Abstract
Knowing the copy number of cellular proteins is critical for understanding cell physiology. By being able to measure the absolute synthesis rates of the majority of cellular proteins, Li et al. (2014) gain insights into key aspects of translation regulation and fundamental principles of cellular strategies to adjust protein synthesis according to the needs.
Accurate accounting is the basis of an efficient economy. In order to understand the rules, trends, and directions of healthy economic growth, one must be able to track the precise amounts of individual products generated, the demand for these goods and the strategies for allocating the resources for their production. In the cell, proteins are the main commodities. They control the majority of cellular activity but their production is very expensive. Knowledge of how much of each protein is made is therefore central to understanding the organization, growth and proliferation of the cell.
As basic as knowing the copy number of individual proteins in the cell may seem, it is a difficult aim to achieve. While whole-cell proteomics and other genome-wide techniques provide useful insights into changes in gene expression under various physiological conditions, estimating the absolute amounts of even limited number of proteins is far more challenging. In this issue of Cell, Li et al. (2014) have succeeded in analyzing the translation output of more than 3,000 E. coli genes and quantify production of more than 95% of the proteins synthesized in fast growing cells.
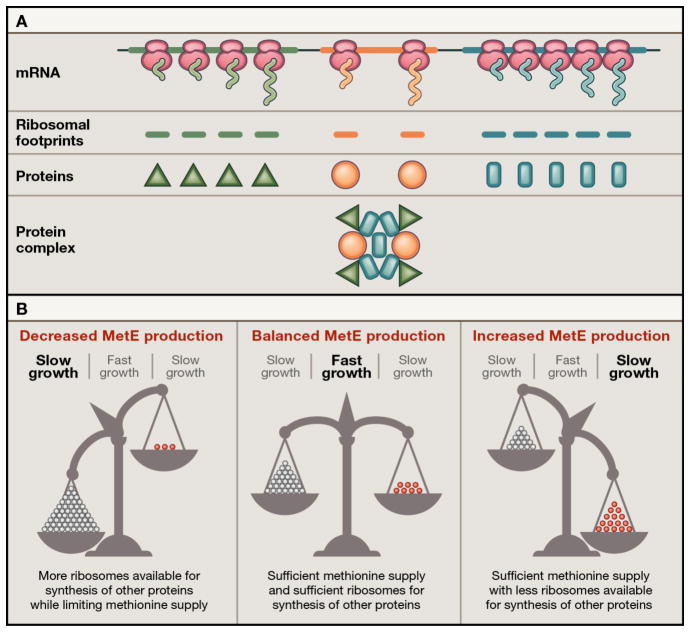
The revolutionary ribosome profiling technique developed by Weissman and colleagues several years ago provides a genome-wide view of translation of individual genes (Ingolia et al., 2009; Li et al., 2012). The method is based on next generation sequencing of the mRNA fragments protected by ribosomes. Each ‘footprint’ represents one translating ribosome, which will most likely generate one protein molecule encoded in the respective gene (Figure 1A). Deep sequencing of the ribosomal footprints hence provides a snapshot of cellular protein synthesis and allows the estimation of the fraction of ribosomes engaged in translation of individual mRNAs, and thus the relative rate of expression of a given gene. Normalizing this by the total protein synthesized during the cell cycle renders the absolute protein synthesis rates, i.e. the number of copies of each protein produced during the cell generation time.
Figure 1. Optimizing the Rates of Protein Synthesis.
(A) Measurement of the absolute rates of protein synthesis shows that the expression of genes in polycistronic operons coding for stable multisubunit protein complexes is usually proportional to the subunit composition of the complex. (B) The protein abundance data reveal that the rate of MetE production is optimized to achieve the most favorable balance between active expression of the MetE enzyme and sufficient translation capacity for synthesizing the rest of the cellular proteins.
By pushing the limits of the technique and achieving an extreme sequencing depth, Li et al. were able to analyze the expression rate of the majority of E. coli genes, including those translated as few as 10 times per generation. This impressively broad and accurate protein accounting makes it possible to explore many aspects of the cellular economy and illuminates the strategies used to achieve secure and fast cell growth at the minimal expense.
A large number of functional complexes in the cell are composed of multiple proteins assembled at a precise stoichiometry. Does the cell produce individual components in a lax fashion and then simply get rid of the unused subunits? Or are the subunits of the complexes synthesized at the ratios that match the need? Of the 64 E. coli protein complexes analyzed by Li et al., 59 adhere to the principle of proportional synthesis, where the production rate of the individual components parallels their stoichiometry in the complex (Figure 1A). However, the genes of bacterial multi-subunit complexes are often organized in operons and thus, their mRNAs are generated in equimolar amounts irrespective of the differential need for the encoded proteins. How then is stoichiometry achieved? Li et al. convincingly show that proportionality of protein production results from tuning the translation rate of individual genes in the polycistronic operons. The principle of proportional synthesis is generally true also for yeast where the production rate of constituents of stable protein complexes has been evolutionarily optimized to be minimally wasteful.
Many other bacterial functional modules, including enzymes involved in the same biochemical pathway, signaling two-components systems and toxin-antitoxin pairs, are also organized in operons. Remarkably, the differential translation rate of individual proteins in such modules is adjusted to match the functional requirements. For example, the authors find that in toxin-antitoxin pairs, the unstable antitoxin is produced more actively than the toxin, even though both genes locate in the same operon, whereas in a two-component signaling complex, the gene of the response regulator is expressed at a higher rate than that of the corresponding kinase. Altogether, the ribosomal profiling census suggests that the translationally coordinated hierarchical expression of functionally related gene products is the key principle for achieving optimal ratios of functional module components.
The knowledge of the copy number of the majority of cellular proteins offers an unprecedented opportunity to examine metabolite flux through biochemical pathways and the logistics of enzyme production at a global level. Li et al. illustrate this facet by analyzing L-methionine biosynthesis under the conditions of limited methionine supply. By correlating the abundance of the enzymes and their catalytic turnover rate, they identify the reaction catalyzed by MetE as the pathway bottleneck. When cells are starved for methionine, the output of the MetE-catalyzed reaction limits the global protein synthesis. Using an analytical model supported with experimental evidence, the authors show that decreasing the abundance of MetE reduces the growth rate due to insufficient methionine production, whereas dedicating more ribosomes to MetE expression diverts too much of the cellular protein synthesis capacity from translation of other essential genes, thereby negatively affecting the growth rate (Figure 1B). This elegant illustration offers a glimpse of how the knowledge of protein abundance can be applied in a much broader way not only for deep exploration of cellular biochemistry but also for biotechnological goals.
New data often necessitate critical re-evaluation of conventional knowledge. The tuning of protein synthesis described by Li et al. is controlled primarily by the frequency of translation initiation. Up until now the rules of initiation in bacteria appeared to be fairly simple. The extent of complementarity between the ribosome binding site in mRNA and the rRNA of the small ribosomal subunit is considered the primary determinant for start codon recognition (Shine and Dalgarno, 1974), whereas the mRNA tertiary structure additionally modulates the efficiency of the ribosome-mRNA interaction (de Smit and van Duin, 1990). Applied to individual genes, these simple rules indeed have certain predictive power and have been able to guide optimization of gene expression (Salis et al., 2009). Strikingly, however, the existing models of translation initiation control largely fail to account for the differences in gene expression rates estimated from the ribosome profiling data. It appears that we are still missing some important factors (mRNA binding proteins? regulatory RNAs?) for the accurate prediction of translation initiation rates in living cells. The newly obtained genome-wide knowledge of absolute rates of gene expression provides fertile ground for in-depth bioinformatics analysis of the underlying principles of translation initiation.
The present study exposes important general principles of gene regulation. However, the data also unmask outliers that do not conform to the common rules. For example, although translation of most cistrons does not show signs of premature translation termination, several genes exhibit an abrupt drop in ribosome density. Such unusual behavior may be indicative of yet unknown translation regulation mechanisms. Another example of non-compliance with the common rule is deviation from proportionality of production of subunits of a small number of stable protein complexes. Do ‘overexpressed’ protein components have some unknown moonlighting functions? Does their rapid turnover play a role in regulation? Exploring these and other odd exceptions may open new doors for better understanding cell biology. We can anticipate that protein accounting, namely the ability to assess the absolute translation rates of cellular polypeptides, will lead to many new discoveries.
Acknowledgments
The work in the authors’ laboratory is supported by the NSF grant MCB 1244455 and NIH grants GM106386 and GM104370.
References
- de Smit MH, van Duin J. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross CA, Weissman JS. Cell. 2014 this issue. [Google Scholar]
- Li GW, Oh E, Weissman JS. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Nature Biotech. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]