Abstract
Allotypes of the natural killer (NK) cell receptor KIR3DL1 vary in both NK cell expression patterns and inhibitory capacity upon binding to their ligands, HLA-B Bw4 molecules, present on target cells. Using a sample size of over 1,500 human immunodeficiency virus (HIV)+ individuals, we show that various distinct allelic combinations of the KIR3DL1 and HLA-B loci significantly and strongly influence both AIDS progression and plasma HIV RNA abundance in a consistent manner. These genetic data correlate very well with previously defined functional differences that distinguish KIR3DL1 allotypes. The various epistatic effects observed here for common, distinct KIR3DL1 and HLA-B Bw4 combinations are unprecedented with regard to any pair of genetic loci in human disease, and indicate that NK cells may have a critical role in the natural history of HIV infection.
NK cells are critical components of the innate immune system that have direct involvement in the antiviral immune response1. NK cells are controlled by many activating and inhibitory receptors2,3, including members of the killer cell immunoglobulin-like receptor (KIR) family. Similarly to other NK cell receptors, KIRs are expressed on T cells as well as on NK cells, affirming their role in both innate and adaptive immunity, but they are distinct from other NK cell receptors in that they are exceptionally diverse and rapidly evolving4. Each KIR locus encodes either an inhibitory or an activating receptor, except for the KIR3DL1 gene, which encodes one common activating allotype, KIR3DS1, and several inhibitory allotypes. The ligands for the inhibitory KIR3DL1 allotypes are HLA-B molecules that contain the Bw4 motif at positions 77–83 (ref. 5), particularly the subset of Bw4 allotypes containing isoleucine at position 80 (Bw4-80I) as opposed to those with threonine (Bw4-80T) at this position5–7. The ligands for KIR3DS1 are not known, but this molecule shares 97% sequence similarity with KIR3DL1 allotypes and may recognize a similar or overlapping set of ligands.
KIRs are expressed in a variegated manner on the NK cell population of a given individual8: only a certain percentage of NK cells express the product of a given KIR gene. Further, mean fluorescence intensity measurements show subtypes of KIR3DL1 to be expressed at different abundances on NK cells9. Recently, it was shown that NK cell inhibitory capacities of individual KIR3DL1 allotypes are closely linked to their abundances on NK cells and to the percentages of cells expressing these molecules within the NK cell population of a given individual10. The functional repercussions of variability at the KIR3DL1 locus provide a logical, operative grouping system for the alleles of this locus: alleles encoding high-expression allotypes (KIR3DL1*h), low-expression allotypes (KIR3DL1*l), and no cell surface expression (*004). KIR3DL1*004 represents an unusual allele as it encodes a protein that is retained within the cell11, a characteristic that may have functional importance. Variation in the affinity of individual KIR3DL1 allotypes for various HLA-B Bw4 allotypes (controlling for expression levels) has also been shown to a limited extent6,10: the KIR3DL1 high-expression allotypes that have been tested show higher affinity for Bw4-80I allotypes than for Bw4-80T, which results in greater inhibition through Bw4-80I recognition.
The most protective HLA class I alleles in terms of HIV disease progression belong to the group of HLA-B Bw4 alleles. In particular, two of these Bw4 alleles, B*27 and B*57 (HLA-B alleles being represented hereinafter by the allele names standing alone), show the greatest protection in several genetic and functional studies (reviewed in ref. 12), the basis of which stems, at least in part, from the immuno-dominant HIV peptides recognized by these allotypes in acquired immune responses13. However, many of the other members of the Bw4 group also tend toward protection relative to the alternative group of HLA-B alleles, Bw6 (ref. 14). As individual allotypes belonging to the Bw4 group recognize distinct peptide motifs15, it is unlikely that common peptide recognition explains the protection conferred by Bw4 as a group; rather, overall Bw4 protection probably has to do with the function of these molecules in the innate immune response as ligands for KIR3DL1 and/or KIR3DS1 (KIR3DL1;3DS1) (ref. 16). Indeed, we have previously shown that the activating KIR3DS1 allele in combination with Bw4-80I associates with protection against HIV disease progression17, as well as against opportunistic infections in HIV+ individuals18. These data raise the possibility that KIR3DS1 may bind one or more of the HLA-B Bw4-80I allotypes on cells infected with HIV-1, directly mediating effector cell killing of the infected target, a parsimonious model in which NK cell activation renders protection against HIV. Given the functional diversity of the KIR3DL1 subtypes in terms of the degree of inhibition they confer, it is possible that they may also differentially affect HIV pathogenesis (apart from the protection conferred by KIR3DS1 + Bw4-80I).
Here we tested the effects of inhibitory KIR3DL1 subtypes in combination with HLA-B allelic groups on HIV disease progression and viral load. These data illustrate a primary role for various distinct combinations of KIR3DL1 and HLA-Bw4 in the innate immune response against HIV. Of note, the highly expressed, highly inhibitory KIR3DL1*h alleles strongly enhance protection conferred by Bw4-80I alleles, including B*57 specifically, an unexpected result given the protective effect of a putatively activating genotype, KIR3DS1 + Bw4-80I, against AIDS progression17. These seemingly contrasting results can be explained logically by a model in which greater dependency on the expression of specific KIR3DL1 + Bw4 receptor-ligand pairs for NK cell inhibition in the resting state (where KIR3DL1*h > KIR3DL1*l) results in more pronounced NK cell responses when that inhibition is abrogated in the face of infection. This model is consistent with recent studies implicating inhibitory receptors for MHC class I in the education and eventual killing capacity of NK cells19–21.
RESULTS
KIR3DL1*004 + Bw4 slows progression to AIDS
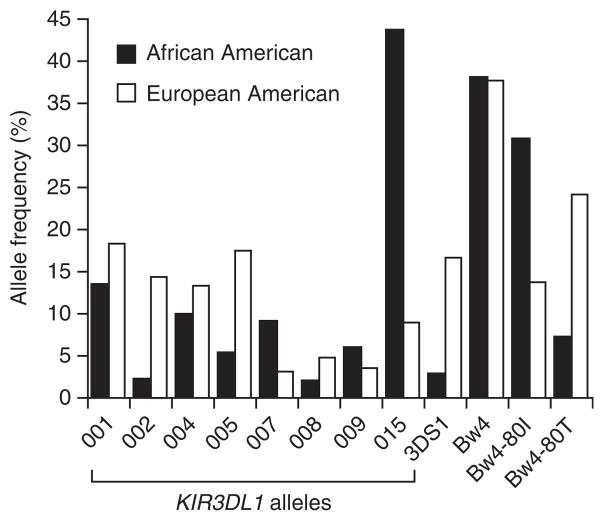
We compared the frequencies of each KIR3DL1 allele in our African American and European American subjects, and the most notable racial differences observed were with KIR3DL1*015 (a highly expressed allele), which was by far the most common allele in African Americans, and, as reported previously, KIR3DS1 (ref. 17) (Fig. 1). As was the case for the KIR3DS1 + Bw4-80I effect17,18, the genetic effects described here were consistent across the two main ethnic groups based on analyses stratified by ancestry (see Methods). All statistics provided in this report were derived from analyses stratified by race.
Figure 1.
Allele frequency of KIR3DL1 alleles and HLA-B Bw4 subtypes. The frequency of each variable is shown for African Americans and European Americans.
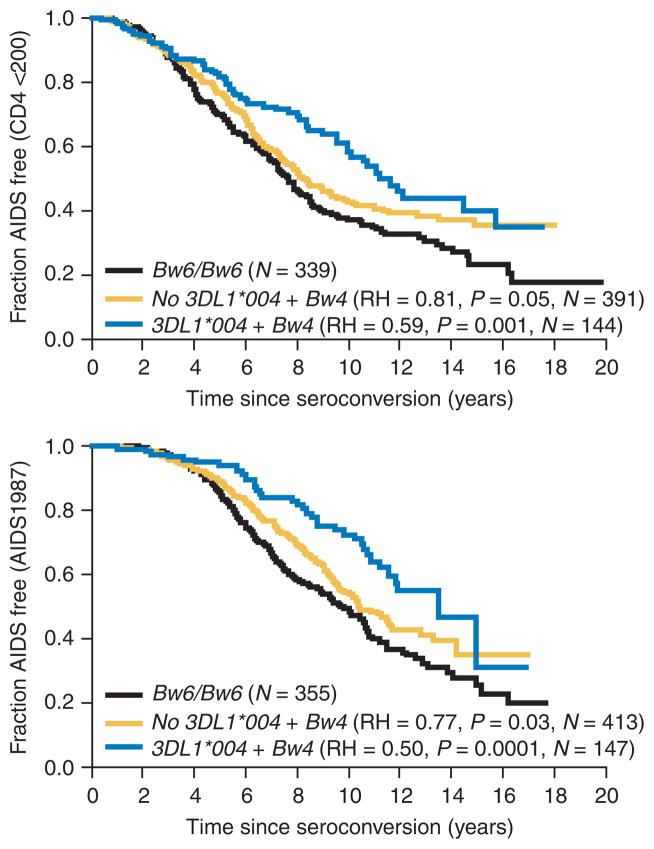
The influence of individual KIR3DL1 alleles on AIDS progression (see Methods for a description of the AIDS outcomes) was tested in a cohort of 915 HIV+ individuals whose date of seroconversion was known within a period of 6 months on average (seroconverters). In order to eliminate confounding by the protective effect of KIR3DS1 + Bw4-80I, individuals with this compound genotype were removed from all analyses described herein. (Our previous study showing protection with this compound genotype17 was performed in this same cohort). HLA-B Bw4 (and not HLA-B Bw6) allotypes serve as ligands for KIR3DL1 subtypes5,7. In Bw6/Bw6 individuals, it does not matter which KIR3DL1 subtype is present because the ligand is absent, so the KIR3DL1 molecule is nonfunctional in these individuals. As the Bw6/Bw6 group does not contain any KIR3DL1 + HLA-B genotype combinations to confound our analyses, we used this group of individuals as a consistent control group for every analysis of KIR3DL1 subtypes, except as noted otherwise. Using Bw6/Bw6 as a consistent reference group allowed direct comparisons of the degrees of effects conferred by distinct KIR3DL1 + Bw4 subtypes. In the presence of Bw4, KIR3DL1*004 showed the most significant protection relative to all other KIR3DL1 alleles (relative hazard (RH) = 0.50 and 0.59, P = 0.0001 and 0.001; the first value in each case refers to the AIDS1987 outcome analysis and the second to the CD4+ T-cell count < 200 cells/mm3 outcome analysis (see Methods), and this convention will apply wherever two statistics are given; Fig. 2 and Supplementary Table 1 online). The q value (the expected proportion of false positives22) for this association (q = 0.00005 and 0.0002) indicates the extreme unlikelihood that this is a false positive discovery (Supplementary Table 1). The protective effect of KIR3DL1*004 was completely dependent on the presence of Bw4, as indicated by the lack of any effect in a comparison of individuals with a Bw6/Bw6 genotype who either had KIR3DL1*004 (Bw6/Bw6 + KIR3DL1*004; N = 86 and 81) or did not (Bw6/Bw6 + no KIR3DL1*004; N = 254 and 246; RH = 1.11 and 0.91, P = 0.56 and 0.59). KIR3DL1*004 is a unique KIR3DL1 allotype in that it is retained within the cell11, a characteristic that has led to its functionality being called into question. Slow progression to AIDS among individuals carrying KIR3DL1*004 + Bw4 raises the possibility that the KIR3DL1*004 molecule may actually bind Bw4 molecules intracellularly with functionally relevant consequences, a possibility that will require further investigation.
Figure 2.
Effect of KIR3DL1*004 + HLA-B Bw4 on AIDS progression. Individuals with KIR3DL1*004 and Bw4 and those lacking this compound genotype were compared with individuals homozygous for HLA-Bw6 for two AIDS outcomes. The subjects were seroconverters of all racial groups in combined cohorts. Relative hazard (RH) and significance (P) are given for Cox proportional model analyses.
Effects of KIR3DL1 allelic groups + Bw4 on AIDS progression
The Bw4 alleles as a group show protection against AIDS progression14, and at least part of this protection can be attributed to two independent KIR + HLA compound genotypes: (i) KIR3DS1 + Bw4-80I (ref. 17) and (ii) KIR3DL1*004 + Bw4 (Supplementary Table 1; Fig. 2). The remaining KIR3DL1 alleles can be divided into one of two groups: the KIR3DL1*h (*001, *002, *008, *015, *009) or KIR3DL1*l (*005, *007) groupings, which were defined previously by KIR3DL1 expression patterns and, in some cases, corresponding inhibitory capacity6,9,10. Furthermore, in terms of intensity of staining and percentage of NK cells expressing the allotype, *h/*h individuals show significant differences from both *h/*l and *l/*l, but *h/*l and *l/*l do not show any observable difference from one another10. To test whether the high-expression alleles differ from the low-expression alleles, individuals were divided into one of two groups: (i) individuals who carried KIR3DL1*h in the absence of KIR3DL1*l (i.e., KIR3DL1 *h/*h or KIR3DL1*h/*004; abbreviated as KIR3DL1*h/*y, where *y = *h or *004), and (ii) individuals with at least one copy of KIR3DL1*l (KIR3DL1*l/*l, KIR3DL1*l/*h, or KIR3DL1*l/*004; abbreviated as KIR3DL1*l/*x, where *x = *l, *h, or *004). (Individuals with KIR3DL1*004 were not excluded from further analyses because this allele was observed at an equal percentage in the *h and *l groups, and the protection conferred by KIR3DL1*004 + Bw4 was not significantly different in the presence of *h versus *l). Although KIR3DL1*h/*y + Bw4 and KIR3DL1*l/*x + Bw4 were each protective compared with Bw6/Bw6, KIR3DL1*h/*y showed a more significant effect (RH = 0.66 and 0.70, P = 0.002 for KIR3DL1*h/*y + Bw4 and RH = 0.79 and 0.77, P = 0.12 and 0.05 for KIR3DL1*l/*x + Bw4; Table 1). Thus, a consistent trend showing more pronounced protection proceeded as follows: KIR3DL1*h/*y + Bw4 > KIR3DL1*l/*x + Bw4 > Bw6/Bw6 (trend P = 0.001 and 0.002).
Table 1.
Effect of distinct allelic combinations of KIR3DL1 and HLA-Bw4 on AIDS progression
| Outcome | Genotype | N | RH | 95% CI | P value | Test for trend | N | P for trend |
|---|---|---|---|---|---|---|---|---|
| CD4 < 200 | 3DL1*h/*y+Bw4 | 306 | 0.70 | 0.56–0.88 | 0.002 | 3DL1*h/*y+Bw4 versus | 306 | 0.002 |
| 3DL1*l/*x +Bw4 | 182 | 0.77 | 0.59–1.00 | 0.05 | 3DL1*l/*x +Bw4 versus | 182 | ||
| Bw6/Bw6 | 327 | |||||||
| 3DL1*h/*y+Bw4-80I | 186 | 0.61 | 0.45–0.81 | 0.001 | 3DL1*h/*y+Bw4-80I versus | 186 | 0.001 | |
| 3DL1*l/*x +Bw4-80I | 87 | 0.89 | 0.64–1.25 | 0.51 | 3DL1*l/*x +Bw4-80I versus | 87 | ||
| Bw6/Bw6 | 327 | |||||||
| 3DL1*h/*y+Bw4-80T | 148 | 0.82 | 0.62–1.08 | 0.15 | 3DL1*l/*x +Bw4-80T versus | 148 | 0.01 | |
| 3DL1*l/*x +Bw4-80T | 113 | 0.68 | 0.50–0.94 | 0.02 | 3DL1*h/*y+Bw4-80T versus | 113 | ||
| Bw6/Bw6 | 327 | |||||||
| AIDS1987 | 3DL1*h/*y+Bw4 | 317 | 0.66 | 0.51–0.86 | 0.002 | 3DL1*h/*y+Bw4 versus | 317 | 0.001 |
| 3DL1*l/*x +Bw4 | 194 | 0.79 | 0.59–1.06 | 0.12 | 3DL1*l/*x +Bw4 versus | 194 | ||
| Bw6/Bw6 | 340 | |||||||
| 3DL1*h/*y+Bw4-80I | 194 | 0.56 | 0.40–0.80 | 0.001 | 3DL1*h/*y+Bw4-80I versus | 194 | 0.001 | |
| 3DL1*l/*x +Bw4-80I | 94 | 0.79 | 0.53–1.19 | 0.26 | 3DL1*l/*x +Bw4-80I versus | 94 | ||
| Bw6/Bw6 | 340 | |||||||
| 3DL1*h/*y+Bw4-80T | 152 | 0.76 | 0.56–1.03 | 0.08 | 3DL1*l/*x +Bw4-80T versus | 152 | 0.05 | |
| 3DL1*l/*x +Bw4-80T | 119 | 0.73 | 0.52–1.04 | 0.08 | 3DL1*h/*y+Bw4-80T versus | 119 | ||
| Bw6/Bw6 | 340 |
P values are based on comparisons between the genotypic variable listed and the Bw6/Bw6 control group. 3DL1*h/*y: KIR3DL1*h/*h or KIR3DL1*h/*004; 3DL1*l/*x: KIR3DL1*l/*l, KIR3DL1*l/*h or KIR3DL1*l/*004. CD4, CD4+ T-cell count (cells/mm3); CI, confidence interval.
KIR3DL1*h/*y + Bw4-80I protects against AIDS progression
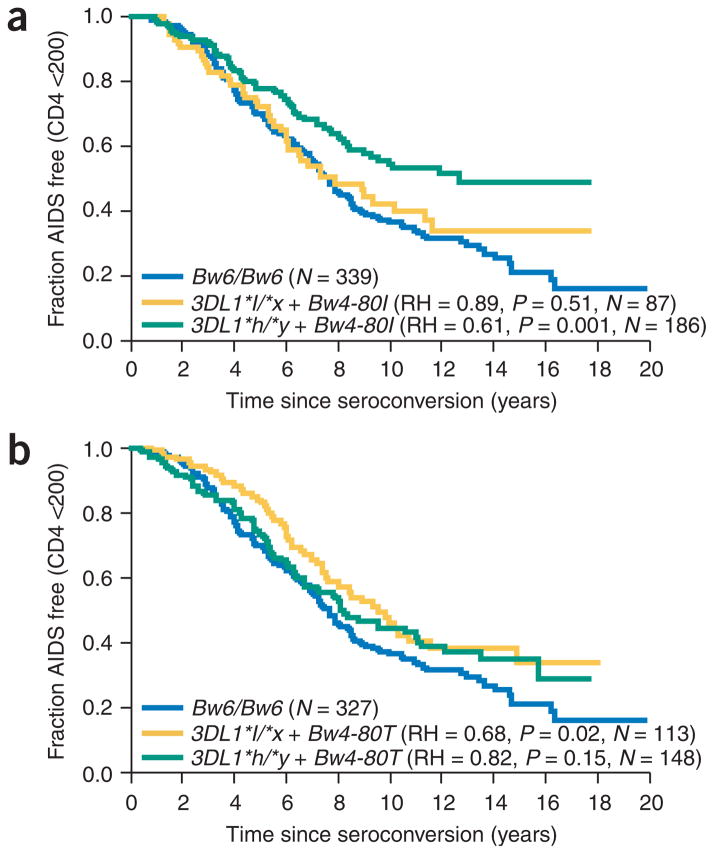
In studies performed to date, HLA-B allotypes with Bw4-80I behave as stronger ligands for KIR3DL1 than do allotypes with Bw4-80T (refs. 6,7). Although this stratification had no effect on KIR3DL1*004 protection (data not shown), the protection conferred by KIR3DL1 *h/*y + Bw4 can be attributed primarily to the subset with KIR3DL1*h/*y + Bw4-80I (RH = 0.56–0.61, P = 0.001; Table 1). In contrast, KIR3DL1*l/*x showed no significant protection in the presence of Bw4-80I, although this KIR + HLA combination did tend toward protection compared with the Bw6/Bw6 group (RH = 0.79 and 0.89; Table 1 and Fig. 3). Indeed, KIR3DL1*h/*y + Bw4-80I was borderline significantly more protective than KIR3DL1*l/*x + Bw4-80I in a stringent, direct comparison of the two groups (RH = 0.63 and 0.66, P = 0.06 and 0.04; Supplementary Table 2 online). KIR3DL1*l/*x did, however, show some protection against CD4+ T-cell count < 200 cells/mm3 (an earlier outcome than AIDS1987, which includes HIV infection plus an AIDS-defining illness) in the presence of Bw4-80T (RH = 0.68, P = 0.02; Table 1 and Fig. 3). Overall, these data support a high-affinity ligand-receptor interaction between Bw4-80I and KIR3DL1*h, and raise the possibility of greater affinity of KIR3DL1*l allotypes for ligands containing Bw4-80T than for those containing Bw4-80I. A significant trend for most protective to least protective proceeded as KIR3DL1*h/*y + Bw4-80I > KIR3DL1*l/*x + Bw4-80I > Bw6/Bw6 (trend P = 0.001; Table 1).
Figure 3.
Effect of KIR3DL1 + HLA-B Bw4 genotypes on progression to CD4+ T-cell count < 200 cells/mm3. (a) KIR3DL1 genotypes with Bw4-80I. (b) KIR3DL1 genotypes with Bw4-80T. Relative hazard (RH) and P values are based on comparisons between the specific genetic variables listed and Bw6/Bw6. The KIR3DL1*h/*y group includes KIR3DL1*h/*h and KIR3DL1*h/*004 individuals, and the KIR3DL1*l/*x group includes KIR3DL1*l/*l, KIR3DL1*l/*h and KIR3DL1*l/*004 individuals.
Synergism between specific Bw4 alleles and KIR3DL1*h and *l
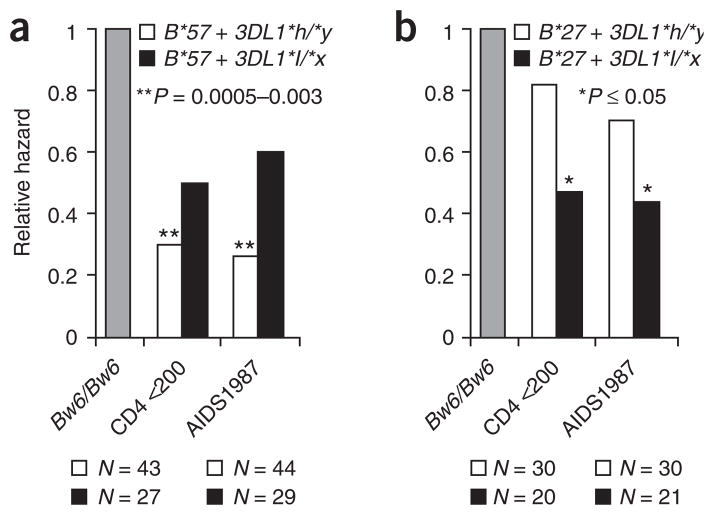
HLA-B*57 and B*27 are the two most protective alleles in our cohorts of HIV-infected individuals23,24, and a sizeable body of literature has indicated that cytotoxic T lymphocytes restricted by these allotypes serve as the basis for their exceptional ability to control HIV (reviewed in ref. 13). All B*57 allelic subtypes encode members of the Bw4-80I group of allotypes, whereas all of the B*27 subtypes in our cohorts, with the exception of six subjects with B*2702, encoded Bw4-80T allotypes. As KIR3DL1*h/*y showed a significant epistatic relationship with Bw4-80I, but not with Bw4-80T, against HIV disease, we tested the hypothesis that KIR3DL1*h/*y would enhance the protective effect of B*57, but not B*27 (with the six individuals with B*2702 removed from the analyses). Using Bw6/Bw6 individuals as the control group in each analysis, both the B*57 + KIR3DL1*h/*y and the B*57 + KIR3DL1*l/*x groups were protective against both AIDS outcomes, but the B*57 effect was enhanced in the presence of KIR3DL1*h/*y as compared with KIR3DL1*l/*x (RH = 0.26 and 0.30, P = 0.003 and 0.0005 for B*57 + KIR3DL1*h/*y; RH = 0.60 and 0.50, P = 0.19 and 0.06 for B*57 + KIR3DL1*l/*x, Fig. 4, Supplementary Table 3 online). The B*57 + KIR3DL1*h/*y genotype conferred stronger protection against AIDS progression than any other host genotype identified to date. The progressive effect of B*57 in the presence of distinct KIR3DL1 groups indicates that the overall protection conferred by B*57 does not stem solely from its function as a cytotoxic T lymphocyte–restricting element in the acquired immune response, but perhaps also as a mediator of innate immunity.
Figure 4.
Synergistic influence of specific HLA-B allotypes with KIR3DL1*h and KIR3DL1*l genotypes on progression to two AIDS outcomes. (a) HLA-B*57; (b)HLA-B*27. Bw6/Bw6 individuals were used as the control group in each analysis. The number of individuals in each category is shown below the bars. The KIR3DL1*h/*y group includes KIR3DL1*h/*h and KIR3DL1*h/*004 individuals, and the KIR3DL1*l/*x group includes KIR3DL1*l/*l, KIR3DL1*l/*h and KIR3DL1*l/*004 individuals.
The group of individuals with B*27-80T (B*2705, N = 60 and B*2709, N = 1), on the other hand, showed borderline significant protection in the presence of KIR3DL1*l/*x and only very moderate, nonsignificant protection in the presence of KIR3DL1*h/*y (RH = 0.44 and 0.47, P = 0.05 and 0.04 for B*27 + KIR3DL1*l/*x; RH = 0.70 and 0.82, P = 0.25 and 0.46 for B*27 + KIR3DL1*h/*y, Fig. 4, Supplementary Table 3). The more prominent protection of B*57 in the presence of KIR3DL1*h/*y than in the presence of KIR3DL1*l/*x accurately parallels the interaction between KIR3DL1*h/*y and Bw4-80I in general, as described above.
Consistent KIR3DL1 + Bw4 effects on plasma HIV RNA
Several KIR3DL1 + Bw4 genotypes showed protection against HIV disease progression relative to the control group Bw6/Bw6, the strongest and most conclusive of which included the following: KIR3DL1*h/*y + Bw4-80I, KIR3DL1*004 + Bw4, KIR3DL1*h/*y + B*57, and to a lesser extent, KIR3DL1*l/*x + B*27. We tested the potential effect of these and related genotypes on MVL measurements that were available for 891 subjects from five studies, 508 of whom were unique to these analyses and were not included in the disease progression analyses. Based on their mean viral load (MVL, given throughout in RNA copies per ml plasma), each individual was categorized into one of three groups: (i) MVL < 2,000, (ii) MVL = 2,000–10,000 or (iii) MVL > 10,000. We then determined the frequency of each compound genotype within each of these groupings and compared it with that of Bw6/Bw6.
First we directly compared the two extreme MVL groupings. The relative frequency of KIR3DL1*h/*y + Bw4-80I was higher than that of Bw6/Bw6 in the MVL <2,000 grouping, whereas the opposite was observed in the MVL >10,000 grouping, and these distributions were significantly different (odds ratio (OR) = 0.24 for KIR3DL1*h/*y + Bw4-80I, P = 6 × 10−8; Table 2). Furthermore, they remained significantly different upon removal of individuals positive for B*57, B*27 and/or KIR3DL1*004 from the KIR3DL1*h/*y + Bw4-80I group (OR = 0.29, P = 5 × 10−4). KIR3DL1*l/*x + Bw4-80I also showed significant protection compared with the Bw6/Bw6 group (OR = 0.43, P = 0.007; Table 2), but the effect was diminished upon removal of B*57, B*27 and/or KIR3DL1*004 (OR = 0.55, P = 0.17). Further, protection conferred by KIR3DL1*l/*x + Bw4-80I was less than that conferred by KIR3DL1*h/*y + Bw4-80I, with borderline significance, in a direct comparison of these two Bw4-80I groupings (OR = 0.58, P = 0.08; Supplementary Table 2). KIR3DL1*004 + Bw4 also showed significant protection relative to the Bw6/Bw6 control group (OR = 0.36, P = 2 × 10−4), and the distributions remained significant upon removal of all individuals with B*57, B*27, and/or KIR3DL1*h/*y + Bw4-80I from the KIR3DL1*004 + Bw4 group (OR = 0.27, P = 5 × 10−5; Table 2). These viral load data concur well with the disease progression data and strongly support protective roles for KIR3DL1*h/*y + Bw4-80I and KIR3DL1*004 + Bw4, even when the samples that overlap between these two groups are removed from the analyses (Supplementary Table 4 online).
Table 2.
Effect of combinations of KIR3DL1 and HLA-B on MVL
| Without covariablesa
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MVL < 2,000 |
MVL > 10,000 |
MVL < 2,000 |
MVL > 10,000 |
MVL < 2,000 |
MVL 2,000– 10,000 |
MVL > 10,000 |
||||||||
| N (%) | N (%) | OR | 95% CI | P value | N (%) | N (%) | OR | 95% CI | P value | N (%) | N (%) | N (%) |
P for trend |
|
| Bw6/Bw6 | 43 (17.3) | 205 (82.7) | 1.00 | 43 (17.3) | 205 (82.7) | 1.00 | 43 (16.0) | 21 (7.8) | 205 (76.2) | |||||
| 3DL1*h/*y + Bw4-80I | 63 (47.7) | 69 (52.3) | 0.24 | 0.14–0.40 | 6 × 10−8 | 22 (42.3) | 30 (57.7) | 0.29 | 0.15–0.59 | 5 × 10−4 | 63 (42.3) | 17 (11.4) | 69 (46.3) | 3 × 10−8 |
| 3DL1*l/*x + Bw4-80I | 25 (32.1) | 53 (67.9) | 0.43 | 0.23–0.79 | 0.007 | 10 (26.3) | 28 (73.7) | 0.55 | 0.24–1.28 | 0.17 | 25 (29.4) | 7 (8.2) | 53 (62.4) | 0.007 |
| 3DL1*004 + Bw4 | 40 (34.2) | 77 (65.8) | 0.36 | 0.21–0.62 | 2 × 10−4 | 25 (41.0) | 36 (59.0) | 0.27 | 0.14–0.50 | 5 × 10−5 | 40 (32.3) | 7 (5.7) | 77 (62.1) | 4 × 10−4 |
| 3DL1*h/*y + B*57 | 39 (69.6) | 17 (30.4) | 0.10 | 0.05–0.19 | 7 × 10−11 | 39 (62.9) | 6 (9.7) | 17 (27.4) | 4 × 10−11 | |||||
| 3DL1*l/*x + B*57 | 10 (37.0) | 17 (63.0) | 0.38 | 0.15–0.95 | 0.04 | 10 (33.3) | 3 (10.0) | 17 (56.7) | 0.02 | |||||
| 3DL1*h/*y + B*27 | 8 (33.3) | 16 (66.7) | 0.37 | 0.14–1.00 | 0.05 | 8 (25.8) | 7 (22.6) | 16 (51.6) | 0.02 | |||||
| 3DL1*l/*x + B*27 | 10 (47.6) | 11 (52.4) | 0.22 | 0.08–0.59 | 0.003 | 10 (41.7) | 3 (12.5) | 11 (45.8) | 0.002 | |||||
P values are based on comparisons between the genotypic variable listed and the Bw6/Bw6 control group. Genotype symbols are as defined in Table 1. CI, confidence interval.
For the Bw4-80I groups, all individuals with KIR3DL1*004, B*57 and B*27 were removed from the analysis. For the KIR3DL1*004 group, all individuals with B*57, B*27 and KIR3DL1*h/*y + Bw4-80I were removed from the analysis.
The individual effects of B*57 (a Bw4-80I allele) and B*27 (a Bw4-80T allele) on HIV MVL closely followed and emphasized the patterns of association observed in the progression analysis (Fig. 4): in comparison with the Bw6/Bw6 group, B*57 showed exceptionally robust protection in the presence of KIR3DL1*h/*y (OR = 0.1, P = 7 × 10−11; Table 2), an effect that was diminished in the presence of KIR3DL1*l/*x (OR = 0.38, P = 0.04), whereas B*27 was more protective in the presence of KIR3DL1*l/*x (OR = 0.22, P = 0.003) than in the presence of KIR3DL1*h/*y (OR = 0.37, P = 0.05). A direct comparison between the B*57 + KIR3DL1*h/*y and the B*57 + KIR3DL1*l/*x effects on viral load indicated a significantly stronger protective effect of B*57 + KIR3DL1*h/*y (OR = 0.23, P = 0.009; Supplementary Table 2). These data indicate that KIR3DL1 allotypes can strongly modulate the protection conferred by B*57 in particular, and imply an important role for B*57 (and possibly B*27) in the innate immune response against HIV.
In a second set of analyses, the differences in frequency trends across the three viral load divisions (MVL < 2,000, MVL = 2,000–10,000, and MVL > 10,000) were compared between individual KIR3DL1 + Bw4 genotypic groups and the Bw6/Bw6 group (Table 2, far right columns). In each comparison, the relative strength and significance of these differences reflected that observed in the comparisons of the two extreme groups (MVL < 2,000 and MVL > 10,000).
DISCUSSION
The functional consequences of allelic variability at the KIR3DL1 locus have been studied in greater detail than that of any other single KIR gene6,10. In this study, we grouped inhibitory KIR3DL1 alleles based on distinct functional characteristics and identified differential protective effects of these allelic groups on HIV disease progression and viral load. KIR3DL1*004 represented the most protective single allele at this locus, and its protection was completely dependent on the presence of HLA-B alleles with the Bw4 motif, the ligands for inhibitory KIR3DL1 allotypes. We found this result surprising, as the KIR3DL1*004 molecule is retained within the cell, offering the possibility that it has a null phenotype. The genetic data presented herein dispel this hypothesis and imply a novel intracellular function for KIR3DL1*004 that directly or indirectly involves the Bw4 ligand. Alternatively, the KIR3DL1*004 molecule may be marking (by linkage disequilibrium) another neighboring locus that confers protection in an additive or synergistic manner with Bw4.
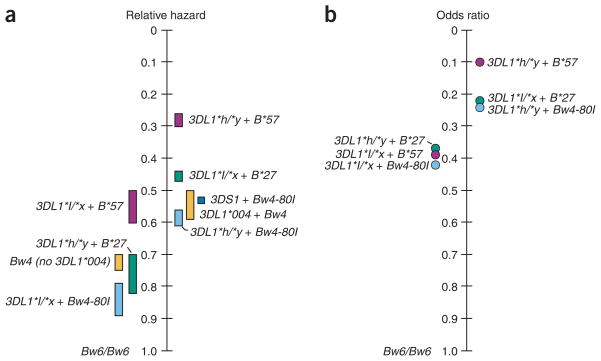
Previous data have indicated a functional distinction between the strongly inhibitory KIR3DL1*h alleles and the weakly inhibitory KIR3DL1*l alleles9,10, and based on data presented herein these groupings also have differential effects in the HIV disease process. Overall, a pattern emerged indicating relatively strong protection through KIR3DL1*h + Bw4-80I, and a weak protective effect through KIR3DL1*l + Bw4-80T. These data were reinforced by parallel conclusions when the individual B*57 (Bw4-80I) and B*27 (Bw4-80T) alleles were examined and shown to be most protective in the presence of KIR3DL1*h/*y and KIR3DL1*l/*x, respectively. Previous functional and genetic studies have also indicated a stronger relationship of some KIR3DL1 allotypes with Bw4-80I than with Bw4-80T (refs. 6,7, 10,17,25), but our genetic data imply greater complexity in the receptor-ligand interactions between these molecules. An analysis of MVL data determined from a group of 891 individuals, only 383 of whom overlapped with the 915 individuals used in the disease progression analyses, showed protective effects of the various KIR3DL1 + HLA-B genotypes strongly consistent with those observed in disease progression analyses. Thus, the influence of these genotypes on disease course occurs early after infection, especially for B*57 + KIR3DL1*h/*y, and may continue throughout the chronic phase of infection. The protection against AIDS progression conferred by various combinations of KIR3DL1 + Bw4 genotypes reported herein represents the first data to support a consequential impact of differential KIR3DL1 allotypic functions on disease outcome of any sort. There was a continuum of synergistic effects of the KIR3DL1 and HLA-B loci (including KIR3DS1 + Bw4-80I) in order by degree of protection (disease progression, Fig. 5a; MVL, Fig. 5b; see also Table 2 and Supplementary Table 5 online) emphasizing the complexity and strength of protection conferred by combinations of these two loci. There is no other pair of loci evaluated thus far that together shows the diversity in protective effects against AIDS (or any other disease) of that observed with KIR3DL1;3DS1 and HLA-Bw4.
Figure 5.
KIR3DL1 + Bw4 continuum of protection. Genotypes are ordered by degree of protection in terms of (a) disease progression and (b) control of viral load. The most protective genotypes are shown on the right of the vertical scale and the alternative (less protective) genotypes are shown on the left. (a) Relative hazard ranges for the two AIDS outcomes (CD4+ T-cell count < 200 cells/mm3 and AIDS1987) relative to the Bw6/Bw6 control group. (b) Odds ratios (dots) of protective genotypes relative to the Bw6/Bw6 control group in terms of their distributions in the < 2,000 versus the > 10,000 MVL groupings. Purple bars and dots, B*57 with KIR3DL1 genotypes; green bars and dots, B*27 with KIR3DL1 genotypes; light blue bars and dots, Bw4-80I with KIR3DL1 genotypes; dark blue bar, Bw4-80I+KIR3DS1; yellow bars, Bw4 with or without KIR3DL1*004.
The activating KIR3DS1 in the presence of Bw4-80I associates with slow progression to AIDS17, lower viral load18, and protection against opportunistic infections during HIV infection18. Based on these data, we proposed that NK cell and/or CD8+ T cell activation through a KIR3DS1–Bw4-80I interaction protects against HIV. Further, activating KIR genotypes may protect against HIV infection26. This model of effector cell activation leading to protection appears incongruent with the protection conferred by inhibitory KIR3DL1 subtypes (particularly those that are expressed at high levels and transmit strong inhibitory signals) in the presence of their class I ligands, as shown herein. However, these findings are actually very compatible given our knowledge of the functional development of NK cells and the role of inhibitory receptors in this development. Broadly accepted models of NK cell repertoire development suggest that NK cells continue to acquire inhibitory receptors until sufficient signals are generated to quench autoreactivity through recognition of autologous class I ligands27–29. The resultant population of NK cells utilizes various KIR + HLA class I pairings that confer various degrees of inhibition to achieve overall nonresponsiveness to self. Therefore, the larger the contribution of a given receptor-ligand pairing to NK cell inhibition under ‘normal’ conditions, the more vigorous an effector cell response will be when the interaction between that inhibitory receptor and its ligand is disrupted. The application of this concept to KIR3DL1 subtypes is particularly apropos, as there are clear distinctions between KIR3DL1*h and KIR3DL1*l subtypes in terms of the level of their expression on a per-cell basis9, the overall frequency of NK cells expressing these subtypes10 and the ability of HLA-Bw4 to increase the frequency of NK cells expressing KIR3DL1*h to a significantly greater extent than KIR3DL1*l10. The activation potential of an NK cell population would also be subject to the affinity between the particular KIR3DL1-Bw4 allotypic pair, an effect implied by the synergism between KIR3DL1*h and B*57 and, alternatively, between KIR3DL1*l and B*27 (Table 2 and Fig. 4).
Notably, this concept of greater inhibition during NK cell development ultimately leading to more vigorous activation under appropriate conditions, such as viral infection, is consistent with data implying inhibitory receptor involvement in the developmental regulation of NK cell responsiveness8,19–21,30,31. These data indicate that weak or missing inhibitory signals during NK cell development result in poor (or complete lack of) activating potential by that NK cell, whereas strong inhibitory signals during NK cell development lead to greater potential for NK cell activation upon interaction with an aberrant target cell. Thus, engagement of highly expressed, highly inhibitory KIR3DL1 allotypes with their ligands may ultimately secure stronger NK cell responses in the event of viral infection, such as HIV, when KIR3DL1-mediated inhibition is suspended.
The complexity of KIR3DL1 is steadily being disentangled by a combination of genetic and functional studies that feed off one another. One allele at this locus, the activating KIR3DS1, has been implicated in resistance to both HIV17 and hepatitis C virus32, and in susceptibility to cervical cancer33. It is now clear that variation among the inhibitory KIR3DL1 subset of alleles also has an influence on HIV disease, and dissection of their differential influence is worth investigating in other diseases—such as cervical neoplasia, in which the presence of Bw4 associates with decreased risk of high-grade cervical lesions and cancer33. Of the three classical HLA class I loci, HLA-B is the most rapidly evolving34 and is also the most consequential in the acquired immune response against HIV from both functional and genetic epidemiological perspectives23,35. Notably, however, among HLA-B alleles, Bw4 alleles show greater protection against HIV than Bw6 alleles, an observation that strongly implicates a defense mechanism involving KIR3DL1, as shown herein. Like HLA-B, the KIR3DL1 locus is also evolving rapidly compared with most other KIR loci36, and in our cohorts it is this locus that shows the strongest effects on HIV disease. It is entirely plausible that coordinated coevolution of these two loci37,38 is driven by pathogens such as HIV, resulting in a primary influence of specific HLA-B + KIR3DL1 allelic combinations in the innate immune response against this virus.
METHODS
Subjects
We used data from a total of 1,496 HIV+ individuals in the studies described. For the disease progression studies, 915 HIV-1–infected subjects for whom the dates of seroconversion were known were derived from four cohorts: the Multicenter AIDS Cohort Study (MACS)39, the Multicenter Hemophilia Cohort Study (MHCS)40, the San Francisco City Clinic Cohort (SFCCC)41 and the AIDS Linked to Intravenous Experience (ALIVE)42. For the viral load studies, 891 subjects for whom viral load data was available were obtained from five cohorts: the MACS, the Swiss HIV Cohort (http://www.shcs.ch), the Study on the Consequences of Protease Inhibitor Era (SCOPE) cohort43, the Massachusetts General Hospital (MGH) Controller Cohort, and the NIAID long term nonprogressor cohort44. There were 383 individuals whose clinical data were used in both the disease progression and the viral load studies. Thus, 508 were used only in the viral load studies and 532 were used only in the disease progression studies. This study was approved by the protocol review office of the US National Cancer Institute institutional review board. Informed consent was obtained at the study sites from all individuals.
HLA genotyping and KIR3DL1 allelic typing
We performed genotyping of the HLA locus following the PCR-SSOP (sequence-specific oligonucleotide probing) typing protocol recommended by the 13th International Histo-compatibility Workshop (http://www.ihwg.org/components/ssopr.htm). For KIR3DL1 subtyping, polymorphic exons 3, 4, 5, 7, 8 and 9 were selectively amplified in four PCR reactions using locus-specific primers (a separate PCR for each of exons 3, 4 and 5, and a fourth PCR for exons 7, 8 and 9 combined). The PCR products were blotted on nylon membranes and hybridized with a panel of 56 sequence-specific oligonucleotide (SSO) probes designed to detect unique sequence motifs of known KIR3DL1 alleles (Supplementary Table 6 online). KIR3DL1 alleles were assigned by the reaction patterns of the SSO probes based on the known KIR3DL1 sequences. Ambiguous SSO typing results were resolved by sequencing analysis.
Plasma HIV RNA measurements
Viral load measurements (HIV RNA copies per ml plasma in the absence of any drug therapy) determined previously from 891 HIV+ individuals were available for analysis. MVL was determined for each individual and categorized into one of three groupings: <2,000, 2,000–10,000, and >10,000. The average number of measurements per person was 7.9 with a range of 1–52 and a total of 7,084 measurements. The ethnic breakdown was as follows: European American (N = 685), African American (N = 136), other (N = 53), unknown (N = 17).
Statistical analysis
All statistics provided were derived from analyses stratified by race. Two AIDS-related outcomes were considered in the survival analysis: (i) a CD4+ T-cell count of <200 cells/mm3 and (ii) progression to AIDS according to the 1987 definition of the US Centers for Disease Control, which includes HIV infection plus an AIDS-defining illness45. In order to eliminate confounding by the previously described protective effect of KIR3DS1 + Bw4-80I, individuals with this compound genotype were removed from all analyses. With the exception of the comparisons listed in Supplementary Table 2, all test variables were compared with those of the Bw6/Bw6 control group.
The influence of individual and groups of KIR3DL1 alleles on AIDS progression was tested using Cox model analyses. We divided KIR3DL1 alleles into high- and low-expressing groups based on the previously defined expression patterns. The high-expressing alleles (KIR3DL1*h) in our cohorts were *001, *002, *008, *015 and *009, and the low-expressing alleles (KIR3DL1*l) were *005 and *007. KIR3DL1*h/*h individuals show significant differences from both KIR3DL1*h/*l and *l/*l, but KIR3DL1*h/*l and *l/*l do not show any observable differences from one another in terms of intensity of staining and percentage of NK cells expressing the allotype10. Furthermore, the high-expression KIR3DL1 alleles are more common than the low-expression alleles, resulting in a small KIR3DL1*l/*l genotypic frequency (13% in our entire dataset, 2.7% of whom had Bw4-80I and 4.7% of whom had Bw4-80T). For these biological and practical reasons, the tests for different effects of high-versus low-expression alleles involved the division of individuals with only high-expression alleles from those with at least one low-expression allele.
We used SAS 9.1 (SAS Institute) for data management and statistical analyses. PROC FREQ was used to compute frequencies on individual variables. PROC LOGISTIC was used for categorical analyses to obtain odds ratios and 95% confidence intervals. PROC LIFETEST and PHREG were used for Kaplan-Meier and Cox model analyses. Statistical significance refers to two-sided P values of <0.05. The q value shown in Supplementary Table 1 is the false discovery rate. The false discovery rate is the expected proportion of false positives when declaring the significance level equal to P; q value estimates were calculated for the effect of individual KIR3DL1 alleles with HLA-Bw4 on AIDS progression using the q value package, v1.1 in the R statistical computing program22.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the US National Cancer Institute, National Institutes of Health (NIH), under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The ALIVE study was supported by US National Institute on Drug Abuse RO1-DA04334. The SCOPE cohort was supported by the NIH (P30 AI027763) and the University of California at San Francisco AIDS Research Institute. The Swiss HIV Cohort study was funded by the Swiss National Science Foundation.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
M.C. designed and supervised the project, and prepared the manuscript; M.P.M. performed KIR genotyping and prepared the manuscript; Y.Q. conducted the data analyses; X.G. performed HLA genotyping; E.Y. contributed to KIR genotyping experiments; P.P., S.G.D, D.W.M. provided intellectual input; S.J.O. provided access to samples and clinical data; E.E.B. participated in data analysis; and J.N.M., F.P., S.C., W.L.S., J.P., J.J.G., S.B., G.D.K., A.T., M.C., B.D.W., and S.G.D. provided clinical samples and data. All authors discussed the results and commented on the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Colucci F, Di Santo JP, Leibson PJ. Natural killer cell activation in mice and men: different triggers for similar weapons? Nat Immunol. 2002;3:807–813. doi: 10.1038/ni0902-807. [DOI] [PubMed] [Google Scholar]
- 4.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 5.Gumperz JE, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 6.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 7.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4- positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valiante NM, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner CM, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 10.Yawata M, et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 12.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 13.Goulder PJ, Watkins DI. HIV and SIV CTL escape: Implications for vaccine design. Nat Rev Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 14.Flores-Villanueva PO, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci USA. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien SJ, Gao X, Carrington M. HLA and AIDS: a cautionary tale. Trends Mol Med. 2001;7:379–381. doi: 10.1016/s1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- 17.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 18.Qi Y, et al. KIR/HLA pleiotropism: Protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2:e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anfossi N, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez NC, et al. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao X, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 24.Gao X, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Vazquez A, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum Immunol. 2005;66:285–289. doi: 10.1016/j.humimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Jennes W, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 27.Dorfman JR, Raulet DH. Acquisition of Ly49 receptor expression by developing natural killer cells. J Exp Med. 1998;187:609–618. doi: 10.1084/jem.187.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Held W, Dorfman JR, Wu MF, Raulet DH. Major histocompatibility complex class I-dependent skewing of the natural killer cell Ly49 receptor repertoire. Eur J Immunol. 1996;26:2286–2292. doi: 10.1002/eji.1830261003. [DOI] [PubMed] [Google Scholar]
- 29.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev. 2006;214:155–160. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 30.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 31.Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006;24:249–257. doi: 10.1016/j.immuni.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 33.Carrington M, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein J, Satta Y, O’hUigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- 35.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 36.Yawata M, Yawata N, Abi-Rached L, Parham P. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol. 2002;22:463–482. [PubMed] [Google Scholar]
- 37.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khakoo SI, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 39.Phair J, et al. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr. 1992;5:490–496. [PubMed] [Google Scholar]
- 40.Goedert JJ, et al. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med. 1989;321:1141–1148. doi: 10.1056/NEJM198910263211701. [DOI] [PubMed] [Google Scholar]
- 41.Buchbinder SP, Katz MH, Hessol NA, O’Malley PM, Holmberg SD. Long-term HIV-1 infection without immunologic progression. AIDS. 1994;8:1123–1128. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 42.Vlahov D, et al. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. J Am Med Assoc. 1998;279:35–40. doi: 10.1001/jama.279.1.35. [DOI] [PubMed] [Google Scholar]
- 43.Emu B, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 45.Council of State and Territorial Epidemiologists; AIDS Program, Center for Infectious Diseases. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. MMWR Morb Mortal Wkly Rep. 1987;36(suppl 1):1S–15S. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.