Abstract
Iron is an essential micronutrient for all eukaryotic organisms because it participates as a redox-active cofactor in many biological processes, including DNA replication and repair. Eukaryotic ribonucleotide reductases (RNRs) are Fe-dependent enzymes that catalyze deoxyribonucleoside diphosphate (dNDP) synthesis. We show here that the levels of the Sml1 protein, a yeast RNR large-subunit inhibitor, specifically decrease in response to both nutritional and genetic Fe deficiencies in a Dun1-dependent but Mec1/Rad53- and Aft1-independent manner. The decline of Sml1 protein levels upon Fe starvation depends on Dun1 forkhead-associated and kinase domains, the 26S proteasome, and the vacuolar proteolytic pathway. Depletion of core components of the mitochondrial iron-sulfur cluster assembly leads to a Dun1-dependent diminution of Sml1 protein levels. The physiological relevance of Sml1 downregulation by Dun1 under low-Fe conditions is highlighted by the synthetic growth defect observed between dun1Δ and fet3Δ fet4Δ mutants, which is rescued by SML1 deletion. Consistent with an increase in RNR function, Rnr1 protein levels are upregulated upon Fe deficiency. Finally, dun1Δ mutants display defects in deoxyribonucleoside triphosphate (dNTP) biosynthesis under low-Fe conditions. Taken together, these results reveal that the Dun1 checkpoint kinase promotes RNR function in response to Fe starvation by stimulating Sml1 protein degradation.
INTRODUCTION
Ribonucleotide reductase (RNR) is an essential enzyme that catalyzes the de novo synthesis of deoxyribonucleoside diphosphates (dNDPs), which are the precursors for DNA replication and repair. Eukaryotic RNRs are comprised of α and β subunits that form an active quaternary structure, (α2)3(β2)m, where m is 1 or 3. α2, referred to as the large or R1 subunit, contains the catalytic and allosteric sites, and β2, known as the small or R2 subunit, harbors a diferric center that is responsible for generating and keeping a tyrosyl radical required for catalysis (reviewed in references 1 to 3). In the budding yeast Saccharomyces cerevisiae, the large R1 subunit is formed by an Rnr1 homodimer and the small R2 subunit is composed of an Rnr2-Rnr4 heterodimer (reviewed in reference 4). Eukaryotic cells tightly control RNR activity to achieve adequate and balanced deoxyribonucleoside triphosphate (dNTP) pools that ensure accurate DNA synthesis and genomic integrity. In response to DNA damage or DNA replication stress or when cells enter S phase of the cell cycle, the yeast Mec1/Rad53/Dun1 checkpoint kinase cascade activates RNR function (reviewed in reference 4). Briefly, genotoxic stress activates Mec1, which phosphorylates and enhances Rad53 kinase activity (5, 6). A diphosphothreonine motif in hyperphosphorylated Rad53 protein is subsequently recognized by Dun1's forkhead-associated (FHA) domain, leading to Rad53-mediated phosphorylation and activation of Dun1 kinase (7–11), which promotes RNR function through multiple mechanisms. One mechanism involves the transcriptional repressor Crt1, which is hyperphosphorylated by Dun1 kinase and released from the promoter regions of RNR2, RNR3, and RNR4 genes, resulting in transcriptional derepression (12). Another dual-checkpoint-dependent mechanism promotes dissociation of Rnr2-Rnr4 from its nuclear anchor protein, Wtm1, and degradation of the Rnr2-Rnr4 nuclear importer protein Dif1, leading to the redistribution of the small R2 subunit from the nucleus to the cytoplasm, where the large R1 subunit resides (13–17). A third mechanism involves the R1 inhibitor Sml1. The R1 active site is oxidized at each turnover cycle and is subsequently regenerated by a cysteine pair (a CX2C motif) located at the R1 carboxyl terminus. Sml1 has been proposed to hinder R1 active-site regeneration based on the observation that it competes with the R1 carboxyl terminus for interactions with the R1 amino-terminal domain, which includes the active site (18–21). During S phase and in response to genotoxic stress, the Mec1/Rad53/Dun1 kinase cascade facilitates Sml1 phosphorylation, ubiquitylation, and degradation by the 26S proteasome, thereby relieving RNR inhibition (22–25). A multimeric complex that includes the E2 ubiquitin-conjugating enzyme Rad6, the E3 ubiquitin ligase Ubr2, and the E2-E3-interacting protein Mub1 mediates the targeted ubiquitylation and degradation of phosphorylated Sml1 (25). Genotoxic stress also increases Rnr1 protein levels by a Rad53-dependent but Dun1-independent mechanism (26).
Iron is an essential element for the vast majority of living organisms and an indispensable cofactor in all eukaryotic RNRs. The extremely low solubility of Fe3+ at physiological pH frequently leads to human Fe deficiency anemia, especially in pregnant women and children. The budding yeast S. cerevisiae has been employed to characterize the strategies that eukaryotic cells use to properly respond to Fe depletion (reviewed in references 27 to 30). Under normal conditions, yeast cells acquire Fe through low-affinity transporters, including Fet4 (31). When Fe becomes scarce, the yeast Aft1 transcription factor activates the expression of a group of genes known as the Fe regulon, which includes high-affinity Fe uptake systems, such as the plasma membrane Ftr1-Fet3 complex, vacuolar Fe mobilization proteins, and the RNA-binding proteins Cth1 and Cth2 (32–39). Importantly, the Aft1 transcription factor does not respond directly to environmental or intracellular Fe levels, but rather to the efficiency of Fe-S cluster (ISC) synthesis in mitochondria (40). Thus, mutants defective in components of the mitochondrial ISC biogenesis core activate Aft1 and constitutively express the Fe regulon regardless of the Fe concentration, whereas no activation is observed in cells defective in the cytosolic ISC assembly pathway (40–42). Furthermore, two functionally redundant ISC-binding monothiol glutaredoxins, Grx3 and Grx4, play a crucial role in directly transmitting the cellular Fe status to the Aft1 transcription factor (43–45). Under Fe-replete conditions, the Grx3-Grx4 proteins interact with the Aft1 protein, inhibiting its function in transcription by promoting its dissociation from its target promoters (45). In the absence of Grx3, Grx4, or its bound ISC or in the presence of AFT1 alleles insensitive to Grx3-Grx4, such as AFT1-1up, the Fe regulon is constitutively active (38, 43, 44). In addition to its role as an Fe sensor, recent studies have demonstrated that Grx3 and Grx4 also participate in intracellular Fe trafficking by facilitating the delivery of Fe to multiple mitochondrial, cytosolic, and nuclear Fe-dependent proteins, including RNR (46, 47).
In response to Fe deficiency, the two Aft1 targets Cth1 and Cth2 facilitate a metabolic remodeling of Fe-dependent pathways by promoting the coordinated degradation of many mRNAs encoding proteins that participate in processes with elevated Fe demands, such as respiration (35, 36, 48–51). More recent results have shown that yeast cells selectively optimize RNR function in response to low Fe availability by activating the redistribution of Rnr2-Rnr4 from the nucleus to the cytoplasm via a mechanism that is independent of the checkpoint kinases Mec1 and Rad53 (52). Instead, Cth1 and Cth2 RNA-binding proteins specifically interact with the WTM1 transcript, leading to its degradation. The resulting decrease of the nuclear anchor Wtm1 protein favors the redistribution of Rnr2-Rnr4 to the cytoplasm, contributing to the optimal synthesis of dNTPs when Fe is scarce (52).
In addition to RNR, Fe is an essential cofactor for important eukaryotic enzymes required for DNA synthesis, such as DNA polymerases (53), and multiple proteins involved in DNA repair. The relevance of appropriately regulating cellular Fe utilization is highlighted by various studies showing that Fe delivery to enzymes in DNA metabolism is critical to avoid nuclear genome instability, a hallmark of cancer and aging (54–57). Despite the direct link between Fe homeostasis and DNA metabolism, little is known about the mechanisms that regulate these enzymes in response to alterations in Fe availability. In this study, we characterize mechanisms that the Dun1 checkpoint kinase utilizes to promote RNR function in response to Fe deficiency. By using a specific Fe2+ chelator and yeast strains defective in Fe acquisition or Fe sensing, we demonstrate that Dun1 specifically promotes the degradation of the Sml1 protein in response to Fe deficiency. Both genetic and biochemical data reveal the details of this regulation and demonstrate its physiological relevance under Fe-deficient conditions.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The yeast strains used in this study are listed in Table 1. Yeast precultures were incubated overnight at 30°C in synthetic complete (SC) medium lacking specific requirements when necessary and reinoculated at an optical density at 600 nm (OD600) of 0.35. To regulate Fe availability, the cells were incubated for 6 to 8 h in SC medium (Fe-sufficient conditions), SC medium supplemented with 100 μM the Fe2+-specific chelator bathophenanthroline disulfonic acid disodium (BPS) (Fe-deficient conditions), or SC medium supplemented with 300 μM ferrous ammonium sulfate (FAS) (Fe excess conditions) before processing. Treatment with either 0.04% methyl methanesulfonate (MMS), 0.2 M hydroxyurea (HU), or 0.2 mg/liter 4-nitroquinoline 1-oxide (4-NQO) was performed during the last 1 or 2 h of SC medium incubation. To determine Sml1 protein degradation dependence on the 26S proteasome, yeast cells from an overnight preculture in SC medium at 25°C were reinoculated in prewarmed SC medium at 37°C. The cells were incubated for 3 h at the restrictive temperature before addition of 100 μM BPS. Then, the cells were incubated for an additional 6 hours at 37°C. To determine Sml1 protein and FET3 mRNA levels in yeast cells expressing members of the ISC synthesis pathway or GRX4 under the control of the GAL1-10 promoter, cells were precultured overnight at 30°C in SC medium containing 2% galactose (SC-Gal medium) instead of glucose, washed twice, and transferred to SC medium for 40 h to repress the expression of the gene of interest. When necessary, cultures were diluted into fresh SC-Gal medium to maintain the exponential growth phase during the whole process. To address genetic interactions between mutants, cells were tested by spot assays on plates and by growth in 96-well plates. For spot assays, yeast cells were grown to exponential phase, spotted in 5-fold serial dilutions on SC medium plates, and incubated at 30°C. For growth in 96-well plates, yeast cells were inoculated at an A600 of 0.1 in 260 μl of liquid SC medium, and the A600 was determined in a Spectrostar Nano absorbance microplate reader (BMG Labtech) every hour for 72 h at 28°C.
TABLE 1.
Yeast strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| SPY386 | BY4741 fet3::URA3 fet4::KanMX4 | 52 |
| SPY122 | BY4741 cth1::KanMX4 cth2::HisMX6 | 35 |
| AXY1928 | BY4741 dun1::KanMX4 fet3::URA3 fet4::KanMX4 | This study |
| SPY350 | BY4741 dun1::KanMX4 | Invitrogen |
| Y300 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 64 |
| DES460 | Y300 TRP1::GAP-RNR1 | 65 |
| DES459 | Y300 TRP1::GAP-RNR1 mec1::HIS3 | 65 |
| DES453 | Y300 TRP1::GAP-RNR1 rad53::HIS3 | 65 |
| MHY307 | Y300 TRP1::GAP-RNR1 dun1::HIS3 | This study |
| YWO0607 | MATa ura3 leu2-3,112 his3-11,15 Gal+ | D. H. Wolf |
| YWO0608 | YWO0607pre1-1 | D. H. Wolf |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 | Invitrogen |
| SPY485 | BY4743 rad6::KanMX4 | Invitrogen |
| SPY487 | BY4743 ubr2::KanMX4 | Invitrogen |
| SPY496 | BY4743 pep4::KanMX4 | Invitrogen |
| SCY037 | MATa GFP-SML1::HIS3 | 11 |
| SCY049 | MATa dun1::URA3 GFP-SML1::HIS3 | 11 |
| SCY0136 | SCY049 DUN1-TAF-S10A::G418 | 11 |
| SCY0137 | SCY049 DUN1-TAF-S139A::G418 | 11 |
| SCY0138 | SCY049 DUN1-TAF-T380A::G418 | 11 |
| SPY349 | BY4741 SML1-GFP::HISMX4 | Invitrogen |
| W303-1A | MATa ura3-1 ade21 trp1-1 his3-11,15 leu2-3,112 | R. Lill |
| Gal-NFS1 | W303-1A pNFS1::GAL1-10–HIS3 | 74 |
| Gal-YAH1 | W303-1A pYAH1::GAL1-10–LEU2 | 75 |
| Gal-NBP35 | W303-1A pNBP35::GAL1-10–HIS3 | 42 |
| Gal-NAR1 | W303-1A pNAR1::GAL1-10–HIS3 | 76 |
| Gal-GRX4 | W303-1A pGRX4::GAL-L-natNT2 grx3::LEU2 | 46 |
| SPY28 | BY4741 aft1::KanMX4 | Invitrogen |
| BY4742 | MATα his3Δ1 leu2 Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| grx3Δ grx4Δ | BY4742 grx3::LEU2 grx4::KanMX4 | 44 |
| AXY1926 | BY4742 dun1::KanMX4 grx3::LEU2 grx4::KanMX4 | This study |
| AXY1988 | BY4741 pGAL1-NFS1-HIS3 | This study |
| AXY2059 | BY4741 pGAL1-NFS1-HIS3 dun1::KanMX4 | This study |
| SPY589 | BY4741 sml1::KanMX4 | Invitrogen |
| AXY2243 | BY4741 sml1::HIS3 fet3::URA3 fet4::KanMX4 | This study |
| AXY2141 | BY4741 sml1::HIS3 dun1::KanMX4 fet3::URA3 fet4::KanMX4 | This study |
| MHY375 | W303-1A sml1::HIS3 | 13 |
| MHY380 | W303-1A sml1::HIS3 rad53::HIS3 | 13 |
| Plasmids | ||
| pRS415 | CEN LEU1 | 77 |
| pRS416 | CEN URA3 | 77 |
| pSP419 | pRS415-CTH2 | 35 |
| pSP476 | pRS416-CTH1 | 35 |
| pMH80 | CEN LEU2 DUN1 | This study |
| pMH62 | 2μm LEU2 DUN1-D328A | This study |
| pSP673 | pRS414-DUN1-FLAG | 9 |
| pSP674 | pRS414-DUN1-R60A-FLAG | 9 |
| pSP675 | pRS414-DUN1-K100A-R102A-FLAG | 9 |
| p413TEF | CEN HIS3 TEFp CYC1t | 58 |
| pSP684 | p413-DUN1(EcoRI) | This study |
| pSP685 | p413-DUN1-R60A | This study |
| pSP686 | p413-DUN1-K100A-R102A | This study |
| pSP692 | p413-DUN1 | This study |
| pSP693 | p413-DUN1-S10A | This study |
| pSP694 | p413-DUN1-S139A | This study |
| pSP695 | p413-DUN1-T380A | This study |
| pAFT1-1up | pRS313-AFT1-1up | J. Kaplan |
Plasmids.
The plasmids used in this study are listed in Table 1. Plasmids pSP673, pSP674, and pSP675 (9) were used as templates to amplify the DUN1 and DUN1-R60A coding sequences and 240 bp from the promoter region, respectively. These fragments were cloned into the p413TEF vector (58), previously digested with SacI and BamHI to remove the TEF promoter, and generated plasmids pSP684, pSP685, and pSP686. A similar strategy was used to obtain plasmids pSP692, pSP693, pSP694, and pSP695, which express DUN1, DUN1-S10A, DUN1-S139A, and DUN1-T380A, respectively, by using genomic DNA from the yeast strains SCY037, SCY0136, SCY0137, and SCY0138 (11), respectively. Other plasmids used in this work have been described previously (Table 1). All PCR amplifications were performed with the Phusion polymerase (Finnzymes), and cloned inserts were sequenced. Escherichia coli strain DH5α was used for the propagation and isolation of plasmids.
Protein analyses.
Total protein extracts were obtained by using the alkali method (59). Equal amounts of protein were resolved in SDS-PAGE gels and transferred onto nitrocellulose membranes. Ponceau staining was used to assess protein transfer. The primary antibodies used in this study included anti-Sml1 (kindly provided by J. Stubbe), anti-Pgk1 (22C5D8; Invitrogen), anti-Dun1 (kindly provided by W. D. Heyer), and anti-Rnr1. Immunoblots were developed with horseradish peroxidase (HRP)-labeled secondary antibodies and the ECL Select Western blotting detection kit (GE Healthcare Life Sciences). Immunoblot images were obtained with an ImageQuant LAS 4000 mini Biomolecular Imager (GE Healthcare Life Sciences). An image representative of at least three independent biological replicates is always shown. Data were processed and specific signals were quantified with ImageQuant TL analysis software (GE Healthcare Life Sciences). Sml1/Pgk1 protein levels were quantified, and the average and standard deviation of at least three independent biological samples is indicated. The values are percentages of that of the wild-type strain in SC medium.
RNA analyses.
Total RNA extraction and cellular mRNA levels were determined as previously described with a few modifications (60). Briefly, 10 to 20 ml of exponentially growing cells was collected by centrifugation, washed with ice-cold water, and frozen at −80°C. Total RNA was extracted from cell pellets using a Millmix 20 bead beater (Tehtnica) in 0.4 ml of LETS buffer (0.1 M LiCl, 0.01 M EDTA, pH 8.0, 0.01 M Tris-HCl, pH 7.4, and 0.2% [wt/vol] SDS), 0.4 ml of phenol (pH 4.5)-chloroform-chloroform-isoamylic alcohol (125:24:1), and 0.3 ml of glass beads. Supernatants were extracted with phenol-chloroform-isoamylic alcohol (125:24:1) and chloroform-isoamylic alcohol (24:1). RNA was precipitated twice, first by adding 2.5 volumes of 96% ethanol and 0.1 volume of 5 M LiCl and second by adding 2.5 volumes of 96% ethanol and 0.1 volume of 3 M sodium acetate. In both cases, RNA was incubated either at −80°C for 3 h or at −20°C overnight. After dissolving in RNase-free MilliQ water, the RNA concentration was determined with a NanoDrop instrument (Thermo Scientific). Then, 2.5 μg of total yeast RNA was treated for 15 min at 25°C with DNase I RNase-free (Roche) according to the manufacturer's protocol. Maxima Reverse Transcriptase (Thermo Scientific) was used to synthesize cDNA from DNase I-treated RNA according to the manufacturer's recommendations. Quantitative real-time PCR was performed in a Light Cycler 480 II (Roche) using the SYBR Premix Ex Taq kit (TaKaRa) for fluorescent labeling. For this purpose, 2.5 μl cDNA was added to each reaction in a final volume of 10 μl. Real-time PCRs using 0.2 μM the corresponding oligonucleotides were performed under the following conditions: 95°C for 10 s, followed by 40 cycles of 10 s at 95°C and 15 s at 55°C. At the end of the amplification cycles, a melting-curve analysis was conducted to verify the specificity of the reaction. A standard curve was made with serial dilutions of the cDNA sample (2 × 10−1, 1 × 10−1, 2 × 10−2, 1 × 10−2, 2 × 10−3, and 1 × 10−3). Primers FET3-qPCR-F (TGACCGTTTTGTCTTCAGGT) and FET3-qPCR-R (TGACCGTTTTGTCTTCAGGT) were used to determine FET3 mRNA levels, whereas primers ACT1-qPCR-F (TCGTTCCAATTTACGCTGGTT) and ACT1-qPCR-R (CGGCCAAATCGATTCTCAA) were used to determine ACT1 mRNA levels. The data and error bars represent the average and standard deviation of three independent biological samples.
dATP and dCTP measurements.
Cells were processed, and dNTPs were determined by a DNA polymerase-based enzymatic assay, as previously described (52, 61).
RESULTS
Sml1 protein levels decrease in response to nutritional and genetic iron deficiencies.
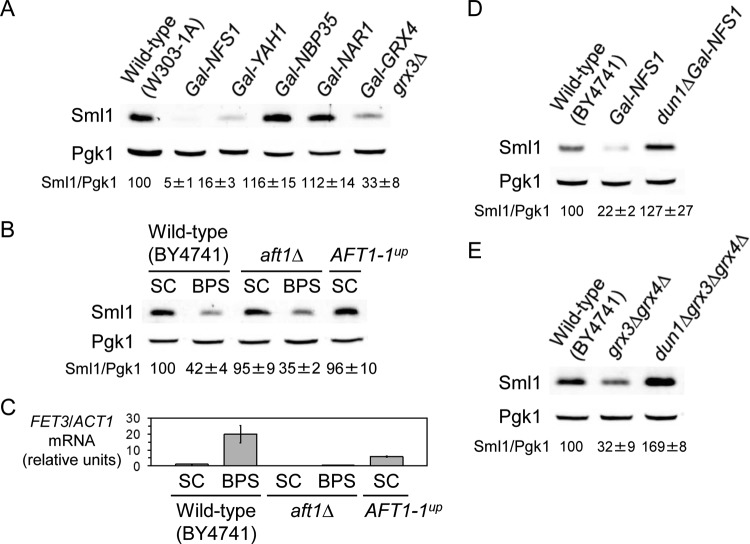
In response to genotoxic stress, Dun1 kinase promotes RNR function through multiple mechanisms, including transcriptional RNR2, RNR3, and RNR4 derepression, redistribution of the small R2 subunit from the nucleus to the cytoplasm, and degradation of the R1 inhibitor Sml1 (4). A recent study has shown that curcumin, a polyphenolic compound, extracted from the Indian spice turmeric, with diverse biological effects, including Fe chelation, promotes the degradation of the large R1 subunit inhibitor Sml1 in budding yeast (62). Therefore, we decided to ascertain whether Sml1 protein abundance is indeed regulated by Fe bioavailability. For this purpose, we grew wild-type cells under either Fe-sufficient (SC medium) or Fe-deficient conditions induced by the addition of the membrane-impermeable Fe2+-specific chelator BPS and compared Sml1 protein levels. As a control, we treated yeast cells with the DNA-damaging agent MMS, which triggered Sml1 protein degradation, as previously reported (Fig. 1A) (22, 23). Notably, Sml1 protein abundance also decreases when BPS is added to the growth medium (Fig. 1A). To further ascertain whether the diminution in Sml1 protein levels was due to Fe deficiency instead of a secondary effect of BPS, we used a strain lacking FET3 and FET4 genes, which are required for high- and low-affinity Fe transport, respectively. As shown in Fig. 1A, fet3Δ fet4Δ cells grown under Fe-sufficient conditions (SC medium) display a reduction in the amount of Sml1 protein compared to wild-type cells grown under the same conditions. Moreover, supplementation of fet3Δ fet4Δ cells with FAS leads to the recovery of Sml1 protein levels to those of wild-type cells under Fe-sufficient conditions (Fig. 1A), consistent with the notion that Sml1 protein decrease is a direct response to Fe deficiency. Taken together, these results demonstrate that the Sml1 protein level falls in response to nutritional and genetic Fe deficiencies.
FIG 1.
Dun1 kinase catalytic activity is required for the diminution of Sml1 protein levels upon genetic and nutritional iron depletion. (A) Sml1 protein abundance decreases in response to nutritional and genetic iron deficiencies. Wild-type BY4741, fet3Δ fet4Δ (SPY386), and cth1Δ cth2Δ (SPY122) yeast strains were grown at 30°C for 6 h in SC medium or SC medium with 100 μM BPS, SC medium with 300 μM FAS, or SC medium with 0.04% MMS added during the last 2 h. Sml1/Pgk1 protein values are shown as percentages of wild-type levels in SC medium (wild-type and fet3Δ fet4Δ strains) or cth1Δ cth2Δ in SC medium (cth1Δ cth2Δ strain). (B) DUN1 is required for Sml1 protein decrease in fet3Δ fet4Δ mutant cells. Yeast fet3Δ fet4Δ (SPY386) and dun1Δ fet3Δ fet4Δ (AXY1928) strains were grown as for panel A. Sml1/Pgk1 protein levels are relative to fet3Δ fet4Δ cells in FAS. (C) Dun1 kinase activity contributes to the drop in Sml1 protein caused by BPS treatment. Yeast dun1Δ (SPY350) cells transformed with plasmid pMH80 (DUN1), pRS416 (dun1Δ), or pMH62 (DUN1-D328A) were grown as for panel A. For each transformant, quantitation of Sml1/Pgk1 protein levels under BPS and MMS conditions are relative to the values obtained in SC medium. In all cases total proteins were extracted, and equal amounts were analyzed by SDS-PAGE. Sml1 and Pgk1 protein levels were determined by immunoblotting with anti-Sml1 and anti-Pgk1 antibodies, respectively. Sml1/Pgk1 protein levels in panels A, B, and C were quantified, and the averages and standard deviations of at least three independent biological replicates are represented. (D) Dun1 and Dun1-D328A protein levels under low-iron conditions. Yeast dun1Δ (SPY350) cells transformed with plasmid pRS416, pMH80 (DUN1), or pMH62 (DUN1-D328A) were grown at 30°C for 6 h in SC medium with 100 μM BPS. Dun1 protein levels were determined by immunoblotting with an anti-Dun1 antibody, and Ponceau staining was used as a loading control.
Dun1 kinase activity, but not Cth1 and Cth2 iron-regulated proteins, is required for Sml1 protein decrease in response to iron deficiency.
Upon Fe limitation, the RNA-binding proteins Cth1 and Cth2 posttranscriptionally activate the downregulation of multiple Fe-dependent processes, including the Wtm1 RNR nuclear anchor (35, 36, 52). To determine whether Cth1 and Cth2 contribute to the decline in Sml1 protein abundance occurring upon Fe depletion, we compared Sml1 protein levels in cth1Δ cth2Δ yeast cells grown under Fe-sufficient (SC medium) or Fe-deficient (BPS) conditions. As shown in Fig. 1A, Fe limitation promotes Sml1 protein diminution in cth1Δ cth2Δ mutant cells as efficiently as in wild-type cells. This result suggests that Sml1 downregulation by low Fe bioavailability is independent of Cth1 and Cth2 proteins.
In response to genotoxic stress, Dun1 protein kinase phosphorylates the Sml1 protein, promoting its ubiquitylation and degradation (22–25). Therefore, we decided to check whether Dun1 plays a role in Sml1 regulation in Fe-deficient cells. For this purpose, we compared Sml1 protein abundance in fet3Δ fet4Δ and dun1Δ fet3Δ fet4Δ mutant cells. As shown in Fig. 1B, dun1Δ fet3Δ fet4Δ cells have a much higher Sml1 protein level than fet3Δ fet4Δ cells under Fe-sufficient conditions (SC medium). Addition of excess Fe (FAS) to the medium removed any difference in Sml1 protein levels between fet3Δ fet4Δ and dun1Δ fet3Δ fet4Δ strains. These results suggest that DUN1 functions in the decrease of Sml1 protein levels observed in cells genetically deficient in Fe uptake. To address whether Dun1 is also required for the Sml1 protein drop described upon nutritional Fe deficiency, we transformed dun1Δ cells with empty vector or the same plasmid expressing wild-type DUN1 under the control of its own promoter (dun1Δ and DUN1 cells, respectively, in Fig. 1C). Yeast transformants were grown under Fe-sufficient, Fe-deficient, and MMS-treated conditions, and Sml1 protein levels were determined. As shown in Fig. 1C, dun1Δ cells expressing the DUN1 gene exhibit a reduction in Sml1 protein abundance upon Fe deficiency or MMS treatment, similar to wild-type cells (Fig. 1A and C). As previously reported (22), cells lacking DUN1 are unable to downregulate Sml1 protein levels upon MMS treatment (Fig. 1C). Importantly, dun1Δ cells do not display any reduction in Sml1 protein levels when grown under low-Fe conditions (Fig. 1C), strongly suggesting that Dun1 is involved in the mechanism that downregulates Sml1 protein levels in response to nutritional Fe deficiency. Previous studies have demonstrated that Dun1 function primarily depends on its capacity to phosphorylate downstream target proteins (23, 63). As such, a D328A substitution in Dun1's active site abolishes Dun1 kinase activity and downstream regulatory functions (23, 63). To ascertain whether Dun1 kinase activity is required for the regulation of Sml1 protein levels during Fe scarcity, we expressed the kinase-dead DUN1-D328A allele in dun1Δ cells and determined Sml1 protein abundance. As shown in Fig. 1C, yeast cells expressing DUN1-D328A are unable to diminish Sml1 protein levels upon Fe scarcity, similar to what happens upon MMS treatment. The failure to downregulate Sml1 was not a consequence of Dun1-D328A protein instability, since its expression levels under low-Fe conditions are similar to those of the wild-type Dun1 protein (Fig. 1D). Taken together, these results strongly suggest that Dun1 kinase activity is required for the downregulation of Sml1 protein abundance in response to Fe deprivation.
The integrity of the Dun1 forkhead-associated domain is important for Sml1 protein diminution upon Fe limitation.
Activation of the DNA damage response depends on the specific interaction between the Dun1 FHA domain and a diphosphothreonine motif in Rad53 checkpoint kinase (8–10). Mutagenesis of FHA domain residues necessary for Dun1 binding to the Rad53 diphosphothreonine motif impairs Dun1 activation and DUN1-dependent responses to DNA damage (9, 11). Mutagenesis to alanine of the Dun1 R60 residue, which is necessary for Dun1 binding to the first phosphothreonine residue in the Rad53 motif, abolishes Dun1 kinase activity, whereas a Dun1 K100A-R102A substitution, which affects Dun1 interaction with the second Rad53 phosphothreonine residue, partially diminishes Dun1 kinase activity (9). To ascertain whether the Dun1 FHA domain functions in Sml1 protein downregulation in response to Fe deficiency, we expressed DUN1-R60A and DUN1-K100A-R102A mutant alleles in a dun1Δ strain and compared its Sml1 protein levels to those in cells lacking DUN1 or expressing wild-type DUN1 (Fig. 2A). As shown in Fig. 2B, both DUN1-R60A- and DUN1-K100A-R102A-expressing cells exhibit a significant defect in Sml1 protein degradation compared to wild-type cells when MMS is added to the growth medium. More importantly, the drop in Sml1 protein caused by Fe deficiency is abolished or significantly impaired in Dun1 R60A or Dun1 K100A-R102A mutants, respectively (Fig. 2B). Both Dun1 R60A and Dun1 K100A-R102A protein concentrations under low-Fe conditions were similar to those of the wild-type Dun1 protein (Fig. 1D and 2D). Collectively, these results suggest that the integrity of the Dun1 FHA domain is required for the Dun1-mediated downregulation of Sml1 protein levels triggered by Fe limitation.
FIG 2.
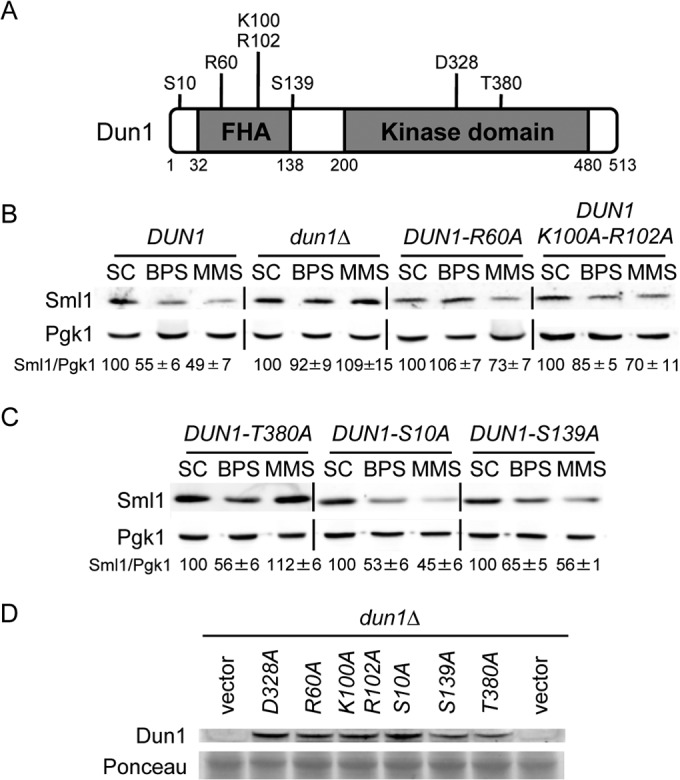
Structure-function analysis of Dun1 protein domains required for Sml1 protein decline in response to iron deficiency. (A) Schematic representation of the most relevant Dun1 domains and amino acid residues. The numbers indicate the amino acid positions. (B) The integrity of the Dun1 FHA domain is essential for the drop in Sml1 protein caused by BPS treatment. Yeast dun1Δ (SPY350) cells transformed with plasmid p413-DUN1(EcoRI) (DUN1), pRS413 (dun1Δ), p413-DUN1-R60A (DUN1-R60A), or p413-DUN1-K100A-R102A (DUN1-K100A-R102A) were grown as described in the legend to Fig. 1. (C) Sml1 protein levels in yeast cells lacking specific Dun1 phosphorylation sites. Yeast dun1Δ (SPY350) cells transformed with plasmid p413-DUN1-T380A (DUN1-T380A), p413-DUN1-S10A (DUN1-S10A), or p413-DUN1-S139A (DUN1-S139A) were grown as described in the legend to Fig. 1. For each transformant in panels B and C, quantitation of Sml1/Pgk1 protein levels under BPS and MMS conditions is relative to the corresponding values obtained in SC medium. A representative image and the average and the standard deviation of three independent biological replicates are shown for each transformant. (D) Mutant Dun1 protein levels under low-iron conditions. Yeast dun1Δ (SPY350) cells transformed with plasmid pRS416, pMH62 (DUN1-D328A) p413-DUN1-R60A (R60A), p413-DUN1-K100A,R102A (K100A-R102A), p413-DUN1-S10A (S10A), p413-DUN1-S139A (S139A), or p413-DUN1-T380A (T380A) were grown at 30°C for 6 h in SC medium with 100 μM BPS. Dun1 protein levels were determined by immunoblotting with an anti-Dun1 antibody, and Ponceau staining was used as a loading control.
The Dun1 T380 residue, a phosphorylation site of Rad53, is not necessary for reduction of Sml1 protein levels in response to Fe deficiency.
In response to genotoxic stress, Rad53 checkpoint kinase activates Dun1 by phosphorylating its T380 residue (11). Thus, DUN1-T380A mutants are unable to activate downstream cellular responses to genotoxic stress, including Sml1 protein degradation (11). To address whether the T380 residue is important to promote the Sml1 protein drop in response to low Fe, we compared Sml1 protein levels in cells lacking DUN1 to those in cells expressing either wild-type DUN1 or DUN1-T380A. As previously reported (11), cells expressing the DUN1-T380A mutant allele are unable to degrade the Sml1 protein upon MMS treatment (Fig. 2C). In contrast, the drop in the Sml1 protein in response to Fe deficiency occurs in DUN1-T380A cells as efficiently as in cells expressing wild-type DUN1 (Fig. 2C). These results strongly suggest that Sml1 protein diminution by Fe limitation is independent of Dun1 T380 phosphorylation by the upstream Rad53 kinase.
To further explore potential Dun1 residues required to promote Sml1 protein decline in response to Fe limitation, we created S10A and S139A substitutions at two previously identified autophosphorylation sites in Dun1 (11). Yeast cells expressing the DUN1-S10A or DUN1-S139A allele show a slight growth defect under genotoxic stress conditions induced by either HU or UV treatment (11). We observe that despite this phenotype, both serine mutants display normal Sml1 protein degradation upon MMS treatment (Fig. 2C). Only cells expressing the DUN1-S139A mutant allele exhibit a slight defect in Sml1 protein decrease in response to Fe deficiency, although the expression levels of Dun1 S139A protein are also reduced (Fig. 2C and D). No defect in Sml1 downregulation is observed for the DUN1-S10A mutant (Fig. 2C).
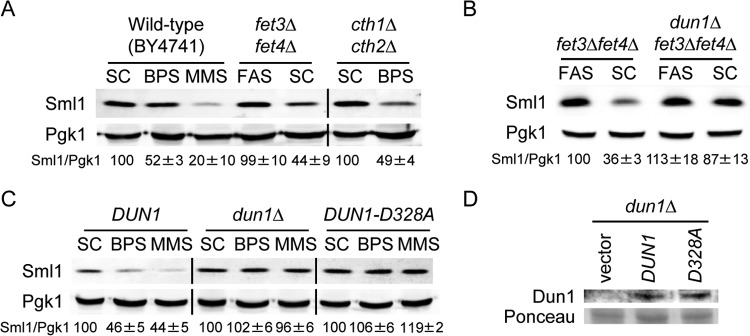
The iron deficiency-induced decline in Sml1 protein levels is independent of the Mec1 and Rad53 checkpoint kinases.
Previous studies have demonstrated that both Mec1 and Rad53 checkpoint kinases are required for the Dun1 activation that leads to Sml1 protein degradation in response to genotoxic stress and S phase (5, 64). To ascertain whether Mec1 and Rad53 participate in Sml1 Fe-mediated regulation, we determined Sml1 protein levels in wild-type, mec1Δ, rad53Δ, and dun1Δ mutant cells under both Fe-sufficient and Fe-deficient conditions. MEC1 and RAD53 are essential genes that can be rescued only by increasing dNTP pools (18, 65). To rescue mec1Δ and rad53Δ lethality, RNR1 was overexpressed under the control of a constitutive promoter in all the strains used in this assay. Although RNR1 overexpression causes an increase in Sml1 protein abundance under normal conditions (references 21 and 22 and data not shown), treatment with either BPS or MMS promotes significant Sml1 downregulation in wild-type cells, which is defective in dun1Δ mutants (Fig. 3). Importantly, in both mec1Δ and rad53Δ yeast strains, Sml1 protein levels diminish as efficiently as in wild-type cells when Fe is scarce. In contrast, MMS-induced Sml1 protein decrease is defective in mec1Δ and rad53Δ mutants compared to wild-type cells (Fig. 3). These results are consistent with the Sml1 downregulation observed in a Dun1-T380A mutant defective in phosphorylation-mediated activation by Rad53 and strongly suggest that, unlike MMS-caused Sml1 degradation, the drop in Sml1 protein produced by low Fe is independent of the Mec1 and Rad53 checkpoint kinases.
FIG 3.
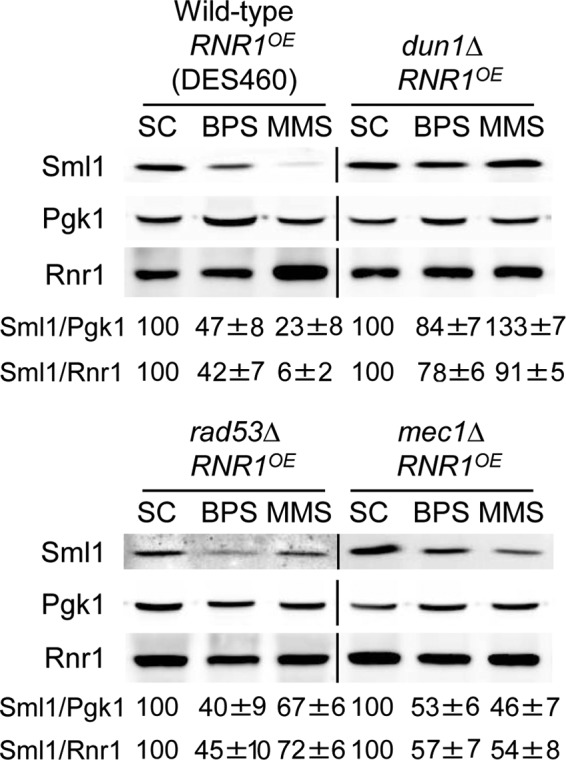
Mec1 and Rad53 checkpoint kinases do not regulate Sml1 protein levels under low-Fe conditions. Wild-type DES460, dun1Δ (MHY307), rad53Δ (DES453), and mec1Δ (DES459) yeast strains overexpressing RNR1 were grown at 30°C for 6 h in SC medium minus tryptophan (SC−trp), SC−trp with 100 μM BPS, and SC−trp with 0.04% MMS added during the last hour. Sml1/Pgk1 and Sml1/Rnr1 protein levels were determined and analyzed as described in the legend to Fig. 1. An anti-Rnr1 antibody was used to determine Rnr1 protein levels. For each strain, Sml1/Pgk1 and Sml1/Rnr1 protein quantitation under BPS and MMS conditions is relative to the values obtained in SC medium. A representative experiment and the average and standard deviation of three independent biological replicates are shown for each strain.
Sml1 protein is degraded by the 26S proteasome in a Rad6-Ubr2 ubiquitin conjugase-ligase complex-dependent manner in response to iron starvation.
In response to DNA damage induced by gamma irradiation, Sml1 protein is degraded by the 26S proteasome, relieving RNR inhibition (25). In order to determine whether the decline in Sml1 protein levels that occurs in response Fe scarcity was also mediated by the 26S proteasome, we compared Sml1 protein amounts in a wild-type strain and the temperature-sensitive pre1-1 mutant, which encodes an essential component of the 26S proteasome (66). Both wild-type and pre1-1 cells were shifted to the restrictive temperature (37°C) for 3 h to inactivate the proteasome in pre1-1 mutant cells. The Fe2+-specific chelator BPS was subsequently added to both cultures to induce Fe deficiency, and Sml1 protein levels were determined at different time points while cells were maintained at the restrictive temperature. As shown in Fig. 4A, the drop in Sml1 protein caused by low Fe is impaired in the pre1-1 strain compared to the wild-type strain, suggesting that the Sml1 protein is degraded by the 26S proteasome in response to Fe deficiency. This result is consistent with the inhibition of Sml1 protein degradation observed when the proteasome inhibitor MG132 is added to curcumin-treated yeast cells (62).
FIG 4.
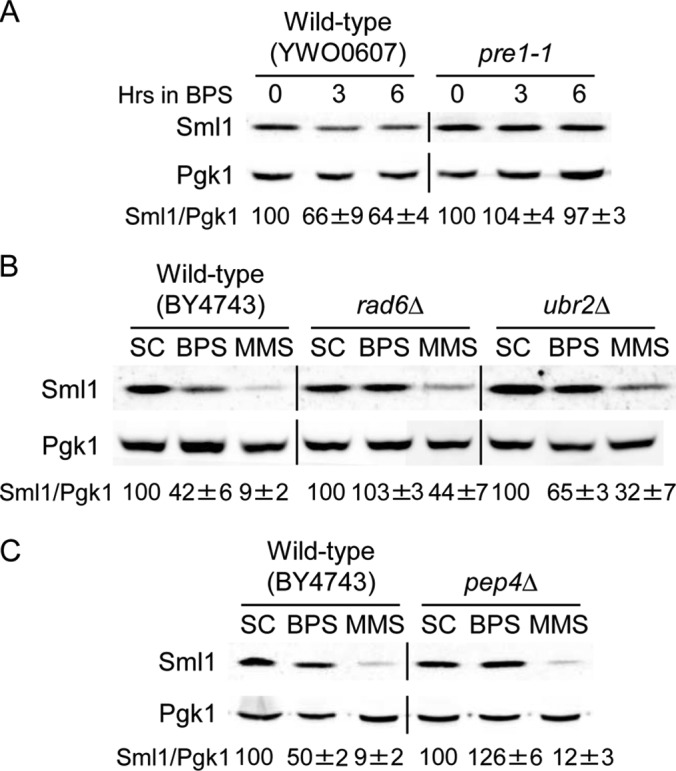
Iron deficiency promotes the 26S proteasomal and vacuolar degradation of Sml1 protein. (A) Wild-type YWO0607 and pre1-1 (YWO0608) yeast strains were grown in SC medium for 3 h at 37°C. Then, 100 μM BPS was added, and the cells were incubated for 6 additional hours at 37°C. (B) Wild-type BY4743, rad6Δ (SPY485), and ubr2Δ (SPY487) yeast strains were grown at 30°C for 6 h in SC medium, SC medium with 100 μM BPS, and SC medium with 0.04% MMS added during the last hour. (C) Wild-type BY4743 and pep4Δ (SPY496) yeast strains were grown at 30°C for 8 h in SC medium, SC medium with 100 μM BPS, and SC medium with 0.04% MMS added during the last hour. In all cases Sml1/Pgk1 protein levels were determined and analyzed as described in the legend to Fig. 1. For each strain, Sml1/Pgk1 protein quantitation under BPS and MMS conditions is relative to the levels obtained in SC medium. A representative experiment and the average and standard deviation of three independent biological replicates are shown.
Sml1 degradation by the 26S proteasome upon gamma irradiation requires the Rad6-Ubr2-Mub1 E2-E3 ubiquitin complex (25). To test whether this ubiquitin ligase complex regulates Sml1 protein degradation during Fe deficiency, we determined Sml1 protein levels in wild-type, rad6Δ, and ubr2Δ yeast cells grown under normal conditions or Fe deficiency and upon MMS treatment. As shown in Fig. 4B, Sml1 protein degradation by MMS treatment is partially defective in rad6Δ and ubr2Δ mutants, indicating that additional mechanisms may contribute to Sml1 regulation by MMS. Iron deficiency importantly diminished Sml1 degradation in both rad6Δ and ubr2Δ mutants (Fig. 4B). These results are consistent with the Rad6-Ubr2 E2-E3 ubiquitin conjugase-ligase complex participating in the degradation of Sml1 protein by the 26S proteasome in response to Fe limitation.
The vacuolar proteinase Pep4 contributes to Sml1 protein turnover in response to iron deficiency.
The degradation of the Sml1 protein by curcumin treatment is inhibited by the addition of the vacuolar proteolysis inhibitor phenylmethylsulfonyl fluoride (PMSF) (62). To further investigate whether the vacuolar proteolytic pathway is involved in the degradation of Sml1 by Fe deficiency, we determined Sml1 protein levels in cells lacking Pep4 vacuolar proteinase A, which is required for the posttranslational maturation of other vacuolar proteases (reviewed in reference 67). Recent results have shown that Sml1 degradation by HU treatment is independent of the Pep4 vacuolar protease (68). Consistent with this result, we observe that PEP4 is not necessary for Sml1 protein turnover upon MMS treatment (Fig. 4C). In contrast, PEP4 is required for Sml1 degradation in response to Fe deficiency, since Sml1 protein is not degraded in the pep4Δ mutant as occurs in wild-type cells (Fig. 4C). Taken together, these results strongly suggest that Fe deficiency promotes the degradation of the Sml1 protein by both the 26S proteasome and the vacuolar proteolytic pathway, whereas DNA damage primarily utilizes the 26S proteasome.
Defects in mitochondrial but not cytosolic iron-sulfur cluster biogenesis promote Sml1 protein depletion.
In budding yeast, the activation of the Fe regulon by the Aft1 transcription factor does not respond directly to cytosolic Fe, but rather to mitochondrial Fe utilization for the biosynthesis of Fe-S clusters (40, 41). To address whether Sml1 protein levels respond to the efficiency of ISC biosynthesis, we used a set of yeast strains that express members of the mitochondrial core or the cytosolic ISC protein assembly machinery under the control of the GAL1 promoter. Cells grown in galactose were shifted to glucose-containing medium to deplete the relevant gene product, and Sml1 protein levels were determined. As shown in Fig. 5A, yeast cells that repress the expression of members of the core mitochondrial ISC synthesis pathway, such as cysteine desulfurase (NFS1) or ferredoxin (YAH1), display a dramatic decrease in Sml1 protein levels even when grown under Fe-sufficient conditions. In contrast, depletion of components of the cytosolic ISC assembly machinery, including NBP35 and NAR1, does not significantly alter Sml1 protein abundance (Fig. 5A). These results indicate that the efficiency of mitochondrial ISC synthesis, but not cytosolic ISC assembly, regulates Sml1 protein levels.
FIG 5.
Identification of genes involved in the regulation of Sml1 protein levels by low iron. (A) Sml1 protein abundance decreases in cells defective in members of the Fe deficiency-sensing pathway. Wild-type W303-1A, Gal-NFS1, Gal-YAH1, Gal-NBP35, Gal-NAR1, and grx3ΔGal-GRX4 yeast strains were grown at 30°C for 40 h in SC medium to repress the expression of the GAL-driven genes. As described in Materials and Methods, the cells were maintained in exponential phase during the whole incubation. (B) Sml1 protein levels in yeast strains with altered Aft1 activity. Wild-type BY4741, aft1Δ, and aft1Δ transformed with plasmid pAFT1-1up were grown at 30°C for 6 h in SC medium or SC medium with 100 μM BPS. (C) FET3 mRNA levels in yeast strains with altered Aft1 activity. Yeast cells were grown as for panel B, total RNA was extracted, and FET3 mRNA levels were determined by quantitative RT-PCR as described in Materials and Methods. FET3 mRNA values were normalized with ACT1 mRNA. (D) Dun1 kinase is required for the Sml1 protein drop observed in Gal-NFS1 cells. Wild-type BY4741, Gal-NFS1 (AXY1988), and dun1ΔGal-NFS1 (AXY2059) cells were grown as described in the legend to panel A. (E) Dun1 kinase is required for the Sml1 protein drop observed in grx3Δ grx4Δ cells. Wild-type BY4741, grx3Δ grx4Δ, and dun1Δ grx3Δ grx4Δ (AXY1926) cells were grown at 30°C in SC medium to exponential growth phase. In panels A, B, D, and E, Sml1/Pgk1 protein levels were determined as described in the legend to Fig. 1 and are relative to the values obtained for the wild-type strain grown in SC medium. In all cases, a representative experiment and the average and standard deviation from at least three independent biological replicates are represented.
The monothiol glutaredoxins Grx3 and Grx4 participate both in the delivery of an Fe sufficiency signal from mitochondrial ISC synthesis to the Fe-sensitive transcription factor Aft1 and in the distribution of intracellular Fe to multiple Fe-requiring proteins (43–47). Depletion of both monothiol glutaredoxins leads to constitutive activation of Aft1 and expression of the Fe regulon (43, 44). Therefore, we decided to address whether cells defective in both monothiol glutaredoxins altered Sml1 protein levels under Fe-replete conditions. For this purpose, we grew a grx3Δ strain expressing a GAL-driven GRX4 gene under glucose-repressing conditions to deplete cells of the Grx4 protein and assessed Sml1 protein abundance. As shown in Fig. 5A, Grx4 depletion leads to reduced Sml1 protein levels under Fe-sufficient conditions. This result suggests that Grx3/4 monothiol glutaredoxins influence the regulation of Sml1 protein levels.
Dun1 kinase, but not the transcription factor Aft1, regulates Sml1 protein degradation in cells defective in ISC synthesis or iron sensing.
The mitochondrial core ISC machinery and the monothiol glutaredoxins Grx3/4, but not the cytosolic ISC assembly components, are required to repress the transcription factor Aft1 under Fe-sufficient conditions to prevent activation of the Fe regulon (40, 41, 43, 44). Moreover, depletion of NFS1, YAH1, or GRX3/4 but not NBP35 or NAR1 activates both the expression of the Fe regulon and Sml1 protein downregulation under Fe-sufficient conditions (Fig. 5A) (40–44). These results suggest that the Aft1 transcription factor could be implicated in Sml1 protein degradation by Fe limitation. To test this hypothesis, we determined Sml1 protein levels in cells lacking AFT1 or expressing the constitutively active allele AFT1-1up (38). We observed that Sml1 protein degradation properly occurs when aft1Δ mutant cells are grown under Fe-deficient conditions (Fig. 5B), despite the lack of activation of the Fe regulon indicated by unchanged FET3 mRNA levels (Fig. 5C). Moreover, constitutive activation of the Fe regulon under Fe-sufficient conditions in the AFT1-1up-expressing cells does not promote Sml1 protein decrease (Fig. 5B and C). Taken together, these results suggest that the Fe-regulated Aft1 transcription factor does not regulate Sml1 protein levels.
We then asked whether the decline in the Sml1 protein concentration observed in mutants defective in mitochondrial ISC synthesis is dependent on Dun1 kinase. For this purpose, wild-type, Gal-NFS1, and dun1Δ Gal-NFS1 yeast cells were grown in galactose and shifted to Fe-sufficient glucose-containing SC medium to switch off the expression of the GAL1-driven NFS1 gene, and Sml1 protein levels were determined. As previously shown (Fig. 5A), depletion of Nfs1 promotes the decrease of Sml1 protein levels (Fig. 5D). Importantly, the drop in Sml1 promoted by NFS1 shutoff does not occur in dun1Δ Gal-NFS1 mutants (Fig. 5D). These results indicate that the decrease in Sml1 protein levels that occurs in cells defective in mitochondrial ISC biosynthesis depends on Dun1 kinase.
Cells depleted of Grx3 and Grx4 also display a drop in the amount of Sml1 protein (Fig. 5A). Thus, we decided to address whether Dun1 kinase regulates the decrease in the Sml1 protein concentration observed for Grx3/4-defective cells. For this purpose, we grew wild-type, grx3Δ grx4Δ, and dun1Δ grx3Δ grx4Δ yeast cells under Fe-sufficient conditions and determined the Sml1 protein levels. As observed for the grx3Δ Gal-GRX4 strain (Fig. 5A), Sml1 protein levels decreased in the grx3Δ grx4Δ mutant strain (Fig. 5E). Importantly, deletion of DUN1 fully abolishes the Sml1 protein drop that occurs in cells lacking GRX3 and GRX4 genes (Fig. 5E). These results indicate that the downregulation of Sml1 protein that occurs in grx3Δ grx4Δ cells depends on Dun1 kinase. Taken together, these data suggest that Dun1 kinase mediates the degradation of Sml1 protein that occurs in yeast cells defective in mitochondrial ISC synthesis or Fe sensing.
Deletion of DUN1 in iron-deficient cells leads to an exacerbated growth defect that is rescued by SML1 deletion.
Since Dun1 kinase is a key regulator of the Fe-dependent enzyme RNR, we decided to explore potential connections between Dun1 and Fe homeostasis. We explored genetic interactions between dun1Δ mutant and fet3Δ fet4Δ cells defective in both high- and low-affinity Fe acquisition. As shown in Fig. 6, the fet3Δ fet4Δ mutant exhibits a significant growth defect compared to wild-type, dun1Δ, and sml1Δ cells in both liquid and solid synthetic complete media. Notably, deletion of the DUN1 gene exacerbates the fet3Δ fet4Δ growth defect, indicating that DUN1 genetically interacts with cells deficient in Fe uptake. Furthermore, deletion of SML1 rescues the synthetic growth defect displayed by the dun1Δ fet3Δ fet4Δ mutant (Fig. 6). Together, these results strongly suggest that Dun1 kinase plays a physiologically important role in cells under Fe deficiency, likely due to its function in downregulating Sml1 protein levels.
FIG 6.
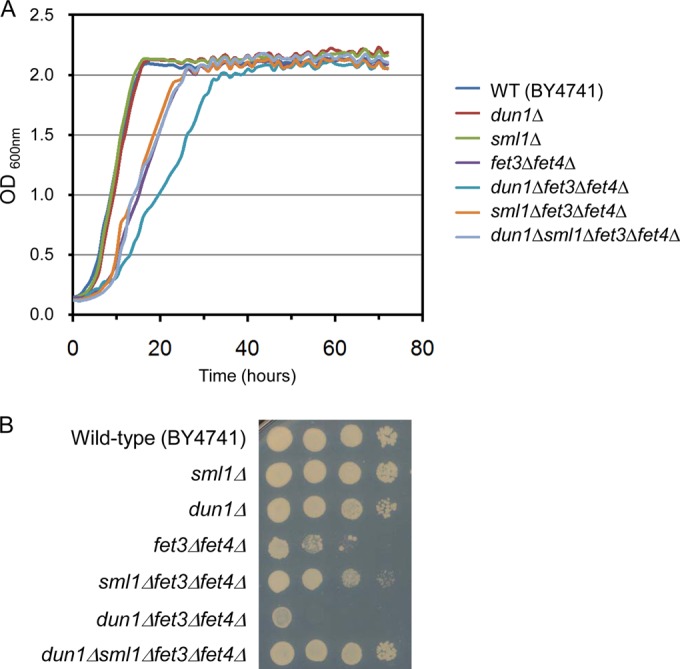
Genetic interactions between dun1Δ, sml1Δ, and fet3Δ fet4Δ yeast mutants. Wild-type (BY4741), dun1Δ (SPY350), sml1Δ (SPY589), fet3Δ fet4Δ (SPY386), dun1Δ fet3Δ fet4Δ (AXY1928), sml1Δ fet3Δ fet4Δ (AXY2243), and dun1Δ sml1Δ fet3Δ fet4Δ (AXY2141) yeast strains were grown to exponential phase and either inoculated in liquid SC medium (3 days at 28°C) with the A600 determined every hour with a Spectrostar Nano absorbance microplate reader (A) or spotted in 5-fold serial dilutions on SC medium plates (2 days at 30°C) (B). An experiment representative of at least three independent biological replicates is shown.
Dun1 kinase is important for dNTP synthesis under iron-deficient conditions.
The main function of Dun1 kinase is to modulate RNR activity. Therefore, the genetic interaction observed between dun1Δ and fet3Δ fet4Δ mutants prompted us to determine whether cells defective in DUN1 displayed alterations in RNR function under Fe-deficient conditions. For this purpose, we grew wild-type and dun1Δ yeast cells under Fe-sufficient (SC medium) and Fe-deficient (BPS) conditions and determined the intracellular dATP and dCTP levels. We observed no significant differences in dATP and dCTP concentrations between wild-type and dun1Δ cells under Fe-replete conditions (Fig. 7). As previously described (52), wild-type cells display an increase in dATP and dCTP after 7 h of incubation in SC medium with 100 μM BPS (Fig. 7). In contrast, no increase in dATP and dCTP levels is observed for dun1Δ cells when Fe bioavailability decreases (Fig. 7). These results strongly suggest that Dun1 kinase contributes to the optimal function of the RNR enzyme in response to Fe deficiency.
FIG 7.
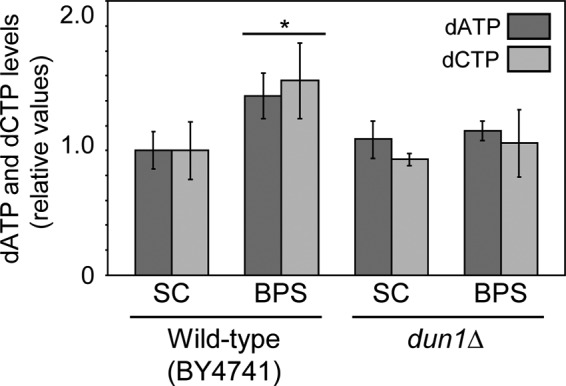
Cells lacking DUN1 display defects in dATP and dCTP synthesis during iron scarcity. Wild-type BY4741 and dun1Δ (SPY350) cells were incubated in SC medium or SC medium with 100 μM BPS for 7 h, and dATP and dCTP levels were determined as described in Materials and Methods. The average and standard deviation from three independent biological replicates are represented. The values are relative to the levels obtained for the wild-type strain grown in SC medium. The asterisk indicates statistically significant (P < 0.05) differences between dNTP levels of wild-type cells grown in BPS and the rest of the conditions assayed.
Rnr1 protein levels increase in response to iron deficiency.
We have shown here that the Dun1-mediated downregulation of Sml1 protein levels is important for dNTP synthesis and growth under Fe-deficient conditions. Given that the Sml1 protein directly binds to the RNR catalytic subunit Rnr1 to inhibit its function, we wanted to know if Rnr1 protein levels change upon Fe deficiency. For this purpose, we grew wild-type cells from two different backgrounds (BY4741 and W303-1A) under Fe-sufficient (SC medium) and Fe-deficient (BPS) conditions and determined the Rnr1 protein levels. As a control, we included a treatment with 4-NQO, a DNA-damaging agent that increases Rnr1 protein levels (26). As shown in Fig. 8A, Rnr1 protein levels increase in response to Fe deficiency, as occurs upon DNA damage.
FIG 8.
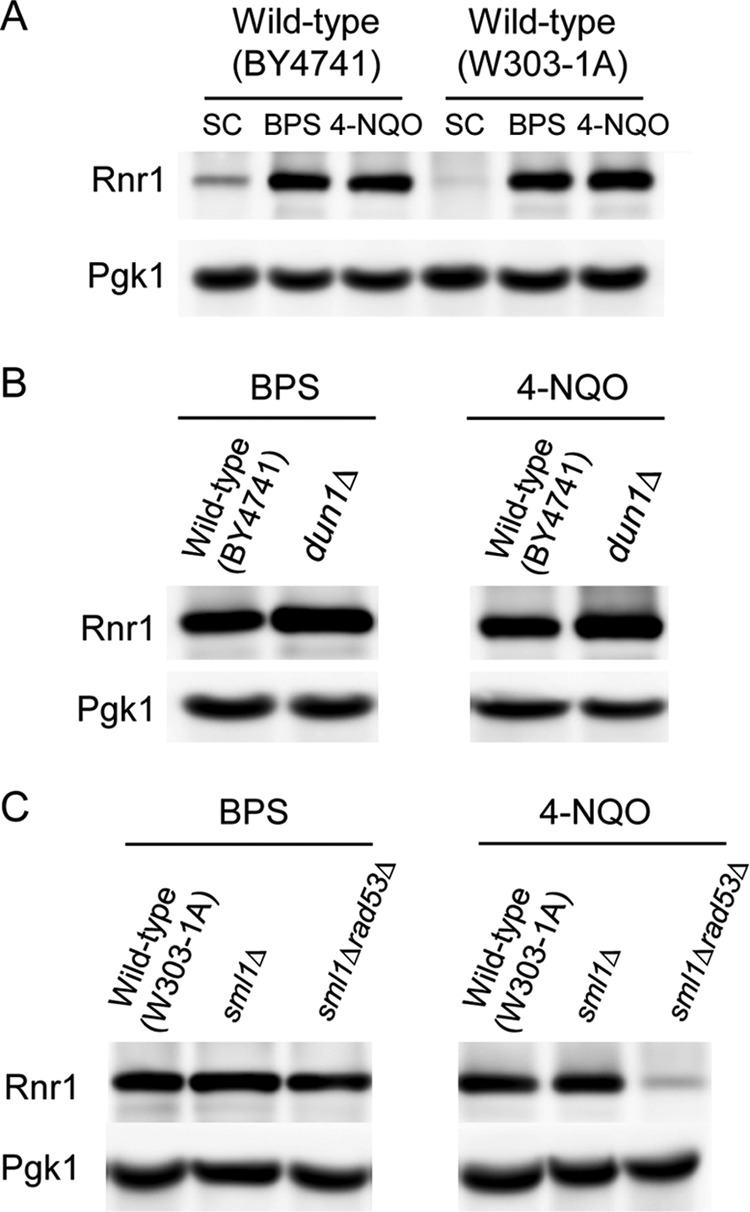
Rnr1 protein levels under iron-deficient conditions. (A) Rnr1 protein levels increase in response to iron deficiency. Wild-type BY4741 and W303-1A strains were grown at 30°C for 6 h in SC medium, SC medium with 100 μM BPS, or SC medium with 0.2 mg/liter 4-NQO added during the last 2 h. (B) Rnr1 protein levels in iron deficiency are not altered in dun1Δ mutant cells. Wild-type (BY4741) and dun1Δ (SPY350) cells were grown as for panel A. (C) Rad53 does not regulate Rnr1 protein levels under iron-deficient conditions. Wild-type (W303-1A), sml1Δ (MHY375), and sml1Δ rad53Δ (MHY380) cells were grown as for panel A. In all cases, Rnr1 protein levels were determined by immunoblotting with an anti-Rnr1 antibody, and Pgk1 protein levels were used as a loading control. An experiment representative of at least three independent biological replicates is shown for each strain.
To ascertain whether the checkpoint kinase cascade controls Rnr1 during Fe deficiency, we determined Rnr1 protein levels in dun1Δ, sml1Δ, and sml1Δ rad53Δ mutant cells and their corresponding wild-type strains grown under Fe-deficient (BPS) and DNA damage (4-NQO) conditions. As previously described (26), Rnr1 expression under 4-NQO requires the wild-type activity of Rad53 but is Dun1 independent (Fig. 8B and C). Importantly, neither dun1Δ nor rad53Δ alters Rnr1 protein levels under low-Fe conditions (Fig. 8B and C). Taken together, these results indicate that yeast cells optimize RNR activity in response to Fe deficiency by coordinately upregulating Rnr1 protein levels and downregulating Sml1 R1 inhibitor, thereby favoring an R1 catalytic subunit in a potentially enhanced active state. Further studies are necessary to decipher the mechanisms that regulate Rnr1 protein abundance under Fe deficiency.
DISCUSSION
In response to genotoxic stress, the activation of the Mec1/Rad53/Dun1 kinase cascade optimizes RNR activity by three simultaneous strategies, including RNR2 and RNR4 transcriptional induction, R2 redistribution to the cytoplasm, and degradation of the R1 inhibitor Sml1. We have previously shown that Fe scarcity promotes the subcellular redistribution of Rnr2 and Rnr4 from the nucleus to the cytoplasm in a mechanism that is Mec1 and Rad53 independent but involves the Cth1/2-mediated turnover of WTM1 mRNA, which encodes an R2 nuclear anchor (52). Since a previous study had shown that curcumin, a component of the Indian spice turmeric with Fe-chelating properties, leads to a decrease of yeast Sml1 protein levels (62), we decided to investigate whether Sml1 degradation was specific to Fe deficiency rather than a consequence of a secondary effect. By using the Fe2+-specific chelator BPS and yeast strains genetically deficient in either Fe uptake (fet3Δ fet4Δ) or Fe sensing (Gal-NFS1, Gal-YAH1, or grx3Δ grx4Δ), we demonstrated that Sml1 protein levels are indeed diminished in response to both nutritional and genetic Fe starvation.
Various genotoxic stresses activate the yeast Mec1/Rad53/Dun1 checkpoint kinase cascade that triggers Sml1 protein ubiquitylation and degradation by the 26S proteasome (22–25). While DNA-damaging agents induce Rad53 and Dun1 hyperphosphorylation, no apparent change in their electrophoretic mobility is observed in response to Fe deficiency (52), suggesting that the Mec1/Rad53/Dun1 kinase cascade is not significantly activated by Fe starvation. Consistent with this notion, we show here that Sml1 protein levels decrease as efficiently in mec1Δ and rad53Δ mutants as in a wild-type strain when Fe is limiting (Fig. 3). Surprisingly, we observed that Dun1 kinase activity is required for the efficient drop in Sml1 protein levels observed in wild-type cells upon Fe limitation (Fig. 1). Furthermore, yeast cells defective in Fe acquisition (the fet3Δ fet4Δ mutant), Fe sensing, or Fe cofactor delivery (Gal-NFS1 and grx3Δ grx4Δ mutants) also display Dun1-dependent downregulation of Sml1 protein levels (Fig. 1 and 5). This Dun1-dependent regulation of Fe deficiency seems to be physiologically relevant, since dun1Δ cells display defects in dATP and dCTP pool levels under low-Fe conditions (Fig. 7), and dun1Δ fet3Δ fet4Δ mutants exhibit a more severe growth defect than dun1Δ and fet3Δ fet4Δ cells (Fig. 6). Furthermore, the effect of DUN1 on fet3Δ fet4Δ mutant growth seems to be primarily mediated by Sml1, since the synthetic growth defect of the dun1Δ fet3Δ fet4Δ mutant is rescued by deletion of SML1 (Fig. 6). Taken together, these observations strongly indicate that Dun1 optimizes RNR function in response to Fe deficiency by stimulating the downregulation of the R1 inhibitor protein Sml1.
We then investigated the mechanisms that yeast cells use to promote Sml1 protein downregulation upon Fe limitation. Our results using pre1-1, rad6Δ, and ubr2Δ mutants are consistent with Sml1 protein being ubiquitylated by the Rad6-Ubr2 ubiquitin conjugase-ligase complex and then degraded by the 26S proteasome when Fe is scarce, as previously described for genotoxic stress (Fig. 4) (25). However, our data for pep4Δ mutants indicate that the vacuolar proteolytic pathway also participates in the degradation of the Sml1 protein when Fe is low, whereas it does not contribute to the regulation induced by MMS treatment (Fig. 4). These results are consistent with recent observations by other groups showing that Sml1 degradation induced by HU occurs in a Pep4-independent manner (68), whereas Sml1 diminution induced by curcumin is defective in the presence of MG132 or PMSF, inhibitors of the proteasomal and vacuolar proteolytic pathways, respectively (62). All these results indicate that both the 26S proteasome and the vacuolar proteolytic pathway contribute to the degradation of the Sml1 protein in response to Fe starvation, whereas the proteasome-dependent pathway is the major mechanism of Sml1 degradation in the DNA damage response.
In response to DNA damage, Rad53 kinase phosphorylates Dun1 at T380, activating its downstream functions, which include Sml1 protein degradation (11). Consistent with a Rad53-independent mechanism, we observed that mutagenesis of Dun1 T380 does not alter Sml1 downregulation by low Fe, while Sml1 degradation by DNA damage is fully abolished (Fig. 2). Several studies have proposed that Rad53-independent pathways activate Dun1 kinase function in yeast (10). For instance, transcriptional activation of SNM1, a gene required for repair of DNA cross-links, in response to DNA damage appears to be dependent on Dun1 but independent of Rad53 kinase (69). Furthermore, Mec1 and Dun1 checkpoint kinases seem to form a Rad53-independent pathway to suppress gross chromosomal rearrangements and silence gene expression in telomeres (70, 71). In these scenarios, Mec1 bypasses the Rad53 requirement to regulate Dun1 kinase. Here, we show that in the case of Fe deficiency-mediated Sml1 protein degradation, Dun1 appears to function independently of Mec1 and Rad53 (Fig. 3).
From yeast to mammals, ISC synthesis seems to play a central role in cellular Fe sensing. In mammals, the Fe-regulatory protein IRP1 controls the expression of multiple genes involved in cellular Fe homeostasis (reviewed in references 72 and 73). Under Fe-replete conditions, IRP1 assembles an Fe-S cluster that confers IRP1 aconitase activity, whereas upon Fe scarcity, IRP1 loses its Fe-S center and its aconitase activity and undergoes extensive conformational changes that enable apo-IRP1 to specifically interact with stem-loop mRNA structures, termed IREs, regulating transcript stability or translation. As detailed above, the activity of Aft1, the most relevant Fe-regulated transcription factor in yeast, also depends on the efficiency of ISC synthesis. Under Fe-sufficient conditions, Grx3-Grx4 proteins bind a 2Fe-2S cluster made in mitochondria and interact with the Aft1 protein, inhibiting its function (43–45). Upon Fe scarcity, the rate of ISC synthesis decreases, allowing Aft1 to activate the Fe deficiency response. To investigate how Fe deficiency is sensed, leading to Dun1-dependent Sml1 degradation, we determined Sml1 protein levels in cells with altered ISC synthesis. Our results indicate that mitochondrial ISC synthesis and Grx3/4 monothiol glutaredoxins are key regulators of Sml1 protein levels (Fig. 5). By using cells with altered Aft1 or Dun1 activity, we show that the mechanism that activates Sml1 turnover when the efficiency of ISC synthesis decreases requires Dun1 but not Aft1 (Fig. 5). Given that the Sml1 drop upon Fe starvation depends on the integrity of the Dun1 FHA domain (Fig. 2), it is possible that a protein other than Rad53 with a diphosphothreonine motif is responsible for transducing the Fe starvation signal to Dun1 kinase. Although further studies are necessary to fully decipher how Dun1 perceives Fe deficiency, the results presented in this work strongly suggest that Dun1 senses the Fe starvation signal by a pathway different from that described for the DNA damage response.
Our studies have highlighted the various strategies that yeast cells employ to optimize RNR function in response to Fe deficiency. First, the RNA-binding proteins Cth1 and Cth2 promote R2 redistribution to the cytoplasm to colocalize with R1 (52), and second, as shown here, the Dun1 kinase promotes the degradation of the R1 inhibitor Sml1. Finally, we revealed that Rnr1 protein levels rise upon Fe deficiency by an unknown mechanism. Further studies should decipher how these strategies are coordinated and whether mammalian cells also regulate RNR function when Fe is scarce.
ACKNOWLEDGMENTS
This work has been supported by a predoctoral fellowship from Conselleria d'Educació de la Generalitat Valenciana to N.S., a predoctoral FPI fellowship from the Spanish Ministry of Economy and Competitiveness to A.M.R., and a postdoctoral JAE-Doc contract from the Spanish Research Council (CSIC) and the European Social Fund to R.D.L. This work has been funded by grant AGL2011-29099 from the Spanish Ministry of Economy and Competitiveness to S.P. and grant CA125574 from the National Institutes of Health to M.H.
We are grateful to members of the Department of Biochemistry and Molecular Biology of the Universitat de Valencia and Systems Biology in Yeasts of Biotechnological Interest in the IATA for technical and scientific assistance. We thank Rodney Rothstein, Stephen Elledge, JoAnne Stubbe, Ulrich Mühlenhoff, Roland Lill, Jerry Kaplan, Wolf-Dietrich Heyer, Dieter Wolf, Huilin Zhou, Jörg Heierhorst, Philip Hieter, and Martin Funk for yeast strains, plasmids, and antibodies.
Footnotes
Published ahead of print 23 June 2014
REFERENCES
- 1.Kolberg M, Strand KR, Graff P, Andersson KK. 2004. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta 1699:1–34. 10.1016/j.bbapap.2004.02.007 [DOI] [PubMed] [Google Scholar]
- 2.Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681–706. 10.1146/annurev.biochem.75.103004.142443 [DOI] [PubMed] [Google Scholar]
- 3.Cotruvo JA, Stubbe J. 2011. Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80:733–767. 10.1146/annurev-biochem-061408-095817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanvisens N, de Llanos R, Puig S. 2013. Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress and iron availability. Biomed. J. 36:51–58. 10.4103/2319-4170.110398 [DOI] [PubMed] [Google Scholar]
- 5.Sun Z, Fay DS, Marini F, Foiani M, Stern DF. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395–406. 10.1101/gad.10.4.395 [DOI] [PubMed] [Google Scholar]
- 6.Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357–360. 10.1126/science.271.5247.357 [DOI] [PubMed] [Google Scholar]
- 7.Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J. 2003. Diverse but overlapping functions of the two forkhead-associated (FHA) domains in Rad53 checkpoint kinase activation. J. Biol. Chem. 278:30421–30424. 10.1074/jbc.C300227200 [DOI] [PubMed] [Google Scholar]
- 8.Lee SJ, Schwartz MF, Duong JK, Stern DF. 2003. Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Mol. Cell. Biol. 23:6300–6314. 10.1128/MCB.23.17.6300-6314.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Yuan C, Hammet A, Mahajan A, Chen ES, Wu MR, Su MI, Heierhorst J, Tsai MD. 2008. Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade. Mol. Cell 30:767–778. 10.1016/j.molcel.2008.05.013 [DOI] [PubMed] [Google Scholar]
- 10.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer WD. 2003. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell. Biol. 23:1441–1452. 10.1128/MCB.23.4.1441-1452.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SH, Smolka MB, Zhou H. 2007. Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 282:986–995. 10.1074/jbc.M609322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang M, Zhou Z, Elledge SJ. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595–605. 10.1016/S0092-8674(00)81601-3 [DOI] [PubMed] [Google Scholar]
- 13.Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. U. S. A. 100:6628–6633. 10.1073/pnas.1131932100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YD, Elledge SJ. 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 20:334–344. 10.1101/gad.1380506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YD, Wang J, Stubbe J, Elledge SJ. 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell 32:70–80. 10.1016/j.molcel.2008.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. 2006. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl. Acad. Sci. U. S. A. 103:1422–1427. 10.1073/pnas.0510516103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Huang M. 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol. Cell. Biol. 28:7156–7167. 10.1128/MCB.01388-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Muller EG, Rothstein R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329–340. 10.1016/S1097-2765(00)80277-4 [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Georgieva B, Chabes A, Domkin V, Ippel JH, Schleucher J, Wijmenga S, Thelander L, Rothstein R. 2000. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol. 20:9076–9083. 10.1128/MCB.20.23.9076-9083.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chabes A, Domkin V, Thelander L. 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274:36679–36683. 10.1074/jbc.274.51.36679 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Yang K, Chen CC, Feser J, Huang M. 2007. Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1. Proc. Natl. Acad. Sci. U. S. A. 104:2217–2222. 10.1073/pnas.0611095104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20:3544–3553. 10.1093/emboj/20.13.3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Rothstein R. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. U. S. A. 99:3746–3751. 10.1073/pnas.062502299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchiki T, Dice LT, Hettich RL, Dealwis C. 2004. Identification of phosphorylation sites on the yeast ribonucleotide reductase inhibitor Sml1. J. Biol. Chem. 279:11293–11303. 10.1074/jbc.M309751200 [DOI] [PubMed] [Google Scholar]
- 25.Andreson BL, Gupta A, Georgieva BP, Rothstein R. 2010. The ribonucleotide reductase inhibitor, Sml1, is sequentially phosphorylated, ubiquitylated and degraded in response to DNA damage. Nucleic Acids Res. 38:6490–6501. 10.1093/nar/gkq552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsaponina O, Barsoum E, Astrom SU, Chabes A. 2011. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 7:e1002061. 10.1371/journal.pgen.1002061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan CD, Kaplan J. 2009. Iron acquisition and transcriptional regulation. Chem. Rev. 109:4536–4552. 10.1021/cr9001676 [DOI] [PubMed] [Google Scholar]
- 28.Philpott CC, Protchenko O. 2008. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7:20–27. 10.1128/EC.00354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanvisens N, Puig S. 2011. Causes and consequences of nutritional iron deficiency in living organisms, p 245–276 In Merkin TC. (ed), Biology of starvation in humans and other organisms. Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 30.Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. 2012. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 1823:1491–1508. 10.1016/j.bbamcr.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 31.Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ. 1994. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269:26092–26099 [PubMed] [Google Scholar]
- 32.Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403–410. 10.1016/0092-8674(94)90346-8 [DOI] [PubMed] [Google Scholar]
- 33.Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552–1557. 10.1126/science.271.5255.1552 [DOI] [PubMed] [Google Scholar]
- 34.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15:1233–1243. 10.1091/mbc.E03-09-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110. 10.1016/j.cell.2004.11.032 [DOI] [PubMed] [Google Scholar]
- 36.Puig S, Vergara SV, Thiele DJ. 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 7:555–564. 10.1016/j.cmet.2008.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutherford JC, Jaron S, Winge DR. 2003. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278:27636–27643. 10.1074/jbc.M300076200 [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi-Iwai Y, Dancis A, Klausner RD. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casas C, Aldea M, Espinet C, Gallego C, Gil R, Herrero E. 1997. The AFT1 transcriptional factor is differentially required for expression of high-affinity iron uptake genes in Saccharomyces cerevisiae. Yeast 13:621–637 [DOI] [PubMed] [Google Scholar]
- 40.Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J. 2004. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279:29513–29518. 10.1074/jbc.M403209200 [DOI] [PubMed] [Google Scholar]
- 41.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280:10135–10140. 10.1074/jbc.M413731200 [DOI] [PubMed] [Google Scholar]
- 42.Hausmann A, Samans B, Lill R, Muhlenhoff U. 2008. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J. Biol. Chem. 283:8318–8330. 10.1074/jbc.M705570200 [DOI] [PubMed] [Google Scholar]
- 43.Pujol-Carrión N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. 2006. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 119:4554–4564. 10.1242/jcs.03229 [DOI] [PubMed] [Google Scholar]
- 44.Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. 2006. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281:17661–17669. 10.1074/jbc.M602165200 [DOI] [PubMed] [Google Scholar]
- 45.Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y. 2012. Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol. Cell. Biol. 32:4998–5008. 10.1128/MCB.00726-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. 2010. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12:373–385. 10.1016/j.cmet.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Liu L, Wu X, An X, Stubbe J, Huang M. 2011. Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 and Dre2 proteins. J. Biol. Chem. 286:41499–41509. 10.1074/jbc.M111.294074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. 2008. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 283:28527–28535. 10.1074/jbc.M804910200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vergara SV, Puig S, Thiele DJ. 2011. Early recruitment of AU-rich element-containing mRNAs determines their cytosolic fate during iron deficiency. Mol. Cell. Biol. 31:417–429. 10.1128/MCB.00754-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prouteau M, Daugeron MC, Seraphin B. 2008. Regulation of ARE transcript 3′ end processing by the yeast Cth2 mRNA decay factor. EMBO J. 27:2966–2976. 10.1038/emboj.2008.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez-Pastor M, Vergara SV, Puig S, Thiele DJ. 2013. Negative feedback regulation of the yeast Cth1 and Cth2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation. Mol. Cell. Biol. 33:2178–2187. 10.1128/MCB.01458-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanvisens N, Bañó MC, Huang M, Puig S. 2011. Regulation of ribonucleotide reductase in response to iron deficiency. Mol. Cell 44:759–769. 10.1016/j.molcel.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. 2012. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8:125–132. 10.1038/nchembio.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. 2009. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137:1247–1258. 10.1016/j.cell.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz de la Loza MC, Gallardo M, Garcia-Rubio ML, Izquierdo A, Herrero E, Aguilera A, Wellinger RE. 2011. Zim17/Tim15 links mitochondrial iron-sulfur cluster biosynthesis to nuclear genome stability. Nucleic Acids Res. 39:6002–6015. 10.1093/nar/gkr193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. 2012. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337:195–199. 10.1126/science.1219723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. 2012. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337:243–245. 10.1126/science.1219664 [DOI] [PubMed] [Google Scholar]
- 58.Mumberg D, Muller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122. 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- 59.Kushnirov VV. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857–860. [DOI] [PubMed] [Google Scholar]
- 60.Garre E, Romero-Santacreu L, Barneo-Munoz M, Miguel A, Perez-Ortin JE, Alepuz P. 2013. Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress. PLoS One 8:e61240. 10.1371/journal.pone.0061240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathews CK, Wheeler LJ. 2009. Measuring DNA precursor pools in mitochondria. Methods Mol. Biol. 554:371–381. 10.1007/978-1-59745-521-3_22 [DOI] [PubMed] [Google Scholar]
- 62.Azad GK, Singh V, Golla U, Tomar RS. 2013. Depletion of cellular iron by curcumin leads to alteration in histone acetylation and degradation of Sml1p in Saccharomyces cerevisiae. PLoS One 8:e59003. 10.1371/journal.pone.0059003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Z, Elledge SJ. 1993. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75:1119–1127. 10.1016/0092-8674(93)90321-G [DOI] [PubMed] [Google Scholar]
- 64.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401–2415. 10.1101/gad.8.20.2401 [DOI] [PubMed] [Google Scholar]
- 65.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956–2970. 10.1101/gad.12.18.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richter-Ruoff B, Wolf DH, Hochstrasser M. 1994. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 354:50–52. 10.1016/0014-5793(94)01085-4 [DOI] [PubMed] [Google Scholar]
- 67.Parr CL, Keates RA, Bryksa BC, Ogawa M, Yada RY. 2007. The structure and function of Saccharomyces cerevisiae proteinase A. Yeast 24:467–480. 10.1002/yea.1485 [DOI] [PubMed] [Google Scholar]
- 68.Baek IJ, Kang HJ, Chang M, Choi ID, Kang CM, Yun CW. 2012. Cadmium inhibits the protein degradation of Sml1 by inhibiting the phosphorylation of Sml1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 424:385–390. 10.1016/j.bbrc.2012.06.103 [DOI] [PubMed] [Google Scholar]
- 69.Wolter R, Siede W, Brendel M. 1996. Regulation of SNM1, an inducible Saccharomyces cerevisiae gene required for repair of DNA cross-links. Mol. Gen. Genet. 250:162–168 [DOI] [PubMed] [Google Scholar]
- 70.Craven RJ, Petes TD. 2000. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:2378–2384. 10.1128/MCB.20.7.2378-2384.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Myung K, Datta A, Kolodner RD. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397–408. 10.1016/S0092-8674(01)00227-6 [DOI] [PubMed] [Google Scholar]
- 72.Rouault TA. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2:406–414. 10.1038/nchembio807 [DOI] [PubMed] [Google Scholar]
- 73.Muckenthaler MU, Galy B, Hentze MW. 2008. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28:197–213. 10.1146/annurev.nutr.28.061807.155521 [DOI] [PubMed] [Google Scholar]
- 74.Muhlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G. 2003. A specific role of the yeast mitochondrial carriers Mrs3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 278:40612–40620. 10.1074/jbc.M307847200 [DOI] [PubMed] [Google Scholar]
- 75.Lange H, Kaut A, Kispal G, Lill R. 2000. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc. Natl. Acad. Sci. U. S. A. 97:1050–1055. 10.1073/pnas.97.3.1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balk J, Pierik AJ, Netz DJ, Muhlenhoff U, Lill R. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23:2105–2115. 10.1038/sj.emboj.7600216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]