Abstract
MDM2 mediates the ubiquitylation and thereby triggers the proteasomal degradation of the tumor suppressor protein p53. However, genetic evidence suggests that MDM2 contributes to multiple regulatory networks independently of p53 degradation. We have now identified the DEAD-box RNA helicase DDX24 as a nucleolar protein that interacts with MDM2. DDX24 was found to bind to the central region of MDM2, resulting in the polyubiquitylation of DDX24 both in vitro and in vivo. Unexpectedly, however, the polyubiquitylation of DDX24 did not elicit its proteasomal degradation but rather promoted its association with preribosomal ribonucleoprotein (pre-rRNP) processing complexes that are required for the early steps of pre-rRNA processing. Consistently with these findings, depletion of DDX24 in cells impaired pre-rRNA processing and resulted both in abrogation of MDM2 function and in consequent p53 stabilization. Our results thus suggest an unexpected role of MDM2 in the nonproteolytic ubiquitylation of DDX24, which may contribute to the regulation of pre-rRNA processing.
INTRODUCTION
MDM2 belongs to the family of RING finger-type ubiquitin ligases (E3s) and functions as a pivotal negative regulator of the tumor suppressor protein p53 (1, 2). MDM2 inhibits p53 function by two distinct mechanisms: it abrogates the transactivation activity of p53 through direct binding to the NH2-terminal region of the protein (3, 4), and it mediates the polyubiquitylation of p53 and thereby targets it for degradation by the 26S proteasome (5–7). On the other hand, p53 binds to the promoter of the MDM2 gene and activates its expression (8, 9), thereby completing a negative-feedback loop that is responsible for strict regulation of p53 function in the absence of stress. Exposure of cells to stress, however, results in downregulation of both the abundance and the activity of MDM2 as well as consequent stabilization and subsequent activation of p53 (1, 2). The mechanisms by which p53 escapes from the inhibitory action of MDM2 differ among cell types and stress signals.
The role of the nucleolus as a stress sensor has recently emerged. Many types of stress signal converge on steps of ribosome biogenesis and thereby activate p53 (10). A group of ribosomal proteins (RPs), including RPL5, RPL11, and RPL23, serve as transmitters of stress signaling. These proteins are released from the nucleolus in response to stress, bind to and inhibit the activity of MDM2 in the nucleoplasm (11), and eventually activate p53 (11–18).
Inhibition of precursor rRNA (pre-rRNA) processing is a key mechanism for induction of nucleolar stress (19). In mammals, the ∼200 genes encoding 47S pre-rRNA, which serves as a precursor for 18S, 5.8S, and 28S rRNAs, are organized in five tandem arrays on the short arms of acrocentric chromosomes. These genes are transcribed by RNA polymerase I (Pol I) in association with the Pol I-specific basal factors SL1 (also known as TAF1B) and UBF (20). The transcribed 47S pre-rRNA undergoes a series of endo- and exonucleolytic cleavages to remove the spacer sequences. In addition, the pre-rRNA is subjected to covalent modifications that include base and ribose methylation as well as uridine isomerization to pseudouridine at specific sites (21, 22). During this process, many assembly factors are associated with the pre-rRNA and its processing products in the form of a preribosomal ribonucleoprotein (pre-rRNP) complex (23). Proteomic characterization of human pre-rRNP complexes has identified >180 assembly factors and 100 small nucleolar ribonucleoprotein particles (snoRNPs) involved in this process (24–27).
DEAD-box RNA helicases constitute a group of such assembly factors, although their precise roles in ribosome biogenesis have not been fully characterized. These enzymes participate in the regulation of essentially all aspects of RNA metabolism, including pre-mRNA splicing, translation, pre-rRNA processing, and mRNA decay. Most of them have been highly conserved through evolution, and many have been shown to contribute to ribosome biogenesis in yeasts (28, 29).
With a proteomics approach, we have now identified proteins that are associated with MDM2 in human cells. Among these MDM2 binding proteins, we focused on DDX24, a nucleolar protein that belongs to the family of DEAD-box RNA helicases. Although MAK5, the Saccharomyces cerevisiae ortholog of human DDX24, was previously found to contribute to the biosynthesis of the 60S ribosome subunit (30), the function of DDX24 in mammals has remained unclear. We found that DDX24 binds to the central acidic region of MDM2 and that such binding promotes the polyubiquitylation of DDX24. However, the polyubiquitylation of DDX24 did not elicit its proteasomal degradation but rather promoted its association with components of pre-rRNP complexes that are required for efficient pre-rRNA processing reactions. Depletion of DDX24 by RNA interference (RNAi) inhibited proper pre-rRNA processing. Our findings thus demonstrate the existence of MDM2-mediated noncanonical polyubiquitylation in human cells.
MATERIALS AND METHODS
Plasmids.
Complementary DNAs encoding wild-type (WT) or mutant forms of human or mouse MDM2 as well as human DDX24, DDX24-ub, p53, p53-ub, ubiquitin, nucleolin (NCL), NIP7, p14ARF, RPL5, RPL11, RPL23, RPL26, and RPS7, each tagged at its NH2 terminus with FLAG, hemagglutinin (HA), Myc, or green fluorescent protein (GFP) epitopes, were subcloned into pcDNA3 (Invitrogen), pRSET (Invitrogen), or pGEX6p (GE Healthcare). A cDNA encoding human ubiquitin, tagged at its NH2 terminus with the HA epitope, was subcloned into pCGN (31). A cDNA for an RNAi-resistant form of human DDX24 was constructed by the introduction of silent mutations into the nucleotide sequence 5′-GCTCGAATCCTTCATAAGAAG-3′ to yield 5′-GCTCGAATaCTTCAcAAaAAG-3′ (where lowercase letters indicate the mutations).
Transfection, immunoprecipitation, and immunoblot analysis.
HCT116 cells were cultured under an atmosphere of 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). Transfection was performed with the FuGENE HD reagent (Roche). The empty pcDNA3 plasmid was included to ensure that cells were transfected with equal amounts of total DNA. Cell lysis and immunoprecipitation were performed as described previously (31), and immunoprecipitates were subjected to immunoblot analysis as described previously (32).
Antibodies.
Antibodies to MDM2 (SMP-14), to p53 (DO-1 or FL-393), and to GFP (sc-8334) as well as control rabbit immunoglobulin G (sc-2027) were obtained from Santa Cruz Biotechnology; antibodies to DDX24 (NB100-2224) were from Novus Biologicals; those to p21 (556430) and to HSP90 (610419) were from BD Biosciences; those to α-tubulin (TU01) and to RPL11 (3A4A7, kindly provided by K. Kawahara) were from Invitrogen; those to RPS6 (5G10) were from Cell Signaling Technology; those to the FLAG epitope (M2) and to the Myc epitope (9E10) were from Sigma; and those to HA (HA11) were from Covance.
Protein identification by LC-MS/MS analysis.
Affinity-purified protein complexes were concentrated by precipitation with chloroform-methanol, fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and stained with silver. Each lane of the stained gel was sliced into equal pieces, and the proteins therein were subjected to in-gel digestion with trypsin. The resulting peptides were dried, dissolved in a mixture of 0.1% trifluoroacetic acid and 2% acetonitrile, and then applied to a Nano flow liquid chromatography (LC) system (Paradigm MS4; Michrom BioResources) equipped with an L-column (C18, 0.15 by 50 mm, particle size of 3 μm; CERI). The peptides were fractionated with a linear gradient of solvent A (2% acetonitrile and 0.1% formic acid in water) and solvent B (90% acetonitrile and 0.1% formic acid in water), with solvent B at 0 to 45% over 20 min, 45 to 95% over 5 min, and 95 to 5% over 1 min at a flow rate of 1 μl/min. Eluted peptides were sprayed directly into a Finnigan LTQ mass spectrometer (Thermo Scientific). Mass spectrometry (MS) and tandem MS (MS/MS) spectra were obtained automatically in a data-dependent scan mode with a dynamic exclusion option. All MS/MS spectra were compared with protein sequences in the International Protein Index (IPI; European Bioinformatics Institute), human version 3.16, with the use of the MASCOT algorithm. Trypsin was selected as the enzyme used, the allowed number of missed cleavages was set at one, and carbamidomethylation of cysteine was selected as a fixed modification. Oxidized methionine and NH2-terminal pyroglutamate were searched as variable modifications. Tolerance of MS/MS ions was 0.8 Da. Assigned high-scoring peptide sequences (MASCOT score of ≥45) were considered for correct identification. If the MASCOT score was <45, the criteria for match acceptance included the following: (i) a peptide sequence length of ≥6, (ii) a MASCOT score for individual peptides of ≥35, (iii) at least three blocks of three successive matches or a block of six successive matches for y or b ions, and (iv) a delta score of ≥15. Identified peptides from independent experiments were integrated and regrouped by IPI accession number. Proteins reproducibly detected in all three independent experiments with HCT116 cells expressing FLAG-tagged mouse MDM2 as the bait were considered MDM2-associated proteins. Functional categorization of identified proteins was performed with the gene ontology (GO) annotations (Gene Ontology database, http://www.geneontology.org).
Protein purification.
For preparation of purified proteins for in vitro assays, glutathione S-transferase (GST) fusion proteins of MDM2, FLAG-DDX24, FLAG-p53, and NIP7 (wild type [WT], A55G, or with a deletion of amino acids 54 to 59 [Δ54–59]) were induced in Rosetta(DE3)/pLys (Novagen) cells at room temperature. The proteins were extracted with buffer BC500 (20 mM Tris-HCl [pH 7.3], 0.2 mM EDTA, 500 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) containing 1% Nonidet P-40 and then purified with glutathione (GSH)-Sepharose (GE Healthcare). If necessary, the GST moiety was removed from the fusion proteins by cleavage with PreScission protease (GE Healthcare). Hexahistidine (His6)-tagged DDX24 was induced in the same manner and purified with nickel-nitrilotriacetic acid (Ni-NTA)–agarose (ProBond purification system; Invitrogen).
In vitro binding assays.
In vitro binding assays were performed as described previously (33). In brief, 1 μg of each GST fusion protein was incubated for 30 min at 37°C with 5 μl of GSH-Sepharose in 200 μl of binding buffer (20 mM HEPES-NaOH [pH 7.5], 150 mM KCl, 10% glycerol, 0.1% Nonidet P-40, 1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM PMSF). Test proteins (100 ng) were then added, and the binding mixtures were incubated first for 30 min at room temperature and then for 1 h at 4°C. The beads were then isolated, washed three times with 0.5 ml of ice-cold binding buffer, resuspended in 20 μl of SDS sample buffer, and boiled. Proteins were separated by SDS-PAGE on a 6% gel and subjected to immunoblot analysis with antibodies to DDX24 or FLAG.
Quantitative RT-PCR analysis.
Total RNA (1 μg) isolated from cells with the use of the Isogen reagent (Nippon Gene) was subjected to reverse transcription (RT) and real-time PCR analysis with primers (forward and reverse, respectively) for human DDX24 (5′-GCTCAGAAACCTGGAGCAGT-3′ and 5′-TAAATCTCCGAGGTACGTGG-3′) and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-GCAAATTCCATGGCACCGT-3′ and 5′-TCGCCCCACTTGATTTTGG-3′), as described previously (34). The amount of DDX24 mRNA was normalized by the corresponding amount of GAPDH mRNA.
Preparation of lentiviral vectors.
Lentiviral vectors were prepared as described previously (35, 36). Gene silencing by RNAi was performed with the lentivirus-based short hairpin RNA (shRNA) expression vector CSII-U6tet. The shRNA-encoding DNA oligonucleotide inserts were generated with the use of the insert design tool for pSilencer vectors (Applied Biosystems). The target sequences were 5′-GGCCAGTATATTATGACTAAA-3′, 5′-GATGATGAGGTATATCAAG-3′, 5′-GCCGGAGCTGAGACTAGATCA-3′, and 5′-GCTCGAATCCTTCATAAGAAG-3′ for MDM2-1, MDM2-2, DDX24-1, and DDX24-2, respectively.
Tetracycline-inducible shRNA expression system.
Cells were infected with a lentivirus encoding TetR (CSII-elongation factor 1 alpha promoter [EF]-TetR-internal ribosome entry site [IRES]-puro) (36) for 2 days. They were then incubated with puromycin (10 μg/ml) for selection and amplification of puromycin-resistant cells. The TetR-expressing cells were then infected with lentiviruses encoding shRNAs. For induction of shRNA expression, the cells were incubated with doxycycline (1 μg/ml) for the times indicated in Fig. 5, 9, and 10, with replenishment of doxycycline after 48 h.
In vivo and in vitro ubiquitylation assays.
For in vivo ubiquitylation assays, cells were transfected with HA-ubiquitin and other expression plasmids as indicated in Fig. 2, 6, and 7. For denaturing immunoprecipitation, cell pellets were lysed in SDS lysis buffer (1% SDS, 5 mM EDTA, 1 mM PMSF, 10 mM 2-mercaptoethanol, DNase I [15 U/ml], aprotinin [10 μg/ml], leupeptin [10 μg/ml]) by consecutive vigorous vortexing and boiling. After the addition of 9 volumes of a solution containing 150 mM NaCl, 0.5% Triton X-100, 50 mM Tris-HCl (pH 7.5), 1 mM PMSF, aprotinin (10 μg/ml), and leupeptin (10 μg/ml), lysates were cleared of debris by centrifugation, and the resulting supernatants were mixed with either anti-FLAG (M2)-agarose affinity gel (Sigma) or protein G (GE Healthcare) and the antibodies indicated above. Ubiquitin conjugates were detected with anti-HA, anti-FLAG, anti-p53, or anti-DDX24.
FIG 2.
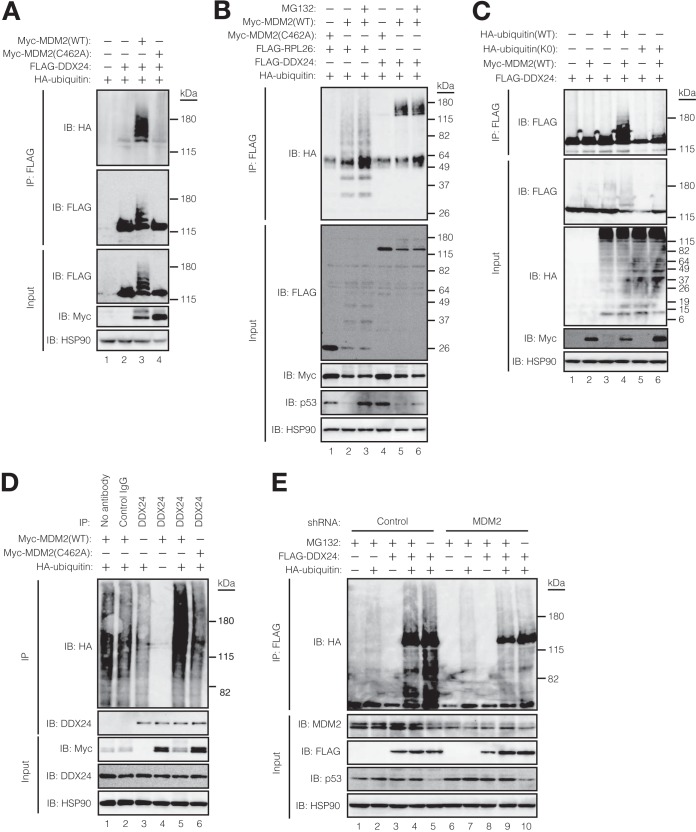
MDM2 promotes DDX24 ubiquitylation in vivo. (A) DDX24 ubiquitylation is upregulated by MDM2 in vivo. HCT116 cells were transfected with the indicated combinations of expression vectors for FLAG-DDX24, Myc epitope-tagged mouse MDM2 (WT or C462A), and HA-ubiquitin. The cells were then subjected to extraction with SDS lysis buffer under denaturing conditions. A portion (15%) of each extract (input) was subjected to immunoblot analysis with the indicated antibodies, whereas the remainder was subjected to immunoprecipitation with anti-FLAG followed by immunoblot analysis with anti-HA and anti-FLAG. (B) MDM2 does not promote the proteasomal degradation of polyubiquitylated DDX24. HCT116 cells were transfected with the indicated combinations of expression plasmids, incubated with or without the proteasome inhibitor MG132 (10 μM) for 3 h, and then subjected to extraction, immunoprecipitation, and immunoblot analysis with the indicated antibodies as in panel A. FLAG-RPL26 was included as a positive control for MDM2-mediated polyubiquitylation. (C) MDM2 induces polyubiquitylation of DDX24 in vivo. HCT116 cells were transfected with the indicated combinations of expression vectors for FLAG-DDX24, Myc-MDM2(WT), and HA-ubiquitin (WT or K0), after which the cells were subjected to extraction, immunoprecipitation, and immunoblot analysis with the antibodies as indicated in panel A. (D) Exogenous MDM2 promotes polyubiquitylation of endogenous DDX24. HCT116 cells were transfected with the indicated combinations of expression vectors for HA-ubiquitin, Myc-MDM2(WT), and Myc-MDM2(C462A), after which the cells were subjected to extraction, immunoprecipitation with anti-DDX24 or control immunoglobulin G (IgG), and immunoblot analysis with the indicated antibodies as for panel A. (E) Knockdown of endogenous MDM2 attenuates the polyubiquitylation of exogenous DDX24. HCT116 cells were infected with lentiviral vectors encoding shRNAs for LacZ (control) or MDM2, transfected with the indicated expression plasmids, and incubated with or without 10 μM MG132 for 3 h. The cells were then subjected to extraction, immunoprecipitation, and immunoblot analysis with the indicated antibodies as described for panel A.
FIG 6.
Ribosomal proteins inhibit MDM2-DDX24 interaction and suppress ubiquitylation of DDX24 by MDM2. (A) Knockdown of DDX24 reduces the extent of p53 ubiquitylation. HCT116 cells infected and treated with doxycycline as described for Fig. 5A were transfected with expression plasmids for the indicated proteins and then subjected to extraction with SDS lysis buffer under denaturing conditions, followed by immunoprecipitation with anti-FLAG. The resulting precipitates, as well as the original cell extracts (input), were subjected to immunoblot analysis with the indicated antibodies. (B) Knockdown of DDX24 does not affect the MDM2-p53 interaction. HCT116 cells infected and treated with doxycycline as described for Fig. 5A were lysed and subjected to immunoprecipitation with anti-p53 or control immunoglobulin G, and the resulting precipitates were subjected to immunoblot analysis with the indicated antibodies (left), followed by quantification of the relative MDM2/p53 band intensity ratio (right). (C) Effects of RPs and DDX24 on MDM2-mediated p53 ubiquitylation. HCT116 cells transfected with the indicated combinations of expression plasmids were subjected to extraction, immunoprecipitation, and immunoblot analysis with the indicated antibodies as described for panel A. (D) Inhibition of MDM2-mediated polyubiquitylation of DDX24 by RPs. HCT116 cells transfected with the indicated combinations of expression vectors were subjected to extraction, immunoprecipitation, and immunoblot analysis with the indicated antibodies as described for panel A. (E) RPs inhibit the MDM2-DDX24 interaction. Lysates of HCT116 cells expressing FLAG-MDM2 and Myc epitope-tagged RPs were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as the original cell lysates (input), were subjected to immunoblot analysis with the indicated antibodies. (F) Depletion of endogenous DDX24 does not suppress actinomycin D-induced p53 activation. HCT116 cells infected and treated with doxycycline as described for Fig. 5A were incubated in the absence (–) or presence (+) of 5 nM actinomycin D (ActD) for 8 h, lysed, and subjected to immunoblot analysis with the indicated antibodies.
FIG 7.
Polyubiquitylation of DDX24 promotes its binding to pre-rRNP complexes. (A) Alignment of ubiquitin-interacting motifs (UIMs) of the indicated human proteins with the region of human NIP7 containing a putative UIM. Positions that are invariant or conserved among many UIMs are shaded black or gray, respectively. (B) In vitro binding of ubiquitylated DDX24 to NIP7. GST-tagged WT or mutant (A55G or Δ54–59) forms of human NIP7 bound to glutathione-conjugated beads were incubated with purified mono- or polyubiquitylated FLAG-DDX24 (generated with methylated or WT ubiquitin, respectively, as described for Fig. 3C). The beads were then precipitated and subjected to immunoblot analysis with anti-FLAG. SDS-PAGE and Coomassie brilliant blue staining of GST and GST fusion proteins used in these assays are also shown. (C) In vivo binding of ubiquitylated DDX24 to NCL. HCT116 cells transfected with the indicated combinations of expression vectors for HA-ubiquitin (WT or K0), MDM2, FLAG-DDX24, and GFP-tagged NCL were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as the original cell lysates, were subjected to immunoblot analysis with the indicated antibodies. (D) In vivo binding of ubiquitylated DDX24 to NIP7. HCT116 cells transfected with the indicated combinations of expression vectors were subjected to immunoprecipitation and immunoblot analysis as described for panel C.
For in vitro ubiquitylation assays, reaction mixtures (20 μl) containing 0.1 μg of E1 (UBE1; LifeSensors), 0.2 μg of the appropriate E2 enzyme (LifeSensors), 1.0 μg of MDM2, 1.0 μg of the appropriate substrate, 20 μg of ubiquitin (Sigma), ubiquitin mutants (LifeSensors), or methylated ubiquitin (37), and reaction buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2 mM ATP, 2 mM dithiothreitol) were incubated at 37°C for 2 h. The reaction was terminated by the addition of SDS sample buffer and boiling, and one-half of the reaction mixture was subjected to immunoblot analysis with anti-FLAG or anti-DDX24.
Ribosome fractionation.
Sucrose density gradient fractionation of ribosomes was performed as described previously (38). Cells were cultured to 50% confluence. After the addition of cycloheximide (100 μg/ml), cells (5 × 107) were lysed in 500 μl of polysome buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 1% Triton X-100, cycloheximide [100 μg/ml]), incubated on ice for 5 min, and centrifuged at 20,000 × g for 10 min at 4°C. The resulting supernatants were applied to a 15 to 47% (wt/vol) sucrose gradient in polysome buffer and centrifuged at 274,000 × g for 4 h at 4°C. Fractionation was evaluated by measurement of absorbance at 254 nm with the use of an EM-1 UV monitor (Bio-Rad). Proteins and RNAs were isolated from the gradient fractions by chloroform-methanol or phenol-chloroform extraction, respectively.
Metabolic labeling of nascent rRNA.
Cells were incubated first for 45 min in phosphate-free DMEM (Life Technologies) supplemented with 10% FBS and doxycycline (1 μg/ml) and then for 45 min in the additional presence of [32P]orthophosphate (15 μCi/ml). The cells were then returned to normal DMEM supplemented with 10% FBS and doxycycline (1 μg/ml) and cultured for 0, 120, or 240 min. Total RNA was extracted from the cells with the use of Isogen and fractionated by electrophoresis on an agarose-formaldehyde gel. The intensities of bands detected by autoradiography were quantitated with the use of ImageJ software (National Institutes of Health).
Northern blot analysis.
Total RNA was extracted from cells with the use of Isogen, and portions (5 μg) of the RNA were fractionated by electrophoresis through a 1% agarose gel containing formaldehyde and MOPS (morpholinepropanesulfonic acid) buffer. The separated RNA molecules were transferred to a Biodyne B nylon filter (Pall) and subjected to hybridization with a 32P-end-labeled DNA oligonucleotide targeted to ITS1 (5′-AAGGGGTCTTTAAACCTCCGCGCCGGAACGCGCTAGGTAC-3′), to ITS2 (5′-CGGGAACTCGGCCCGAGCCGGCTCTCTCTTTCCCTCTCCG-3′), or to 5.8S rRNA (5′-GCGTTCGAAGTGTCGATGATCAATGTGTCCTGCAATTCAC-3′) (38). The intensity of bands detected by autoradiography was quantitated with the use of ImageQuant TL software (GE Healthcare).
Immunoprecipitation analysis of rRNA association with DDX24.
HCT116 cells expressing FLAG-DDX24 were subjected to immunoprecipitation with anti-FLAG. The rRNA species that coimmunoprecipitated with DDX24 were then identified by Northern blotting with the ITS1 probe as described above. The intensity of bands detected by autoradiography was quantitated with the use of ImageQuant TL software (GE Healthcare).
Immunofluorescence analysis.
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 10 min, permeabilized with 0.1% saponin, and exposed to 5% bovine serum albumin before being immunostained with anti-HA and Alexa Fluor 488-conjugated goat antibodies to mouse immunoglobulin G (Invitrogen). Samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vector Laboratories). Images were acquired with a Zeiss LSM 510 laser-scanning inverted confocal microscope.
RESULTS
DDX24 interacts with the central region of MDM2.
To identify proteins that are physically associated with MDM2 in cells, we transiently transfected HCT116 cells with an expression vector for FLAG epitope-tagged mouse MDM2 and then subjected cell lysates to immunoprecipitation with anti-FLAG. Immunoprecipitated proteins were eluted from the antibody-coated beads with the FLAG peptide and subjected to SDS-PAGE. Mass spectrometry (MS) of the fractionated proteins identified 163 molecules that were detected in each of three replicate experiments. Functional categorization of these MDM2-associated proteins with GO annotations revealed that a substantial proportion (42 proteins) were related to nucleolar localization (Table 1), consistent with the previous observation that MDM2 binds to certain nucleolar proteins (39–42). Among the identified MDM2-interacting proteins, we focused on the DEAD-box RNA helicase DDX24, given that several other nucleolar DEAD-box proteins were known to participate in ribosome biogenesis (28, 29).
TABLE 1.
Proteins identified by LC-MS/MS analysis of tryptic peptides derived from endogenous proteins captured by FLAG-MDM2 expressed in HCT116 cells
| Protein function(s) and name | UniProt accession no. | Yeast ortholog | Domain(s)a | No. of peptides |
|---|---|---|---|---|
| DNA repair | ||||
| HMGB1 | P09429 | NHP6B | HMG box | 4 |
| HMGB2 | P26583 | NHP6B | HMG box | 3 |
| XRCC5 | P13010 | YKU80 | Leucine zipper, Ku | 2 |
| Protein and RNA metabolism | ||||
| RPL11 | P62913 | RPL11A | 10 | |
| RPL3 | P39023 | RPL3 | 4 | |
| RPL9 | P32969 | RPL9B | 3 | |
| RPS10 | P46783 | RPS10A | 3 | |
| RPS14 | P62263 | RPS14A | 4 | |
| RPS19 | P39019 | RPS19A | 3 | |
| RPS2 | P15880 | RPS2 | S5 DRBM | 6 |
| RPS25 | P62851 | RPS25A | 3 | |
| RPS3A | P61247 | RPS1A | 9 | |
| RPS6 | P62753 | RPS6A | 7 | |
| RPS7 | P62081 | RPS7A | 8 | |
| RPS9 | P46781 | RPS9A | S4 RNA binding | 5 |
| RNA metabolism, splicing, processing | ||||
| DDX24 | Q9GZR7 | MAK5 | Helicase | 12 |
| WDR77 | Q9BQA1 | PFS2 | WD repeats | 14 |
| HNRNPF | P52597 | RRM | 4 | |
| HNRNPH1 | P31943 | RRM | 6 | |
| HNRNPK | P61978 | PBP2 | RGG box, KH | 6 |
| RBM10 | Q6PKH5 | RRM, G patch, zinc finger | 18 | |
| SNRNP200 | O75643 | BRR2 | Helicase, SEC63, coiled coil | 8 |
| GRWD1 | Q9BQ67 | RRB1 | WD repeats | 4 |
| NCL | P19338 | NSR1 | RRM | 9 |
| TRMT10C | Q7L0Y3 | TRM10 | Coiled coil | 7 |
| Protein metabolism | ||||
| CCT2 | P78371 | CCT2 | 10 | |
| CCT4 | P50991 | CCT4 | 7 | |
| CCT5 | P48643 | CCT5 | 6 | |
| PSMC2 | P35998 | RPT1 | 2 | |
| TCP1 | P17987 | TCP1 | 10 | |
| Lipid metabolism: HADHA | P40939 | EHD3 | 30 | |
| Signal transduction, phosphorylation, ubiquitylation | ||||
| NPM1 | P06748 | 6 | ||
| TP53 | P04637 | DNA binding | 9 | |
| YWHAB | P31946 | BMH2 | 6 | |
| YWHAQ | P27348 | BMH2 | 6 | |
| CDK9 | P50750 | CTK1 | Protein kinase | 4 |
| STK38 | Q15208 | CBK1 | Protein kinase, AGC kinase | 14 |
| STK38L | B4E3J8 | CBK1 | Protein kinase, AGC kinase | 8 |
| PIP4K2C | Q8TBX8 | MSS4 | PIPK | 4 |
| TRIM21 | P19474 | B30.2/SPRY, zinc finger, coiled coil | 9 | |
| Regulation of transcription: THRAP3 | Q9Y2W1 | 10 | ||
| Cell adhesion: EZR | P15311 | FERM | 3 |
HMG, high mobility group; DRBM, double-stranded RNA binding motif; RRM, RNA recognition motif; KH, K homology; PIPK, phosphatidylinositol phosphatase kinases; FERM, four-point-one, ezrin, radixin, moesin.
To confirm the interaction between MDM2 and DDX24 in mammalian cells, we transfected HCT116 cells with expression vectors both for FLAG-tagged human DDX24 and for HA-tagged mouse MDM2 and then subjected cell lysates to immunoprecipitation with anti-FLAG. Immunoblot analysis of the resulting precipitates with anti-HA and anti-FLAG revealed the specific interaction between FLAG-DDX24 and HA-MDM2 (Fig. 1A). Endogenous DDX24 was also present together with FLAG-MDM2 in immunoprecipitates prepared with anti-FLAG (Fig. 1B). To examine whether DDX24 and MDM2 interact directly in vitro, we performed a pulldown assay. Recombinant human MDM2 tagged with GST at its NH2 terminus was produced in bacteria and tested for its ability to bind to recombinant His6-tagged human DDX24 also produced in bacteria (Fig. 1C). GST-MDM2 indeed interacted with His6-DDX24 in vitro, whereas GST alone did not, consistent with our observations in vivo. Furthermore, a series of deletion mutants of MDM2 (Fig. 1D) that were tagged with FLAG at their NH2 termini were expressed in HCT116 cells and subjected to a coimmunoprecipitation assay for endogenous DDX24. All MDM2 constructs that contained the region encompassing amino acids 221 to 272 were found to associate with DDX24, whereas those lacking this region did not (Fig. 1E), suggesting that this region is essential for DDX24 binding. This region is located within the central acidic domain of MDM2, which is required for its ability to target p53 for degradation and also overlaps sequences that are required for interaction with a variety of regulatory factors, including the tumor suppressor ARF as well as multiple ribosomal or nucleolar proteins (43).
FIG 1.
Identification of DDX24 as a protein that interacts with MDM2. (A) Exogenous MDM2 interacts with exogenous DDX24 in vivo. Lysates of HCT116 cells transfected with vectors for HA-tagged mouse MDM2 and FLAG-tagged human DDX24 (or the empty vector) were subjected to immunoprecipitation (IP) with anti-FLAG, and the resulting precipitates, as well as the original cell lysates (input), were subjected to immunoblot analysis (IB) with antibodies to HA and to FLAG. (B) Exogenous MDM2 interacts with endogenous DDX24 in vivo. Lysates of HCT116 cells expressing FLAG-tagged mouse MDM2 were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as the original cell lysates, were subjected to immunoblot analysis with antibodies to DDX24 and to FLAG. (C) MDM2 interacts with DDX24 in vitro. Bacterially expressed recombinant GST or GST-tagged human MDM2 bound to glutathione-conjugated beads was incubated with His6-tagged human DDX24. The beads were then precipitated, with half being subjected to immunoblot analysis with anti-DDX24. The other half was subjected to SDS-PAGE followed by staining with Coomassie brilliant blue (CBB) to visualize the recombinant proteins. (D) Schematic representation of WT and deletion mutants of mouse MDM2. The mutants contained a C462A substitution that abolishes E3 activity. The ability of the mutants to bind to DDX24 or to p53, as determined in panel E, is indicated, as are the regions of MDM2 found to be required for such binding. NLS, nuclear localization sequence; NES, nuclear export sequence. (E) DDX24 preferentially binds to the acidic domain of MDM2. Lysates of HCT116 cells expressing FLAG-tagged deletion mutants of MDM2 shown in panel D were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as the original cell lysates, were subjected to immunoblot analysis with antibodies to DDX24, to p53, to FLAG, or to HSP90 (loading control).
DDX24 is polyubiquitylated by MDM2 but not degraded by the proteasome.
Given that MDM2 is an E3 ubiquitin ligase that targets p53 for degradation, we hypothesized that DDX24 is also a substrate of this enzyme. To test this possibility, we transfected HCT116 cells with vectors for FLAG-DDX24 and HA-ubiquitin together with a vector for Myc epitope-tagged WT MDM2 or an E3 activity-defective mutant in which Cys462 is replaced with Ala (C462A). Expression of Myc-MDM2(WT), but not that of Myc-MDM2(C462A), resulted in the appearance of a slowly migrating ladder of bands reactive with both anti-HA and anti-FLAG, suggesting that MDM2 markedly increased the extent of FLAG-DDX24 ubiquitylation (Fig. 2A and B). The abundance of Myc-MDM2(WT) was lower than that of Myc-MDM2(C462A), most likely as a result of the autoubiquitylation and rapid degradation of the former protein. The higher-molecular-weight conjugates detected in the presence of HA-ubiquitin were not observed with an HA-tagged lysine-less (K0) mutant of ubiquitin (Fig. 2C), suggesting that DDX24 was polyubiquitylated. We next confirmed that forced expression of WT MDM2, but not that of the C462A mutant, also increased the ubiquitylation of endogenous DDX24 (Fig. 2D). Conversely, shRNA-mediated depletion of endogenous MDM2 resulted in a marked decrease in the level of FLAG-DDX24 ubiquitylation (Fig. 2E). We also examined whether DDX24 is a direct substrate of MDM2 in vitro. MDM2 and GST- and FLAG-tagged DDX24 were expressed in bacteria, purified, and subsequently applied to ubiquitylation assays (Fig. 3A). Only in the presence of all the reaction components, including UBE1 (E1), UBE2D2 (E2), MDM2 (E3), ubiquitin, and DDX24 (substrate), was MDM2 able to ubiquitylate DDX24 directly. These results indicated that MDM2 serves as an E3 ligase for DDX24.
FIG 3.
MDM2 mediates polyubiquitylation of DDX24 in vitro. (A) In vitro ubiquitylation of DDX24 by MDM2. Recombinant human MDM2 was assayed for ubiquitylation activity with GST-FLAG-DDX24 as the substrate in the absence (–) or presence (+) of the indicated reaction mixture components. Each reaction was stopped after 2 h at 37°C by the addition of SDS sample buffer, and the mixture was subjected to immunoblot analysis with anti-FLAG. The positions of unmodified GST-FLAG-DDX24 and of GST-FLAG-DDX24 conjugated with ubiquitin (Ub) are indicated. (B) In vitro ubiquitylation assays with a panel of representative E2s. Recombinant human MDM2 was assayed for ubiquitylation activity in the presence of E1 (UBE1), ubiquitin, and a panel of E2s and with GST-FLAG-p53 (left) or GST-FLAG-DDX24 (right) as the substrate. Each reaction mixture was subjected to immunoblot analysis with antibodies to FLAG (left) or to DDX24 (right). −, no E2. (C) MDM2 mediates polyubiquitylation of DDX24 in vitro. Recombinant human MDM2 was assayed for ubiquitylation activity in the presence of E1 (UBE1), E2 (UBE2D), and either WT or methylated (Me) ubiquitin and with GST-FLAG-p53 (left) or GST-FLAG-DDX24 (right) as the substrate. Each reaction mixture was subjected to immunoblot analysis with anti-FLAG. The positions of unmodified, monoubiquitylated (MonoUb), and polyubiquitylated (PolyUb) substrates are indicated. (D) MDM2 and UBE2D mediate DDX24 polyubiquitylation with all single-lysine-substitution mutants of ubiquitin. Recombinant human MDM2 was assayed for ubiquitylation activity in the presence of E1 (UBE1), E2 (UBE2D), and either WT ubiquitin, ubiquitin mutants (in which the lysine residue at position 6, 11, 27, 29, 33, 48, or 63 was replaced with arginine), or methylated (Me) ubiquitin and with GST-FLAG-p53 (left) or GST-FLAG-DDX24 (right) as the substrate. Each reaction mixture was analyzed as described for panel B.
E3 ligases of the RING finger family rely on a ubiquitin-conjugating enzyme (E2) for the transfer of ubiquitin to substrate proteins. E2s are largely responsible for selection of the lysine residue to which the ubiquitin chain will be attached, and they therefore directly control the fate of the substrate (44, 45). We examined which E2s are able to mediate the conjugation of ubiquitin to DDX24 in cooperation with MDM2. A panel of E2s was tested for the ability to support the MDM2-mediated ubiquitylation of DDX24 as well as that of p53 as a positive control (46) (Fig. 3B). We found that both p53 and DDX24 were ubiquitylated by MDM2 in the presence of UBE2L3 or members of the UBE2D or UBE2E family, suggesting that MDM2 has a broad range of functional E2 partners for ubiquitylation of DDX24 and p53. In the presence of methylated ubiquitin, which can be conjugated to substrates but cannot serve as a site for further ubiquitin conjugation (37), p53 underwent oligoubiquitylation (Fig. 3C), suggesting that monoubiquitylation occurred on several lysine residues, as previously described (46). In contrast, only a monoubiquitylated form of DDX24 was detected in the presence of methylated ubiquitin (Fig. 3C), suggesting that MDM2 mediates DDX24 polyubiquitylation on a single lysine residue.
We next determined the type of isopeptide linkage in the polyubiquitin chain formed on DDX24 with the use of ubiquitin mutants in which the lysine residues at position 6, 11, 27, 29, 33, 48, or 63 are individually replaced with arginine. All seven ubiquitin mutants supported the polyubiquitylation of DDX24 and p53 in cooperation with UBE2D2 and MDM2 (Fig. 3D), suggesting that MDM2 has the ability to form polyubiquitin chains containing all possible isopeptide linkages and does not show any preference for lysine linkages in vitro.
Given that MDM2 targets p53, HIPK2, and RPL26 for proteasomal degradation (47, 48), we examined whether MDM2-mediated polyubiquitylation of DDX24 affects the stability of the protein. Treatment of HCT116 cells with the proteasome inhibitor MG132 did not affect the abundance of endogenous DDX24, whereas it resulted in a marked increase in the amount of endogenous p53 (Fig. 4A). Consistently with this finding, MG132 did not affect the level of polyubiquitylated DDX24 (Fig. 2B and E). Furthermore, whereas expression of FLAG-MDM2(WT) in HCT116 cells induced the degradation of p53 in a concentration-dependent manner, it had no effect on DDX24 abundance (Fig. 4B). In contrast, expression of FLAG-MDM2(C462A) resulted in an increase in the steady-state level of endogenous p53, suggestive of a dominant negative effect of this mutant on p53 stability. The half-life of p53 determined with a cycloheximide chase assay was reduced by expression of FLAG-MDM2(WT) but increased by that of FLAG-MDM2(C462A) (Fig. 4C). In contrast, DDX24 was found to be a highly stable protein, with a half-life of >15 h, and its stability was not affected by expression of either FLAG-MDM2(WT) or FLAG-MDM2(C462A). We also examined the effect of shRNA-mediated depletion of endogenous MDM2 on the steady-state level of endogenous DDX24 (Fig. 4D). Whereas depletion of MDM2 in HCT116 cells resulted in the accumulation of p53 in proportion to the extent of depletion, it had no effect on the amount of DDX24. A cycloheximide chase assay confirmed that the stability of DDX24 was not affected by depletion of MDM2 (Fig. 4E). Collectively, these data indicated that MDM2-mediated polyubiquitylation of DDX24 does not target the protein for proteasomal degradation.
FIG 4.
MDM2-mediated polyubiquitylation does not target DDX24 for proteasomal degradation. (A) MG132 treatment does not affect the abundance of endogenous DDX24. HCT116 cells were incubated in the absence (–) or presence (+) of 10 μM MG132 for 4.5 h, after which cell lysates were subjected to immunoblot analysis with the indicated antibodies. (B) Overexpression of MDM2 does not reduce the steady-state level of endogenous DDX24. HCT116 cells were transfected (or not) with various amounts (1×, 2×, or 4×) of an expression vector for FLAG-tagged WT or C462A mutant forms of mouse MDM2 (or with the empty vector), after which cell lysates were subjected to immunoblot analysis with the indicated antibodies. (C) Overexpression of MDM2 does not promote DDX24 turnover. HCT116 cells transfected as described for panel B and incubated with cycloheximide (CHX; 100 μg/ml) for the indicated times were subjected to immunoblot analysis of DDX24, p53, FLAG, and α-tubulin (loading control) (top) as well as to subsequent quantification of the amount of DDX24 and p53 (bottom). The half-life of p53 in the transfected cells is indicated. (D) RNAi-mediated depletion of MDM2 does not result in accumulation of DDX24. HCT116 cells were infected (or not) with lentiviral vectors encoding shRNAs specific for LacZ (control) or MDM2 (MDM2-1 or MDM2-2), after which cell lysates were subjected to immunoblot analysis with the indicated antibodies. (E) Ablation of MDM2 does not affect the stability of DDX24. HCT116 cells infected as described for panel D and incubated with cycloheximide (100 μg/ml) for the indicated times were subjected to immunoblot analysis of DDX24, p53, and MDM2 (top), followed by quantification of the amount of DDX24 and p53 (bottom). The half-lives of DDX24 and p53 in the infected cells are indicated.
DDX24 is essential for proper pre-rRNA processing.
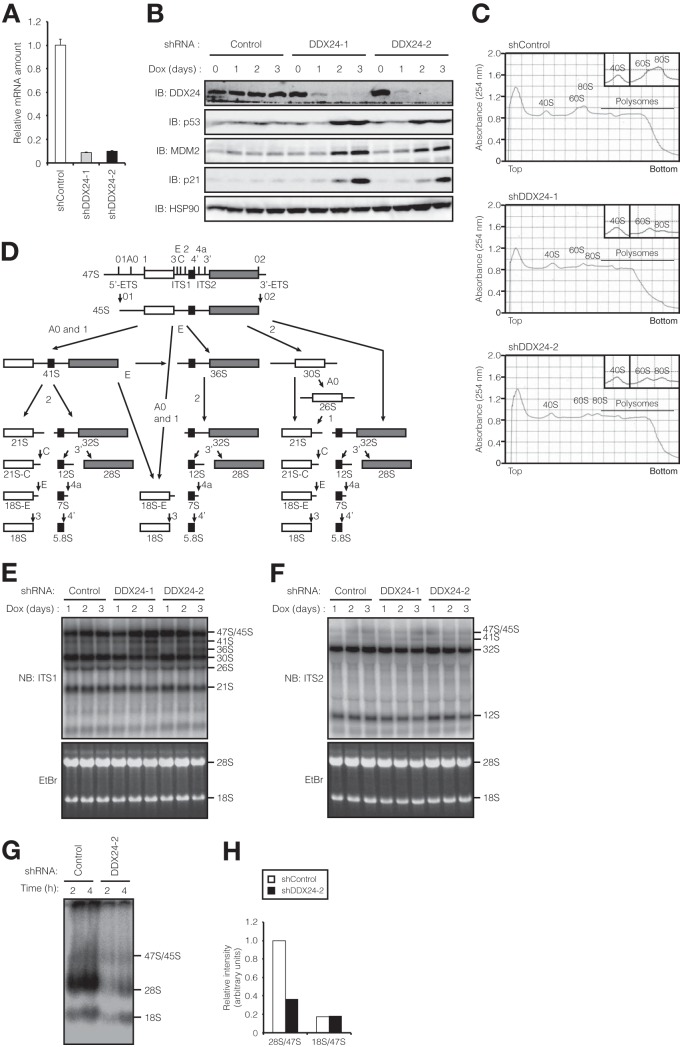
To uncover the role of DDX24, we depleted HCT116 cells of this protein with the use of a tetracycline-induced shRNA expression system (36). Quantitative RT-PCR analysis revealed that the amount of DDX24 mRNA was reduced by up to 95% after shRNA induction by treatment of the cells for 3 days with doxycycline (Fig. 5A). Immunoblot analysis also showed a substantial reduction in the abundance of DDX24 protein as early as 1 day after the onset of doxycycline treatment (Fig. 5B). MAK5, the yeast ortholog of human DDX24, was previously shown to be required for biosynthesis of the 60S ribosomal subunit (30). To examine whether depletion of DDX24 affected the translation machinery in human cells, we subjected extracts prepared from control or DDX24-depleted HCT116 cells to sucrose density gradient centrifugation in order to generate polysome profiles. DDX24-depleted cells manifested a marked reduction in the abundance of the 60S ribosomal subunit as well as an overall reduction in the extent of ribosome synthesis (Fig. 5C), with the latter effect likely being secondary to the 60S subunit deficiency. We next examined whether DDX24 contributes to pre-rRNA processing by monitoring the steady-state levels of processing intermediates in DDX24-depleted cells with the use of Northern blot analysis. The boundaries of most pre-rRNA intermediates relevant to this work have already been mapped (22, 49) (Fig. 5D). Sequences in the internal transcribed spacer 1 (ITS1) and ITS2 sites located immediately upstream of cleavage sites 2 and 3′, respectively, were used as probes. Depletion of DDX24 reduced the abundance of 32S, 30S, 21S, and 12S pre-rRNAs and slightly increased that of 41S and 36S pre-rRNAs, whereas the amount of 47S/45S precursor forms remained unaffected (Fig. 5E and F). A reduction in the amount of 30S pre-rRNA and its downstream products was previously shown to occur in situations in which processing at site 2 is defective (22, 49). The slight accumulation of the 41S and 36S pre-rRNAs was reproducible and is consistent with slower processing of site 2 in DDX24-depleted cells, suggesting that DDX24 is required for processing of ITS1 sites. To obtain a more quantitative and dynamic view of the role of DDX24 in pre-rRNA processing, we measured the effects of DDX24 depletion on the accumulation of rRNA species labeled with 32P in vivo. As with our results indicative of a defect in processing of ITS1 site 2, the rate of 28S rRNA production was greatly reduced in DDX24-depleted cells compared with that in control cells, whereas the effect on 18S rRNA synthesis was minimal (Fig. 5G and H). A block in the 18S rRNA pathway due to impaired production of pre-18S intermediates (30S and 21S pre-rRNAs) observed in DDX24-depleted cells is likely compensated for by the production of another pre-18S rRNA intermediate, 18S-E, that can also be generated from 45S pre-rRNA by alternative cleavage at sites A0, 1, and E (22, 49) (Fig. 5D). Collectively, these results suggested that DDX24 plays a role in the proper processing of pre-rRNA.
FIG 5.
Role for DDX24 in regulation of pre-rRNA processing. (A) Quantification of DDX24 mRNA knockdown. Tetracycline repressor (TetR)-expressing HCT116 cells were infected with a lentiviral vector encoding shRNAs specific for LacZ (shControl) or DDX24 (shDDX24-1 or shDDX24-2) and were then incubated with doxycycline (Dox; 1 μg/ml) for 72 h. Total RNA was then extracted from the cells and subjected to RT and real-time PCR analysis of DDX24 mRNA. Data are means ± standard deviations (SD) of results from three independent experiments. (B) Time course of DDX24 protein knockdown in HCT116 cells. Lysates of cells infected as described for panel A and exposed to doxycycline for the indicated times were subjected to immunoblot analysis with the indicated antibodies. (C) Reduced 60S ribosomal subunit levels in DDX24-depleted cells. Extracts of HCT116 cells infected and treated with doxycycline as described for panel A were subjected to sucrose density gradient centrifugation. The absorbance of RNA in the gradient fractions was monitored at 254 nm. The region of each gradient containing 40S, 60S, and 80S subunits is shown expanded in the insets. (D) Schematic representation of human pre-rRNA processing pathways. ETS, external transcribed spacer. (E and F) Impaired pre-rRNA processing in DDX24-depleted cells. HCT116 cells infected and treated with doxycycline as described for panel B were subjected to Northern blot analysis (NB) with probes complementary to sequences immediately upstream of ITS1 site 2 (E) or of ITS2 site 3′ (F). The ethidium bromide (EtBr)-stained gels are also shown. (G) DDX24 depletion inhibits 28S rRNA accumulation. HCT116 cells infected and treated with doxycycline as described for panel A were pulse-labeled with [32P]orthophosphate and then incubated in normal medium for 2 or 4 h. Total RNA was then isolated from the cells and subjected to agarose gel electrophoresis followed by autoradiography. (H) Ratio of the amount of 28S or 18S rRNA to that of 47S pre-rRNA for cells at the 4-h time point in panel G. Values are expressed relative to the 28S/47S ratio for control cells.
Upregulation of p53, MDM2, and p21 was apparent in DDX24-depleted cells (Fig. 5B), suggesting that DDX24 deficiency might prevent p53 degradation by attenuating MDM2-mediated ubiquitylation. To examine this possibility, we monitored the effect of DDX24 depletion on the extent of MDM2-mediated ubiquitylation of p53. Depletion of DDX24 indeed greatly reduced the extent of p53 ubiquitylation (Fig. 6A) without abrogating the interaction between MDM2 and p53 (Fig. 6B), suggesting that DDX24 depletion may give rise to nucleolar stress and consequent inhibition of MDM2 activity through binding of RPs (10, 19). We found that not only MDM2-mediated p53 ubiquitylation (14, 16–18) (Fig. 6C) but also DDX24 ubiquitylation (Fig. 6D) was impaired by RPs. The modes of inhibition appeared to differ, however. RPs formed a complex with MDM2-p53, whereas they inhibited the interaction between MDM2 and DDX24 (Fig. 6E). Nucleolar stress induced by a low concentration (5 nM) of actinomycin D was previously shown to facilitate the binding of RPs to MDM2, and knockdown of these RPs attenuated this interaction as well as p53 accumulation (15–17). Depletion of DDX24 did not affect the actinomycin D-induced accumulation of p53 (Fig. 6F), however, suggesting that DDX24 does not influence the MDM2-p53 axis during nucleolar stress but rather acts downstream of the RP-MDM2 pathway.
Polyubiquitylation of DDX24 by MDM2 promotes its interaction with pre-rRNP complexes.
One of the functions of nonproteolytic ubiquitylation is the recruitment of proteins that harbor ubiquitin binding domains (UBDs) to ubiquitin or polyubiquitin chains (50, 51). Several lines of evidence suggest that human DDX24, nucleolin (NCL), and NIP7 function in association during ribosome biogenesis as part of the same pre-rRNP complexes. These proteins were thus shown to copurify with the processing factor parvulin (Par14) and ribosomal protein S19 (RPS19) (24–27). NCL plays a role in the processing of pre-rRNA (52), and NIP7 is required for processing of ITS1 sites in pre-rRNA (38, 53).
These various observations led us to hypothesize that components of pre-rRNP complexes harboring UBDs might recognize the polyubiquitin chain formed on DDX24 by MDM2. We found that NIP7 harbors a motif similar to a sequence known as the ubiquitin-interacting motif (UIM), which is conserved among members of a specific class of UBDs (Fig. 7A). We thus tested whether the putative UIM in NIP7 shows any preference for monoubiquitylated or polyubiquitylated DDX24. Mono- or polyubiquitylated FLAG-DDX24 generated by MDM2 in vitro in the presence of methylated or WT ubiquitin, respectively, was incubated with bacterially expressed and purified GST-NIP7, and binding was tested with a pulldown assay (Fig. 7B). Although NIP7 associated with both mono- and polyubiquitylated DDX24, it showed a preference for ubiquitin polymers attached to DDX24 and did not interact with nonubiquitylated DDX24. The binding of NIP7 to both mono- and polyubiquitylated DDX24 was markedly reduced in extent when the conserved alanine residue located in the putative UIM at position 55 was replaced with glycine (A55G). Furthermore, the binding was almost completely attenuated when the UIM core region (residues 54 to 59) of NIP7 was deleted (Δ54–59). These results thus suggested that the UIM of NIP7 confers affinity for a polyubiquitin chain formed on DDX24 by MDM2.
To assess the importance of DDX24 polyubiquitylation for pre-rRNP complex formation in vivo, we investigated whether ubiquitylated DDX24 interacts with NCL and NIP7 in HCT116 cells. Coimmunoprecipitation experiments revealed that the interaction of DDX24 with NCL (Fig. 7C) or NIP7 (Fig. 7D) was promoted by MDM2-mediated polyubiquitylation. In contrast, the binding of NCL or NIP7 to DDX24 was greatly attenuated in the presence of the K0 mutant of ubiquitin instead of the WT protein, consistent with the notion that the polyubiquitin chain formed on DDX24 is required for its efficient interaction with NCL or NIP7. The interaction between DDX24 and NIP7 was dependent on the UIM of NIP7 (Fig. 7D), consistent with the results of our in vitro binding assays.
We next examined the possible effect of polyubiquitylation on the binding of DDX24 to rRNA. Complex formation by rRNA species and DDX24 was assessed by immunoprecipitation and Northern blot analysis. We detected a substantial increase in the extent of DDX24 binding to both 30S and 21S pre-rRNAs in cells overexpressing both MDM2 and ubiquitin (Fig. 8A and B). Immunoblot analysis of the distribution of DDX24 among sucrose density gradient fractions also revealed that heavily polyubiquitylated DDX24 cosedimented with preribosomal particles, although a significant amount of Myc-MDM2 and HA-ubiquitin formed a substantial proportion of the polyubiquitylated DDX24 in the soluble fractions (Fig. 8C). To further clarify whether polyubiquitylated DDX24 associates with preribosomal particles, we analyzed pooled pre-40S and 60S subunit-containing fractions from the first gradient on a second gradient. Polyubiquitylated DDX24 again cosedimented at least in part with the preribosomal particles (Fig. 8D), suggesting that polyubiquitylated DDX24 interacts with preribosomal complexes during ribosome synthesis.
FIG 8.
Polyubiquitylated DDX24 associates with pre-rRNA molecules and preribosomal particles. (A) MDM2 promotes the binding of DDX24 to pre-rRNAs. HCT116 cells transfected with the indicated combinations of expression vectors were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as the original cell lysates (input), were subjected to Northern blot analysis with a probe complementary to a sequence upstream of ITS1 site 2. (B) Quantification of coprecipitated pre-rRNAs in experiments similar to those whose results are shown in panel A. The intensities of the pre-rRNA bands (lanes 7 to 10) were quantified and normalized by that of the 30S or 21S bands in lane 7. Data are means ± SD from three independent experiments. (C and D) Polyubiquitylated DDX24 is incorporated into preribosomal particles. (C) Extracts of HCT116 cells expressing FLAG-DDX24, Myc-MDM2, and HA-ubiquitin were subjected to sucrose density gradient centrifugation. Twenty fractions were collected, and protein and RNA molecules extracted from each fraction were subjected to immunoblot analysis with the indicated antibodies or to electrophoresis on a 1% agarose gel and then staining with ethidium bromide, respectively. The fractions containing soluble proteins, monosomes, and polysomes are indicated at the top. (D) Fractions containing pre-40S and pre-60S subunits (fractions 4 to 10) for the gradient shown in panel C were pooled and subjected to a second density gradient centrifugation. Each of the resulting fractions was subjected to immunoblot analysis with the indicated antibodies or to electrophoresis on a 1% agarose gel, followed by staining with ethidium bromide, as described for panel C.
To evaluate the role of ubiquitylated DDX24 in pre-rRNA processing, we monitored the pattern and time course of the appearance of pre-rRNAs and mature 5.8S rRNA after inactivation of both MDM2 and DDX24 or of MDM2 alone. We found that, in both instances, the abundances of 41S, 32S, 30S, 21S, and 12S pre-rRNAs as well as that of 5.8S rRNA correlated well with those observed after depletion of DDX24 alone (Fig. 9), suggesting that pre-rRNA processing is influenced by MDM2-dependent ubiquitylation of DDX24. Although we attempted to elucidate the role of DDX24 ubiquitylation by generating DDX24 mutants that cannot be ubiquitylated, a series of DDX24 mutants in which a cluster of lysines was replaced with arginines still underwent ubiquitylation, probably as a result of an “escape” of ubiquitylation from the original site to alternative neighboring sites. Instead, we generated a mutant form of DDX24 (DDX24-ub) that mimics the ubiquitylated form of the protein by fusion of a ubiquitin moiety to its COOH terminus (Fig. 10A). Direct fusion of a ubiquitin moiety to p53 was previously shown to alter the subcellular distribution of p53 in a manner similar to that apparent after its posttranslational monoubiquitylation (54). Indeed, immunostaining revealed a cytoplasmic localization for p53-ub, whereas p53, DDX24, and DDX24-ub were localized predominantly to the nucleus (Fig. 10B). After verification by immunoblot analysis that DDX24-ub gave rise to both mono- and polyubiquitylated forms in HCT116 cells (Fig. 10C), we expressed the mutant protein in cells subjected to depletion of both MDM2 and DDX24. Quantitation of pre-rRNA transcripts after 3 days of doxycycline treatment showed that DDX24-ub expression prevented the abnormality in the abundance of 30S pre-rRNA and that of its downstream product, 21S pre-rRNA, observed in the double-knockdown cells (Fig. 10D to I). Although it is unclear how well expression of the DDX24-ub construct mimics the physiological situation, these results suggest that ubiquitylated DDX24 likely contributes to pre-rRNA processing. This rescue of the steady-state levels of pre-rRNA processing intermediates was specific for the ubiquitylated protein, given that expression of an RNAi-resistant form of DDX24 alone did not show this effect (Fig. 10G and I).
FIG 9.
Similarity of pre-rRNA processing phenotypes after depletion of DDX24, MDM2, or both proteins. (A and B) TetR-expressing HCT116 cells were infected with lentiviral vectors for the indicated shRNAs and incubated with doxycycline (1 μg/ml) for the indicated times, after which RNA and protein molecules were extracted from the cells and subjected to Northern blot analysis with ITS1, ITS2, or 5.8S rRNA probes or to immunoblot analysis with the indicated antibodies. (C to J) Quantification of pre-rRNA and rRNA levels after doxycycline treatment for 3 days in Northern blots similar to those shown in panels A and B. Data are expressed relative to the corresponding value for cells expressing control shRNA and are means ± SD from three independent experiments. NS, not significant; *, P < 0.05; **, P < 0.01 (two-tailed Student's t test).
FIG 10.
Role of DDX24 ubiquitylation in pre-rRNA processing. (A) Schematic representation of DDX24-ub and p53-ub, which contain one copy of the human ubiquitin sequence fused to the COOH terminus of the respective human protein. (B) In-frame fusion of ubiquitin does not affect the nuclear localization of DDX24. HCT116 cells expressing the indicated HA-tagged proteins were subjected to immunofluorescence staining with anti-HA (green). Nuclei were also stained with DAPI (blue). Bar, 20 μm. (C) Immunoblot analysis of the DDX24-ub fusion protein. HCT116 cells expressing the indicated HA-tagged proteins were lysed and subjected to immunoblot analysis with the indicated antibodies. (D) Expression of DDX24-ub corrects the pre-rRNA processing defects of MDM2 and DDX24 double-knockdown cells. TetR-expressing HCT116 cells infected with the indicated lentiviral shRNA vectors were transfected with expression plasmids for RNAi-resistant forms of FLAG-tagged DDX24 or DDX24-ub in the presence of doxycycline for the indicated times. The cells were then subjected to Northern blot analysis and to immunoblot analysis as described for Fig. 9A. (E to I) Quantification of pre-rRNA levels after doxycycline treatment for 3 days in Northern blots similar to that in panel D. Data are expressed relative to the corresponding value for cells expressing control shRNA and transfected with the empty vector and are means ± SD from three independent experiments. NS, not significant; *, P < 0.05; **, P < 0.01 (two-tailed Student's t test).
DISCUSSION
We have shown that MDM2 interacts with a member of the DEAD-box family of ATP-dependent RNA helicases. DDX24 binds to the central acidic region of MDM2, and this binding promotes polyubiquitylation of DDX24. Unexpectedly, however, the polyubiquitylation of DDX24 does not elicit its degradation by the proteasome. Instead, the polyubiquitin chain formed on DDX24 promotes interaction with NIP7, a component of pre-rRNP processing complexes, through the UIM domain of the latter protein. DDX24 was found to be required for the early steps of pre-rRNA processing, as is NIP7 (38).
While our study was in progress, DDX24 was found to be required for pre-rRNA processing in HeLa cells, with its depletion in these cells resulting in a reduction in the amounts of 47/45S, 30S, 21S, and 18S-E pre-rRNAs as well as in the accumulation of 26S pre-rRNA (55). The accumulation of 26S pre-rRNA is thought to occur in situations in which the processing at site 1 that gives rise to the 5′ end of 18S rRNA becomes defective and the maturation of 18S rRNA and 40S subunit synthesis are thus impaired, consistent with the observed reduction in the amounts of 21S and 18S-E intermediates. On the other hand, with relation to 60S subunit synthesis, the observed reduction in the amount of 30S pre-rRNA is suggestive of a defect in ITS1 site 2 processing, but the analysis was not sufficient to provide a clear answer in this regard. Collectively, the results of this previous study thus suggest that DDX24 contributes to the processing of site 1 as well as possibly to that of site 2. Our results show that DDX24 is required for the processing of ITS1 site 2 (Fig. 5D to F), for maturation of 28S rRNA (Fig. 5G and H), and for 60S subunit synthesis (Fig. 5C), findings that are not contradictory to those of the previous study. However, we did not observe a reduction in the amount of 18S rRNA (Fig. 5G and H) or in the extent of 40S subunit synthesis (Fig. 5C) in DDX24-depleted cells. In addition, we did not detect a substantial reduction in the amount of 47S/45S pre-rRNA in normally cycling HCT116 cells depleted of DDX24. These differences between the results of the two studies may be attributable to the difference in cell lines or culture conditions, with the operation of alternative pre-rRNA processing pathways or the kinetics of certain cleavage reactions possibly being dependent on cell type, physiological state, or developmental stage (22). Regardless of their differences, however, both studies identify DDX24 as an important regulator of pre-rRNA processing, and this finding is not unique to a single cell line.
MDM2 appears to play two concomitant roles in the regulation of rRNA biosynthesis. First, it promotes the degradation of p53, resulting in increased transcription of 47S pre-rRNA by Pol I (20). Second, MDM2 might contribute to the efficient processing of pre-rRNA intermediates to mature rRNAs partly through modification of DDX24. Impairment of such processing results in nucleolar stress and inhibits MDM2 activity, leading to the arrest both of 47S pre-rRNA transcription through p53 stabilization and of pre-rRNA processing through attenuation of the DDX24 polyubiquitylation. We found that DDX24 overexpression resulted in moderate attenuation of MDM2-p53 interaction (data not shown), whereas it did not affect the level of p53 polyubiquitylation (Fig. 6C). Furthermore, depletion of DDX24 did not affect actinomycin D-induced disruption of the MDM2-p53 interaction and consequent p53 stabilization (Fig. 6F). Although the regulatory mechanisms of DDX24 expression and the biological relevance of its ability to affect the MDM2-p53 interaction remain unknown, this system may constitute a checkpoint to ensure the proper production of rRNA.
Although MDM2 catalyzes polyubiquitylation of p53 and DDX24, it also mediates monoubiquitylation of histones H2A and H2B (56), dihydrofolate reductase (57), and the transcription factor FOXO4 (58) through direct interaction with these proteins. The choice between mono- and polyubiquitylation might be determined by the conformational restriction of the particular substrate-MDM2 complex or by the involvement of monoubiquitylation-specific E2s, such as UBE2W and UBE2T (59). The precise mechanism by which the different types of ubiquitin chain are formed by a single E3 enzyme remains to be uncovered, however.
How does MDM2 catalyze both degradation-associated polyubiquitylation (for p53) and degradation-independent polyubiquitylation (for DDX24)? This capability is likely explained by a difference in the topology of the ubiquitin chains attached to the substrates. Evidence suggests that K11-, K29-, and K48-linked chains are targeting signals for the proteasome (60), and at least four consecutive ubiquitin molecules in the case of a K48-linked chain are necessary for recognition by the proteasome (44, 61). Given that MDM2 is capable of catalyzing the formation of polyubiquitin chains on both DDX24 and p53, which contain all seven possible isopeptide linkages, the polyubiquitin chain conjugated to DDX24 likely includes a mixture of linkages that is not recognized by the proteasome. This in vitro finding is consistent with previous observations that several physiologically important E3s, including a U-box E3 (CHIP) and RING finger E3s (MuRF1 and MDM2), form polyubiquitin chains that contain all seven types of isopeptide linkage and which are also forked (Lys6 plus Lys11, Lys27 plus Lys29, and Lys29 plus Lys33) (62). One possible explanation for the different fates of polyubiquitylated proteins is that cells contain additional auxiliary factors, such as S5a (also known as RPN10), that block the formation of nondegradable polyubiquitin chains to facilitate substrate degradation (62–64). We speculate that DDX24 might escape from these factors by physical or spatial sequestration, resulting in a fate different from that of other substrates, such as p53.
The biochemical reactions underlying ribosome synthesis are mediated by many assembly factors that associate transiently with the pre-rRNP complexes. A variety of complexes are formed during the assembly of RPs, with their composition changing in a hierarchical and stepwise manner (23, 24). There are three major types of complex with regard to RP content (27). The first type contains RPs of both the 40S and 60S subunits but with a higher ratio of 40S proteins, such as the complexes isolated by affinity purification of RPS19 (25). The second type contains RPs of both the 40S and 60S subunits but with a higher ratio of 60S proteins, such as the complexes described for NCL, nucleophosmin, Par14, and NOP56 (26, 65–68). The third type is highly enriched in 60S RPs and includes SBDS and ISG20L2 (69, 70). DDX24 and NIP7 are present specifically together in the RPS19 and Par14 complexes (27), consistent with our observations, suggesting the importance of DDX24 polyubiquitylation in this association. Given that the proposed role of DEAD-box proteins in ribosome biogenesis lies in the remodeling of rRNA-rRNA and rRNA-snoRNA duplexes as well as of the many RNA binding proteins associated with preribosomal particles during assembly, the association of polyubiquitylated DDX24 with pre-rRNP complexes suggests the importance of such remodeling functions in these complexes. Although the precise function of NIP7 remains to be determined, we speculate that the association between the polyubiquitin chain attached to DDX24 and the UIM of NIP7 plays a role in the proper deployment of these factors during the formation of a variety of pre-rRNP complexes. NCL is thought to function as a scaffold for many proteins that participate in rRNA processing at different stages of ribosome biosynthesis (71, 72), and we found that it copurifies selectively with polyubiquitylated DDX24. This observation also supports the possibility that DDX24 polyubiquitylation determines the composition of pre-rRNP complexes. In yeast, the pre-60S processing factor NSA1 is present in both polyubiquitylated and sumoylated forms, with polyubiquitylation and sumoylation of this protein appearing to regulate its dissociation from preribosomes induced by the AAA-type ATPase RIX7 (73–75). In the course of the present study, we noticed that NIP7 is also mono- or diubiquitylated by unknown enzymes in HCT116 cells (unpublished data). These observations suggest that a number of processing factors are modified by nonproteolytic ubiquitylation (50, 60), which may play a role similar to that of sumoylation, which is necessary for efficient ribosome biogenesis and export (76).
ACKNOWLEDGMENTS
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. T.Y. was also supported by a research fellowship of the Japan Society for the Promotion of Science for Young Scientists.
We thank Masatoshi Kitagawa for HCT116 cells; Hiroyuki Miyoshi, Hiromitsu Nakauchi, Yohei Kobayashi, and Shin Yonehara for vectors; Naoko Nishimura, Kayoko Tsunematsu, Koji Oyamada, Emiko Koba, and Mizuho Oda for technical assistance; Akane Ohta for help with preparation of the manuscript; and Katsuyoshi Mihara, Shigeki Furuya, and Kohichi Kawahara for both technical assistance and helpful discussion.
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.Brooks CL, Gu W. 2006. p53 ubiquitination: Mdm2 and beyond. Mol. Cell 21:307–315. 10.1016/j.molcel.2006.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marine JC, Lozano G. 2010. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 17:93–102. 10.1038/cdd.2009.68 [DOI] [PubMed] [Google Scholar]
- 3.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237–1245. 10.1016/0092-8674(92)90644-R [DOI] [PubMed] [Google Scholar]
- 4.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857–860. 10.1038/362857a0 [DOI] [PubMed] [Google Scholar]
- 5.Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299. 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- 6.Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27. 10.1016/S0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- 7.Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387:299–303. 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- 8.Barak Y, Juven T, Haffner R, Oren M. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juven T, Barak Y, Zauberman A, George DL, Oren M. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8:3411–3416 [PubMed] [Google Scholar]
- 10.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol. Cell 40:216–227. 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Lu H. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16:369–377. 10.1016/j.ccr.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. 1994. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 14:7414–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577–587. 10.1016/S1535-6108(03)00134-X [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23:8902–8912. 10.1128/MCB.23.23.8902-8912.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhat KP, Itahana K, Jin A, Zhang Y. 2004. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 23:2402–2412. 10.1038/sj.emboj.7600247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai MS, Lu H. 2004. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279:44475–44482. 10.1074/jbc.M403722200 [DOI] [PubMed] [Google Scholar]
- 17.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. 2004. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24:7654–7668. 10.1128/MCB.24.17.7654-7668.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin A, Itahana K, O'Keefe K, Zhang Y. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24:7669–7680. 10.1128/MCB.24.17.7669-7680.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deisenroth C, Zhang Y. 2010. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 29:4253–4260. 10.1038/onc.2010.189 [DOI] [PubMed] [Google Scholar]
- 20.Grummt I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17:1691–1702. 10.1101/gad.1098503R [DOI] [PubMed] [Google Scholar]
- 21.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359. 10.1007/s00018-008-8027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullineux ST, Lafontaine DL. 2012. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie 94:1521–1532. 10.1016/j.biochi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 23.Staley JP, Woolford JL., Jr 2009. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 21:109–118. 10.1016/j.ceb.2009.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi N, Yanagida M, Fujiyama S, Hayano T, Isobe T. 2003. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22:287–317. 10.1002/mas.10057 [DOI] [PubMed] [Google Scholar]
- 25.Orru S, Aspesi A, Armiraglio M, Caterino M, Loreni F, Ruoppolo M, Santoro C, Dianzani I. 2007. Analysis of the ribosomal protein S19 interactome. Mol. Cell. Proteomics 6:382–393. 10.1074/mcp.M600156-MCP200 [DOI] [PubMed] [Google Scholar]
- 26.Fujiyama-Nakamura S, Yoshikawa H, Homma K, Hayano T, Tsujimura-Takahashi T, Izumikawa K, Ishikawa H, Miyazawa N, Yanagida M, Miura Y, Shinkawa T, Yamauchi Y, Isobe T, Takahashi N. 2009. Parvulin (Par14), a peptidyl-prolyl cis-trans isomerase, is a novel rRNA processing factor that evolved in the metazoan lineage. Mol. Cell. Proteomics 8:1552–1565. 10.1074/mcp.M900147-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simabuco FM, Morello LG, Aragao AZ, Paes Leme AF, Zanchin NI. 2012. Proteomic characterization of the human FTSJ3 preribosomal complexes. J. Proteome Res. 11:3112–3126. 10.1021/pr201106n [DOI] [PubMed] [Google Scholar]
- 28.Bleichert F, Baserga SJ. 2007. The long unwinding road of RNA helicases. Mol. Cell 27:339–352. 10.1016/j.molcel.2007.07.014 [DOI] [PubMed] [Google Scholar]
- 29.Linder P, Jankowsky E. 2011. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12:505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- 30.Zagulski M, Kressler D, Becam AM, Rytka J, Herbert CJ. 2003. Mak5p, which is required for the maintenance of the M1 dsRNA virus, is encoded by the yeast ORF YBR142w and is involved in the biogenesis of the 60S subunit of the ribosome. Mol. Genet. Genomics 270:216–224. 10.1007/s00438-003-0913-4 [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama KI, Nakayama K. 1999. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 18:2401–2410. 10.1093/emboj/18.9.2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. 2003. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 100:10231–10236. 10.1073/pnas.1831009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, Kunkel TA, van Harn T, Xia B, Correll M, Quackenbush J, Livingston DM, Gygi SP, Sicinski P. 2011. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 474:230–234. 10.1038/nature10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125. 10.1038/sj.emboj.7600217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahara-Hanaoka S, Sudo K, Ema H, Miyoshi H, Nakauchi H. 2002. Lentiviral vector-mediated transduction of murine CD34(–) hematopoietic stem cells. Exp. Hematol. 30:11–17. 10.1016/S0301-472X(01)00761-5 [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi Y, Yonehara S. 2009. Novel cell death by downregulation of eEF1A1 expression in tetraploids. Cell Death Differ. 16:139–150. 10.1038/cdd.2008.136 [DOI] [PubMed] [Google Scholar]
- 37.Hershko A, Heller H. 1985. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128:1079–1086. 10.1016/0006-291X(85)91050-2 [DOI] [PubMed] [Google Scholar]
- 38.Morello LG, Hesling C, Coltri PP, Castilho BA, Rimokh R, Zanchin NI. 2011. The NIP7 protein is required for accurate pre-rRNA processing in human cells. Nucleic Acids Res. 39:648–665. 10.1093/nar/gkq758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klibanov SA, O'Hagan HM, Ljungman M. 2001. Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J. Cell Sci. 114:1867–1873 [DOI] [PubMed] [Google Scholar]
- 40.Latonen L, Kurki S, Pitkanen K, Laiho M. 2003. p53 and MDM2 are regulated by PI-3-kinases on multiple levels under stress induced by UV radiation and proteasome dysfunction. Cell. Signal. 15:95–102. 10.1016/S0898-6568(02)00044-X [DOI] [PubMed] [Google Scholar]
- 41.Dai MS, Sun XX, Lu H. 2008. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 28:4365–4376. 10.1128/MCB.01662-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lessard F, Stefanovsky V, Tremblay MG, Moss T. 2012. The cellular abundance of the essential transcription termination factor TTF-I regulates ribosome biogenesis and is determined by MDM2 ubiquitinylation. Nucleic Acids Res. 40:5357–5367. 10.1093/nar/gks198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manfredi JJ. 2010. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 24:1580–1589. 10.1101/gad.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye Y, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10:755–764. 10.1038/nrm2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Wijk SJ, Timmers HT. 2010. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24:981–993. 10.1096/fj.09-136259 [DOI] [PubMed] [Google Scholar]
- 46.Shloush J, Vlassov JE, Engson I, Duan S, Saridakis V, Dhe-Paganon S, Raught B, Sheng Y, Arrowsmith CH. 2011. Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2. J. Biol. Chem. 286:4796–4808. 10.1074/jbc.M110.157669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S. 2007. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell 25:739–750. 10.1016/j.molcel.2007.02.008 [DOI] [PubMed] [Google Scholar]
- 48.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. 2008. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32:180–189. 10.1016/j.molcel.2008.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJ, Watkins NJ. 2013. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 200:577–588. 10.1083/jcb.201207131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen ZJ, Sun LJ. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275–286. 10.1016/j.molcel.2009.01.014 [DOI] [PubMed] [Google Scholar]
- 51.Trempe JF. 2011. Reading the ubiquitin postal code. Curr. Opin. Struct. Biol. 21:792–801. 10.1016/j.sbi.2011.09.009 [DOI] [PubMed] [Google Scholar]
- 52.Ginisty H, Amalric F, Bouvet P. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476–1486. 10.1093/emboj/17.5.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanchin NI, Goldfarb DS. 1999. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302:1972–1975. 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- 55.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol. Cell 51:539–551. 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 56.Minsky N, Oren M. 2004. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell 16:631–639. 10.1016/j.molcel.2004.10.016 [DOI] [PubMed] [Google Scholar]
- 57.Maguire M, Nield PC, Devling T, Jenkins RE, Park BK, Polanski R, Vlatkovic N, Boyd MT. 2008. MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Res. 68:3232–3242. 10.1158/0008-5472.CAN-07-5271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenkman AB, de Keizer PL, van den Broek NJ, Jochemsen AG, Burgering BM. 2008. Mdm2 induces mono-ubiquitination of FOXO4. PLoS One 3:e2819. 10.1371/journal.pone.0002819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 60.Kulathu Y, Komander D. 2012. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13:508–523. 10.1038/nrm3394 [DOI] [PubMed] [Google Scholar]
- 61.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94–102. 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282:17375–17386. 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- 63.Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. 2009. S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 28:1867–1877. 10.1038/emboj.2009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ziv I, Kleifeld O, Glickman M. 2009. Nonconformity in ubiquitin compliance. EMBO J. 28:1825–1827. 10.1038/emboj.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanagida M, Shimamoto A, Nishikawa K, Furuichi Y, Isobe T, Takahashi N. 2001. Isolation and proteomic characterization of the major proteins of the nucleolin-binding ribonucleoprotein complexes. Proteomics 1:1390–1404. [DOI] [PubMed] [Google Scholar]
- 66.Hayano T, Yanagida M, Yamauchi Y, Shinkawa T, Isobe T, Takahashi N. 2003. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 278:34309–34319. 10.1074/jbc.M304304200 [DOI] [PubMed] [Google Scholar]
- 67.Lindstrom MS, Zhang Y. 2008. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem. 283:15568–15576. 10.1074/jbc.M801151200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD. 2008. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell. Biol. 28:7050–7065. 10.1128/MCB.01548-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coute Y, Kindbeiter K, Belin S, Dieckmann R, Duret L, Bezin L, Sanchez JC, Diaz JJ. 2008. ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol. Cell. Proteomics 7:546–559. 10.1074/mcp.M700510-MCP200 [DOI] [PubMed] [Google Scholar]
- 70.Ball HL, Zhang B, Riches JJ, Gandhi R, Li J, Rommens JM, Myers JS. 2009. Shwachman-Bodian Diamond syndrome is a multi-functional protein implicated in cellular stress responses. Hum. Mol. Genet. 18:3684–3695. 10.1093/hmg/ddp316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379–390. 10.1016/0092-8674(89)90241-9 [DOI] [PubMed] [Google Scholar]
- 72.Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. 1998. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 273:19025–19029. 10.1074/jbc.273.30.19025 [DOI] [PubMed] [Google Scholar]
- 73.Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E. 2006. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7:1311–1321. 10.1111/j.1600-0854.2006.00471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kressler D, Roser D, Pertschy B, Hurt E. 2008. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 181:935–944. 10.1083/jcb.200801181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strunk BS, Karbstein K. 2009. Powering through ribosome assembly. RNA 15:2083–2104. 10.1261/rna.1792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finkbeiner E, Haindl M, Muller S. 2011. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30:1067–1078. 10.1038/emboj.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]