Abstract
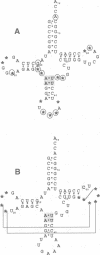
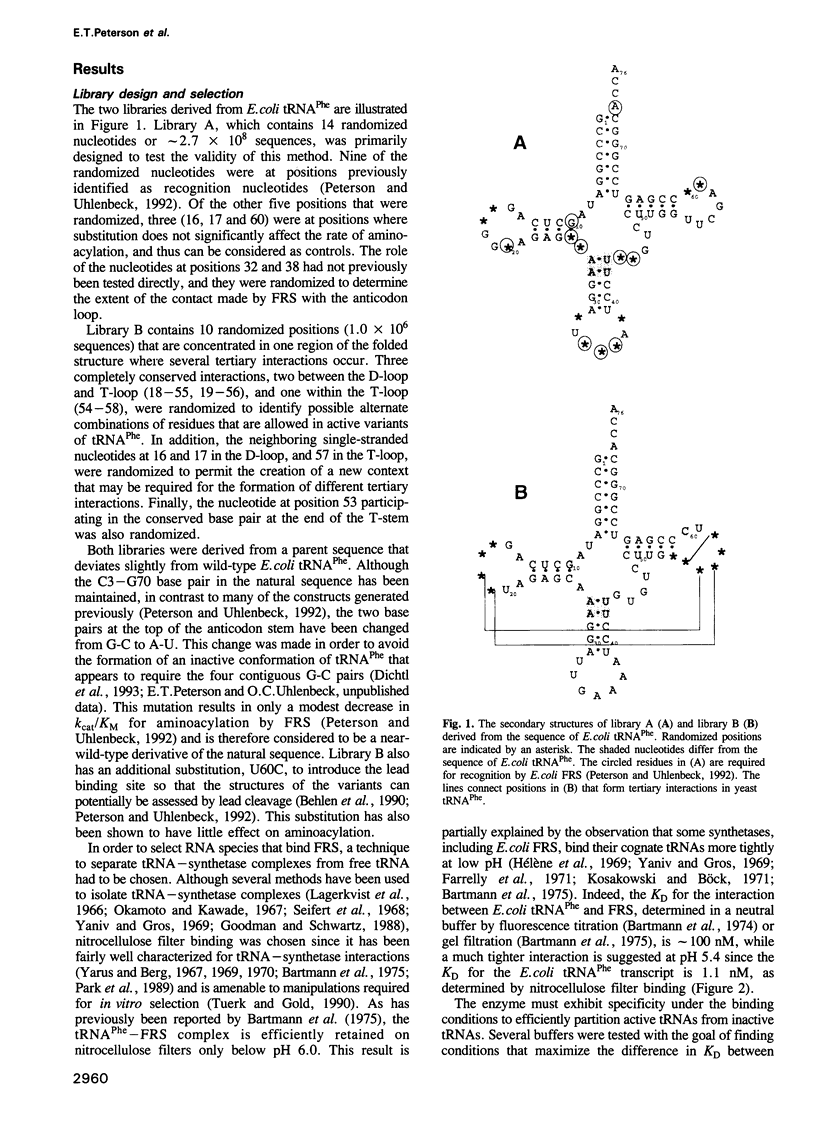
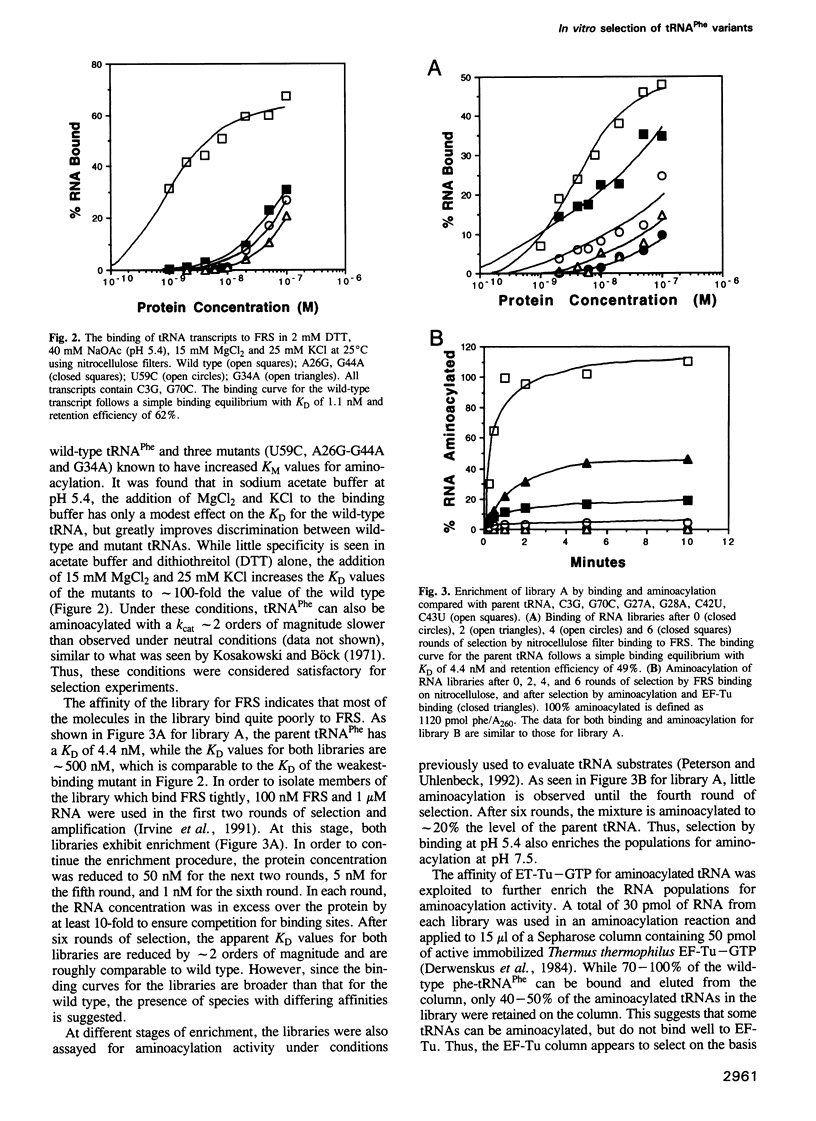
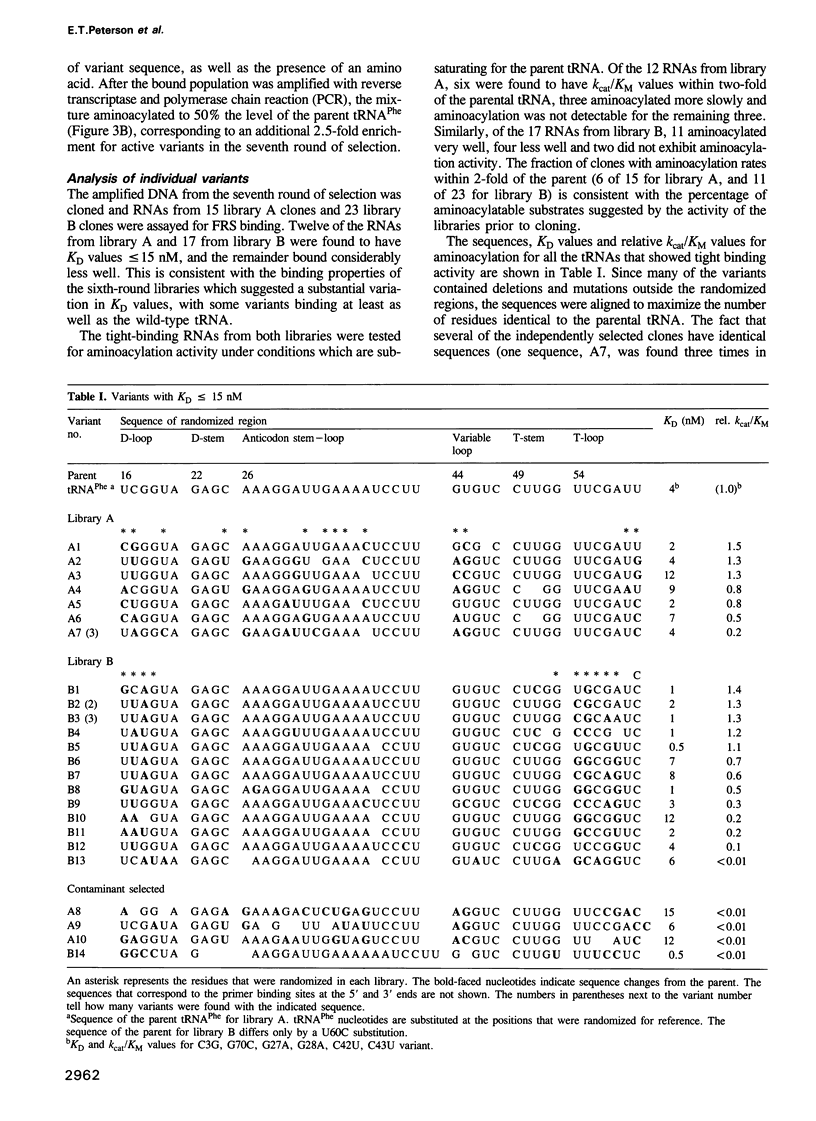
In vitro selection was used to isolate active Escherichia coli tRNA(Phe) variants from randomized libraries. Functional tRNAs were first selected by multiple rounds of binding to Escherichia coli phenylalanyl-tRNA synthetase. These variants were then aminoacylated and selected for affinity to elongation factor-Tu. By randomizing potential recognition nucleotides, the importance of residues U20, G34, A35, A36 and U59, previously identified to be required for specific recognition by E. coli phenylalanyl-tRNA synthetase (FRS), was confirmed. However, the sequences of several active variants imply that the wild-type tertiary interactions G10-C25-U45 and A26-G44 are not required for recognition, as previously suggested. Selection of functional tRNAs from a second library randomized at positions normally involved in conserved tertiary interactions revealed new combinations of nucleotides at these positions, suggesting the presence of novel tertiary interactions. In both libraries, active sequences containing deletions were isolated. Taken together, it is clear that FRS is active with substrates having an unexpectedly broad sequence diversity. Finally, the potency of this method is illustrated by the identification of a second class of variants that was isolated by virtue of the presence of an impurity in the FRS preparation.
Full text
PDF
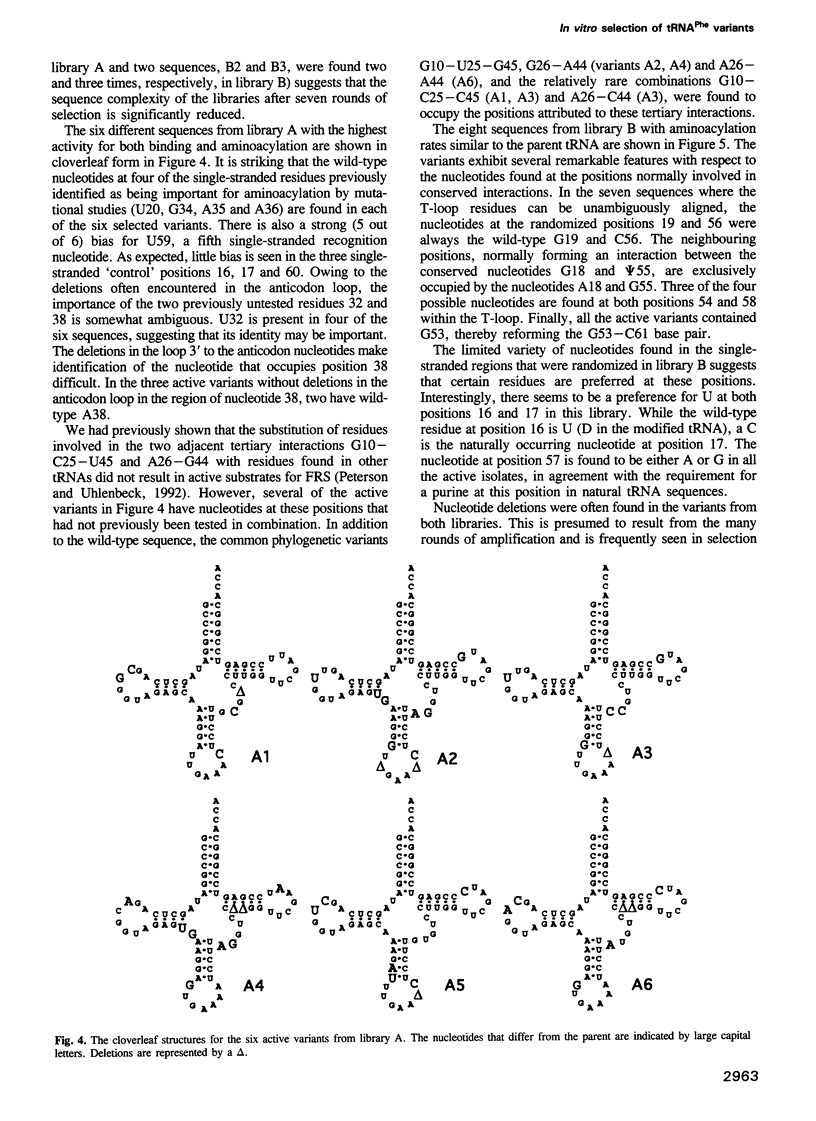
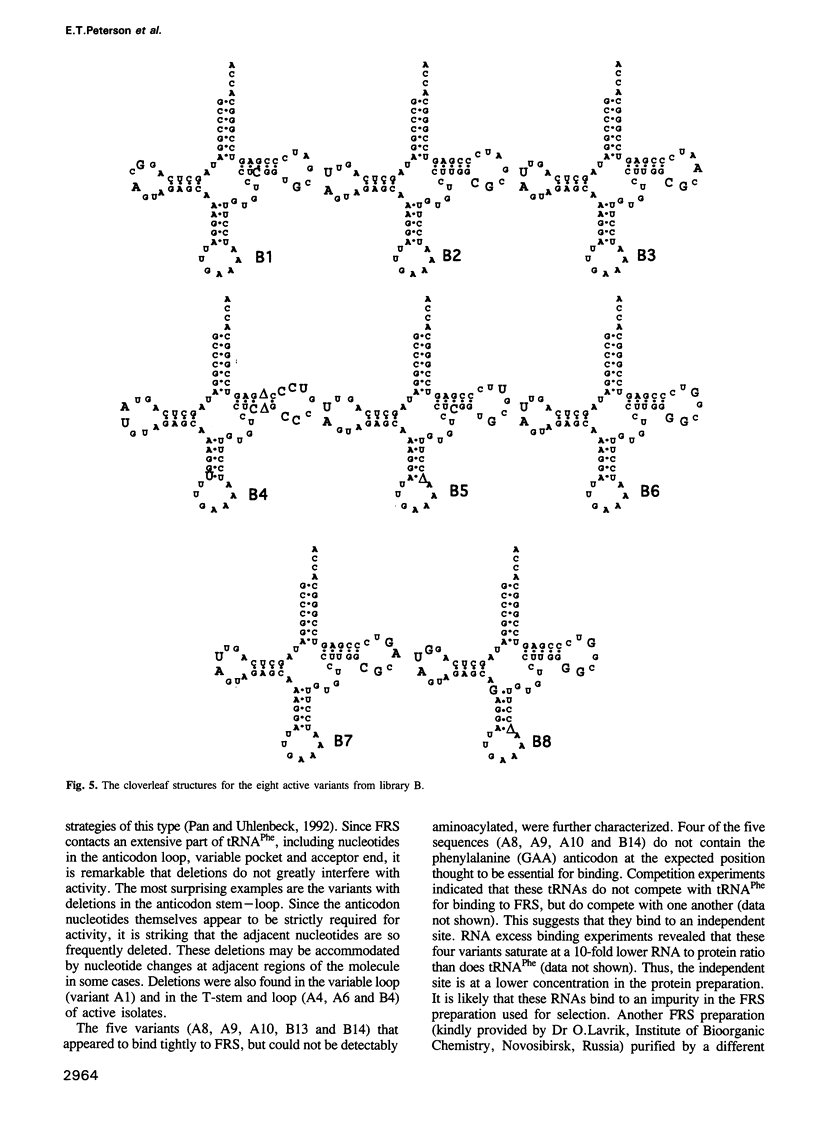



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartel D. P., Zapp M. L., Green M. R., Szostak J. W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991 Nov 1;67(3):529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- Bartmann P., Hanke T., Hammer-Raber B., Holler E. Equilibrium analysis of L-Phe-tRNA Phe complexes with L-phenylalanyl transfer ribonucleic acid synthetase of Escherichia coli K 10. Biochemistry. 1974 Sep 24;13(20):4171–4175. doi: 10.1021/bi00717a016. [DOI] [PubMed] [Google Scholar]
- Bartmann P., Hanke T., Holler E. Active site stoichiometry of L-phenylalanine: tRNA ligase from Escherichia coli K(-10). J Biol Chem. 1975 Oct 10;250(19):7668–7674. [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Derwenskus K. H., Fischer W., Sprinzl M. Isolation of tRNA isoacceptors by affinity chromatography on immobilized bacterial elongation factor Tu. Anal Biochem. 1984 Jan;136(1):161–167. doi: 10.1016/0003-2697(84)90318-x. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Pan T., DiRenzo A. B., Uhlenbeck O. C. Replacement of RNA hairpins by in vitro selected tetranucleotides. Nucleic Acids Res. 1993 Feb 11;21(3):531–535. doi: 10.1093/nar/21.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly J. G., Longworth J. W., Stulberg M. P. The interaction of phenylalanyl transfer ribonucleic acid synthetase and phenylalanine transfer ribonucleic acid. Complex formation and resulting fluorescence quenching. J Biol Chem. 1971 Mar 10;246(5):1266–1270. [PubMed] [Google Scholar]
- Goodman R., Schwartz I. Kinetic analysis of an E.coli phenylalanine-tRNA synthetase mutant. Nucleic Acids Res. 1988 Aug 11;16(15):7477–7486. doi: 10.1093/nar/16.15.7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayase Y., Jahn M., Rogers M. J., Sylvers L. A., Koizumi M., Inoue H., Ohtsuka E., Söll D. Recognition of bases in Escherichia coli tRNA(Gln) by glutaminyl-tRNA synthetase: a complete identity set. EMBO J. 1992 Nov;11(11):4159–4165. doi: 10.1002/j.1460-2075.1992.tb05509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Novel transfer RNAs that are active in Escherichia coli. Biochemistry. 1992 May 5;31(17):4157–4160. doi: 10.1021/bi00132a001. [DOI] [PubMed] [Google Scholar]
- Hélene C., Brun F., Yaniv M. Fluorescence study of interactions between valyl- t RNA synthetase and valine-specific tRNA's from Escherichia coli. Biochem Biophys Res Commun. 1969 Oct 22;37(3):393–398. doi: 10.1016/0006-291x(69)90927-9. [DOI] [PubMed] [Google Scholar]
- Irvine D., Tuerk C., Gold L. SELEXION. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J Mol Biol. 1991 Dec 5;222(3):739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- Kosakowski H. M., Böck A. Substrate complexes of phenylalanyl-tRNA synthetase from Escherichia coli. Eur J Biochem. 1971 Dec 22;24(1):190–200. doi: 10.1111/j.1432-1033.1971.tb19670.x. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U., Rymo L., Waldenström J. Structure and function of transfer ribonucleic acid. II. Enzyme-substrate complexes with valyl ribonucleic acid synthetase from yeast. J Biol Chem. 1966 Nov 25;241(22):5391–5400. [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Kawade Y. Electrophoretic separation of complexes of aminoacyl-tRNA synthetase and transfer RNA. Biochim Biophys Acta. 1967;145(3):613–620. doi: 10.1016/0005-2787(67)90120-7. [DOI] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+. Biochemistry. 1992 Apr 28;31(16):3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Park S. J., Hou Y. M., Schimmel P. A single base pair affects binding and catalytic parameters in the molecular recognition of a transfer RNA. Biochemistry. 1989 Mar 21;28(6):2740–2746. doi: 10.1021/bi00432a056. [DOI] [PubMed] [Google Scholar]
- Peterson E. T., Uhlenbeck O. C. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1992 Oct 27;31(42):10380–10389. doi: 10.1021/bi00157a028. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Pütz J., Florentz C., Giegé R. Influence of tRNA tertiary structure and stability on aminoacylation by yeast aspartyl-tRNA synthetase. Nucleic Acids Res. 1993 Jan 11;21(1):41–49. doi: 10.1093/nar/21.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Romby P., Carbon P., Westhof E., Ehresmann C., Ebel J. P., Ehresmann B., Giegé R. Importance of conserved residues for the conformation of the T-loop in tRNAs. J Biomol Struct Dyn. 1987 Dec;5(3):669–687. doi: 10.1080/07391102.1987.10506419. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Behlen L. S., DiRenzo A. B., Uhlenbeck O. C. Recognition of yeast tRNA(Phe) by its cognate yeast phenylalanyl-tRNA synthetase: an analysis of specificity. Biochemistry. 1992 May 5;31(17):4161–4167. doi: 10.1021/bi00132a002. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry. 1990 Mar 13;29(10):2523–2532. doi: 10.1021/bi00462a014. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D., Gold L., Platt T. Selective enrichment of RNA species for tight binding to Escherichia coli rho factor. FASEB J. 1993 Jan;7(1):201–207. doi: 10.1096/fasebj.7.1.7678562. [DOI] [PubMed] [Google Scholar]
- Schneider D., Tuerk C., Gold L. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J Mol Biol. 1992 Dec 5;228(3):862–869. doi: 10.1016/0022-2836(92)90870-p. [DOI] [PubMed] [Google Scholar]
- Seifert W., Nass G., Zillig W. Electrophoretic separation of tRNA-bound leucyl-tRNA synthetase from Escherichia coli extracts. J Mol Biol. 1968 Apr 28;33(2):507–511. doi: 10.1016/0022-2836(68)90208-8. [DOI] [PubMed] [Google Scholar]
- Tsai D. E., Harper D. S., Keene J. D. U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 1991 Sep 25;19(18):4931–4936. doi: 10.1093/nar/19.18.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai D. E., Kenan D. J., Keene J. D. In vitro selection of an RNA epitope immunologically cross-reactive with a peptide. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8864–8868. doi: 10.1073/pnas.89.19.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Tuerk C., MacDougal S., Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M., Gros F. Studies on valyl-tRNA synthetase and tRNA from Escherichia coli. II. Interaction between valyl-tRNA synthetase and valine acceptor tRNA. J Mol Biol. 1969 Aug 28;44(1):17–30. doi: 10.1016/0022-2836(69)90402-1. [DOI] [PubMed] [Google Scholar]
- Yarus M., Berg P. On the properties and utility of a membrane filter assay in the study of isoleucyl-tRNA synthetase. Anal Biochem. 1970 Jun;35(2):450–465. doi: 10.1016/0003-2697(70)90207-1. [DOI] [PubMed] [Google Scholar]
- Yarus M., Berg P. Recognition of tRNA by aminoacyl tRNA synthetases. J Mol Biol. 1967 Sep 28;28(3):479–490. doi: 10.1016/s0022-2836(67)80098-6. [DOI] [PubMed] [Google Scholar]
- Yarus M., Berg P. Recognition of tRNA by isoleucyl-tRNA synthetase. Effect of substrates on the dynamics of tRNA-enzyme interaction. J Mol Biol. 1969 Jun 14;42(2):171–189. doi: 10.1016/0022-2836(69)90037-0. [DOI] [PubMed] [Google Scholar]