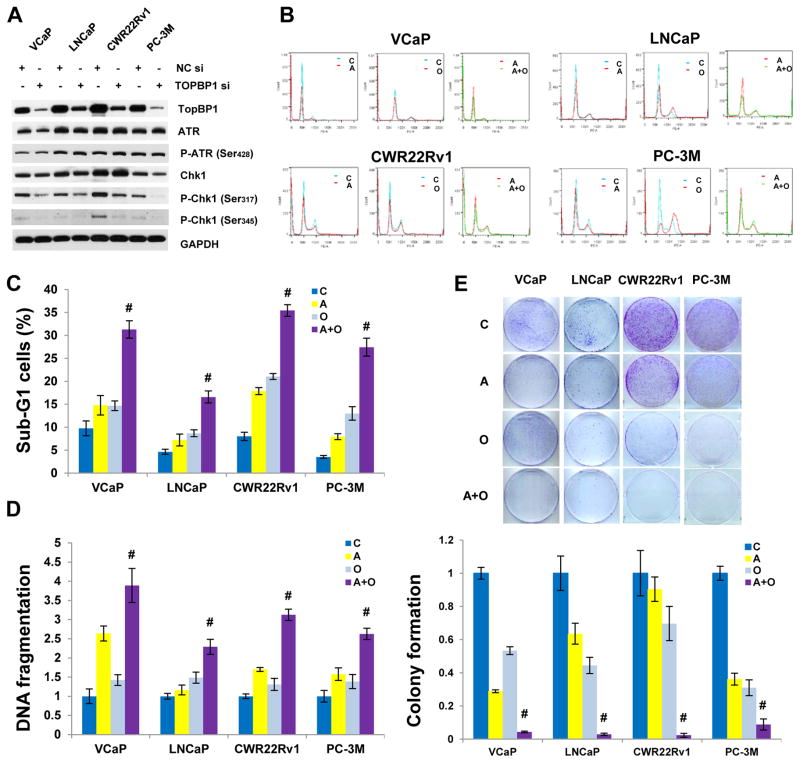
As reported above, TOPBP1 is one of important target of AR and c-Myb and is a crucial activator of ATR-Chk1 signaling pathway. Silencing of TOPBP1 by siRNA led to markedly reduced Chk1 phosphorylation at both Ser317 and Ser345 (Fig. 6A). Since c-Myb and TopBP1 are not druggable, targeting downstream Chk1 becomes a viable strategy. Cell cycle analysis showed that Chk1/2 inhibitor AZD 7762 (AZD) abrogated cell cycle checkpoints leading to the reduction of G1 and G2/M cells and the increase of sub-G1 cells; the combination of AZD and OLA further reduced G1 cells and increased sub-G1 cells (Fig. 6B) with synergistic cytotoxicity (Fig. 6C), DNA fragmentation (Fig. 6D) and suppression of colony growth (Fig. 6E) in both AR positive and AR negative PCa cells (also see Supplementary Table S4).
Fig. 6. Chk1 inhibition synergizes with PARP inhibition to increase cytotoxicity and DNA fragmentation and suppress cologenic growth in PCa cells.
(A) Effect of TopBP1 on ATR-Chk1 signaling pathway. (B) Representative cell cycle profiles. (C) Cell cycle analysis. PCa cells were treated with Chk1 inhibitor AZD7762 (200 nM) or PARP inhibitor olaparib (10 μM for VCaP and LNCaP and 5 μM for CWR22Rv1 and PC-3M) for 48 h. C, vehicle control (DMSO); A, AZD7762; O, olaparib; A+O, AZD7762+olaparib. (D) DNA fragmentation assay using a Cell Death ELISA Kit (Roche, Mannheim, Germany). Experimental conditions are the same as cell cycle analysis. (E) Colony assay. The data are presented as mean ± standard deviation in (C) and (D), as mean ± standard error in (E). # indicates synergistic effect.
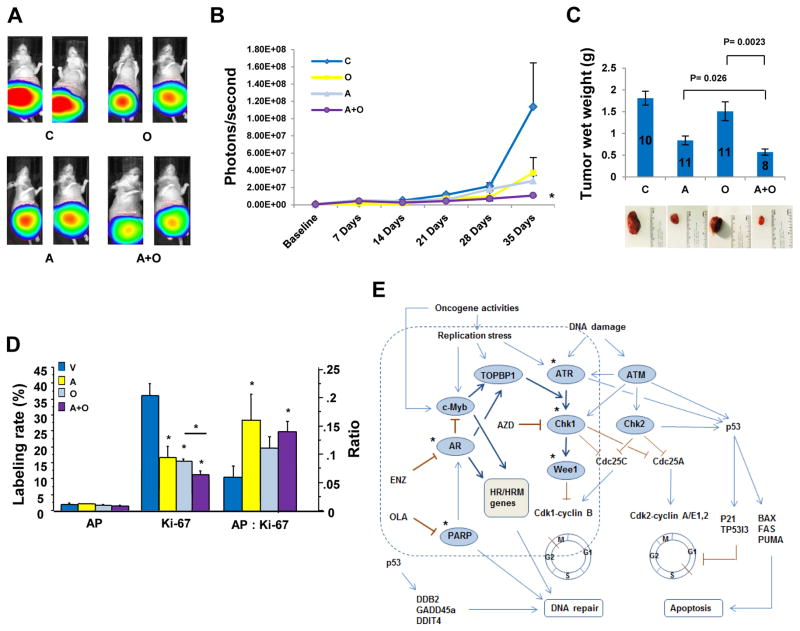
To move one step towards potential clinical applications, we confirmed our in vitro findings in vivo using a VCaP orthotopic xenograft prostate cancer model. VCaP is derived from a vertebral metastatic lesion of a patient with hormone refractory prostate cancer (28) and has been used in preclinical studies with PARP inhibitors (23, 29). Our in vivo data showed that OLA or AZD alone inhibited tumor growth, but the combination of AZD and OLA significantly increased the therapeutic effect (Fig. 7, A–C). IHC analysis of day 35 tumors showed significantly reduced cell proliferation (Ki67 staining) and an increased ratio of apoptosis to cell proliferation (AP: Ki67) in the combination of AZD and OLA (Fig. 7D). Low frequency of TUNEL-positive cells may result from selection of anti-apoptotic cells at the end of drug treatment.
Fig. 7. Chk1 inhibition synergizes with PARP inhibition to suppress xenograft tumor growth in vivo.
C, vehicle control; A, AZD7762; O, olaparib; A+O, AZD7762+olaparib. (A) Representative luminescence images. (B) Quantitative luminescence data showing xenograft tumor growth. Photon signals in combination group were significantly smaller than those in AZD alone (P= 0.0459), but not statistically significant compared to OLA alone due to bigger signal variation in OLA group. (C) Tumor wet weight. Numbers in the bar graph are number of mice in each group. Error bars in (B and C) are standard errors. (D) IHC analysis of apoptosis (TUNEL), cell proliferation (Ki67 staining) and ratio of apoptosis to cell proliferation (AP: Ki67) in the tumors treated with C (n= 8), O (n= 8), A (n= 7) or A+O (n= 7). *: P values <0.05. (E) Proposed AR and c-Myb coregulated DDR signaling pathway. In AR-positive, androgen-sensitive prostate cancer, AR and c-Myb coregulate TOPBP1, BRCA1 and other their common DDR target genes, promoting ATR-Chk1 signaling and DNA repair. In AR negative PCa or upon ADT or the impairment of AR signaling, MYB is derepressed and predominately in charge of DDR gene regulation. * indicates druggable therapeutic targets. PARP and Chk1 are two important DDR gene targets.
Overall, this study identified a novel mechanism by which c-Myb is activated by AR inhibition or impairment of AR signaling; generated an AR- and c-Myb regulated DDR gene signature that significantly correlate with metastasis, hormone resistance, recurrence, and shorter overall survival in PCa patients; and developed synergistic therapeutic approaches for targeting PARP and c-Myb-TopBp1-ATR-Chk1 pathway in AR inhibition-resistant prostate cancer (Fig. 7E).
DISCUSSION
This study identifies a novel mechanism of AR regulation of MYB in PCa by which MYB is transcriptionally repressed by AR in hormone-naïve PCa cells, but is derepressed by ADT or impairment of AR signaling. We also demonstrated that MYB silencing induced PCa cell death; inhibited PCa cell proliferation, motility and clonal growth in vitro; and suppressed PCa growth and spontaneous metastasis in vivo, indicating a crucial role for c-Myb in PCa cell survival and malignant progression. Our microarray data revealed that c-Myb and AR regulate strikingly similar bioprocesses including a substantial subset of DDR target genes. AR- and c-Myb-regulated DDR gene signature positively correlated with metastasis, castration resistance, recurrence, and reduced overall survival in PCa patients. Silencing of MYB or its target DDR genes BRCA1 or TOPBP1 generated synergistic effects with the PARP inhibitor OLA. Targeting the c-Myb-TopBp1-ATR-Chk1 pathway by using the Chk1 inhibitor AZD mimicked the synergistic effects observed with the combination of MYB, BRCA1 or TOPBP1 silencing and OLA. Although the combination of AZD and OLA was studied in pancreatic and mammary cell lines (30, 31), this study is the first in vitro and in vivo demonstration of AZD synergy with OLA in PCa. Our results establish a new mechanistic framework for understanding the progression of PCa to a castration- and drug-resistant state, and the roles of c-Myb and its DDR target genes in this process. The DDR gene signature derived from this study may function as predictive biomarkers for the efficacy of second-generation AR inhibitors or second-line therapeutic options in patients whose PCa continues to progress on these agents. The finding of synergistic effect from the combination of Chk1 inhibition and OLA provides strong rationale for clinical applications.
The novel finding of AR regulation of MYB is important, given the critical role of AR in the normal prostate and PCa and the role of c-Myb as an oncogenic transcription factor with diverse functions. We emphasize that AR suppression of c-Myb occurs only at physiologic concentrations of androgen (≥1 nM) and not at castration concentrations (≤10 pM) (Fig. 1I). This explains why high abundance of AR and c-Myb co-exist in metastatic PCa and CRPC and together promote PCa progression. The finding of overlapping regulatory functions of AR and c-Myb with regard to DDR genes in PCa is novel and interesting given the transition of oncogenic signaling control in PCa during the development of CRPC (2). Our data support a model in which (i) AR and c-Myb co-regulate DDR in hormone-naïve PCa, with AR functioning as the dominant regulator in most cases; (ii) derepression of c-Myb in CRPC and in AR negative PCa increases its capacity to promote DDR genes and therefore can replace AR for the dominant role in promoting PCa cell growth, survival, castration resistance, and drug resistance vis-à-vis DDR gene upregulation. Additional studies are warranted to further test this model and to assign a phenotypic profile(s) to the transition from AR-dominant to c-Myb-dominant regulation of DDR expression, including CRPC in which both AR and c-Myb activities may play important roles in DDR upregulation.
Prior to this study, information on biological function of c-Myb in PCa was very limited. There was only one report that showed c-Myb overexpression promoted proliferation, survival, motility and invasion in PCa cells in vitro (14). Our study provides the first line of evidence for a substantial impact of c-Myb on the tumor growth and metastasis in vivo. Furthermore, our study for the first time reveals a crucial role of c-Myb in regulation of DDR to promote DNA repair and therefore enhance cancer cell survival.
An important finding in this study is the identification of TOPBP1 as one of the AR- and c-Myb-regulated DDR genes, since TopBp1 is an essential activator for the ATR-Chk1 signaling pathway that plays a critical role in both cell cycle control and DNA repair. While c-Myb and TopBP1 are not druggable targets, their abundances in clinical samples may function as biomarkers to predict therapeutic resistance to OLA and targeting Chk1 is a clinically viable strategy. Mechanistically, synergy of AZD+OLA may be the consequence of targeting two crucial DDR pathways simultaneously. PARP inhibition induces cell death and causes accumulation of DNA-unrepaired cells at G2, while abrogation of cell cycle checkpoints by a Chk1 inhibitor pushes unrepaired DNA-damaged cells to premature mitosis and mitotic catastrophe-induced cell death. Another possibility may be, as reported, that Chk1 inhibition led to reduction of Rad51 Thr309 phosphorylation by Chk1, resulting in HR deficiency (25, 32). Future studies into mechanistic details of AZD+OLA synergy are warranted.
During the preparation of this manuscript, two publications reported AR regulation of DDR gene expression programs to govern DDA repair and cancer cell survival. Goodwin et al. reported that AR activities are induced by DNA damage, and promote DNA repair and cell survival though regulation of its key target gene DNAPKcs (33). Polkinhorn et al. reported that PCa treated with ionizing radiation plus androgen demonstrated enhanced DNA repair and decreased DNA damage while AR inhibition caused increased DNA damage and decreased clonogenic survival (34). Our current study supports the role of AR in regulation of DDR and promotion of PCa cell survival. It reveals that MYB is derepressed by ADT or impairment of AR signaling pathway; that c-Myb and AR co-regulate a substantial set of DDR genes in PCa; and that c-Myb plays a dominant role in the maintenance high abundances of these DDR genes and supports cell survival in AR positive, castration-resistant or AR-negative, androgen-independent PCa.
Overall, this study has established a genetic relationship between AR and c-Myb and between AR and c-Myb and their DDR target genes. The DDR gene–derived signature based on our results may provide prognostic and/or predictive biomarkers for second-generation AR inhibitors and second-line therapies for patients with CRPC. We have demonstrated synergistic therapeutic effects of MYB, TOBBP1 and BRCA1 silencing with OLA in PCa cells, and have identified AZD+OLA as a synergistic combination for advanced PCa regardless AR status. Although AZD7762 clinical trials were recently terminated due to cardiotoxicity, the scientific concept of the combination of a Chk1 inhibitor and OLA remains a viable clinical strategy. Future work is warranted to develop and evaluate the DDR gene signature as biomarkers and to test a Chk1 inhibitor and OLA combination strategy in clinical settings for advanced PCa.
MATERIALS AND METHODS
Cell lines
VCaP, LNCaP, DU145, CWR22Rv1, and PC-3M cell lines were validated by short tandem repeat DNA fingerprinting with the AmpFℓSTR Identifiler kit (Applied Biosystems) in MD Anderson’s Characterized Cell Line Core Facility.
Patient derived xenografts
The MD Anderson Cancer Center prostate cancer patient derived xenografts (PDXs) were developed as previously described (5, 35) with the support of the Prostate Cancer Foundation (Santa Monica, CA) and the David H. Koch Center for Applied Research in Genitourinary Cancers at The University of Texas MD Anderson Cancer Center (Houston, TX). Tissue samples for PDX development were obtained during radical prostatectomies, bone surgeries to palliate skeletal complications, or biopsies of symptomatic lesions under approved by the Institutional Review Board of The University of Texas M.D. Anderson Cancer Center. The information regarding PDXs is summarized in supplementary Table S2.
Immunohistochemistry
Twenty-seven human PCa radical prostatectomy specimens with a pathologic stage of pT2 and pathologic differentiation patterns with Gleason scores of 6 (n=11), 7 (n=9), 8 (n=6), or 9 (n=1) were obtained with informed consent from patients and used to analyze c-Myb and DDR genes. The patients had received no treatment prior to surgery. Additionally, nine bone biopsies from the patients who had various hormonal therapies were included (supplementary Table S1). Immunostaining results were scored according to the function of the staining intensity (from 0 [negative] to 3 [strong]) and the extent of positive staining of the cancerous area (1 = <10%; 2 = 10–50%; 3 = >50%). The immunostaining scores were 0 (grade 0), 1 (grades 1–3), 2 (grades 4–6), or 3 (grade >6). In addition, c-Myb (Epitomics, cat#1792-1) and AR (Santa Cruz, sc-816) immunostainings were also performed and scored on tissue microarray slides composed of 19 tumor xenografts generated from different histologic types of PCa (see Supplementary Table S2). Other antibodies used for IHC: BRCA1 (Biocare Medical, CM345C), XRCC3 (Abcam, ab20254), TopBP1 (Abcam, ab2402), TUNEL KIT (Millipore, S7101), Ki-67 (Santa Cruz, sc-15402).
Probes and primers for qRT-PCR
Taqman probe and primers from Applied Biosystems: Hu-B-actin: cat#4326315E. Taqman probe and primers from Integrated DNA Technologies: MYB forward: 5′CATGTTCCATACCCTGTAGCG3′, c-MYB reverse: 5′TTCTCGGTTGACATTAGGAGC3′, MYB probe: 5′TTATAGTGTCTCTGAATGGCTGCGGC3′. Primer sequences for AR and c-Myb regulated DDR genes are listed in supplementary Table S5.
Antibodies for Western blotting analysis
AR (sc-816) from Santa Cruz; c-Myb (ab109127), BRCA1 (ab16780) and TopBP1 (ab2402) from Abcam; GAPDH (#21188), ATR (#2790), P-ATR (Ser428) (#2853), Chk1 (#2360), P-Chk1 (Ser317) (#12302), P-Chk1 (Ser345) (#2348) from Cell Signaling.
siRNA transfection
PCa cells were seeded at desire density (VCAP, 1.0 × 106; LNCaP, 5 × 105; CWR22Rv1, 3 × 105; PC-3M, 2 × 105 in 6-well plates or 1/5 numbers in 24-well plates or 1/30 numbers in 96-well plates). On next day, cells were untransfected, mock transfected, or transfected with 20 nM siMYB or negative control siRNA (siNC). RNA and protein were prepared 48 hours after siRNA transfection.
Construction of MYB promoter reporters
A BAC (bacterial artificial chromosome) clone (RP11-32B1) containing the human MYB locus was purchased from CHORI Bacpac Resources (Oakland, CA). PCR was performed using the BAC clone RP11-32B1 as template to amplify human MYB promoter sequences (−1481 to +489) with forward primer: 5′CCCAGTCAGCAGAAGTCTCAAA3′ and reverse primer: 5′GCAGCTACTAAACAATCCAGCA3′. The PCR product was cloned into the pGL3-basic luciferase reporter vector (Promega) SmaI site to obtain MYB pro 1.2k-ex1-int1-Luc. Then the 875 nt from 5′ end of promoter was deleted using SacI sites to generate c. The intron 1 was further deleted with EcoRI and BglII to get MYB pro 325nt-ex1-Luc. Exon1 was deleted by PstI and EcoRI from MYB pro 325nt-ex1-int1-Luc to produce MYB pro 324nt-int1-Luc. MYB pro 324nt-Luc was obtained by PstI and BglII deletion of exon 1 and intron 1 MYB pro 325nt-ex1-int1-Luc. All the constructs were confirmed by DNA sequencing.
ChIP assays
ChIP assays were performed using a Millipore ChIP kit (Millipore, 17-295). The input and immunoprecipitated DNA were subjected to PCR using primers corresponding to the −496 to −352 base pairs of the MYB promoter (forward: 5′-AGCGGGGTTTGCTCAGGAAA-3′; reverse: 5′-GGGTCGCCGCTCCCATT-3′). The antibodies (ChIP grade): AR (Abcam, ab74272), LSD1 (Abcam, ab17721), trimethyl-histone H3K4 (Abcam, ab8580), trimethyl-histone H3K27 (Millipore, 07-449), acetyl-histone H3K27 (Millipore, 07-360), RNA polymerase II (Santa Cruz, sc-900X), and normal rabbit immunoglobulin G (Santa Cruz, sc-2025 for mouse and sc-2027 for rabbit). Results were presented as mean ± standard deviation for replicate samples.
Wound healing assay
Twenty-four hours after siRNA transfection, a straight longitudinal incision was made on the monolayer of cells using a pipet tip. After the removal of existing medium, fresh medium was added and the cells were incubated for 24 hours (CWR22Rv1 and PC-3M), or 48 hours (LNCaP), or 72 hours (VCaP). Cells were then stained with HEMA3 (Biochemical Sciences, Cat#122-911) and the number of cells migrating into the clear area were counted and imaged with a microscope using NIS-Elements AR2.30 software (Nikon).
Boyden chamber assay
Twenty-four hours after siRNA transfection, cells were trypsinized and single cell suspension was prepared. Cells were seeded into each Falcon cell culture insert (8.0 μm pore size, 24 well format, Becton Dickinson Labware, 353097) at desired density (VCAP, 1.5 × 104; LNCaP, 1 × 104; CWR22Rv1, 5 × 103; PC-3M, 3 × 103) with 300 μl of serum-free medium and with 700 μl complete medium in outer well. After 16–24 hour incubation, the medium and cells inside the insert were carefully removed and cells transmigrated onto the outer membrane of the insert were stained, counted and imaged as described in wound healing assay.
Colony assay
For siRNA transfected cells: PCa cells were untransfected, mock transfected, or transfected with 5 nM siMYB or siNC. Twenty-four hours after siRNA transfection, cells were trypsinized and reseeded into 6-well plates at low density (VCAP, 1.0 × 105; LNCaP, 2 × 104; CWR22Rv1, 1 × 104; PC-3M, 5 × 103). Cells were grown for 1–2 weeks for colony formation. Colonies were fixed with cold methanol for 30 min, stained with 0.5 % crystal violet in 25% methanol for 30 min, and counted and imaged as described in wound healing assay. For drug treated PCa cells: PCa cells were seeded into 6-well plates at low density as described above and grown for 48 hours. Then cells were treated with AZD7762 (AZD) (Selleckchem) (100 nM for VCaP, 200 nM for LNCaP, CWR22Rv1 and PC-3M) or olaparib (OLA) (Selleckchem) (10 μM for VCaP and LNCaP, 5 μM for CWR22Rv1 and PC-3M) or combination of AZD and OLA. Medium containing drug or vehicle control (DMSO) was renewed every 3 days. Colonies were stained, counted and imaged as described above.
MYB shRNA xenograft tumor model
Stable PC-3M xenograft lines were generated by the transduction of a lentivirus expressing shMYB or shNC and a lentivirus expressing bioluminescent luciferase. The PC-3M cells (1.0 × 106) were injected into the prostates of athymic nude mice. Tumor growth was monitored weekly by bioluminescent images. Mice were sacrificed and the tumor wet weights were determined 28 days after cancer cell injection. Experiments were performed in accordance of animals’ care guidelines.
Gene expression microarray analysis
Significance Analysis of Microarrays software (Stanford University) was used to identify specific gene sets that are regulated by siRNAs targeting AR, or MYB. Gene ontology enrichment analysis using Ingenuity Pathway Analysis (Ingenuity Systems) was used to determine AR, or c-Myb top-regulated biologic processes and AR- and/or c-Myb–regulated DDR genes. Microarray data were deposited in the GEO database (accession number GSE49287).
Flow cytometry analysis
PCa cells were treated with 10 μM OLA or vehicle control DMSO 24 h after siRNA transfection. Fourty-eight h after OLA treatment, cells were harvested and stained with propidium iodide and then analyzed on FACSCanto II (BD Biosciences). Cell cycle profiles and quantitative data were obtained using FlowJo software (Tree Star, Inc., Ashland, OR). For AZD and OLA combination experiments, PCa cells were treated with 200 nM AZD or OLA (10 μM for VCaP and LNCaP and 5 μM for CWR22Rv1 and PC-3M) or combination or vehicle control for 48 hours.
Correlation analysis of gene signature
Six prostate-specific gene expression microarray datasets were obtained from the GEO database. Three datasets (GSE32269, GSE2443, and GSE25136) include raw data, from which we extracted an expression profile using the RMA method of the Bioconductor Affy package (R version 2.15.2). The remaining three datasets (GSE28680, GSE6811, and GSE16560) contained expression profiles that we used directly. All expression profiles were further normalized using quartile methods, and multiple probes corresponding to the same gene were collapsed using the MaxMean method of the weighted correlation network analysis (WGCNA) package (Bioconductor). For each sample in every dataset, we calculated signature scores for our DDR-gene signature using the single sample gene set enrichment analysis method in the gene set variation analysis (GSVA) package (Bioconductor). For datasets that did not contain every gene in a signature, we used a reduced signature containing all the signature genes that were present. With GSE16560, we evaluated the prognostic power of each signature by classifying patients into low-scoring and high-scoring groups depending on the median of their signature scores, and we compared survival information between the two groups using the log-rank test.
DNA fragmentation assay
DNA fragmentation assay was performed using a Cell Death Detection ELISA Kit (Roche, Mannheim, Germany) according to manufacturer’s portocol. The assay is a photometric enzyme-immunoassay for the qualitative and quantitative in vitro determination of cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) after induced cell death.
Orthotopic xenograft prostate cancer model for AZD and OLA combination
VCaP cells were transduced with lentivirus stably expressing luciferase. Aliquots of 3.5 × 106 VCaP-luciferase cells in 15 μL PBS were injected directly into the right lobe of the dorsolateral prostate in athymic nude male mice (Taconic Farm, Hudson, NY) to induce orthotopic tumors. The tumors were allowed to grow for 14 days before treatment. The experimental groups received intraperitoneal injections of 40 mg/kg OLA 5 days per week; 25 mg/kg AZD twice a day every 3 days; or combination of these two drugs for 35 days. Tumor size was monitored by measuring the luminescence signal using the IVIS 200 Series (PerkinElmer, Wellesley, MA) and mice were sacrificed after 5 weeks of treatment and the tumors were collected and weighed.
Statistical analysis
For data with a non-normal distribution or data with a small sample size, a non-parametric method, Wilcoxon rank sum test was used. This covers qRT-PCR data, luciferase assay, ChIP assay, MTS assay, wound healing assay, Boyden chamber assay, cell cycle analysis and DNA fragmentation assay as well as TUNEL and Ki-67 immunohistochemical analyses. ANOVA with desired pair-wise comparison was used in the comparison of single drug vs combination of two drugs. Synergism was determined using two-way ANOVA test (36).
Supplementary Material
Fig. S1. Correlation analysis of MYB and AR mRNA expression in prostate cancer.
Fig. S2. MYB mRNA expression under different conditions that impair AR signaling.
Fig. S3. qRT-PCR validation of AR- and c-Myb-coregulated DDR genes.
Table S1. Clinical treatment associated with PCa bone biopsies
Table S2. Patient derived xenografts
Table S3. Combinatorial effects of DDR gene silencing and OLA on PCa cells.
Table S4. Synergism determination
Table S5. Primer sequences for AR- and c-Myb-regulated DDR genes
Table S6. siRNA and shRNA sequences
One Sentence Summary.
This study identified a novel mechanism of c-Myb activation by androgen deprivation therapy or impairment of AR signaling which led to identification of a therapy resistance pathway defined by an AR- and c-Myb-regulated DNA damage response (DDR) gene signature; the study also developed synergistic therapeutic approaches for targeting poly (ADP-ribose) polymerase and the c-Myb-TopBP1-ATR-Chk1 signaling pathway in castration-resistant prostate cancer.
Acknowledgments
We thank Sarah Bronson and Dawn Chalaire for editing the manuscript.
Funding: This work was supported in part by National Cancer Institute grant R0150588 (to T.C.T.); National Cancer Institute grant P50140388, the Prostate Cancer Specialized Program of Research Excellence at The University of Texas MD Anderson Cancer Center; the National Institutes of Health through MD Anderson’s Cancer Center Support Grant, CA16672; and Tony’s Prostate Cancer Research Foundation.
Footnotes
Supplementary references: References (37–39) in the reference list.
Author contributions: TC Thompson and L Li conceived and designed the study and wrote the paper. L Li prepared samples for gene expression microarray, performed Western blotting analysisand flow cytometric experiments. W Chang performed gene expression microarray analysis, gene signature correlation analysis and flow cytometric experiments. G Yang conducted immunohistochemistry and tissue microarray analysis. C Ren performed q-RT-PCR, luciferase reporter assay and ChIP assay. S Park, T Karantanos, S Karanika and J Wang designed and conducted xenograft model experiments for AZD and OLA synergy studies. L Li and J Yin contributed to wound healing assays, Boyden chamber assays and colony assays. PK Shah conducted AR and c-Myb correlation analysis using published data and our own microarray data. S Park, H Takahiro, M Dobashi and W Zhang conducted MYB shRNA xenograft mouse model experiments. T Karantanos and J Wang performed DNA fragmentation assays. J-S Lee and H-S Lee conducted gene expression microarray experiments. B Broom performed gene signature correlation analysis and synergism determination. L Xiao contributed to statistical analysis. E Efstathiou, SN Maity, AM Aparicio, EM Li Ning Tapia, P Troncoso, and N Navone contributed to pathological analysis of human PCa samples and the establishment of patient tissue xenograft lines. PG Corn contributed to interpretation of data and manuscript preparation. All authors contributed to data analysis.
Competing interests: The authors declare no competing financial interest.
REFERENCES AND NOTES
- 1.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 2.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013 doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Nelson PS, Liu XS, Brown M, Balk SP. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan X, Balk SP. Mechanisms mediating androgen receptor reactivation after castration. Urol Oncol. 2009;27:36–41. doi: 10.1016/j.urolonc.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzelepi V, Zhang J, Lu JF, Kleb B, Wu G, Wan X, Hoang A, Efstathiou E, Sircar K, Navone NM, Troncoso P, Liang S, Logothetis CJ, Maity SN, Aparicio AM. Modeling a lethal prostate cancer variant with small-cell carcinoma features. Clinical Cancer Research. 2012;18:666–677. doi: 10.1158/1078-0432.CCR-11-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberti C. Taxane- and epothilone-based chemotherapy: from molecule cargo cytoskeletal logistics to management of castration-resistant prostate carcinoma. Eur Rev Med Pharmacol Sci. 2013;17:1658–1664. [PubMed] [Google Scholar]
- 7.Rescigno P, Buonerba C, Bellmunt J, Sonpavde G, De Placido S, Di Lorenzo G. New perspectives in the therapy of castration resistant prostate cancer. Curr Drug Targets. 2012;13:1676–1686. doi: 10.2174/138945012803529956. [DOI] [PubMed] [Google Scholar]
- 8.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 9.Stenman G, Andersson MK, Andren Y. New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle. 2010;9:2986–2995. doi: 10.4161/cc.9.15.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards J, Krishna NS, Grigor KM, Bartlett JM. Androgen receptor gene amplification and protein expression in hormone refractory prostate cancer. Br J Cancer. 2003;89:552–556. doi: 10.1038/sj.bjc.6601127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanno B, Sesti F, Cesi V, Bossi G, Ferrari-Amorotti G, Bussolari R, Tirindelli D, Calabretta B, Raschella G. Expression of Slug is regulated by c-Myb and is required for invasion and bone marrow homing of cancer cells of different origin. J Biol Chem. 2010;285:29434–29445. doi: 10.1074/jbc.M109.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knopfova L, Benes P, Pekarcikova L, Hermanova M, Masarik M, Pernicova Z, Soucek K, Smarda J. c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Mol Cancer. 2012;11:15. doi: 10.1186/1476-4598-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Yang G, Ren C, Tanimoto R, Hirayama T, Wang J, Hawke D, Kim SM, Lee JS, Goltsov AA, Park S, Ittmann MM, Troncoso P, Thompson TC. Glioma pathogenesis-related protein 1 induces prostate cancer cell death through Hsc70-mediated suppression of AURKA and TPX2. Mol Oncol. 2013;7:484–496. doi: 10.1016/j.molonc.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava SK, Bhardwaj A, Singh S, Arora S, McClellan S, Grizzle WE, Reed E, Singh AP. Myb overexpression overrides androgen depletion-induced cell cycle arrest and apoptosis in prostate cancer cells, and confers aggressive malignant traits: potential role in castration resistance. Carcinogenesis. 2012;33:1149–1157. doi: 10.1093/carcin/bgs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 17.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 19.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wang F, Tang T, Guo C. The role of PARP1 in the DNA damage response and its application in tumor therapy. Front Med. 2012;6:156–164. doi: 10.1007/s11684-012-0197-3. [DOI] [PubMed] [Google Scholar]
- 21.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellisen LW. PARP inhibitors in cancer therapy: promise, progress, and puzzles. Cancer Cell. 2011;19:165–167. doi: 10.1016/j.ccr.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, Palanisamy N, Siddiqui J, Yan W, Cao X, Mehra R, Sabolch A, Basrur V, Lonigro RJ, Yang J, Tomlins SA, Maher CA, Elenitoba-Johnson KS, Hussain M, Navone NM, Pienta KJ, Varambally S, Feng FY, Chinnaiyan AM. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson R, Eastman A. The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design. British journal of clinical pharmacology. 2013;76:358–369. doi: 10.1111/bcp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai C, Wang H, He HH, Chen S, He L, Ma F, Mucci L, Wang Q, Fiore C, Sowalsky AG, Loda M, Liu XS, Brown M, Balk SP, Yuan X. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest. 2013;123:1109–1122. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korenchuk S, Lehr JE, MCL, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 29.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, Liu F, Planck JL, Ravindranathan P, Chinnaiyan AM, McCue P, Gomella LG, Raj GV, Dicker AP, Brody JR, Pascal JM, Centenera MM, Butler LM, Tilley WD, Feng FY, Knudsen KE. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vance S, Liu E, Zhao L, Parsels JD, Parsels LA, Brown JL, Maybaum J, Lawrence TS, Morgan MA. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle. 2011;10:4321–4329. doi: 10.4161/cc.10.24.18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Booth L, Cruickshanks N, Ridder T, Dai Y, Grant S, Dent P. PARP and CHK inhibitors interact to cause DNA damage and cell death in mammary carcinoma cells. Cancer Biol Ther. 2013;14:458–465. doi: 10.4161/cbt.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 33.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, Ma T, Den RB, Dicker AP, Feng FY, Knudsen KE. A Hormone-DNA Repair Circuit Governs the Response to Genotoxic Insult. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, Arora VK, Yen WF, Cai L, Zheng D, Carver BS, Chen Y, Watson PA, Shah NP, Fujisawa S, Goglia AG, Gopalan A, Hieronymus H, Wongvipat J, Scardino PT, Zelefsky MJ, Jasin M, Chaudhuri J, Powell SN, Sawyers CL. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navone NM, Olive M, Ozen M, Davis R, Troncoso P, Tu SM, Johnston D, Pollack A, Pathak S, von Eschenbach AC, Logothetis CJ. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clinical Cancer Research. 1997;3:2493–2500. [PubMed] [Google Scholar]
- 36.Slinker BK. The statistics of synergism. Journal of molecular and cellular cardiology. 1998;30:723–731. doi: 10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
- 37.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ, Barrette T, Everett J, Siddiqui J, Kunju LP, Navone N, Araujo JC, Troncoso P, Logothetis CJ, Innis JW, Smith DC, Lao CD, Kim SY, Roberts JS, Gruber SB, Pienta KJ, Talpaz M, Chinnaiyan AM. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sircar K, Huang H, Hu L, Cogdell D, Dhillon J, Tzelepi V, Efstathiou E, Koumakpayi IH, Saad F, Luo D, Bismar TA, Aparicio A, Troncoso P, Navone N, Zhang W. Integrative molecular profiling reveals asparagine synthetase is a target in castration-resistant prostate cancer. Am J Pathol. 2012;180:895–903. doi: 10.1016/j.ajpath.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aparicio A, Tzelepi V, Araujo JC, Guo CC, Liang S, Troncoso P, Logothetis CJ, Navone NM, Maity SN. Neuroendocrine prostate cancer xenografts with large-cell and small-cell features derived from a single patient’s tumor: morphological, immunohistochemical, and gene expression profiles. The Prostate. 2011;71:846–856. doi: 10.1002/pros.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Correlation analysis of MYB and AR mRNA expression in prostate cancer.
Fig. S2. MYB mRNA expression under different conditions that impair AR signaling.
Fig. S3. qRT-PCR validation of AR- and c-Myb-coregulated DDR genes.
Table S1. Clinical treatment associated with PCa bone biopsies
Table S2. Patient derived xenografts
Table S3. Combinatorial effects of DDR gene silencing and OLA on PCa cells.
Table S4. Synergism determination
Table S5. Primer sequences for AR- and c-Myb-regulated DDR genes
Table S6. siRNA and shRNA sequences