Summary
The term innate immunity typically refers to a quick but nonspecific host defense response against invading pathogens. The innate immune system comprises particular immune cell populations, epithelial barriers, and numerous secretory mediators including cytokines, chemokines, and defense peptides. Innate immune cells are also now recognized to play important contributing roles in cancer and pathological inflammatory conditions. Innate immunity relies on rapid signal transduction elicited upon pathogen recognition via pattern recognition receptors (PRRs) and cell:cell communication conducted by soluble mediators, including cytokines. A majority of cytokines involved in innate immune signaling use a molecular cascade encompassing receptor-associated Jak protein tyrosine kinases and STAT (signal transducer and activator of transcription) transcriptional regulators. Here, we focus on roles for STAT proteins in three major innate immune subsets: neutrophils, macrophages, and dendritic cells (DCs). While knowledge in this area is only now emerging, understanding the molecular regulation of these cell types is necessary for developing new approaches to treat human disorders such as inflammatory conditions, autoimmunity, and cancer.
Keywords: Neutrophils, macrophages, dendritic cells, cytokines, STAT proteins
Introduction
A classic view of the innate immune system includes specific hematopoietic lineages that are dedicated to rapid pathogen clearance. These cell types specialize in phagocytosis and recognizing pattern molecules that exist or are produced by microbial invaders and necrotic cells (e.g. bacterial lipopolysaccharide, extracellular DNA). The association of pattern molecules with surface receptors (PRRs) on innate immune cells elicits rapid intracellular signal transduction. This culminates in numerous responses, which are designed to alert and educate the host immune system (1). One of the most crucial responses initiated by PRR signaling is synthesis and secretion of cytokine mediators. Cytokines elicit stress hematopoietic responses to combat infection (e.g. emergency granulopoiesis) and guide the generation of appropriate adaptive immune subsets (2-5). Our understanding of innate immunity has broadened, in part due to the recognition that epithelial cells and other molecular messengers (e.g. anti-microbial peptides) have critical roles in immune activation. These topics have been covered in excellent reviews (6-10). Accordingly, here, we focus on the classic innate immune subsets and the roles for cytokine signaling via signal transducer and activators of transcription (STAT) proteins in their regulation.
Many of the cytokines involved in innate immunity are categorized as interacting with class I or class II receptors, which comprise a large gene family (11, 12). Class I receptor cytokines are 4-α-helical bundle proteins that are typically involved in stimulating proliferation, differentiation, and function of innate and adaptive immune populations (11). These include granulocyte colony-stimulating factor [(G-CSF) also known as colony-stimulating factor 3 (CSF3)], granulocyte-macrophage colony-stimulating factor [(GM-CSF) also known as (CSF2)], interleukin-3 (IL-3), IL-5, IL-6, and IL-11. These proteins regulate hematopoiesis by signaling through high affinity cell surface receptors that share common subunits (e.g. IL-3Rβ for IL-3, IL-5, and GM-CSF; gp130 for IL-6, IL-11) (13, 14). Other important class I receptor cytokines require the common γ chain (γc), which was first identified as a crucial component of the IL-2 receptor (15). The γc-activating cytokines are key regulators of B and T-lymphocyte development; accordingly, mutation of γc or its selective Jak kinase (Jak3) results in severe immunodeficiency in mice and humans (15, 16). The Jak-STAT cascade is a major intracellular signaling response elicited by class I cytokine receptors, although it is important to note that other signaling modules such as the mitogen-activated protein kinase (MAPK) and phosphoinositol 3-kinase (PI3K) pathways are also stimulated. Typically, in the Jak-STAT pathway, the activated kinase phosphorylates tyrosine residues within the cytokine receptor intracellular domain. These phosphorylated tyrosines provide docking sites for the SH2-domain of STAT proteins. Cytoplasmically localized STATs associate with the receptor and undergo Jak-mediated tyrosine phosphorylation, which leads to their hetero- or homodimerization via SH2-phosphotyrosine interactions. The dimerized STATs are competent to accumulate in the nucleus, bind DNA and regulate expression of cytokine-responsive genes. This signaling cascade results in direct and rapid changes in gene expression upon cytokine stimulation (17-19).
Cytokines that interact with class II receptors include interferons (IFNs), which are further subdivided into type I (e.g. IFN-α, IFN-β), II (IFN-γ) and III (IFN-γ) IFNs, as well as IL-10 and IL-10-related molecules (e.g. IL-20, IL-22) (12). These cytokines also bind via high affinity heteromeric receptors that activate the Jak-STAT cascade (19, 20). In addition, other important protein mediators regulate innate immunity through receptor tyrosine kinases (RTKs). These include the major DC and macrophage growth factors FMS-like tyrosine kinase 3 ligand (Flt3L) and macrophage colony-stimulating factor [(M CSF) also known as CSF1] (21, 22). Interestingly, the Flt3L and M-CSF receptors stimulate STAT protein activation; this may occur by direct receptor-mediated activation via the kinase function intrinsic to the RTK intracellular domain (23, 24). Here, we discuss the developmental origins of neutrophils, macrophages and DCs, with a focus on the growth factors and STAT-mediated mechanisms that regulate these critical innate immune populations.
Neutrophil development and granulopoietic cytokines
Neutrophils (also known as neutrophilic granulocytes) are the most numerous immune cells in bone marrow and blood. These short-lived cells usually undergo apoptosis after performing their host-defense activities, which include secretion of proteases, reactive oxygen intermediates, antimicrobial peptides and neutrophil extracellular “traps” (NETs) (25). Activated neutrophils also release cytokines that orchestrate subsequent adaptive immune responses. Like other leukocytes, neutrophils develop from hematopoietic stem cells (HSCs) through defined progenitor populations, each with less developmental potential than its immediate precursor. Neutrophil progenitors include hematopoietic multipotent progenitors (MPP), common myeloid progenitors (CMP) and granulocyte-macrophage progenitors (GMPs), as well as morphologically defined precursor populations (e.g., myeloblasts, promyelocytes, myelocytes, metamyelocytes) (25). In adults, neutrophils undergo their main development stages within the bone marrow environment, where a large reserve pool of mature cells is retained for rapid release during systemic infection.
G-CSF is the principal cytokine regulator of granulopoiesis, stimulating neutrophil production and mobilization from bone marrow. G-CSF is present constitutively at low levels in the circulation, and is rapidly induced during infection (3, 26-28). G-CSF-null (Csf3−/−) mice develop chronic neutropenia, characterized by an approximate 50% reduction in granulocytic precursors and a more severe reduction (~ 70%) in circulating neutrophils (29). A similar phenotype was observed in mice carrying null or targeted mutations of the G-CSF receptor (G-CSFR) (e.g. Csf3r−/− mice) (30-32), highlighting the importance of signals elicited by engagement of G-CSF/G-CSFR in normal granulopoiesis. G-CSF regulates proliferation and survival of granulocytic progenitors and differentiated granulocytes (33, 34). Of note, G-CSF itself is not chemotactic, yet GCSF signaling through the G-CSFR is required for efficient migration of neutrophils from bone marrow in steady state conditions (34-36). While G-CSF-G-CSFR signaling is critical for maintaining normal circulating neutrophil numbers, the incomplete loss of neutrophils in Csf3−/− or Csf3r−/− mice indicates compensatory mechanisms can support neutrophil production.
Other cytokines, including IL-6, GM-CSF, and Flt3L, were identified as independent regulators of granulopoiesis, albeit to a lesser extent than G-CSF. For example, administration of recombinant IL-6 stimulated multiple hematopoietic progenitor subsets to expand in number and increased circulating neutrophil levels (37). Important genetic evidence was obtained for the role of IL-6 when mice with disruption of both Il6 and Csf3r genes (Il6−/− Csf3r−/− mice) were found to have more severe neutropenia compared to Csf3r−/− animals (38). Similarly, mice lacking G-CSF and GM-CSF have fewer neutrophils versus mice lacking either gene alone (39). Furthermore, cytokines such as stem cell factor (SCF) or Flt3L synergize with G-CSF in enhancing mobilization of hematopoietic progenitors and neutrophils, indicating important roles for these factors in granulopoiesis at progenitor stages and terminal neutrophil differentiation (40, 41).
Roles for STATs in neutrophil development and function
Activation of G-CSFR by G-CSF binding induces trans-phosphorylation of the receptor-associated Jak1 and Jak2 proteins, which subsequently stimulate several signal cascades. Among these, G-CSFR-mediated STAT activation is robust and rapid; therefore, significant effort has gone toward understanding whether and how STATs regulate granulopoiesis. Experiments in cell culture systems ex vivo demonstrated that G-CSF activated STAT1, STAT3, and STAT5, with STAT3 as the dominant response pathway (42-44). Accordingly, STAT3 was shown to regulate G-CSF-induced development of neutrophil morphology, as well as expression of the integrin Mac-1 (CD11b/CD18), the enzyme myeloperoxidase (MPO) and the secondary granule proteins lactoferrin and 24p3 lipocalin in tissue culture studies with granulocytic cell lines (45-47). These results led to the idea that STAT3 would be a crucial, positive regulator of granulopoiesis. Surprisingly, however, several groups showed using cre-loxP-mediated conditional Stat3 deletion that Stat3-deficiency in the hematopoietic system leads to neutrophilia (48-51). These Stat3-deficient models have an approximate threefold increase in circulating neutrophil numbers and a modest enhancement of mature neutrophils in bone marrow. Significantly, mice with MXcre-induced or hematopoietic-specific (Tie2cre-mediated) Stat3-deficiency contain relatively normal numbers of granulocyte progenitors and precursor populations in bone marrow, including lineage-Sca-1+ c-kit+ (LSK) cells, G-CSF- and GM-CSF-responsive colony-forming cells, promyelocytes, myelocytes, and metamyelocytes (48, 50-52). These results collectively indicate STAT3 is dispensable for early stages of granulopoiesis, yet has an important role in restraining peripheral neutrophil numbers.
The negative role of STAT3 in granulopoiesis appears to be due in part to STAT3-dependent induction of SOCS3, a member of the SOCS protein family that inhibits cytokine signaling responses (53) (Fig. 1). SOCS proteins suppress Jak kinase function, promote proteasome-mediated degradation of cytokine signaling components, and serve as competitive inhibitors that block STAT binding to cytokine receptors (53). The murine Socs3 promoter contains 2 consensus STATx binding sites, which suggested Socs3 was an important STAT-regulated gene (54). Accordingly, STAT3-activating cytokines such as IL-6 or leukemia-inhibitory factor (LIF) stimulate STAT3 interaction at the Socs3 promoter in vivo, leading to rapid induction of Socs3 transcription (54, 55). Subsequent studies revealed Socs3 as one of the most highly induced genes following STAT3 activation (56); thus, Socs3 expression is often used as a readout for STAT3 signaling. SOCS3 dampens STAT3 signal transduction from the G-CSFR and, therefore, SOCS3 plays a critical role in suppressing the signaling cascade that activates its expression, consistent with its autoregulatory function (54, 57) (Fig. 1). Hematopoietic-restricted Socs3 ablation leads to neutrophilia (57). Following G-CSF treatment, hematopoietic Socs3-deficient mice exhibit elevated neutrophilia and pathological neutrophil-mediated inflammation, indicating SOCS3 negatively regulates G-CSF-G-CSFR functions in vivo (57). These results support the idea that STAT3-directed induction of SOCS3 is critical for dampening neutrophil production, in line with observations of elevated neutrophil numbers in hematopoietic Stat3- or Socs3-deficient mice (48, 57). However, the onset of neutrophilia in hematopoietic Socs3-deficient mice is delayed compared to hematopoietic-restricted Stat3-deficient animals (49, 51, 52, 57). This may be due to differences in G-CSFR signaling in each model and/or STAT3-mediated control of additional target genes in granulopoiesis, a subject covered in more detail below.
Fig. 1. Roles for STAT3 in G-CSFR signal transduction.
G-CSF-activated STAT3 induces genes involved in G-CSFR signal termination (Socs3), emergency granulopoiesis (Cebpb, c-myc) and neutrophil migration (Cxcl2, Il8rb).
How does G-CSF promote the generation of Stat3-deficient granulocytes? The inability of Stat3-deficient granulocytes to activate SOCS3 (or STAT3) in response to G-CSF results in enhanced activation of other G-CSFR-induced pathways including augmented STAT1 and Erk1/2 signaling (48). Importantly, both STAT1 and Erk1/2 signaling responses promote granulocyte proliferation and delay neutrophil apoptosis; the latter is accomplished by induction of antiapoptotic proteins (48, 50, 58, 59). Thus, STAT3-SOCS3 signaling elicited by G-CSFR is crucial for maintaining neutrophil homeostasis, yet other G-CSFR signaling responses stimulate granulopoiesis in the absence of STAT3 and SOCS3.
Socs3-deficient myeloid cells show elevated STAT3 activation but not enhanced MAPK signaling in response to G-CSF (57), suggesting STAT3 may also stimulate genes that positively regulate granulopoiesis. Support for this concept was obtained through studies of mice that express a truncated isoform of the G-CSFR that contains a mutation (Csf3rd715F) that attenuates STAT3 and STAT5 signaling, engineered by targeted gene replacement at the Csf3r locus (32). Csf3rd715F animals display significant fewer metamyelocytes and mature neutrophils in homeostasis, while their immediate precursors, myelocytes and promyelocytes, are increased. Furthermore, Csf3rd715F mice fail to adequately induce peripheral neutrophil numbers following G-CSF administration or bacterial challenge (32). Granulocytic progenitors from Csf3rd715F mice exhibit suppressed proliferative and differentiation responses to G-CSF. These functions can be rescued by reconstitution with constitutively active STAT3. By contrast, dominant-negative STAT3 abrogated granulocytic cell expansion and differentiation in response to G-CSF ex vivo (32). These data support the idea that STAT3 has complex roles in granulopoiesis involving negative and positive regulatory functions.
Using hematopoietic Stat3-deficent mice, we found STAT3 is required to mediate G-CSF-driven ‘emergency’ granulopoiesis (51), a response that occurs during systemic infection and is mimicked by clinical application of recombinant G-CSF cytokine (60, 61). Significantly, we showed G-CSF-driven emergency granulopoiesis is independent of SOCS3 (51), indicating additional STAT3 target genes are involved. We discovered STAT3 controls the ability of LSK and immature granulocyte progenitor populations to increase in number during G-CSF treatment (51, 52). Moreover, STAT3 is required for hematopoietic progenitor proliferation elicited by recombinant G-CSF or bacterial infection (52), most likely explaining its role in regulating progenitor amounts during emergency granulopoiesis. STAT3 acts by directly inducing expression of the transcriptional regulator C/EBPβ, which is necessary for emergency but not steady state granulopoiesis (62). Furthermore, STAT3 works in collaboration with C/EBPβ to induce c-Myc expression (52) (Fig. 1), contributing to sustained progenitor proliferation during emergency conditions. Interestingly, the induction of c-Myc is mediated by direct and indirect STAT3-mediated mechanisms. These include direct interaction of STAT3 and C/EBPβ with the c-Myc promoter, and indirect suppression of C/EBPα interaction at the c-Myc promoter (52). C/EBPα is a key regulator of steady state granulopoiesis. Its principal actions include controlling the expression important granulocyte genes, inducing cell cycle arrest in developing granulocytic progenitors, including c-Myc repression, which enables terminal neutrophil differentiation (63, 64). In addition, other transcriptional regulators such as PU.1 and Gfi1 have important roles in granulopoiesis at progenitor as well as late differentiation stages (65-67). However, STAT3 is unique in its ability to link extracellular cytokine cues elicited during infection (e.g. G-CSF) with the transcriptional machinery that drives granulocytic progenitor proliferation needed to sustain emergency granulopoiesis (Fig. 1).
An additional and often overlooked aspect of emergency granulopoiesis is the rapid release of mature neutrophils from the bone marrow reserve, a phenomenon termed neutrophil mobilization. We found STAT3 controls important neutrophil functions required for mobilization including chemoattractant-stimulated actin reorganization and G-CSF-responsive transcription of Cxcl2 (encoding MIP2) and Il8rb (encoding CXCR2, a receptor for MIP-2) (51, 68, 69)(Fig. 1). Accordingly, STAT3 is necessary for acute neutrophil mobilization elicited by G-CSF or MIP-2 (51, 68). However, the amount of mature neutrophils accumulates in circulation during prolonged G-CSF treatment in hematopoietic Stat3-deficient mice (51), suggesting alternative or compensatory migration pathways are elicited in the absence of STAT3. Furthermore, STAT3 is required for chemotaxis of mature neutrophils toward CXCR2 ligands including MIP-2 or KC (51), indicating STAT3 involvement in neutrophil responses that are necessary to localize to sites of infection or inflammation (25). These data point to a potential mechanism that might lead to elevated circulating neutrophil amounts in hematopoietic Stat3-deficient mice. The requirement for STAT3 in neutrophil chemotaxis may cause defective tissue margination, contributing to neutrophil accumulation in blood. Hence, neutrophilia in hematopoietic Stat3-deficient mice may be due to combined effects of the failure to activate the negative regulator SOCS3 and impaired neutrophil trafficking (i.e., tissue margination) (3, 48, 51, 57, 68). Importantly, the human immunodeficiency condition Hyper-immunoglobulin E syndrome (HIES), associated with inactivating mutations in one allele of STAT3, is reported to show defects in neutrophil chemotaxis (70). Much remains to be learned about neutrophil function in HIES, however these date suggest specific STAT3 functions may be conserved in humans. In summary, STAT3 has numerous, complex and non-redundant roles in regulating steady state and emergency granulopoiesis including SOCS3 induction, G-CSF-responsive progenitor proliferation and neutrophil mobilization (Fig. 1).
Macrophage origin and growth factors
Macrophages are tissue-resident phagocytic cells that specialize in clearance of dead cells and ingestion of invading pathogens. Macrophages are an important phagocytic component of the reticuloendothelial system (or mononuclear phagocytic system). Macrophage populations are highly diverse in adult organs and include subsets such as splenic marginal zone macrophages, liver Kupffer cells and brain microglia. Macrophage heterogeneity is increasingly recognized as a critical cellular component of inflammatory and neoplastic diseases (71). Conditioned by their residential environment, macrophages gain distinct functions that enable them to participate in tissue homeostasis and host defense (71, 72). Unlike most immune cells, macrophages derived during embryogenesis can persist into adulthood and maintain a tissue-specific reservoir through local proliferation. For example, the microglia population in adult brain is established prenatally and derives primarily from primitive progenitors found in the ectoderm of the yolk sac (73). Under homeostatic conditions and in response to inflammation, embryonically derived microglia are maintained locally without replenishment from circulating precursor cells. However, blood monocytes or myeloid precursors can contribute to the microglia pool under certain conditions, such as central nervous system (CNS) inflammation induced by irradiation or complete lack of embryonic microglia due to genetic PU.1 ablation (74-77).
Lineage tracing studies have shown that a majority of F4/80 high macrophages in other tissues, such as spleen (red pulp macrophage), pancreas, liver (Kupffer cells), and lung (alveolar macrophage), also arise from yolk sac progenitors. Development of these populations does not require the adult hematopoietic regulator c-Myb (78, 79), consistent with their origin from embryonic precursors and not adult bone marrow HSCs. Alternatively, F4/80low tissue macrophages are thought to arise from bone marrow-derived monocytes and are continuously replenished by hematopoietic progenitors (78, 79). More recently, using parabiosis and fate-mapping approaches, macrophages in lung, spleen, peritoneum, and bone marrow were shown to undergo stochastic self-renewal by proliferating in situ, in homeostasis and after nongenotoxic ablation (80). These new findings argue against the contribution of circulating monocytes to tissue-resident F4/80high macrophages, although some studies indicate monocytes may act as a source of tissue macrophages under certain experimental settings (81-84). By contrast, all resident macrophages in mouse intestine were shown to arise from blood monocytes in steady state conditions (85). Tumor-infiltrating and atherosclerotic-plaque macrophages also appear to have monocyte origin (84, 86, 87). Thus, it appears that the majority of tissue-resident macrophages in homeostasis, with the exception of intestinal populations, are distinct from macrophages that arise during inflammation or pathological conditions.
Regardless of their diversity in anatomical location, morphology, and origin, the development of most if not all macrophage populations requires activation of colony-stimulating factor 1-receptor (CSF-1R), a high affinity receptor for M-CSF. Targeted inactivation of murine Csf1r results in a severe deficiency of blood monocytes and tissue macrophages (e.g. microglia), as well as impaired bone resorption associated with reduced osteoclasts, which are derived from bone macrophages (73, 88, 89). Furthermore, blockade of CSF-1R signals by CSF-1R-neutralizating antibodies or tyrosine kinase inhibitors also eliminated or reduced mononuclear phagocytic cell populations (90-94), confirming an essential role for CSF-1R-mediated signals in macrophage development. Depletion of tissue-resident macrophages is more profound in Csf1r−/− mice versus animals carrying a nonsense point mutation in Csf1 (Csf1op/op mice), the gene encoding M-CSF/CSF-1 (88, 95). For example, microglia are present in Csf1op/op mice, but not Csf1r−/− animals. These results suggested the presence of an alternative ligand(s) for CSF-1R, leading to identification of IL-34 as a second CSF-1R ligand. Interestingly, IL-34 is required exclusively for the development of microglia, but not other tissue macrophage subsets (96), results that are consistent with the presence of microglia in Csf1op/op mice. Additional cytokines, such as GM-CSF and IL-4, have been shown to maintain or expand tissue macrophage populations (94, 97), further reflecting the complexity of this innate immune subset and its regulatory control mechanisms.
STAT proteins in macrophage polarization
Although M-CSF and CSF-1R have well established roles in macrophage development, very little information exists about the intracellular signaling cascades they utilize to direct macrophage generation. Engagement of M-CSFR stimulates activation of multiple signaling pathways, including Tyk2, STAT1, STAT3, MAPK, and PI3K (23, 98-100). Bone marrow-derived macrophages lacking expression of the PI3K p85α subunit show impaired proliferation and migration in response to M-CSF or GM-CSF stimulation (101). These results suggest the potential for PI3K signals to mediate macrophage development, although it remains unclear whether bone marrow-derived macrophages reflect native macrophage populations found in homeostatic conditions. While certain transcription factors are known to control macrophage generation, including PU.1 and IRF8 (65, 66, 102-108), the precise mechanisms that link extracellular M-CSF or IL-34 with intracellular cascades that drive macrophage development remain unclear.
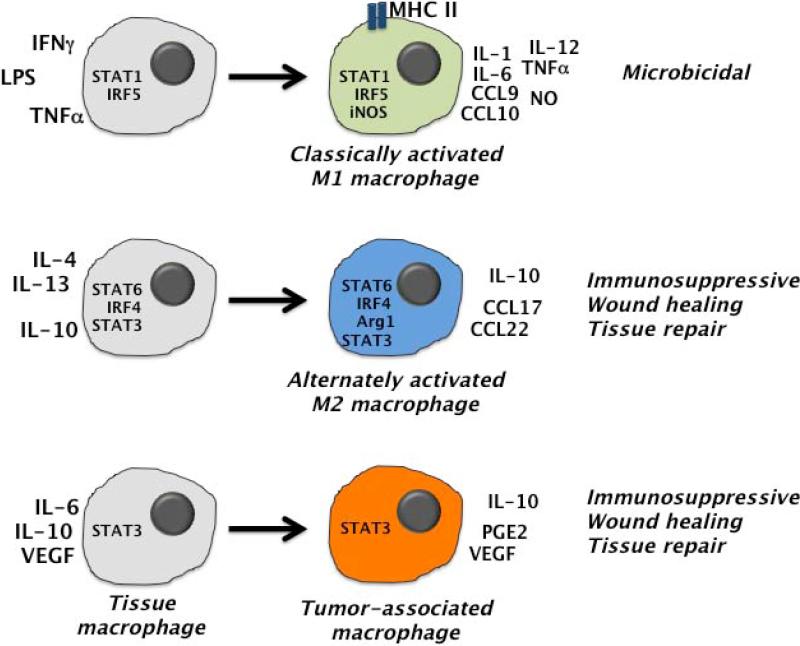
Extracellular factors also have crucial roles in macrophages by regulating functional distinctions between differentiated populations. For example, macrophages have been subdivided into two major types based on their functional differences: the ‘classically activated’ M1 macrophage subset and the ‘alternatively activated’ M2 macrophage population, although subsets with intermediate or additional phenotypes also exist (72, 109, 110) (Fig. 2). PRR agonists and cytokines that are encountered in infection and inflammatory environments control M1 and M2 macrophage polarization (72, 109-111). This process resembles CD4+ T-cell differentiation (e.g. Th1, Th2, Th17 polarization), which is influenced by local cytokine cues (4). In macrophage polarization, stimulation with the Toll-like receptor 4 (TLR4) antagonist LPS, the cytokine IFN-γ or TNF-α can drive the M1 phenotype (110, 112-114). M1 macrophages produce high levels of proinflammatory cytokines (e.g. TNFα, IL-1, IL-6, IL-12) and Th1-attracting chemokines (e.g. CXCL9, CXCL10), and thus are efficient in eliciting Th1-type immune responses (110, 115). M1 macrophages also have enhanced antimicrobial capability, as revealed by elevated inducible nitric oxide synthase (iNOS) and nitric oxide (NO) production (110, 116). The classic M1 traits include enhanced microbicidal activity, induction of MHC class II and antigen-presenting function, and IL-12 production (109, 110). These features endow M1 macrophages with effective bacterial killing activity and induction of appropriate adaptive immunity, thus the M1 population is broadly considered to be proficient at mediating host defense responses (Fig. 2).
Fig. 2. Signals mediating macrophage polarization.
(Left) Extracellular and intracellular factors involved in regulating the polarization of tissue macrophages. (Right) Polarized macrophage populations (M1, M2, TAM) are illustrated, showing intracellular signaling molecules, canonical soluble factors and major biological functions.
Since IFN-γ has a key role in M1 macrophage polarization, it is of little surprise that Jak1, Jak2, and STAT1-mediated responses are involved, as these are the major signaling molecules elicited by this cytokine (117). IFN-γ-activated STAT1 homodimers induce transcription of classic M1 macrophage genes (e.g. iNOS, IL-12 and CIITA) by direct binding to the gamma-activated sequence (GAS, a STAT consensus binding element) in their proximal promoters (117). In the case of LPS stimulation, engagement of TLR4 induces IFN-β synthesis, which signals via an autocrine manner to induce IFN-stimulated gene factor 3 (ISGF3) (20, 118, 119). ISGF3 is a heteromeric complex comprising STAT1, STAT2, and IRF9. The activated ISGF3 complex translocates into the nucleus, where it binds interferon-stimulated response elements (ISREs) in the promoter regions of target genes (20). While ISREs have been found in the promoter regions for the genes encoding iNOS, IL-12 and CIITA, evidence in support of a direct interaction between ISGF3 and these regions is lacking. By contrast, LPS was reported to stimulate STAT1 activation and recruitment to iNOS GAS element, although this was detected at a delayed time point compared to IFN-γ-induced STAT1 interaction (2 h versus 30 min), suggesting that new protein synthesis, i.e. IFN-β, was also involved in LPS-mediated iNOS upregulation (118, 120, 121). To date, the precise mechanisms that are employed by LPS/TLR4 to activate M1 polarization remain unclear; however, STAT1 is well established as cytokine-responsive factor involved in M1 induction (Fig. 2).
By contrast, macrophages that develop in the presence of Th2 cytokines such as IL–4, IL-10, and IL-13 are classified as M2 (122-125) (Fig. 2). M2 macrophages express high levels of IL-10, arginase 1 (Arg1) and the chemokines CCL17 and CCL22 (126-129). They are thought to act as important regulators in tissue repair (i.e. enhancing repair mechanisms), Th2-mediated allergic and parasitic immunity, and tumor development (109, 110, 124). The induction of the M2 phenotype depends on STAT6, a signaling molecule activated by IL-4 and IL-13 (130). IL-4- or IL-13-mediated activation of STAT6 not only directly upregulates expression of Arg1, one of the most specific M2 macrophage markers, but also induces other key transcription factors (i.e. PPAR-γ and C/EBPβ) that synergize with STAT6 to regulate M2-specific genes (131-133). Conversely, IL-10 inhibits STAT1 phosphorylation and signals predominantly through STAT3, which stimulates Arg1 upregulation in mycobacteria-infected macrophages (56, 132, 134). STAT3 also has a well-established role in suppressing production of proinflammatory cytokines in macrophages and neutrophils, and curbing lethal intestinal inflammation mediated by these cells in vivo (114, 135). This may relate to its ability to interact physically with the NF-κB p65 and p50 subunits, and thereby act as a dominant-negative inhibitor of NF-κB signaling to the M1 macrophage hallmark gene iNOS (136-139). Furthermore, the interferon regulatory factors IRF5 and IRF4 have been shown to specifically direct polarization into M1 or M2 macrophages, respectively (Fig. 2) (72, 140, 141); however, it is not clear whether STAT signals are involved in IRF regulation during macrophage polarization.
Collectively, STATs play crucial roles in determining macrophage functional polarization (72) (Fig. 2). The balance of signal strength and crosstalk between STAT1-or STAT3/STAT6-mediated pathways appear to be key factors leading to M1 or M2-related gene activation and subsequent macrophage polarization (72, 110). In addition, stimulation of human macrophages with GM-CSF or M-CSF promotes monocyte differentiation into M2- or M1-polarized phenotypes (142, 143). STAT5 appears to be involved in GM-CSF-mediated macrophage differentiation (144), while signals downstream of M-CSF are less investigated.
STATs in tumor-associated macrophages and myeloid-derived suppressor cells
An additional, specialized macrophage population is now recognized in cancer: tumor-associated macrophages (TAMs) (Fig. 2). TAMs are typically defined by the cell surface profile Ly6C+ MHC class II+ CX3CR1+ CCR2+ CD62L+ in mouse, and CD14+ HLA-DR+ CD115+ CD312+ CD16+ in human. TAMs have gained increasing attention due to their immunosuppressive role during tumorigenesis (111, 145, 146). TAMs appear to be highly related to another immune-suppressing population in tumors, myeloid-derived suppressor cells (MDSCs), which are generally divided into granulocytic CD11b+ Ly6G+ Ly6Clo and monocytic CD11b+ Ly6G− Ly6Chi subpopulations in mouse (147, 148). Accordingly, MDSCs represent a heterogeneous population of myeloid cells, many of which are incompletely terminally differentiated. Because of their high functional similarities and potential overlap in developmental origins, TAMs and MDSCs are often discussed together. In most established tumors, TAMs exhibit phenotypes synonymous with M2 macrophages (145, 146)(Fig. 2). This includes low IL-12 and MHC class II expression, reduced antimicrobial capability, and elevated production of angiogenic and immunosuppressive factors (e.g. VEGF, prostaglandin E2, IL-10). For example, during carcinogen-induced lung tumor development, infiltrating macrophages appear to switch from an M1 to M2 phenotype (149). However, variations in TAM characteristics have been reported in different tumor models, such as mice bearing D1-DMBA-3 mammary tumors, which contain myeloid cells with low expression of macrophage markers, impaired inflammatory function, and present neither M1 or M2 features (150).
Constitutive activation of STAT3 in cancer has been recognized as a crucial signal that promotes tumor growth, angiogenesis and tumor-mediated immunosuppression (151). Importantly, activated nuclear STAT3 has been detected in not only in malignant cells, but also in tumor-associated stromal and immune cells, including TAMs and MDSCs (152). Both tumor and immune cells can produce STAT3-activating factors, such as IL-6, IL-10 and VEGF, which are then able to induce STAT3 signaling in an autocrine as well as paracrine manner. Conditional ablation of STAT3 in hematopoietic cells using the Mx1-cre-loxP system resulted in greater anti-tumor immune responses, thus suppressing tumor progression (151). Similarly, myeloid cell-restricted disruption of Stat3 using the LysMcre model showed enhanced Th1 responses associated with hyperactivity of M1-like macrophages (135), which may exert anti-tumor immunity. Moreover, STAT3 blockade in MDSCs inhibited Arg1 activity and abrogated their suppressor function (153). These data point to STAT3 as a critical immunosuppressive signal in tumor-associated immune subsets including TAMs and MDSCs. It was reported in various tumor models that STAT3 in MDSCs mediated the upregulation of the myeloid-related protein S100A9 and NADPH oxidase, which are critical in MDSC generation and mediating their suppressive function (154, 155). In addition, tumor-activated STAT3 and protein kinase C βII counter-regulate each other and block the differentiation of monocytes into a DC-like population (156). Thus, the role for STAT3 in tumor-associated immune populations is complex, involving effects on myeloid differentiation as well as expression of tumor-regulatory genes.
IFN-γ and STAT1, in contrast, serve important immune surveillance functions against tumors, as evidenced by the enhanced sensitivity to carcinogen-induced tumor growth in mice deficient in IFNγR or STAT1 (157). However, TAMs were shown to express high levels of activated STAT1, and TAMs isolated from mouse EL-4 lymphomas mediated T cell suppressive function via a STAT1-dependent, yet STAT3-and STAT6-independent manner (158, 159). Along the same lines, genetic disruption of STAT1 or delivery of IFN-γ-neutralizing antibodies significantly enhanced IL-12-mediated cytotoxic T-cell (CTL) activity and restrained tumor progression (160). Furthermore, lack of SOCS1 leads to spontaneous development of colorectal carcinomas, which are associated with STAT1 hyperactivity (161). Taken together, these data support the idea that STAT1-dependent signals in TAMs may promote carcinogenesis by suppressing T-cell activities.
Origin and growth factors for dendritic cells
Dendritic cells (DCs) are a heterogeneous immune population that appear closely related yet discrete from macrophages. DCs display distinct phenotypic and functional features based on their anatomical location and developmental origin, a diversity shared with macrophages. However, DCs lack typical macrophage markers, exhibit potent antigen-presenting function and have progenitor/precursors distinct from macrophages (162, 163). The classical or conventional DCs, which are frequently referred to as cDCs, were first identified in splenocyte cultures in mid-1970s by virtue of their unique morphology, including the presence of dendrites or branched cellular protrusions that mediate lymphocyte contacts (164, 165). It is now well established that numerous subsets of DCs reside in lymphoid and nonlymphoid organs. Murine DCs share the common marker CD11c and comprise the splenic-resident CD11b− CD8α+ (CD8α+ DCs), CD11b+ CD4+ (CD4+ DCs) and CD11b+ CD4− CD8α− (CD4− CD8α− DCs) populations; nonlymphoid organ or tissue CD11b− CD103+ DCs (CD103+ DCs); lymph node resident CD8α+ and CD8α− subsets; epidermal Langerhans’ cells (langerin+); intestinal CD103+ and CD11b+ subsets; and plasmacytoid DCs, which are found in circulation, bone marrow and spleen in homeostasis (166). The majority of these DC subsets are able to acquire, process and present exogenous or cell-associated antigens to prime adaptive lymphocyte responses. Recently, CD8α+ and CD103+ DCs have gained attention, due to their superior ability to secrete IL-12 and cross-present exogenous antigen on MHC class I molecules (167-170). Due to these activities, CD8α+ and CD103+ DCs are indispensably required to elicit efficient CD8+ T-cell responses against viral infections, tumors, or vaccines (168, 171-173).
By contrast, pDCs are unique in their plasma cell-like morphology and ability to secrete abundant amounts of type I IFN upon viral infection (174). Due to this exclusive role, pDCs were predicted to participate in mediating anti-viral responses, a role that has been supported by recent evidence in pDC-deficient mice (175). Moreover, pDCs undergo a maturation process following TLR stimulation in which they acquire antigen-presenting function and the ability to modulate T-cell responses (176, 177). In addition, pDCs mediate immune tolerance to dietary and tumor antigens, contribute to wound healing and participate in the pathogenesis of experimentally induced lupus (178-181).
Langerhans’ cells in the epidermis resemble tissue macrophages in terms of ontogeny and developmental cues; yet appear closely related to DCs by virtue of certain phenotypic and functional properties such as the expression of DC hallmark molecule CD11c and antigen presentation capability. Consequently, the classification of Langerhans’ cells as DC versus macrophage has been extensively debated. Recent analysis of the ImmGen Consortium Project shows migrating Langerhans’ cells acquire DC signature genes such as Flt3 and Zbtb46 (182, 183), suggesting close relationship to DCs. However, Langerhans’ cells arise exclusively from embryonic precursors under steady state and require the cytokines IL-34 and TGF-β for their development (89, 96, 184, 185). The embryonic origin and IL-34 dependence is reminiscent of macrophage/microglia populations (discussed above). Langerhans’ cells appear before birth in the epidermis, where they maintain the resident population throughout adult life via self-renewal. Under inflammatory settings, however, blood-borne precursors may contribute to Langerhans’ cell repopulation (186-189).
Despite their obvious differences in function, the pDC and conventional DC populations, with the exception of Langerhans’ cells, derive from bone marrow resident hematopoietic progenitors that express the tyrosine kinase receptor Flt3 (190). Common lymphoid progenitors (CLPs) and CMPs contain Flt3+ subsets, which are able to generate DCs upon adoptive transfer in vivo, while Flt3− CMPs and Flt3− CLPs lack this ability (190). This evidence was among the first to suggest that the DC lineages required Flt3 but development was not restricted to the classic myeloid or lymphoid branches of the hematopoietic system. Recent work has established that Flt3+ CLPs and Flt3+ CMPs have unique developmental potential, with Flt3+ CLPs more proficient at generating pDCs compared to Flt3+ CMPs (191-193). DC development proceeds through committed progenitors for the mononuclear phagocyte lineages, the macrophage/DC progenitor (MDP) population, which has lost the potential to generate granulocytes and thus is placed developmentally downstream of the GMP stage (162, 194-196). MDPs give rise to common DC progenitors (CDP), which generate conventional DCs and pDCs but not macrophage populations (197, 198). CDPs further develop into pre-cDCs, a restricted precursor for conventional DCs (162). Pre-DCs exit the bone marrow and migrate to lymphoid and nonlymphoid tissues, where they give rise to the myriad of distinct conventional DC subsets. By contrast, pDCs arise from CDPs through as yet undefined precursors and complete their terminal differentiation within the bone marrow (162, 197, 198). The restricted development of DCs from Flt3+ progenitor populations is entirely consistent with data that indicates the cytokine Flt3 ligand (Flt3L) is a nonredundant growth factor required for DC homeostasis. Flt3L is produced by numerous cell types including endothelial cells, stromal cells, LSK bone marrow cells and activated T cells (199-201). By contrast, Flt3 expression is highly restricted to early hematopoietic progenitors (HSC, LMPP), DC progenitors (MDP, CDP), pre-cDCs and mature DC subsets (196, 202). Mice that lack either Flt3L or Flt3 show a profound deficiency in DCs, including conventional and pDC lineages, and have reduced amounts of bone marrow DC progenitors (203-205). Administration of recombinant Flt3L or Flt3L overexpression in transgenic mice markedly boosts pDC and conventional DC numbers (e.g., CD8α+, CD4+ and CD103+ DCs) in vivo (206-211). Moreover, Flt3L drives murine DC generation from bone marrow progenitors, or human DC generation from peripheral blood (212-214).
GM-CSF is well known as a myeloid cell growth factor, regulating many aspects of neutrophil, macrophage, and DC maturation and function (215). GM-CSF is commonly used to generate large amounts of conventional DCs ex vivo for experimental as well as clinical use (214, 216-218). GM-CSF also has a specific and unique role in repressing pDC development ex vivo and in vivo (210, 214, 219). Injection of recombinant GM-CSF cytokine or hydrodynamic gene transfer with GM-CSF-encoding plasmid markedly enhances the production of numerous conventional DC populations in lymphoid and nonlymphoid organs, including CD8α+ DCs, CD8α− CD4− DCs and CD103+ DCs (210, 220-222). Surprisingly, however, mice lacking GM-CSF (Csf2−/−) or the GM-CSF receptor β chain (Csf2rb−/−), which is required for GM-CSF receptor signaling, showed only minor defects in lymphoid organ DC populations (205, 220). These data initially suggested GM-CSF was dispensable for DC generation under steady state conditions. However, a better recognition of specific nonlymphoid organ DC populations led to the recent finding that GM-CSF is uniquely required for the accumulation and survival of CD103+ DCs in the dermis, intestine and lung (89, 205, 223). These results indicate that GM-CSF plays a distinct role in regulating DC accrual in nonlymphoid versus lymphoid organs. This function has been postulated to be due to differences in homeostatic GM-CSF amounts in these organs (89, 205, 223), an interesting concept that requires further study.
Function of STATs in DC subset diversification
STAT3 and STAT5 have been reported to be activated upon Flt3 receptor engagement by Flt3L or in cells that express a constitutively activated mutant of Flt3 associated with human leukemia (219, 224). Significantly, however, lin− Flt3+ DC progenitors respond to Flt3L by uniquely stimulating STAT3, without detectable STAT5 activation (219). Accordingly, several groups including our own have demonstrated the importance of STAT3 in DC development using conditional Stat3-deficient mice. This approach has been necessary as genome-wide Stat3 deletion leads to embryonic lethality (225). Conditional deletion of STAT3 in hematopoietic cells (Tie2/Tek-cre Stat3f/f mice) was reported to abrogate Flt3L-dependent DC generation in vitro and in vivo, deplete the total CD11c+ population, and reduce the proportion of DC precursors without affecting HSC amounts (226). Using a similar hematopoietic-specific Stat3-deficient model, we found that pDCs were uniquely sensitive to Stat3-deficiency while conventional CD8α+, CD4+, and CD8α− CD4− DCs in spleen and tissue CD103+ DCs were present in normal amounts under homeostatic conditions (177, 210). In agreement with prior work, we found Flt3L-driven production of conventional CD8α+, CD4+ and CD8α− CD4− DCs in spleen, as well as pDCs in bone marrow, spleen and tissue, required STAT3 (210, 226). Moreover, studies of mice with DC-restricted Stat3-deficiency (CD11c cre) support the concept that pDCs but not conventional DC lineages are uniquely dependent upon STAT3 in steady state conditions (177, 210, 227). Interestingly, however, Flt3L-responsive generation of the nonlymphoid organ CD103+ DC population does not require STAT3 (210). Since this subset showed an approximate 10-fold expansion during Flt3L treatment (210), the results highlight the importance of understanding roles for other signaling pathways elicited by Flt3L. For example, Flt3L has been reported to stimulate MAPK signaling, yet little is understood about the MAPK pathway in DC development. Furthermore, it remains unclear whether Flt3L-driven expansion of CD103+ DCs is mediated by cell autonomous signals that are activated by Flt3 directly in CD103+ DCs or their immediate precursors, or by paracrine effects within the tissues that are induced by Flt3L and act indirectly to stimulate CD103+ DC amounts.
By investigating cellular and molecular responses elicited by STAT3, our laboratory has obtained evidence indicating that STAT3 functions by regulating several aspects of DC development. Using ex vivo cultures with bone marrow lin− Flt3+ DC progenitors from hematopoietic Stat3-deficient mice, we found STAT3 is necessary for Flt3L-dependent progenitor proliferation (219). By contrast, STAT3 is dispensable for progenitor proliferation in response to GM-CSF (219). We also discovered that Flt3L stimulates C/EBPβ expression (210). This suggests the possibility that DC progenitor proliferative responses to Flt3L could be mediated by STAT3-dependent activation of C/EBPβ, similar to the mechanisms used by G-CSF and STAT3 to induce granulocytic progenitor cell growth (52) (Fig. 1). While this model remains to be tested, the requirement for STAT3 in Flt3L-driven progenitor proliferation is likely to explain the broad role for STAT3 in mediating expansion of the conventional and pDC lineages in response to elevated levels of circulating Flt3L (210, 219, 226).
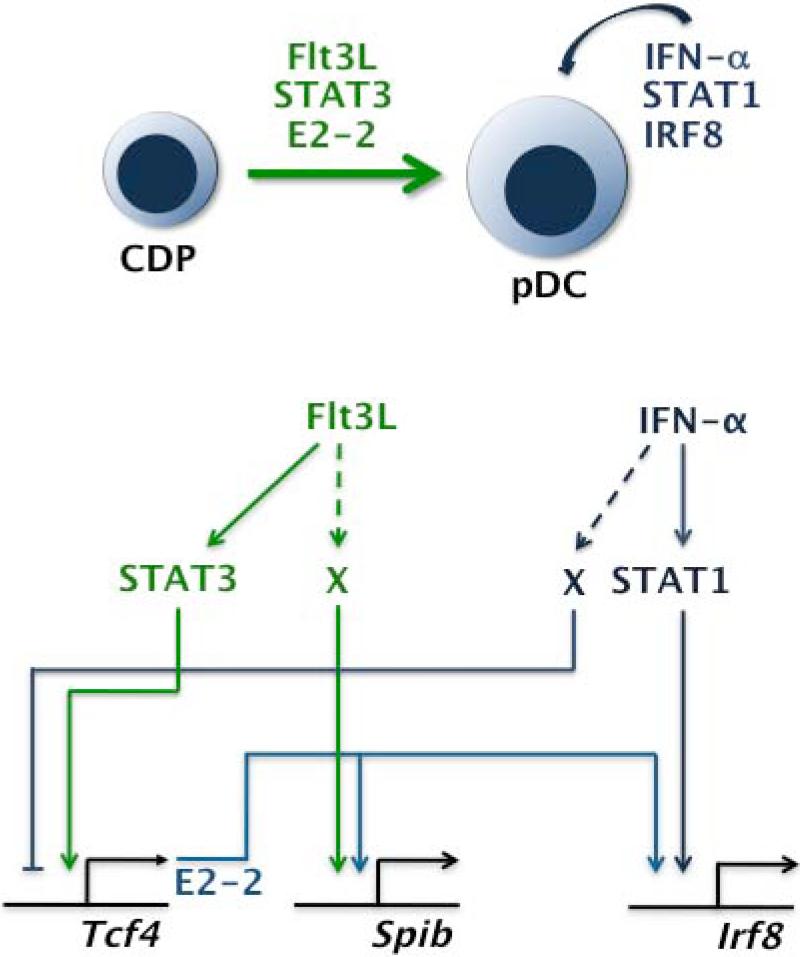
The unique function that our laboratory and others identified for STAT3 in regulating pDC development was reminiscent of studies focusing on the transcriptional regulator E2-2. E2-2 is a basic helix-loop-helix (bHLH) transcription factor that binds E box consensus sites to regulate gene expression (228). Deletion of Tcf4, the gene encoding E2-2, leads to a profound deficiency in pDCs but not other DC lineages (229), demonstrating its fundamental importance in pDC development. E2-2 controls several genes that are uniquely expressed in pDCs and/or required for pDC function, including IRF7, TLR7 and TLR9 (229-231). Furthermore, continued expression of E2-2 is required to maintain pDC cellular identity, as conditional deletion in differentiated pDCs causes them to acquire conventional DC-like traits (230, 231). We found that Flt3L stimulates Tcf4 expression in CDPs, with Tcf4 induction specifically dependent on STAT3 (210). Using gene reporter assays with wildtype and mutated STAT3 isoforms, we showed that STAT3 transcriptional activity is required to stimulate Tcf4 expression in response to Flt3L. Moreover, Flt3L-activated STAT3 is recruited to the Tcf4 promoter in vivo (210). These data collectively indicate that Tcf4 is a direct STAT3 target gene in DC progenitors and suggest STAT3-mediated Tcf4 induction is important for promoting pDC development from the progenitor population (Fig. 3). Therefore, STAT3 not only regulates proliferative responses of DC progenitors upon Flt3L engagement, but also controls expression of the pDC-specific regulator E2-2 (210, 219). It is highly likely that STAT3 regulates additional target genes in DC progenitors and pDCs; genome-wide gene profiling and STAT3 chromatin immunoprecipitation experiments in both populations will be needed for a better understanding of the function of STAT3 in DC development.
Fig. 3. Model for pDC transcriptional regulation by Flt3L- and IFN-α-responsive STATs.
Cytokine-STAT signals involved in pDC development and function, showing STAT target genes (Tcf4, Irf8) and effects of E2-2 on the pDC transcriptional network. X = unknown factor(s) predicted to participate in cytokine-responsive regulation of Spib and Irf8.
Since GM-CSF-responsive progenitor proliferation and GM-CSF-mediated expansion of conventional DC subsets in vivo is independent of STAT3 (210, 219), we undertook experiments to investigate the role of STAT5 in DCs. STAT5 is a key signaling protein activated upon GM-CSF engagement with its receptor. Following ligand binding, the GM-CSF receptor activates its β-chain-associated Jak2 kinase. This leads to receptor tyrosine phosphorylation, followed by the recruitment and tyrosine phosphorylation of STAT5A and STAT5B (referred to herein as STAT5). Mice with genome-wide deletion of Stat5a and Stat5b (Stat5−/− mice) die at birth (232); therefore, we used bone marrow chimeric mice reconstituted with Stat5−/− fetal liver cells to investigate the role of STAT5 in DCs. These experiments showed that Stat5−/− chimeric mice had reduced proportions and absolute numbers of donor-derived bone marrow CD11c+ CD11b− DCs relative to Stat5-sufficient controls. By contrast, we did not observe an effect on CD11c+ CD11b+ DCs (a population comprises both CD4+ and CD8α−CD4-DCs), suggesting STAT5 regulates specific conventional DC populations (219). More strikingly, we found that STAT5 inhibited pDC generation following bone marrow reconstitution. The absolute number and frequency of donor-derived pDCs was higher in chimeras reconstituted with Stat5−/− progenitors versus Stat5-sufficient controls (219). Prior studies had shown that GM-CSF inhibits pDC generation in ex vivo culture systems (214). Using Stat5−/− fetal liver progenitors, we confirmed that STAT5 mediated the suppressive effect of GM-CSF on pDC generation (219). Thus, collectively, these experiments pointed to critical roles for STAT5 in blocking pDC development and promoting generation of CD11b− DCs.
To further investigate STAT5 function in DCs, we utilized mice with hematopoietic-restricted Stat5-deficiency. These animals were obtained by breeding Tie2 cre transgenic mice with animals carrying the floxed Stat5a Stat5b locus (termed herein Stat5f/f) (232). This approach provides an advantage to bone marrow chimeras reconstituted with Stat5−/− fetal liver cells since Stat5 deletion occurs in adult (not fetal) hematopoietic progenitors. Moreover, the Stat5−/− bone marrow chimeras show dramatically reduced spleen cell numbers, which likely reflects the importance of STAT5 in lymphopoiesis (233), yet confounds analysis of splenic DC populations. Accordingly, we compared the proportion and absolute number of bone marrow, splenic and tissue DC populations in hematopoietic Stat5-deficient mice and Stat5-sufficient animals in steady state conditions and following GM-CSF treatment. We confirmed that STAT5 has a critical role in suppressing pDC development in homeostasis and upon exposure to GM-CSF (210). Interestingly, we determined that splenic conventional DC subsets including CD8α+, CD4+, and CD8α− CD4− DCs were unaffected in hematopoietic Stat5-deficient mice. Taken in light of their lack of dependence on STAT3 (210), the data collectively suggest that alternate intracellular signaling cascades are important for the homeostatic generation of splenic CD8α+, CD4+ and CD8α− CD4− DCs. By contrast, STAT5 plays a critical role in the nonlymphoid CD103+ DC population. This subset was significantly reduced in hematopoietic Stat5-deficient mice in steady state, and failed to expand in these animals during GM-CSF treatment (210). These data agree with the selective dependence of CD103+ DCs on GM-CSF (89, 205, 223). Thus, CD103+ DC development is regulated by GM-CSF and STAT5, while this signaling cascade inhibits pDC generation (89, 205, 210, 219, 223, 234, 235). GM-CSF is stimulated during infection, most notably during responses to bacterial pathogens (26); therefore, the distinct regulation of DC subsets by GM-CSF may indicate that cytokines tailor antigen-presenting populations for effective pathogen removal, a possibility that requires further investigation.
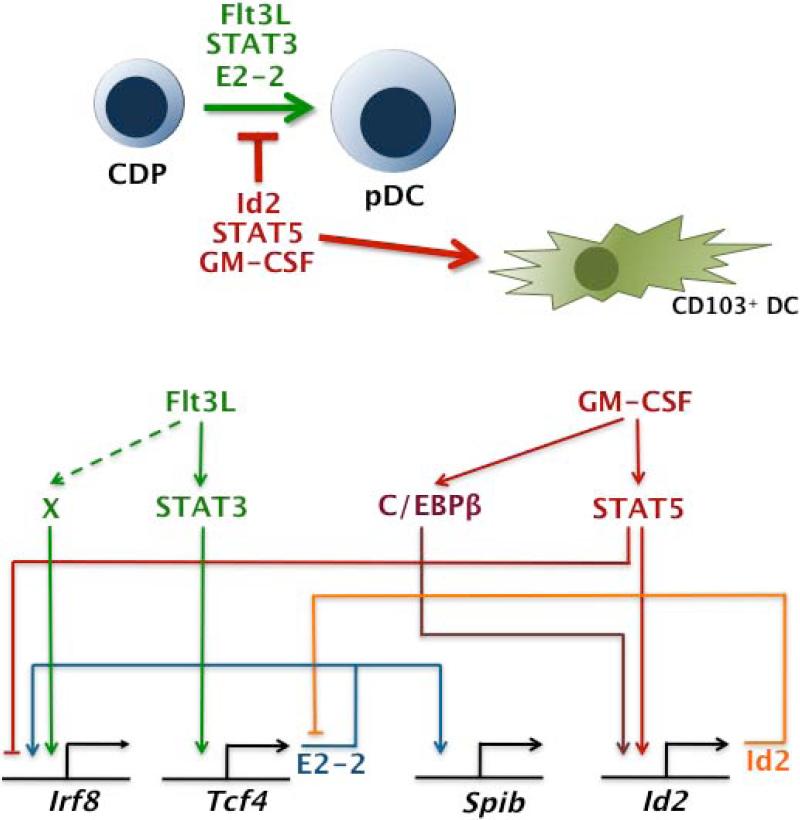
We noted that the unique role for STAT5 in suppressing pDCs and stimulating CD103+ DCs was similar to the function of the transcriptional regulator Id2 in DCs. Id2 is a HLH protein that lacks a basic DNA binding domain (228). Thus, Id2 is able to heterodimerize with bHLH proteins and interfere with their DNA binding function (228). Significantly, Id2 interacts with the pDC regulator E2-2 and blocks its transcriptional activity (236). Furthermore, Id2 is expressed at high amounts in conventional DCs but not pDCs (237). These results collectively suggested Id2 as a negative regulator of the pDC lineage and a positive factor for conventional DCs, responses that could be achieved by E2-2 blockade. In agreement with this model, pDC amounts and type I IFN production are increased in Id2−/− mice, while CD103+ DCs and CD8α+ DCs were significantly reduced (237, 238). Therefore, Id2 controls the production of pDC and conventional DC lineages in an opposing manner (inhibition vs. promotion), although it remains unclear whether and how Id2 interferes with E2-2 or other bHLH proteins during DC development to affect this outcome.
Because STAT5 and Id2 showed similar roles in negatively controlling pDC generation and promoting CD103+ DC development, we investigated the possibility that Id2 was directly controlled by STAT5. Using molecular techniques including chromatin immunoprecipitations and gene expression analyses, we found that GM-CSF-activated STAT5 bound the Id2 promoter in vivo and upregulated Id2 expression in CDPs (210) (Fig. 4). These results suggest a model in which GM-CSF-responsive STAT5 and Flt3L-responsive STAT3 signaling in CDPs influences the expression of the key DC transcriptional regulators Id2 and E2-2 (210, 234, 235) (Fig. 4). The developmental outcome most likely depends on the timing and amplitude of STAT-mediated transcriptional responses, as well as competitive interactions between the protein products of STAT-dependent genes (Fig. 4).
Fig. 4. Model for DC transcriptional regulation by Flt3L- and GM-CSF responsive STATs.
Roles for Flt3L-STAT3 and GM-CSF-STAT5 in DC development, including effects of E2-2 and Id2 on the DC transcriptional network.
In addition, we identified Irf8 as a STAT5-repressed gene in DC progenitors (219). IRF8 is required for the generation and function of pDCs (239, 240). Significantly, IRF8 is also important for development of the conventional CD8α+ and CD103+ DC subsets (237, 239, 241-243). We found that GM-CSF-activated STAT5 interacts directly with the Irf8 proximal promoter to inhibit its transcription (Fig. 4) (219). These data are consistent with the negative role for STAT5 in pDCs, and suggest it is mediated by suppression of Irf8 in addition to induction of Id2. However, because IRF8 is a key positive factor for the CD8α+ and CD103+ DC subsets, it is not yet clear why STAT5-mediated suppression of Irf8 is insufficient to block their development. It is possible that individual DC lineages have distinct requirements for the amount of IRF8 expressed, with pDCs requiring higher IRF8 amounts versus conventional DCs. Alternatively, dedicated progenitor populations may have different needs for IRF8 or encounter discrete microenvironments with or without the ability to elicit GM-CSF-STAT5 signaling.
Few studies to date have investigated roles for STAT proteins in regulating DC function. DC-restricted Stat3-deficient mice develop chronic intestinal inflammation, accompanied by elevated levels of circulating pro-inflammatory cytokines (227). Bone marrow-derived DCs (i.e. DCs generated from bone marrow in GM-CSF cultures) show enhanced pro-inflammatory gene expression upon TLR stimulation (227). These results are consistent with the anti-inflammatory activity of STAT3 (114, 135), although additional studies are needed to evaluate this possibility and define the underlying mechanisms involved. By contrast, DC-restricted deletion of Stat5 led to a deficiency in DC maturation induced by thymic stromal lymphopoietin (TSLP), a potent mediator of Th2 and allergic responses (244). Stat5-deficient DCs showed reduced upregulation of the CD80 and CD86 costimulatory molecules, impaired production of the lymphocyte chemoattractant CCL17 and a decreased ability to induce CD4+ Th2 development ex vivo (244). Moreover, DC-restricted STAT5 expression was critical for induction of Th2-mediated immunity at sites of environmental contact (e.g. skin, lungs) (244). These results point to an essential and non-redundant function for STAT5 in transmitting TSLP signals within DCs to affect adaptive immunity.
STAT1 is well known as a key signal transducer for type I, II, and III IFNs (20). Accordingly, a positive feedback loop of type I IFN receptor signaling via STAT1 is important for robust production of type I IFNs in pDCs following TLR stimulation (245, 246). Furthermore, IFNs act on pDCs and conventional DCs to upregulate MHC and costimulatory molecules, modify cytokine production profiles, and affect antigen presentation; thus, STAT1 expression in DCs is important for modulating the nature of DC-elicited immune responses (177, 247-250). Apart from its role in stimulating DC function, IFN-α appears to have a unique ability to regulate pDC generation and affect the outcome of pDC-mediated Th responses (177). High circulating amounts of IFN-α induce a STAT1-dependent increase in pDCs as well as the pDC-regulatory factor IRF8 (Fig. 3), responses that are accompanied by elevated amounts of lin− Flt3+ DC progenitors (177). The latter result is consistent with the recent identification of type I (and type II) IFNs as proliferation-inducing factors for HSCs (251, 252). In homeostatic conditions, type I IFN and STAT1 are selectively required for accrual of pDCs in Peyer's Patches, lymphoid organs that are adjacent to the intestine (177). Moreover, IFN-α synergizes with Flt3L to facilitate the proliferation and survival of CLPs, and promote their differentiation into pDCs (253). Interestingly, pDCs that are developed in IFN-α-containing cultures show reduced Tcf4 expression and an enhanced propensity to induce Th17 cells versus Flt3L-derived pDCs (177), suggesting type I IFN exposure skews pDC functional responses. By contrast, the conventional DC lineages are repressed by type I IFNs or viral infection (a potent inducer of type I IFNs), an effect that has been reported to require STAT1 or STAT2, respectively (177, 254). Collectively, these results suggest type I IFNs have complimentary effects on pDC and conventional DC development (stimulatory or inhibitory, respectively), in addition to their well established role in promoting maturation of differentiated DCs. It will now be important to understand how type I IFN signals in DCs direct appropriate immune responses or participate in disease development.
Summary and concluding comments
While the field has made major strides in understanding the roles for growth factors and transcriptional regulators in innate immune development, much remains to be learned about the pathways that control progenitor proliferation, differentiation decisions, and functional responses. Specifically, we still lack information on other signaling cascades that are directly downstream of Flt3 in DCs, and their role in mediating DC generation. We also have only rudimentary knowledge of the functions for STAT proteins in mature innate immune cells, as well as the precise mechanisms by which STAT proteins link extracellular cytokine signals with transcriptional responses that affect developmental decisions or functional outcomes. These topics are critical to investigate further as differentiated innate immune cells and potentially their circulating precursors encounter microenvironments rich in cytokines that will elicit STAT activation. Indeed, it is expected that STATs will participate in many aspects of innate immune function, including immunosuppressive roles in tumor microenvironments and pro-inflammatory functions in immune disease. Harnessing the power of these cell populations is critical for improvements in human immunotherapy, and understanding STAT protein function will be a key factor in learning how to regulate subsequent anti-tumor immune responses.
Acknowledgements
We thank Drs. Yong-Jun Liu and Huiyuan Zhang for discussion and insight. Our laboratory research was supported by NIH grants AI073587 and AI098099; an investigator-initiated Preclinical Research Agreement with Amgen Inc, an Institutional Research grant from MD Anderson, and pilot grants from the MD Anderson Center for Inflammation and Cancer, the MD Anderson Stem Cell and Developmental Biology Center and the MD Anderson Center for Cancer Epigenetics (to SSW). Dr. Haiyan Li was supported by an R.E. “Bob” Smith Fellowship and a pilot grant from the MD Anderson Center for Inflammation and Cancer.
Footnotes
The authors have no conflicts to report.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 3.Panopoulos AD, Watowich SS. Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and ‘emergency’ hematopoiesis. Cytokine. 2008;42:277–288. doi: 10.1016/j.cyto.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 6.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 7.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nature reviews Immunology. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 8.Peters BM, Shirtliff ME, Jabra-Rizk MA. Antimicrobial peptides: primeval molecules or future drugs? PLoS Pathogens. 2010;6:e1001067. doi: 10.1371/journal.ppat.1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirota JA, Knight DA. Human airway epithelial cell innate immunity: relevance to asthma. Curr Opin Immunol. 2012;24:740–746. doi: 10.1016/j.coi.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 11.Bazan JF. Haemopoietic receptors and helical cytokines. Immunol Today. 1990;11:350–354. doi: 10.1016/0167-5699(90)90139-z. [DOI] [PubMed] [Google Scholar]
- 12.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagley CJ, Woodcock JM, Stomski FC, Lopez AF. The structural and functional basis of cytokine receptor activation: lessons from the common beta subunit of the granulocyte-macrophage colony-stimulating factor, interleukin-3 (IL-3), and IL-5 receptors. Blood. 1997;89:1471–1482. [PubMed] [Google Scholar]
- 14.Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 15.Sugamura K, et al. The interleukin-2 receptor gamma chain: its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu Rev Immunol. 1996;14:179–205. doi: 10.1146/annurev.immunol.14.1.179. [DOI] [PubMed] [Google Scholar]
- 16.Pesu M, Candotti F, Husa M, Hofmann SR, Notarangelo LD, O'Shea JJ. Jak3, severe combined immunodeficiency, and a new class of immunosuppressive drugs. Immunol Rev. 2005;203:127–142. doi: 10.1111/j.0105-2896.2005.00220.x. [DOI] [PubMed] [Google Scholar]
- 17.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 18.Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 19.Stark GR, Darnell JE., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 21.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 22.Hume DA, MacDonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 2012;119:1810–1820. doi: 10.1182/blood-2011-09-379214. [DOI] [PubMed] [Google Scholar]
- 23.Novak U, et al. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86:2948–2956. [PubMed] [Google Scholar]
- 24.Zhang S, Broxmeyer HE. Flt3 ligand induces tyrosine phosphorylation of gab1 and gab2 and their association with shp-2, grb2, and PI3 kinase. Biochem Biophys Res Commun. 2000;277:195–199. doi: 10.1006/bbrc.2000.3662. [DOI] [PubMed] [Google Scholar]
- 25.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Cheers C, Haigh AM, Kelso A, Metcalf D, Stanley ER, Young AM. Production of colony-stimulating factors (CSFs) during infection: separate determinations of macrophage-, granulocyte-, granulocyte-macrophage-, and multi-CSFs. Infect Immun. 1988;56:247–251. doi: 10.1128/iai.56.1.247-251.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watari K, et al. Serum granulocyte colony-stimulating factor levels in healthy volunteers and patients with various disorders as estimated by enzyme immunoassay. Blood. 1989;73:117–122. [PubMed] [Google Scholar]
- 28.Kawakami M, et al. Levels of serum granulocyte colony-stimulating factor in patients with infections. Blood. 1990;76:1962–1964. [PubMed] [Google Scholar]
- 29.Lieschke GJ, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 30.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 31.Semerad CL, Poursine-Laurent J, Liu F, Link DC. A role for G-CSF receptor signaling in the regulation of hematopoietic cell function but not lineage commitment or differentiation. Immunity. 1999;11:153–161. doi: 10.1016/s1074-7613(00)80090-4. [DOI] [PubMed] [Google Scholar]
- 32.McLemore ML, et al. STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity. 2001;14:193–204. doi: 10.1016/s1074-7613(01)00101-7. [DOI] [PubMed] [Google Scholar]
- 33.Lord BI, Molineux G, Pojda Z, Souza LM, Mermod JJ, Dexter TM. Myeloid cell kinetics in mice treated with recombinant interleukin-3, granulocyte colony-stimulating factor (CSF), or granulocyte-macrophage CSF in vivo. Blood. 1991;77:2154–2159. [PubMed] [Google Scholar]
- 34.Basu S, Hodgson G, Katz M, Dunn AR. Evaluation of role of G-CSF in the production, survival, and release of neutrophils from bone marrow into circulation. Blood. 2002;100:854–861. doi: 10.1182/blood.v100.3.854. [DOI] [PubMed] [Google Scholar]
- 35.Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. 2002;17:413–423. doi: 10.1016/s1074-7613(02)00424-7. [DOI] [PubMed] [Google Scholar]
- 36.Wengner AM, Pitchford SC, Furze RC, Rankin SM. The coordinated action of G CSF and ELR + CXC chemokines in neutrophil mobilization during acute inflammation. Blood. 2008;111:42–49. doi: 10.1182/blood-2007-07-099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pojda Z, Tsuboi A. In vivo effects of human recombinant interleukin 6 on hemopoietic stem and progenitor cells and circulating blood cells in normal mice. Exp Hematol. 1990;18:1034–1037. [PubMed] [Google Scholar]
- 38.Liu F, Poursine-Laurent J, Wu HY, Link DC. Interleukin-6 and the granulocyte colony-stimulating factor receptor are major independent regulators of granulopoiesis in vivo but are not required for lineage commitment or terminal differentiation. Blood. 1997;90:2583–2590. [PubMed] [Google Scholar]
- 39.Seymour JF, Lieschke GJ, Grail D, Quilici C, Hodgson G, Dunn AR. Mice lacking both granulocyte colony-stimulating factor (CSF) and granulocyte-macrophage CSF have impaired reproductive capacity, perturbed neonatal granulopoiesis, lung disease, amyloidosis, and reduced long-term survival. Blood. 1997;90:3037–3049. [PubMed] [Google Scholar]
- 40.Molineux G, Migdalska A, Szmitkowski M, Zsebo K, Dexter TM. The effects on hematopoiesis of recombinant stem cell factor (ligand for c-kit) administered in vivo to mice either alone or in combination with granulocyte colony-stimulating factor. Blood. 1991;78:961–966. [PubMed] [Google Scholar]
- 41.Molineux G, McCrea C, Yan XQ, Kerzic P, McNiece I. Flt-3 ligand synergizes with granulocyte colony-stimulating factor to increase neutrophil numbers and to mobilize peripheral blood stem cells with long-term repopulating potential. Blood. 1997;89:3998–4004. [PubMed] [Google Scholar]
- 42.Tian SS, Lamb P, Seidel HM, Stein RB, Rosen J. Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood. 1994;84:1760–1764. [PubMed] [Google Scholar]
- 43.Tian SS, Tapley P, Sincich C, Stein RB, Rosen J, Lamb P. Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood. 1996;88:4435–4444. [PubMed] [Google Scholar]
- 44.Dong F, et al. Stimulation of Stat5 by granulocyte colony-stimulating factor (G CSF) is modulated by two distinct cytoplasmic regions of the G-CSF receptor. J Immunol. 1998;161:6503–6509. [PubMed] [Google Scholar]
- 45.Shimozaki K, Nakajima K, Hirano T, Nagata S. Involvement of STAT3 in the granulocyte colony-stimulating factor-induced differentiation of myeloid cells. The J Biol Chem. 1997;272:25184–25189. doi: 10.1074/jbc.272.40.25184. [DOI] [PubMed] [Google Scholar]
- 46.Panopoulos AD, Bartos D, Zhang L, Watowich SS. Control of myeloid-specific integrin alpha Mbeta 2 (CD11b/CD18) expression by cytokines is regulated by Stat3-dependent activation of PU.1. J Biol Chem. 2002;277:19001–19007. doi: 10.1074/jbc.M112271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Arcasoy MO, Watowich SS, Forget BG. Cytokine signals through STAT3 promote expression of granulocyte secondary granule proteins in 32D cells. Exp Hematol. 2005;33:308–317. doi: 10.1016/j.exphem.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CK, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 49.Welte T, et al. STAT3 deletion during hematopoiesis causes Crohn's disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci USA. 2003;100:1879–1884. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamezaki K, et al. Roles of Stat3 and ERK in G-CSF signaling. Stem Cells. 2005;23:252–263. doi: 10.1634/stemcells.2004-0173a. [DOI] [PubMed] [Google Scholar]
- 51.Panopoulos AD, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108:3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010;116:2462–2471. doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- 54.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc Natl Acad Sci USA. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L, et al. IL-6 signaling via the STAT3/SOCS3 pathway: functional analysis of the conserved STAT3 N-domain. Mol Cell Biochem. 2006;288:179–189. doi: 10.1007/s11010-006-9137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 57.Croker BA, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 58.McLoughlin RM, et al. Interplay between IFN-gamma and IL-6 signaling governs neutrophil trafficking and apoptosis during acute inflammation. J Clin Invest. 2003;112:598–607. doi: 10.1172/JCI17129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hakkim A, et al. Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nat Chem Biol. 2011;7:75–77. doi: 10.1038/nchembio.496. [DOI] [PubMed] [Google Scholar]
- 60.Zhan Y, Lieschke GJ, Grail D, Dunn AR, Cheers C. Essential roles for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF in the sustained hematopoietic response of Listeria monocytogenes-infected mice. Blood. 1998;91:863–869. [PubMed] [Google Scholar]
- 61.Crawford J, Caserta C, Roila F, Group EGW. Hematopoietic growth factors: ESMO Clinical Practice Guidelines for the applications. Ann Oncol. 2010;21(Suppl):v248–251. doi: 10.1093/annonc/mdq195. [DOI] [PubMed] [Google Scholar]
- 62.Hirai H, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. at Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 63.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 64.Johansen LM, et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 66.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 67.Hock H, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen-Jackson H, Panopoulos AD, Zhang H, Li HS, Watowich SS. STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood. 2010;115:3354–3363. doi: 10.1182/blood-2009-08-240317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen-Jackson HT, Li HS, Zhang H, Ohashi E, Watowich SS. G-CSF-activated STAT3 enhances production of the chemokine MIP-2 in bone marrow neutrophils. J Leuk Biol. 2012;92:1215–1225. doi: 10.1189/jlb.0312126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paslin D, Norman ME. Atopic dermatitis and impaired neutrophil chemotaxis in Job's syndrome. Arch Dermatol. 1977;113:801–805. [PubMed] [Google Scholar]
- 71.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 73.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beers DR, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 76.Mildner A, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 77.Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11:775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- 78.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 79.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parwaresch MR, Wacker HH. Origin and kinetics of resident tissue macrophages. Parabiosis studies with radiolabelled leucocytes. Cell Tissue Kinetics. 1984;17:25–39. doi: 10.1111/j.1365-2184.1984.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 82.Wacker HH, Radzun HJ, Parwaresch MR. Kinetics of Kupffer cells as shown by parabiosis and combined autoradiographic/immunohistochemical analysis. Virchows Archiv B. 1986;51:71–78. doi: 10.1007/BF02899017. [DOI] [PubMed] [Google Scholar]
- 83.Landsman L, Jung S. Lung macrophages serve as obligatory intermediate between blood monocytes and alveolar macrophages. J Immunol. 2007;179:3488–3494. doi: 10.4049/jimmunol.179.6.3488. [DOI] [PubMed] [Google Scholar]
- 84.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bain CC, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ingersoll MA, Platt AM, Potteaux S, Randolph GJ. Monocyte trafficking in acute and chronic inflammation. Trends Immunol. 2011;32:470–477. doi: 10.1016/j.it.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dai XM, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 89.Greter M, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36:1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Conway JG, et al. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc Natl Acad Sci USA. 2005;102:16078–16083. doi: 10.1073/pnas.0502000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Irvine KM, Burns CJ, Wilks AF, Su S, Hume DA, Sweet MJ. A CSF-1 receptor kinase inhibitor targets effector functions and inhibits pro-inflammatory cytokine production from murine macrophage populations. FASEB J. 2006;20:1921–1923. doi: 10.1096/fj.06-5848fje. [DOI] [PubMed] [Google Scholar]
- 92.Ohno H, et al. A c-fms tyrosine kinase inhibitor, Ki20227, suppresses osteoclast differentiation and osteolytic bone destruction in a bone metastasis model. Mol Cancer Ther. 2006;5:2634–2643. doi: 10.1158/1535-7163.MCT-05-0313. [DOI] [PubMed] [Google Scholar]
- 93.MacDonald KP, et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue-and tumor-associated macrophages but does not inhibit inflammation. Blood. 2010;116:3955–3963. doi: 10.1182/blood-2010-02-266296. [DOI] [PubMed] [Google Scholar]
- 94.Hashimoto D, Miller J, Merad M. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 2011;35:323–335. doi: 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoshida H, et al. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yusoff P, Hamilton JA, Nolan RD, Phillips WA. Haematopoietic colony stimulating factors CSF-1 and GM-CSF increase phosphatidylinositol 3-kinase activity in murine bone marrow-derived macrophages. Growth factors. 1994;10:181–192. doi: 10.3109/08977199409000236. [DOI] [PubMed] [Google Scholar]
- 99.Buscher D, Hipskind RA, Krautwald S, Reimann T, Baccarini M. Ras-dependent and -independent pathways target the mitogen-activated protein kinase network in macrophages. Mol Cell Biol. 1995;15:466–475. doi: 10.1128/mcb.15.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jaworowski A, Christy E, Yusoff P, Byrne R, Hamilton JA. Differences in the kinetics of activation of protein kinases and extracellular signal-related protein kinase 1 in colony-stimulating factor 1-stimulated and lipopolysaccharide-stimulated macrophages. Biochem J. 1996;320:1011–1016. doi: 10.1042/bj3201011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Munugalavadla V, Borneo J, Ingram DA, Kapur R. p85alpha subunit of class IA PI-3 kinase is crucial for macrophage growth and migration. Blood. 2005;106:103–109. doi: 10.1182/blood-2004-10-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olson MC, et al. PU. 1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity. 1995;3:703–714. doi: 10.1016/1074-7613(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 103.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 104.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 106.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 107.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi CF, Li Z, Raffeld M, Wang H, Kovalchuk AL, Morse HC., 3rd. Differential expression of IRF8 in subsets of macrophages and dendritic cells and effects of IRF8 deficiency on splenic B cell and macrophage compartments. Immunol Res. 2009;45:62–74. doi: 10.1007/s12026-008-8032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 110.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 112.Mackaness GB. Cellular immunity and the parasite. Adv Exp Med Biol. 1977;93:65–73. doi: 10.1007/978-1-4615-8855-9_5. [DOI] [PubMed] [Google Scholar]
- 113.Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. JJ Immunol. 1988;141:2407–2412. [PubMed] [Google Scholar]
- 114.O'Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mantovani A. The chemokine system: redundancy for robust outputs. Immunol Today. 1999;20:254–257. doi: 10.1016/s0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 116.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Ann Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 117.Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 118.Toshchakov V, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 119.Platanias LC. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 120.Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J Biol Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 121.Ohmori Y, Hamilton TA. Requirement for STAT1 in LPS-induced gene expression in macrophages. J Leuk Biol. 2001;69:598–604. [PubMed] [Google Scholar]
- 122.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goerdt S, Orfanos CE. Other functions, other genes: alternative activation of antigen-presenting cells. Immunity. 1999;10:137–142. doi: 10.1016/s1074-7613(00)80014-x. [DOI] [PubMed] [Google Scholar]
- 124.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 125.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 126.Fenton MJ, Buras JA, Donnelly RP. IL-4 reciprocally regulates IL-1 and IL-1 receptor antagonist expression in human monocytes. J Immunol. 1992;149:1283–1288. [PubMed] [Google Scholar]
- 127.Schebesch C, et al. Alternatively activated macrophages actively inhibit proliferation of peripheral blood lymphocytes and CD4+ T cells in vitro. Immunology. 1997;92:478–486. doi: 10.1046/j.1365-2567.1997.00371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol. 1998;160:5347–5354. [PubMed] [Google Scholar]
- 129.Bonecchi R, et al. Divergent effects of interleukin-4 and interferon-gamma on macrophage-derived chemokine production: an amplification circuit of polarized T helper 2 responses. Blood. 1998;92:2668–2671. [PubMed] [Google Scholar]
- 130.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 131.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest. 2007;117:1155–1166. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qualls JE, et al. Arginine usage in mycobacteria-infected macrophages depends on autocrine-paracrine cytokine signaling. Science signaling. 2010;3:ra62. doi: 10.1126/scisignal.2000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.El Kasmi KC, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ito S, et al. Interleukin-10 inhibits expression of both interferon alpha-and interferon gamma-induced genes by suppressing tyrosine phosphorylation of STAT1. Blood. 1999;93:1456–1463. [PubMed] [Google Scholar]
- 135.Takeda K, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 136.Battle TE, Frank DA. The role of STATs in apoptosis. Curr Mol Med. 2002;2:381–392. doi: 10.2174/1566524023362456. [DOI] [PubMed] [Google Scholar]
- 137.Yu Z, Zhang W, Kone BC. Signal transducers and activators of transcription 3 (STAT3) inhibits transcription of the inducible nitric oxide synthase gene by interacting with nuclear factor kappaB. Biochem J. 2002;367:97–105. doi: 10.1042/BJ20020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yoshida Y, et al. Interleukin 1 activates STAT3/nuclear factor-kappaB cross-talk via a unique TRAF6-and p65-dependent mechanism. J Biol Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- 139.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krausgruber T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 141.Satoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 142.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 143.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 144.Lehtonen A, Matikainen S, Miettinen M, Julkunen I. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J Leuk Biol. 2002;71:511–519. [PubMed] [Google Scholar]
- 145.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 146.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4:376–389. [PMC free article] [PubMed] [Google Scholar]
- 147.Movahedi K, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 148.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zaynagetdinov R, et al. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J Immunol. 2011;187:5703–5711. doi: 10.4049/jimmunol.1100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Torroella-Kouri M, et al. Identification of a subpopulation of macrophages in mammary tumor-bearing mice that are neither M1 nor M2 and are less differentiated. Cancer Res. 2009;69:4800–4809. doi: 10.1158/0008-5472.CAN-08-3427. [DOI] [PubMed] [Google Scholar]
- 151.Kortylewski M, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 152.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 153.Vasquez-Dunddel D, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123:1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Corzo CA, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Farren MR, et al. Tumor-Induced STAT3 Signaling in Myeloid Cells Impairs Dendritic Cell Generation by Decreasing PKCbetaII Abundance. Sci Signal. 2014;7:ra16. doi: 10.1126/scisignal.2004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaplan DH, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Biswas SK, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation). Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 159.Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol. 2005;174:4880–4891. doi: 10.4049/jimmunol.174.8.4880. [DOI] [PubMed] [Google Scholar]
- 160.Torrero MN, Xia X, Henk W, Yu S, Li S. Stat1 deficiency in the host enhances interleukin-12-mediated tumor regression. Cancer Res. 2006;66:4461–4467. doi: 10.1158/0008-5472.CAN-05-3554. [DOI] [PubMed] [Google Scholar]
- 161.Hanada T, et al. IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J Exp Med. 2006;203:1391–1397. doi: 10.1084/jem.20060436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hettinger J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14:821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 164.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. II. Functional properties in vitro. J Exp Med. 1974;139:380–397. doi: 10.1084/jem.139.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Merad M, Manz MG. Dendritic cell homeostasis. Blood. 2009;113:3418–3427. doi: 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Iyoda T, et al. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J Exp Med. 2002;195:1289–1302. doi: 10.1084/jem.20020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bedoui S, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol. 2009;10:488–495. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- 170.Henri S, et al. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. The J Exp Med. 2010;207:189–206. doi: 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fuertes MB, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li L, et al. Cross-dressed CD8alpha+/CD103+ dendritic cells prime CD8+ T cells following vaccination. Proc Natl Acad Sci USA. 2012;109:12716–12721. doi: 10.1073/pnas.1203468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Helft J, et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 2012;122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 175.Cervantes-Barragan L, et al. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc Natl Acad Sci USA. 2012;109:3012–3017. doi: 10.1073/pnas.1117359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kadowaki N, Antonenko S, Lau JY, Liu YJ. Natural interferon alpha/beta-producing cells link innate and adaptive immunity. J Exp Med. 2000;192:219–226. doi: 10.1084/jem.192.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li HS, et al. Cell-intrinsic role for IFN-alpha-STAT1 signals in regulating murine Peyer patch plasmacytoid dendritic cells and conditioning an inflammatory response. Blood. 2011;118:3879–3889. doi: 10.1182/blood-2011-04-349761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Goubier A, et al. Plasmacytoid dendritic cells mediate oral tolerance. Immunity. 2008;29:464–475. doi: 10.1016/j.immuni.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gregorio J, et al. Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J Exp Med. 2010;207:2921–2930. doi: 10.1084/jem.20101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Conrad C, et al. Plasmacytoid dendritic cells promote immunosuppression in ovarian cancer via ICOS costimulation of Foxp3(+) T-regulatory cells. Cancer Res. 2012;72:5240–5249. doi: 10.1158/0008-5472.CAN-12-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Di Domizio J, et al. Nucleic acid-containing amyloid fibrils potently induce type I interferon and stimulate systemic autoimmunity. Proc Natl Acad Sci USA. 2012;109:14550–14555. doi: 10.1073/pnas.1206923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Randolph G, Merad M. Reply to: “Can DCs be distinguished from macrophages by molecular signatures?”. Nat Immunol. 2013;14:189–190. doi: 10.1038/ni.2517. [DOI] [PubMed] [Google Scholar]
- 184.Borkowski TA, Letterio JJ, Farr AG, Udey MC. A role for endogenous transforming growth factor beta 1 in Langerhans cell biology: the skin of transforming growth factor beta 1 null mice is devoid of epidermal Langerhans cells. J Exp Med. 1996;184:2417–2422. doi: 10.1084/jem.184.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ginhoux F, Merad M. Ontogeny and homeostasis of Langerhans cells. Immunol Cell Biol. 2010;88:387–392. doi: 10.1038/icb.2010.38. [DOI] [PubMed] [Google Scholar]
- 186.Merad M, et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Merad M, et al. Depletion of host Langerhans cells before transplantation of donor alloreactive T cells prevents skin graft-versus-host disease. Nat Med. 2004;10:510–517. doi: 10.1038/nm1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol. 2008;8:935–947. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- 189.Ghigo C, et al. Multicolor fate mapping of Langerhans cell homeostasis. J Exp Med. 2013;210:1657–1664. doi: 10.1084/jem.20130403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.D'Amico A, Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med. 2003;198:293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu L, D'Amico A, Hochrein H, O'Keeffe M, Shortman K, Lucas K. Development of thymic and splenic dendritic cell populations from different hemopoietic precursors. Blood. 2001;98:3376–3382. doi: 10.1182/blood.v98.12.3376. [DOI] [PubMed] [Google Scholar]
- 192.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 193.Sathe P, Vremec D, Wu L, Corcoran L, Shortman K. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood. 2013;121:11–19. doi: 10.1182/blood-2012-02-413336. [DOI] [PubMed] [Google Scholar]
- 194.Fogg DK, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 195.Liu K, Nussenzweig MC. Origin and development of dendritic cells. Immunol Rev. 2010;234:45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 196.Schmid MA, Kingston D, Boddupalli S, Manz MG. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev. 2010;234:32–44. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 197.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 198.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 199.McClanahan T, et al. Biochemical and genetic characterization of multiple splice variants of the Flt3 ligand. Blood. 1996;88:3371–3382. [PubMed] [Google Scholar]
- 200.Solanilla A, et al. Expression of Flt3-ligand by the endothelial cell. Leukemia. 2000;14:153–162. doi: 10.1038/sj.leu.2401635. [DOI] [PubMed] [Google Scholar]
- 201.Sitnicka E, Bryder D, Theilgaard-Monch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–472. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 202.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 204.Waskow C, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 2009;114:835–843. doi: 10.1182/blood-2009-02-206318. [DOI] [PubMed] [Google Scholar]
- 206.Maraskovsky E, et al. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J Exp Med. 1996;184:1953–1962. doi: 10.1084/jem.184.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Maraskovsky E, et al. In vivo generation of human dendritic cell subsets by Flt3 ligand. Blood. 2000;96:878–884. [PubMed] [Google Scholar]
- 208.Fong L, et al. Altered peptide ligand vaccination with Flt3 ligand expanded dendritic cells for tumor immunotherapy. Proc Natl Acad Sci USA. 2001;98:8809–8814. doi: 10.1073/pnas.141226398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Manfra DJ, Chen SC, Jensen KK, Fine JS, Wiekowski MT, Lira SA. Conditional expression of murine Flt3 ligand leads to expansion of multiple dendritic cell subsets in peripheral blood and tissues of transgenic mice. J Immunol. 2003;170:2843–2852. doi: 10.4049/jimmunol.170.6.2843. [DOI] [PubMed] [Google Scholar]
- 210.Li HS, et al. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 2012;120:4363–4373. doi: 10.1182/blood-2012-07-441311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tsapogas P, et al. In vivo evidence for an instructive role of fms-like tyrosine kinase-3 (FLT3) ligand in hematopoietic development. Haematologica. 2014;99:638–646. doi: 10.3324/haematol.2013.089482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hubert P, et al. In vitro propagated dendritic cells from patients with human-papilloma virus-associated preneoplastic lesions of the uterine cervix: use of Flt3 ligand. Cancer Immunol Immunother. 1998;47:81–89. doi: 10.1007/s002620050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Blom B, Ho S, Antonenko S, Liu YJ. Generation of interferon alpha-producing predendritic cell (Pre-DC)2 from human CD34(+) hematopoietic stem cells. J Exp Med. 2000;192:1785–1796. doi: 10.1084/jem.192.12.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gilliet M, et al. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med. 2002;195:953–958. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hamilton JA. GM-CSF in inflammation and autoimmunity. Trends Immunol. 2002;23:403–408. doi: 10.1016/s1471-4906(02)02260-3. [DOI] [PubMed] [Google Scholar]
- 216.Inaba K, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Caux C, et al. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to GM-CSF+TNF alpha. J Exp Med. 1996;184:695–706. doi: 10.1084/jem.184.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28:509–520. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Vremec D, Lieschke GJ, Dunn AR, Robb L, Metcalf D, Shortman K. The influence of granulocyte/macrophage colony-stimulating factor on dendritic cell levels in mouse lymphoid organs. Eur J Immunol. 1997;27:40–44. doi: 10.1002/eji.1830270107. [DOI] [PubMed] [Google Scholar]
- 221.Daro E, et al. Polyethylene glycol-modified GM-CSF expands CD11b(high)CD11c(high) but notCD11b(low)CD11c(high) murine dendritic cells in vivo: a comparative analysis with Flt3 ligand. J Immunol. 2000;165:49–58. doi: 10.4049/jimmunol.165.1.49. [DOI] [PubMed] [Google Scholar]
- 222.O'Keeffe M, et al. Effects of administration of progenipoietin 1, Flt-3 ligand, granulocyte colony-stimulating factor, and pegylated granulocyte-macrophage colony-stimulating factor on dendritic cell subsets in mice. Blood. 2002;99:2122–2130. doi: 10.1182/blood.v99.6.2122. [DOI] [PubMed] [Google Scholar]
- 223.King IL, Kroenke MA, Segal BM. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J Exp Med. 2010;207:953–961. doi: 10.1084/jem.20091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Choudhary C, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 225.Takeda K, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19:903–912. doi: 10.1016/s1074-7613(03)00332-7. [DOI] [PubMed] [Google Scholar]
- 227.Melillo JA, et al. Dendritic cell (DC)-specific targeting reveals Stat3 as a negative regulator of DC function. J Immunol. 2010;184:2638–2645. doi: 10.4049/jimmunol.0902960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 229.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yao Z, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci USA. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Li H, Watowich S. A STATus report on dendritic cell development. J Leukoc Biol. 2012;92:445–459. doi: 10.1189/jlb.0212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li HS, Watowich SS. Diversification of dendritic cell subsets: emerging roles for STAT proteins. JakSTAT. 2013;2:e25112. doi: 10.4161/jkst.25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Liu YP, Burleigh D, Durning M, Hudson L, Chiu IM, Golos TG. Id2 is a primary partner for the E2-2 basic helix-loop-helix transcription factor in the human placenta. Mol Cell Endocrinol. 2004;222:83–91. doi: 10.1016/j.mce.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 237.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hacker C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 239.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 240.Schiavoni G, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tailor P, Tamura T, Morse HC, 3rd, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bell BD, et al. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat Immunol. 2013;14:364–371. doi: 10.1038/ni.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Takauji R, et al. CpG-DNA-induced IFN-alpha production involves p38 MAPK-dependent STAT1 phosphorylation in human plasmacytoid dendritic cell precursors. J Leuk Biol. 2002;72:1011–1019. [PubMed] [Google Scholar]
- 246.Prakash A, Smith E, Lee CK, Levy DE. Tissue-specific positive feedback requirements for production of type I interferon following virus infection. Biol Chem. 2005;280:18651–18657. doi: 10.1074/jbc.M501289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Johnson LM, Scott P. STAT1 expression in dendritic cells, but not T cells, is required for immunity to Leishmania major. Journal of immunology. 2007;178:7259–7266. doi: 10.4049/jimmunol.178.11.7259. [DOI] [PubMed] [Google Scholar]
- 248.Pilz A, et al. Dendritic cells require STAT-1 phosphorylated at its transactivating domain for the induction of peptide-specific CTL. J Immunol. 2009;183:2286–2293. doi: 10.4049/jimmunol.0901383. [DOI] [PubMed] [Google Scholar]
- 249.Diamond MS, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kernbauer E, et al. Conditional Stat1 ablation reveals the importance of interferon signaling for immunity to Listeria monocytogenes infection. PLoS Pathogens. 2012;8:e1002763. doi: 10.1371/journal.ppat.1002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 252.Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Chen YL, Chen TT, Pai LM, Wesoly J, Bluyssen HA, Lee CK. A type I IFN-Flt3 ligand axis augments plasmacytoid dendritic cell development from common lymphoid progenitors. J Exp Med. 2013;210:2515–2522. doi: 10.1084/jem.20130536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]