Abstract
Histone H3 methylation at R17 and R26 recently emerged as a novel epigenetic mechanism regulating pluripotency in mouse embryos. Blastomeres of four-cell embryos with high H3 methylation at these sites show unrestricted potential, whereas those with lower levels cannot support development when aggregated in chimeras of like cells. Increasing histone H3 methylation, through expression of coactivator-associated-protein-arginine-methyltransferase 1 (CARM1) in embryos, elevates expression of key pluripotency genes and directs cells to the pluripotent inner cell mass. We demonstrate CARM1 is also required for the self-renewal and pluripotency of embryonic stem (ES) cells. In ES cells, CARM1 depletion downregulates pluripotency genes leading to their differentiation. CARM1 associates with Oct4/Pou5f1 and Sox2 promoters that display detectable levels of R17/26 histone H3 methylation. In CARM1 overexpressing ES cells, histone H3 arginine methylation is also at the Nanog promoter to which CARM1 now associates. Such cells express Nanog at elevated levels and delay their response to differentiation signals. Thus, like in four-cell embryo blastomeres, histone H3 arginine methylation by CARM1 in ES cells allows epigenetic modulation of pluripotency.
Keywords: CARM1, ES cells, pluripotency, differentiation, chromatin, arginine histone methylation
Introduction
Development of the mouse embryo from the fertilized egg leads progressively to the establishment of two lineages of cells by the blastocyst stage. One of these lineages, the inner cell mass (ICM), retains pluripotency and provides progenitor cells for the future body of the animal. Pluripotent embryonic stem (ES) cells are derived from this lineage. The other cell lineage differentiates into trophectoderm, an extra-embryonic tissue [1]. It has been recently shown that differences in specific epigenetic modifications between early blastomeres contribute to the first steps in diversification of these two lineages [2]. It was found that blastomeres as early as at the four-cell stage could be heterogeneous in the extent of their pluripotency and consequently in their fate. Thus, although some show unrestricted potential, others tend to give rise to the trophectoderm lineage and cannot support full development when aggregated in chimeras of like cells [1]. This decreased potency is, at least in part, reversible because these cells can fully contribute to development when surrounded by cells with full potential [1]. The four-cell blastomeres with reduced developmental potential have lower levels of histone H3 arginine 17 and 26 methylation [2]. Conversely, cells with higher levels of this epigenetic modification are predisposed to contribute to pluripotent ICM. That this correlation is causal was suggested by the elevation of expression of coactivator-associated-protein-arginine-methyltransferase 1 (CARM1) in individual blastomeres, leading to increased histone methylation, resulting in higher expression of pluripotency transcription factors such as Nanog and Sox2 and direction of progeny to the ICM [2]. Together these observations have pointed to the importance of arginine methylation in modulating the pluripotent properties of blastomeres; although all cells are initially pluripotent, reduced CARM1 activity can tip the balance and predispose cells to differentiate. This raises the question of whether CARM1 might have a similar role in modulating pluripotency of ES cells.
Transcription factors Oct4, Sox2, and Nanog are critical to maintain ES cell pluripotency and suppress their differentiation [3]. Nanog blocks ES cell differentiation into extra-embryonic ectoderm [4, 5]; reduction of Oct4 levels induces trophectoderm differentiation [6]; and reduced expression of Sox2 results in multiple lineages with trophectoderm predominating [7-9]. The unique dynamic and plastic chromatin architecture of ES cells [10] allows their chromatin to be easily modified and be more accessible to transcription factors. Epigenetic modifications are important in regulating gene expression in this open chromatin environment. Generally, methylation of histone H3 at lysine 27 (H3K27me3) marks repressed genes, whereas H3K4me3 is associated with active genes [11]. Recent studies indicate that a large number of genes important for development harbor both marks and thus have so-called, bivalent domains [12-14]. Interestingly, many of these marks are co-occupied by master factors Oct4, Sox2, and Nanog [14, 15]. So far, most studies of epigenetic modifications of histones in ES cells have focused on lysine methylases such as Ezh2, a component of the polycomb complex that methylates H3K27 to repress differentiation-specific genes and maintain pluripotency of ES cells [14, 16, 17].
We now find that pluripotent ES cells have CARM1 associated with the Oct4 and Sox2 promoters, which display arginine methylation of histone H3. When CARM1 is depleted, ES cells enter differentiation programs. Conversely, ES cells expressing elevated levels of CARM1 are more resistant to differentiation signals than wild-type ES cells. Such cells with increased CARM1 and detectable arginine methylation of histone H3 at the Nanog promoter respond to differentiation signals by prolonged and elevated expression of Nanog. Our study highlights, for the first time, the importance of histone arginine methylation in the epigenetic regulation of pluripotency in ES cells. It appears that like in early blastomeres, CARM1 provides a means of modulating the transition from the pluripotent to the differentiated state.
Materials And Methods
Cell Culture
HM1 ES cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, http://www.invitrogen.com), supplemented with 15% ES qualified fetal bovine serum (FBS; Gibco), 0.055 mM β-mercaptoethanol (Gibco), 2 mM L-glutamine, 0.1 mM MEM nonessential amino acid, 5,000 units/ml penicillin/streptomycin, and 1,000 units/ml of leukemia inhibitory factor (LIF) (Chemicon, Temecula, CA, http://www.chemicon.com). Differentiation was induced in media supplemented with 0.1 μM retinoic acid (RA) in the absence of LIF or simply lacking LIF. Alkaline phosphatase staining used a kit from Chemicon.
RNAi Design and Transfection
Nineteen-basepair sequences of CARM1 (5′-GTACACAGTGAACTTCTTA-3′ or 5′-GCACCTATAATCTCAGCAG-3′) were cloned into pSUPER.neo (Oligoengine Inc., Seattle, U.S.A., http://www.oligoengine.com) to express short hairpin RNA for RNAi 1 or 2. BLAST searches ensured no significant sequence similarity with other genes. After transfections with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com), the cells were selected by neomycin (150 μg/ml) for 3 days. The cells were then harvested for RNA extraction and Western blotting.
Generation of an RNAi-Resistant CARM1 Expression Construct
The CARM1 RNAi-immune construct was generated using site-directed mutagenesis kit (Stratagene, La Jolla, CA, http://www.stratagene.com). The CARM1 RNAi target site was destroyed by five DNA point mutations without causing changes of the encoded amino acid sequence. The following primers were used: forward: GGCCAAATCTGTCAAATACACAGTCAACTTCCTGGAAGCCAAAGAAGGC; reverse: GCCTTCTTTGGCTTCCAGGAAGTTGACTGTGTATTTGACAGATTTGGCC.
RNA Extraction, Reverse Transcription, and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated using TRI Reagent (Sigma-Aldrich Co., St. Louis, U.S.A., http://www.sigmaaldric.com) and purified with the RNAeasy Mini Kit (Qiagen, Hilden, Germany, http://www1.qiagen.com) after DNase (Ambion, Austin, TX, http://www.ambion.com) treatment. Reverse transcription used SuperScript III Kit (Invitrogen). Quantitative polymerase chain reaction in real time used an ABI PRISM 7000 Sequence Detection System and SYBR Green Master Mix. Gene expression levels were normalized toβ-actin. Each primer set gave a single product of the correct size.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was as described previously [15]. Crosslinking was with 1% formaldehyde for 10 minutes followed by neutralization with 0.125 M glycine. Chromatin (approximately 400-500 bp after sonication) was immunoprecipitated using: α-CARM1 (Upstate 07-080; Upstate, Charlottesville, VA, http://www.upstate.com); α-histone H3R17di-me (Abcam ab8284; Abcam, Cambridge, U.K., http://www.abcam.com), α-histone H3R26di-me (Abcam ab8047); the amount of antibody used in each immunoprecipitation and its ratio to magnetic protein G beads used in the assay was empirically determined to yield the highest fold enrichments. Fold enrichment was calculated by determining apparent IP efficiency (ratios of ChIP enriched DNA over input sample) and normalized to the level observed at a control region [14] defined as 1.0. SDs were calculated from biological replicates based on triplicate real-time polymerase chain reaction measurements of DNA.
Generation of CARM1 Overexpressing Stable Embryonic Stem Cell Line
CARM1 was cloned into pCAGIPpuro (kind gift of Ian Chambers) and transfected into HM1 cells. After 1 week of puromycin selection (1.5 μg/ml), ES colonies were picked and replated. CARM1 overexpression was confirmed by Western blot.
Construction of Chimeras with CARM1 Overexpressing Embryonic Stem Cells
CARM1 overexpressing ES cells were trypsinized for 1 to 2 minutes and washed with phosphate-buffered saline. Cells were dispersed by pipetting and resuspended in ES medium. Mouse eight-cell embryos were recovered from spontaneously ovulating F1 (CBA/C57Bl) females mated with EGFP transgenic males and zona pellucidae removed by treatment with Acid Tyrode’s solution as described before [1]. Individual ES cell clumps were placed into a depression in the culture dish and a single embryo added to each clump. Such aggregates were cultured in KSOM medium at 37 °C until the blastocyst stage as described before [1]. Chimeras were examined on a Zeiss LSM 510 (Carl Zeiss Inc., Oberkochen, Germany, http://www.ziess.com) confocal microscope to assess the contribution of host and donor cells.
Protein Extraction and Western Blotting
Protein extracts were obtained using cell lysis buffer (50 mM Tris-HCl pH 8.0; 150 mM NaCl; 2 mM EDTA; 1% NP-40) with protease inhibitor (Roche Diagnostics, Basel, Switzerland, http://www.roche-applied-science.com). Cell lysates were cleared at 10,000 g for 15 minutes at 4 °C. Protein concentrations were determined using Bradford reagent assay (Bio-Rad, Hercules, CA, http://www.bio-rad.com). Proteins were separated by SDS–PAGE, transferred to immunoblot PVDF membrane (Bio-Rad), and probed with specific primary antibodies and appropriate horseradish peroxidase-conjugated antibodies. Signals were detected using SuperSignal West Pico Chemiluminescent substrate (Pierce, Rockford, IL, http://www.piercenet.com). The following antibodies were used for Western blotting: α-Oct4 (Abcam ab19857); α-Nanog (Santa Cruz sc33760; Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com); α-α-tubulin (Sigma T9026); and α-β-actin (Abcam ab8226).
Microarray Hybridizations
ES cells were transfected, as described previously, with plasmid expressing shRNA targeted against either CARM1 or GFP control. After 72 hours growth in selection media, total RNA was isolated and 500 ng amplified using the Illumina Total Prep RNA Amplification Kit (Ambion) according to the manufacturer’s instructions. The biotinylated cRNA samples (1500 ng per sample) were applied to Illumina Mouse WG-6 v1.1 Expression BeadChips and hybridized overnight at 58 °C. Chips were washed, detected, and scanned according to the manufacturer’s instructions. Triplicate samples were used to compare transcript expression differences between CARM1-targeted RNAi ES cells and control treated (GFP-targeted) ES cells.
Microarray Analysis
Data from the triplicate samples were quantile-normalized [18] and analyzed using the “Bioconductor” (http://www.bioconductor.org) “Lumi” (http://www.bioconductor.org/packages/2.0/bioc/html/lumi.html) and “Limma” packages [19]. Data were P value-adjusted [20] to yield a sorted list of differentially expression genes. Genes that displayed significant (P < 0.005) fold expression changes >1.5 or <0.6 between the replicate arrays were identified (according to Loh et al. [21]; supporting information 1). Log2 transformed fold gene expression differences in each of the three replicate experiments were also subjected to hierarchical clustering using the “Cluster” program set to the “Euclidean Distance” similarity metric and “Average Linkage” clustering functions [22]. The results were visualized using “Treeview” [22]. Discreet clusters of differentially expressed genes were then interrogated for overrepresentation of “Gene Ontology” (GO) terms contained within the MGI- M. musculus “Biological Process” database (P < 0.005) using the Princeton University Generic GO term finder web interface [23].
Results
CARM1 Depletion Directs Embryonic Stem Cells to Differentiate
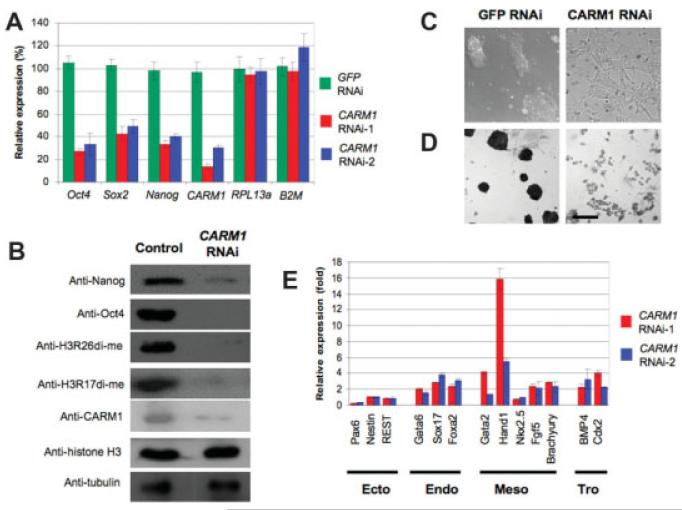
The correlation of levels of histone H3 arginine 17 and 26 methylation, catalyzed by CARM1, with extent of pluripotency of mouse embryo blastomeres [2], has raised the question of whether CARM1 might have a similar role in modulating pluripotency of ES cells. In a first series of experiments to examine CARM1 function in ES cells, we decided to knock down CARM1 transcript levels using constructs expressing two independent CARM1 shRNAs (Fig. 1A). We found that these constructs led to a dramatic decrease in CARM1 protein levels in comparison to control cells (Fig. 1B). Consistent downregulation of CARM1 led to a reduction in histone H3 methylation on arginines 26 and 17 (Fig. 1B). ES cells in which CARM1 was downregulated no longer formed distinct alkaline phosphatase positive colonies characteristic of ES cell cultures. Instead they adopted a variety of morphologies characteristic of differentiating cells (Fig. 1C, D). To test whether the observed phenotype was specific, we cotransfected plasmids expressing the most efficient CARM1 shRNA and RNAi-resistant CARM1 mRNA. This did not result in ES cell differentiation or loss of ES cell colony morphology, thus rescuing the CARM1 RNAi phenotype (supporting information Fig. 1).
Figure 1. Coactivator-associated-protein-arginine-methyltransferase 1 (CARM1) is required to maintain embryonic stem (ES) cell pluripotency.
(A) Transfection of CARM1 shRNA downregulates Nanog, Sox2, and Oct4 mRNA levels. RNA was extracted 3 days after transfection and quantified by real-time polymerase chain reaction. Mean values of the indicated transcript levels plotted as percentages relative to those after transfection of empty vector control. Sample was assayed in duplicate and normalized to endogenous β-actin. (B) CARM1 knockdown results in decreased Oct4, Nanog, and CARM1 protein revealed by Western blotting and of methylation of histone H3 at R26 and R17. α-tubulin and α-histone H3 are loading controls. (C) Cell differentiation after CARM1 depletion but not after transfection of GFP RNAi plasmid. Note stellate cells after CARM1 RNAi. Scale bar: 50 μM. (D) Reduced alkaline phosphatase staining indicating ES cell differentiation caused by CARM1 depletion compared to transfection GFP control. Scale bar: 50 μM. (E) CARM1 depletion directs ES cells to express indicated endodermal (endo); mesodermal (meso), and trophectodermal (Tro) marker genes assessed by real-time polymerase chain reaction. Mean levels (after 3 days) expressed relative to the vector control (onefold) and normalized to β-actin expression levels.
We then asked whether downregulation of CARM1 in ES cells would be associated with changes in the levels of expression of the transcription factors such as Oct4, Sox2, and Nanog that are critical to maintain ES cell pluripotency and suppress their differentiation. We found that after 3 days of CARM1 knockdown, ES cells showed a reduction in Oct4, Sox2, and Nanog mRNAs and proteins (Fig. 1A, B), whereas nontarget GFP RNAi did not result in changes of transcript levels (Fig. 1A). Although reductions in transcriptional activity are likely to be widespread after CARM1 knockdown, CARM1 depletion did not uniformly affect all genes because transcripts of the housekeeping genes RPL13A and B2M were not reduced (Fig. 1A). Together, these results suggest that CARM1 is required to maintain expression of key transcription factors, Oct4, Sox2, and Nanog, necessary for ES pluripotency.
It is known that Oct4 and Sox2 act synergistically on the enhancers of their own genes to provide a positive feedback loop regulating their expression [24, 25] and also regulate Nanog expression [26, 27]. It could be expected that reduced expression of this pluripotency network of genes would result in increased expression of marker genes for specific differentiated lineages. Indeed, quantitative real-time polymerase chain reaction after CARM1 RNAi revealed the expression of two trophectoderm markers, BMP4 and Cdx2 was increased by 2.3- and 4.1-fold, respectively; of three endoderm markers, Gata6, Sox17, and FoxA2 expression increased by 2-, 2.8-, and 2.4-fold, respectively; the mesoderm marker Gata2 increased 4.1-fold, whereas Hand1 was dramatically upregulated by up to 15.9-fold and Nkx2.5 showed no significant change in expression level. However, expression levels of three ectoderm markers showed no significant change (Fig. 1E).
Taken together these results indicate that after CARM1 downregulation concomitant with the loss of expression of pluripotency transcription factors, ES cells initiate differentiation into trophectoderm, endoderm, and mesoderm cells.
Downregulation of CARM1 Causes Increased Expression of Developmental Genes and Reduced Expression of Pluripotency Genes
To better understand how CARM1 depletion results in ES cell differentiation, we wanted to survey the global gene expression changes occurring at the mRNA level. To this end, we performed whole genome cDNA microarray hybridizations comparing the transcriptome from ES cells with either CARM1 or GFP (negative control) specific RNAi. We found that 1,187 genes were upregulated (>1.5-fold) and 587 transcripts were downregulated (<0.6-fold), indicative of large-scale changes within the transcriptome on CARM1 depletion (supporting information 1). To examine whether we could define discreet clusters of common gene regulation that could provide a functional insight between the detected gene expression differences, we performed a hierarchical clustering of our microarray data. This analysis yielded six discernable clusters of commonly regulated genes with three each associated with up- and downregulation (Fig. 2A). Each gene cluster (a list of the genes in each cluster is given in supporting information 2) was then interrogated for overrepresented GO) terms relating to “biological process” (summary findings in Fig. 2A and full GO results in supporting information 3). Overall downregulated gene clusters were associated with a lack of self-renewal capacity. This is reinforced by the presence of known ES cell specific genes or those associated with pluripotency in the downregulated gene clusters (Fig. 2B). For example, the two most highly downregulated clusters contain the pluripotent triad of transcription factors (Nanog, 1.97-fold; Oct4, 2.18-fold; and Sox2, 1.69-fold) as well as the histone H3-lysine 27 methyltransferase, Ezh2 (1.97-fold), an essential gene contributing to ES cell-specific bivalent domains of chromatin structure [13]. Upregulated clusters were highly enriched for terms relating to “development” and “differentiation.” When we compared this data set with one from ES cells grown without LIF or in the presence of RA [28], we found strong overlap. This was particularly striking in, although not confined to, cluster 5. This cluster contained genes that consistently displayed the highest levels of gene upregulation after CARM1 depletion, marking them out as the most sensitive to CARM1 regulation (supporting information 2 and 3). The overrepresentation of developmentally related GO terms in upregulated gene clusters is consistent with the observed differentiation of ES cells (Fig. 1). It also highlights that cell differentiation programs that would ordinarily be repressed in ES cells expressing CARM1 become aberrantly activated on CARM1 depletion. Examination of the upregulated gene clusters revealed markers of the major germ layers (Fig. 2B) further emphasizing this point. Interestingly, in addition to developmentally related GO terms, we also observed terms relating to “apoptosis” in upregulated gene clusters. Similar findings were made in studies of RA-induced ES cell differentiation [28]. In summary, our global expression analysis shows that on CARM1 depletion, not only are cell differentiation programs aberrantly activated, but those that regulate ES cell integrity per se are also downregulated. It is unlikely that CARM1 directly regulates all the gene expression changes observed but rather a key subset of genes. On CARM1 depletion, it is these genes that become misregulated that then invoke the large-scale changes in the transcriptome. Thus, analysis of the global gene expression changes indicates CARM1 is an important player in regulation of ES cell pluripotency.
Figure 2. Gene expression microarray profiling of changes in ES cells after coactivator-associated-protein-arginine-methyltransferase 1 (CARM1) RNAi.
Gene expression changes associated with targeted knockdown of CARM1 mRNA in embryonic stem (ES) cells were determined by expression microarray analysis (see supporting information for complete data). (A) Log2 transformed fold gene expression differences, from each of three replicates, were subject to hierarchical clustering (see “Methods”) and are shown in classical thumbnail-dendogram format (downregulated and upregulated genes shown as green and red tiles, respectively). Six discernable clusters of differential gene expression were identified representing downregulated (clusters 1, 2, and 3) and upregulated genes (clusters 4, 5, and 6). The genes within each cluster were interrogated for overrepresentation of “Gene Ontology” (GO) terms relating to “biological process.” Examples of significantly enriched GO terms, together with associated P values, are shown for each of the six clusters. The asterisk (*) denotes a significant term associated with both down- and upregulated clusters and the section mark (§) indicates a term found in other clusters that nonetheless show the same trend in gene regulation, i.e., up or down. (B) Examples of specific gene regulation across the triplicate samples taken as individual tiles from the thumbnail-dendogram. Genes were selected on the basis of their known roles in either pluripotency and ES cells (downregulated genes) or their roles in differentiation to distinct germ layers (upregulated). The cluster from which each regulated gene was derived is indicated. All genes exhibit fold expression changes <0.6 (downregulated) or >1.5 (upregulated) across the triplicates (P < 0.005).
CARM1 Binds to the Oct4 and Sox2 Promoters
The reduction in Oct4, Sox2, and Nanog transcript levels after CARM1 downregulation could suggest that all three genes may be direct targets for CARM1 regulation. To investigate this possibility, we performed ChIP on undifferentiated ES cell chromatin using a CARM1 antibody. Real-time polymerase chain reaction using specific primers located in the proximal region of the Oct4 promoter revealed significant enrichment of these DNA fragments in the anti-CARM1 ChIP (Fig. 3A) relative to polymerase chain reaction of intergeneic sequences. Oct4 sequences could not be detected in a CARM1 ChIP after CARM1 RNAi (supporting information Fig. 2). This indicates a direct interaction between CARM1 and the Oct4 promoter and suggests that CARM1 may be involved in activating or maintaining Oct4 expression in ES cells. We then asked whether histone H3 showed methylation at arginines 17 and 26 by performing ChIP with antibodies that recognize these modifications. As expected, if CARM1 was active at the Oct4 promoter, we found the promoter fragments clearly enriched in both antihistone H3R17di-me and antihistone H3R26di-me ChIP DNA (Fig. 3A). After CARM1 RNAi, these marks were also removed (supporting information Fig. 2). This suggests that CARM1 can facilitate Oct4 expression through the modification of histone H3R17 and H3R26 on its promoter.
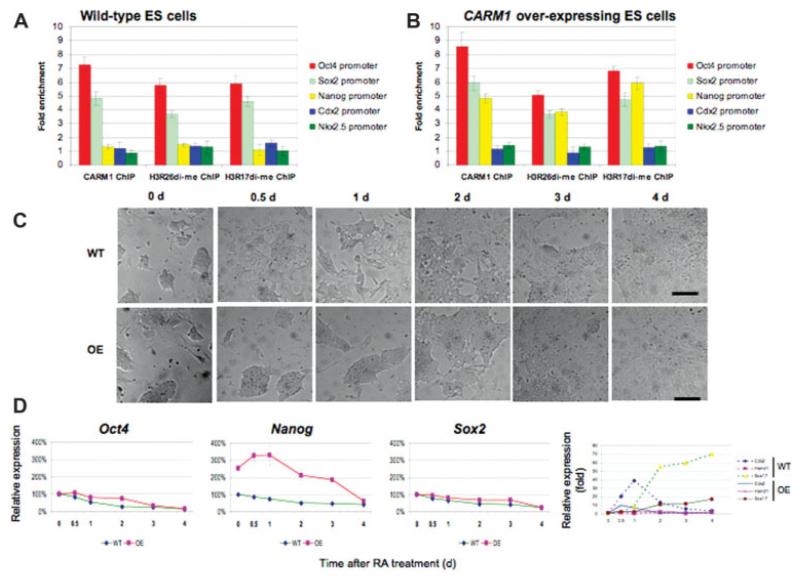
Figure 3. Coactivator-associated-protein-arginine-methyltransferase 1 (CARM1) overexpression leads embryonic stem (ES) cells to resist retinoic acid (RA)-induced ES cell differentiation.
Chromatin immunoprecipitation (ChIP) was performed on sonicated chromatin from wild-type ES cells (A) and CARM1 overexpressing cells (B) using anti-CARM1, antihistone H3R26di-me, and antihistone H3R17di-me antibodies. Immunoprecipitated DNA was analyzed by quantitative real-time polymerase chain reaction with primers from within 1-kb region of transcriptional start site of the investigated gene. Fold enrichments were calculated from the apparent IP efficiency (ratio of ChIP enriched DNA over input) and normalized to the level at a control region defined as 1.0 for a given extract from a specific cell line. This allows comparisons of fold enrichment to be made for a given extract but does not permit quantitative comparisons between cell lines. Error bars represent SD and were calculated from biological replicates (performed in triplicate) of the quantitative polymerase chain reaction data. (C) Differentiation of CARM1 overexpressing cells is delayed on retinoic acid treatment relative to wild-type ES cells. ES cells were treated with 0.1 μM retinoic acid in the absence of leukemia inhibitory factor (LIF) and observed after 0.5, 1, 2, 3, and 4 days of RA treatment. Scale bar: 50 μM. (D) Quantification of transcript level changes of Oct4, Sox2, Nanog, and three lineage marker genes (Cdx2, Hand1, and Sox17) on RA treatment of wild-type and CARM1 overexpressing ES cells. Gene expression levels were analyzed by real-time polymerase chain reaction. Relative expression given as percentages or fold relative to untreated control cells at day 0 and normalized to endogenous β-actin level.
Similarly, we could detect enrichment of Sox2 promoter sequences in the anti-CARM1 ChIP in wild-type ES cells. Sox2 promoter fragments also showed substantial arginine methylation at R17 and R26 (Fig. 3A). Paralleling the Oct4 data, no Sox2-derived sequences could be detected in anti-CARM1 or either antihistone arginine methylation ChIP DNAs after CARM1-specific RNAi (supporting information Fig. 2). The presence of histone H3 R26 and R17 methylation at the Oct4 and Sox2 promoters, albeit at different levels, is indicative of functional CARM1 recruitment at these loci. In contrast, the Nanog promoter did not show any detectable enrichment (with the same three antibodies) in wild-type ES cells, implying a lack of CARM1 occupancy at this locus (Fig. 3A). This strongly resembled the profile obtained for genes important for differentiation of mesoderm (Nkx2.5) or trophectoderm (Cdx2) that are not expressed in pluripotent cells. Thus, downregulation of Oct4 and Sox2 after CARM1 RNAi appears to be a direct consequence of reduced recruitment of CARM1 protein (and its enzymatic activity) at their promoters while downregulation of Nanog to be secondary to the loss of binding of Oct4 and Sox2 proteins to its own enhancer.
Embryonic Stem Cells Overexpressing CARM1 are More Resistant to Differentiation
It has been shown that elevation of CARM1 expression in the mouse embryo elevates expression of several pluripotency genes and directs cells to ICM [2]. Thus, we wondered whether CARM1 might have a similar effect on the pluripotent state in ES cells. To test this, we constructed transformed ES cell lines stably expressing elevated levels of CARM1 (supporting information Fig. 3A, B). We found that the resulting ES cell line had typical ES cell morphologies (Fig. 3C) and was Alkaline phosphatase (AP)-positive (not shown). Interestingly, when these CARM1 overexpressing ES cells were aggregated with eight-cell host embryos, they showed preferential contribution to ICM and formed uniform and coherent clones (supporting information Fig. 4). They also formed colonies with similar efficiency to wild-type ES cells (supporting information Fig. 5). The expression level of Oct4 in this CARM1 overexpression line was comparable to control ES cells (1.1- and 1.2-fold respective elevation of transcript and protein). However, the expression of Nanog was elevated to 2.3- and 2.2-fold for both mRNA and protein (supporting information Fig. 3A, 3B). We also compared the binding of CARM1 with the promoter regions of pluripotency marker genes such as Oct4, Sox2, and Nanog or the differentiation marker genes, Cdx2 and Nkx2.5, by ChIP (Fig. 3B). This showed that like in the wild-type ES cells, CARM1 associated with both Oct4 and Sox2 promoters in ES cells expressing elevated CARM1 levels. Interestingly, in contrast to parental ES cells, we also detected CARM1 binding to the Nanog promoter in ES cells expressing elevated CARM1 (Fig. 3B). In neither parental nor CARM1 overexpressing ES cells was CARM1 associated with the Cdx2 and Nkx2.5 promoters. We also tested whether histone H3 was modified on arginines 17 and 26 on the promoters of these five genes. Whereas wild-type ES cells showed enrichment of these modifications only on Oct4 and Sox2, CARM1 overexpressing cells also showed an enrichment of Nanog promoter sequences in both antihistone H3R17di-me and H3R26di-me ChIP DNA (Fig. 3B). Therefore, these results suggest that the elevated CARM1 levels lead to functional CARM1 recruitment and the appearance of histone H3 arginine methylation at the Nanog promoter.
We then wanted to assay the functional consequences of elevated CARM1 expression in ES cells and specifically test whether this might affect their response to a differentiation signal. Our first test was to treat CARM1 overexpressing ES cells with 0.1 μM RA in the absence of LIF (normally used to maintain pluripotency). We found CARM1 overexpressing cells maintained the characteristic appearance of ES cells for as long as 2 days of treatment in contrast to wild-type ES cells that began to show characteristic differentiation morphologies after as little as 0.5 day of RA treatment (Fig. 3C). Thus, it appears that CARM1 upregulation makes ES cells resist RA-induced differentiation, although eventually they do respond to the hormone.
We considered how the ability to delay differentiation might be reflected in expression level changes of Oct4, Sox2, and Nanog on RA treatment. We found that whereas in wild-type ES cells, Oct4, Nanog, and Sox2 transcript levels showed a gradual decline after RA treatment, in the CARM1 over-expressing cells, the expression dynamics of these gene was different. Oct4 and Sox2 levels declined with a similar, if slightly delayed, time course, whereas Nanog levels, in contrast, increased (Fig. 3D). Moreover, this transient increase was maintained for 24 hours before declining to similar levels to wild-type after 4 days (Fig. 3D). This increase was also reflected in levels of Nanog protein (supporting information Fig. 3C). Thus, it appears that ES cells expressing 10-fold elevated CARM1 do respond to a differentiation signal, but it is considerably delayed for a period during which Nanog expression persists at a higher level. This is reminiscent of the consequences of CARM1 overexpression in mouse blastomeres that leads to elevated levels of Nanog and the contribution of the cells preferentially to ICM [2].
Because CARM1 is known to positively regulate expression of genes responsive to nuclear hormone receptors [29] and thus may affect genes under the control of the RA receptor, we also wanted to examine the ability of cells overexpressing CARM1 to respond to an alternative differentiation signal, the removal of LIF from the medium. Comparable to the effects of RA treatment, we found the morphology of control ES cells indicated differentiation as early as 12 hours after LIF withdrawal. Once again, ES cells with elevated CARM1 were retarded in their differentiation and only developed apparent differentiated cell morphologies 48 hours after LIF withdrawal. Like with RA treatment, LIF withdrawal led to progressive diminution of Oct4 and Sox2 levels in wild-type and CARM1 overexpressing cells with some delay in the latter. Nanog expression was further elevated in the CARM1 overexpressing cells before declining after a similar time course to that seen in RA treatment (supporting information Fig. 6). Thus, CARM1 overexpression, its associated occupancy, and histone H3 arginine modification of the Nanog promoter appear to prime cells so that they respond to a differentiation signal by increasing Nanog expression. Because Nanog is a pluripotency related transcription factor, its extended and elevated expression could contribute to the retarded response to the differentiation signal [30].
Discussion
Epigenetic modifications are beginning to emerge as important ways to regulate gene expression in the plastic chromatin of ES cells [10-14]. Our present study adds to this by showing that CARM1, a histone arginine methyltransferase, contributes to maintaining ES cell pluripotency. CARM1 appears to play its role, at least in part, by sustaining Oct4 and Sox2 activity through arginine methylation of histone H3 at their promoters. Loss of CARM1 leads to the downregulation of Oct4 and Sox2. Because Oct4 and Sox2 act together to enhance Nanog expression [26, 27], their downregulation would be sufficient to account for the downregulation of Nanog and the direction of ES cells to differentiate into various lineages. Thus, once CARM1 levels fall beneath a critical threshold, ES cells begin to differentiate under conditions in which they would otherwise continue to renew as pluripotent cells. This result points to CARM1 as a pluripotency related gene in mouse ES cells.
Our results are consistent with findings that progeny of blastomeres with naturally higher levels of histone H3 R26/17me at the four-cell stage tend to contribute more cells to the pluripotent ICM and to show greater extent of developmental potency than progeny of blastomeres with very low levels of these modifications [1, 2]. Thus, by affecting levels of arginine methylation, CARM1 could influence developmental pathways of cells that are otherwise similarly pluripotent. When the zygotic expression of CARM1 is disrupted, this results in small embryos that die perinatally [31]. This suggests that there are both maternal and zygotic contributions of CARM1 to embryonic development. Thus CARM1−/− embryos will inherit a dowry of maternal CARM1 protein [2] that, in turn, would ensure development of the embryo beyond the stage at which it relies on the pluripotency of ICM and epiblast. It would be interesting to determine, in future studies, whether in mice in which maternal CARM1 is eliminated, the formation and size of the ICM/epiblast population is affected.
CARM1 is a well-characterized histone H3 methylase and many reports detail its role in the epigenetic regulation of transcription (reviewed [29]). It is important, however, to also highlight that CARM1 has been documented to methylate nonhistone proteins. These include components of histone acetyl transferase complexes resulting in altered composition and function [32-34] and a number of RNA-binding proteins involved in transcription elongation, splicing, and translation [35-37]. Although we cannot exclude regulation at other levels, our finding that CARM1 is recruited to Oct4, Sox2, and Nanog promoters (the last after CARM1 overexpression) and is associated with R26 and R17 methylation (that is then lost after CARM1 depletion) strongly suggests that CARM1 achieves the effects we describe, at least in part through modification of chromatin at the promoters of key pluripotency genes. As a transcriptional coactivator, CARM1 is considered to affect a broad number of genes. Nevertheless, there must be some specificity to its activity in pluripotent cells because the microarray analysis reported here identifies genes whose expression is decreased, genes that are unaffected, and genes whose expression increases sgyrt CARM1 downregulation. ChIP indicates that these differential effects reflect a requirement for methylation of histone H3 at R17 and R26 to activate expression of Oct4 and Sox2 but not for expression of mesoderm (Nkx2.5) or trophectoderm (Cdx2) genes. This provides the first demonstration that epigenetic modifications mediated by CARM1 are important to regulate expression of specific gene sets in ES cell self-renewal and pluripotency.
The resistance to differentiation of ES cells expressing elevated CARM1 mirrors the effects of its overexpression in blastomeres that express elevated levels of Nanog and contribute progeny preferentially to ICM [2]. In ES cells engineered to express elevated levels of CARM1, this methyltransferase now becomes recruited to the Nanog promoter. This gives a modest 2.5-fold increase in Nanog expression unless the cells meet signals for differentiation when Nanog expression is prolonged and elevated a further 1.5-fold and the differentiation process is accordingly delayed. However, ES cells expressing elevated CARM1, and Nanog, can still differentiate if they receive a signal to do so, although differentiation is considerably delayed. Thus, the effects of CARM1 in sustaining expression of pluripotency genes can be overridden. Importantly, CARM1 overexpressing cells do not display any proliferative advantage over the wild-type cells, but rather resist differentiation. It is of interest that in both embryos and ES cells, the effects of increased CARM1 expression are mediated, at least in part, through Nanog, a protein now proposed as unessential for pluripotency but acting rather by resisting or reversing alternative gene expression programs in pluripotent cells [30]. In many ways, CARM1 seems to act in a not dissimilar manner. Our model suggests that in both early embryos and ES cells, CARM1 behaves not as an absolute switch, but more as a modulator of the expression of genes governing the pluripotent state (Fig. 4).
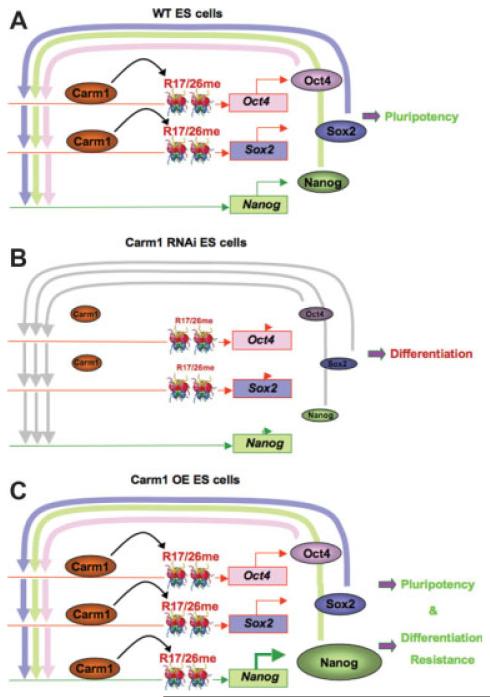
Figure 4. Model for the modulation of pluripotency by coactivator-associated-protein-arginine-methyltransferase 1 (CARM1).
CARM1 activity is required for embryonic stem (ES) cell pluripotency because cells differentiate after its downregulation (A versus B). Pluripotency is maintained in wild-type cells in part as a result of arginine methylation of histone H3 by CARM1 at the Oct4 and Sox2 promoters. The Oct4 and Sox2 proteins together act on their own enhancers and the Nanog enhancer. Nanog expression tips the balance towards maintaining the pluripotent state Overexpression of CARM1 (C) now leads to arginine methylation of histone H3 at the Nanog promoter elevating Nanog expression, causing resistance to differentiation. Abbreviations: OE, CARM1-overexpressing cells; WT, wild-type.
Supplementary Material
Acknowledgments
MZG is grateful to the Wellcome Trust for a Senior Research Fellowship and associated program grant that supported this work. We are grateful to Ian Chambers for cell lines and advice and Paul Robson for valuable comments.
Footnotes
Disclosure Of Potential Conflicts Of Interest: The authors indicate no potential conflict of interest.
References
- 1.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, et al. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132:479–490. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- 2.Torres-Padilla ME, Parfitt DE, Kouzarides T, et al. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 4.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 5.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 6.Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 7.Avilion AA, Nicolis SK, Pevny LH, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanova N, Dobrin R, Lu R, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 9.Masui S, Nakatake Y, Toyooka Y, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 10.Meshorer E, Yellajoshula D, George E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 14.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 15.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 16.O’Carroll D, Erhardt S, Pagani M, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 21.Loh YH, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle EI, Weng S, Gollub J, et al. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okumura-Nakanishi S, Saito M, Niwa H, et al. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 25.Chew JL, Loh YH, Zhang W, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda T, Tada M, Kubota H, et al. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodda DJ, Chew JL, Lim LH, et al. Transcriptional regulation of Nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 28.Sharova LV, Sharov AA, Piao Y, et al. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 30.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 31.Yadav N, Lee J, Kim J, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YH, Coonrod SA, Kraus WL, et al. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naeem H, Cheng D, Zhao Q, et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Q, Yi P, Wong J, et al. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng D, Cote J, Shaaban S, et al. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Park S, Kilburn B, et al. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 37.Ohkura N, Takahashi M, Yaguchi H, et al. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J Biol Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.