Abstract
Transcriptional regulatory mechanisms likely contribute to the etiology of inflammatory bowel disease (IBD), as genetic variants associated with the disease are disproportionately found at regulatory elements. However, the transcription factors regulating colonic inflammation are unclear. To identify these transcription factors, we mapped epigenomic changes in the colonic epithelium upon inflammation. Epigenetic marks at transcriptional regulatory elements responded dynamically to inflammation and indicated a shift in epithelial transcriptional factor networks. Active enhancer chromatin structure at regulatory regions bound by the transcription factor hepatocyte nuclear factor 4α (HNF4A) was reduced during colitis. In agreement, upon an inflammatory stimulus, HNF4A was downregulated and showed a reduced ability to bind chromatin. Genetic variants that confer a predisposition to IBD map to HNF4A binding sites in the human colon cell line CaCo2, suggesting impaired HNF4A binding could underlie genetic susceptibility to IBD. Despite reduced HNF4A binding during inflammation, a temporal knockout model revealed HNF4A still actively protects against inflammatory phenotypes and promotes immune regulatory gene expression in the inflamed colonic epithelium. These findings highlight the potential for HNF4A agonists as IBD therapeutics.
INTRODUCTION
The colonic epithelium is an integral component in inflammatory bowel disease (IBD) pathology, as compromised epithelial integrity permits increased interaction between the gut immune system and luminal antigens. However, the colonic epithelium is not merely a passive barrier against luminal microbes; active epithelial roles include antigen presentation, adaptive and innate immune regulation, and antimicrobial peptide production, among others (1–3). A detailed molecular understanding of the epithelium's role in IBD and how the epithelium responds to an inflammatory insult could offer therapeutic alternatives or innovations to current treatments.
Transcriptional regulatory networks serve as the interface between the extracellular environment and genome regulation. Defining how the regulatory networks of the epithelium respond to inflammation could provide important insights into the role of the epithelium in IBD. Transcriptional regulatory networks can be inferred from a cell's epigenome, which is a collection of epigenomic marks, typically a histone posttranslational modification that is associated with a particular genome function. Transcriptional enhancer epigenomic marks are strong predictors of cellular identity and gene expression (4, 5). Nucleosomes containing histone 3, lysine 27 acetylation (H3K27ac) can be used to identify regions that have distal regulatory activity (transcriptional enhancers), flank chromatin-accessible transcription factor binding regions, and are predictive of active transcription in a condition-specific manner (4, 6, 7). Changes in H3K27ac levels and DNA accessibility predict changes in transcription factor occupancy (8, 9); dynamic enhancer chromatin structures can thus serve as a discovery tool to identify shifts in transcription factor regulatory networks induced by disease.
To identify the transcriptional networks impacted in a mouse model of colitis, we profiled the genome-wide H3K27ac levels before and during colonic inflammation. Epigenomic profiling of the colonic epithelium revealed a redistribution of enhancer activity upon inflammation. Notably, the genomic regions losing the enhancer chromatin conformation were enriched in DNA motifs recognized by the hepatocyte nuclear factor 4α (HNF4A) transcription factor, suggesting that HNF4A chromatin binding activity may be compromised in the inflamed epithelium, and prompted a detailed investigation into HNF4A gene regulation and function during colitis. A genome-scale analysis of HNF4A binding revealed diminished interactions between HNF4A and chromatin in the inflamed condition. Direct transcriptional targets of HNF4A were identified in the inflamed colon and included immune regulatory target genes which were reduced in the absence of HNF4A. HNF4A binding events were overrepresented for human genetic variants associated with increased IBD risk. Finally, we provide evidence that HNF4A plays an active role in suppressing colitis during an inflammatory bout. Together, our study resolves the temporal window in which HNF4A suppresses colonic inflammation and for the first time provides direct regulatory targets of HNF4A in this tissue and in the inflamed state. These insights into the molecular mechanisms of HNF4A during colonic inflammation should bolster efforts to restore HNF4A activity as a therapeutic approach for acute colitis.
MATERIALS AND METHODS
Mice.
C57BL/6J mice, 12 to 14 weeks old, were housed under specific-pathogen-free conditions according to protocol 11-017, which was approved by the Institutional Animal Care and Use Committee of Rutgers University. All tissues were collected between 12:00 and 14:00 to avoid circadian variability.
Experimental colitis and isolation of colon epithelial cells.
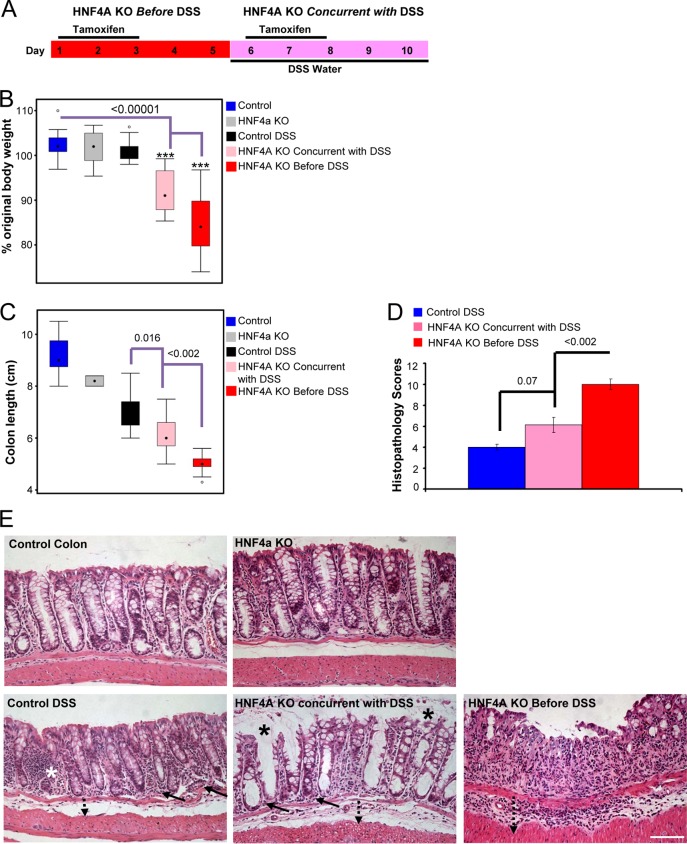
Mixed genders of mice were used to study the effects of dextran sodium sulfate (DSS)-induced inflammation on weight loss, colon length, and histopathology of the distal colon in control and HNF4A knockout (KO) mice before and concurrent with DSS treatment (see Fig. 6, below). DSS colitis was induced with 3% DSS (molecular weight, 36,000 to 50,000; MP Biomedicals, Solon, OH) solution in drinking water for up to 7 days. Daily changes in body weight were assessed. Water consumption was monitored daily to confirm the amounts of DSS consumed, as reported previously (10). Control mice were littermates given normal water and housed and processed identically. Tamoxifen-induced HNF4A knockout mice were induced by intraperitoneal injection of tamoxifen, 2 mg/day for 3 days; HNF4A KO before DSS treatment was achieved by 3 consecutive days of tamoxifen injections followed by a 2-day rest before 5 days of DSS treatment; HNF4A KO concurrent with genetic ablation was achieved by 3 consecutive days of tamoxifen injection concurrent with the start of the 5-day treatment with DSS. All control animals received vehicle-only treatments with sunflower oil and/or normal water (instead of DSS-containing water) in parallel with tamoxifen injections (see Fig. 6A, below).
FIG 6.
HNF4A plays a protective role both before and concurrently with DSS-induced colitis. (A) Strategy to inactivate HNF4A before introducing the inflammatory stimulus or concurrently with the inflammatory stimulus. After 5 days of DSS treatment, mice were measured for body weight loss (n ≥ 17 mice per genotype) (B) and colon length shortening (n = 2 to 15 mice per genotype) (C), and histolopathology scores were determined (n = 8 mice per genotype) (D). (E) Representative histology from each treatment, upon which the scores in panel D were derived. Increased inflammation in the lamina propria (solid arrow), basal crypt damage (white star), and increased submucosal space (dotted arrow) were observed in control-treated mice. Mice with HNF4A KO induced after DSS showed more patent, vacuolar crypts (black star) and some compromise of the surface epithelium and crypt architecture. Complete erosion of the surface epithelium and entire crypt loss was evident in HNF4A KO mice before DSS treatment. Bar, 100 μm. Histopathology score criteria are detailed in Materials and Methods. An ANOVA and Tukey's HSD were performed to calculate significance.
The histolopathology scores below in Fig. 6D were calculated by using metrics defined previously (11) for distal colon after 5 days of DSS treatment and followed three criteria for scoring: severity of inflammation, crypt damage, and ulceration. Possible severity of inflammation scores were 0 (rare inflammatory cells in lamina propria), 1 (increased number of inflammatory cells in lamina propria), 2 (confluent inflammatory cells extended to submucosa), and 3 as the maximum (transmural extension of inflammatory infiltrate). Crypt damage scores were 0 (intact crypt), 1 (loss of basal one-third of crypt), 2 (loss of basal two-thirds of crypt), 3 (entire crypt loss), 4 (change in epithelial surface with erosion), or 5, the maximum (confluent erosion). Ulceration was scored from 0 (absence of ulcer), 1 (1 or 2 foci of ulceration), 2 (3 or 4 foci of ulceration), or 3, the maximum (extensive erosion). Histopathological examination was performed 5 days post-DSS treatment for 8 control mice (5 males and 3 females), 8 mice in the HNF4A KO concurrent with DSS treatment group (4 males and 4 females), and 8 mice in the HNF4A KO before DSS treatment group (3 males and 5 females). Mice used for weight loss analysis (see Fig. 6B) in each set were as follows: control (29 mice, 12 males and 17 females), control DSS (24 mice, 9 males and 15 females), HNF4A KO (22 mice, 16 males and 6 females), HNF4A KO concurrent with DSS (22 mice, 12 males and 10 females), HNF4A KO before DSS (19 mice, 9 males and 10 females). Mice used for obtaining colon length data (the entire colon) (see Fig. 6C) were as follows: control (7 mice, 4 males and 3 females), HNF4A KO (2 males), control DSS (18 mice, 5 males and 13 females), HNF4A KO before DSS (14 mice, 5 males and 9 females), HNF4A KO concurrent with DSS (15 mice, 6 males and 9 females).
For chromatin immunoprecipitation and DNA sequencing (ChIP-seq) analysis, colon epithelial cells were isolated by using a scraping method with little modification (12). Briefly, distal colon segments were opened in ice-cold phosphate-buffered saline (PBS) and scraped by using a glass slide; the mucosal scrapings were resuspended in ice-cold PBS, and remaining tissue submucosa, muscular, and serosa tissues were analyzed histologically to confirm the differentiated nature of colon epithelial cells in mucosal scrapings. Two washes and low-speed centrifugation runs (150 × g) were used to reduce blood cell contamination from the epithelial preparations. Mixed genders of mice were used for ChIP-seq analysis: 3 mice (2 males and 1 female) per experimental set (HNF4A and H3K27ac ChIP-seq) were pooled for the control colon group and 8 DSS-treated mice (5 females and 3 males) per experimental set (HNF4A and H3K27ac) were pooled for ChIP-seq for the inflamed colon group. Equal amounts of total chromatin input (measured using Picogreen [Life Technologies]) were used for control and DSS-treated colon specimens: 80 μg chromatin for each HNF4A ChIP and 60 μg chromatin per H3K27ac ChIP.
Quantitative RT-PCR, immunoblotting, immunofluorescence, and immunohistochemistry.
Total RNA was extracted from respective samples (colon scrapings) by using TRIzol (Ambion). Five micrograms of total RNA from control and DSS-inflamed colon was used to purify poly(A) mRNA with the NEBNext poly(A) mRNA magnetic isolation module (E7490S/L; New England Biolabs). cDNA was synthesized from purified Poly(A) mRNA by using oligo(dT) primers (SuperScript III first-strand synthesis kit; Invitrogen). Quantitative reverse transcription-PCR (qRT-PCR) was performed using an ABI Prism 7900HT machine (Applied Biosystems, Foster City, CA). Results were normalized to hypoxanthine phosphoribosyltransferase expression and wild-type, untreated control animals. Primers sequences are available upon request. The antibodies for Western blotting and immunohistochemistry were as follows: rabbit anti-HNF4A (1:1,000; sc-6556; Santa Cruz Biotechnoloy), mouse anti-β-actin (1:2,000; sc-47778; Santa Cruz Biotechnology), rabbit anti-RelB (1:1,000 for Western blotting and 1:500 for immunohistochemistry; 4922; Cell Signaling). Donkey anti-rabbit IgG–horseradish peroxidase (HRP; 1:2,000; NA934V) and sheep anti-mouse IgG–HRP (1:2,000, NXA931) were from GE Healthcare, and biotinylated anti-rabbit IgG (1:200; BA-1000) was from Vector Laboratories. For immunoblotting, colonic epithelial scrapings were collected as detailed above and sonicated in cold tissue lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM EDTA, 1 mM NaVO4, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% Triton X-100, plus 1× Roche Complete protease inhibitor cocktail and 1× Roche phosphatase inhibitor cocktail) and then left on ice for 20 min. The supernatants were collected after centrifugation at 12,000 × g for 10 min at 4°C and stored at −80°C for further use. For Western blotting, 50 μg of whole cell lysate was heat denatured in 5× SDS sample buffer, separated by 10% SDS-PAGE, and transferred onto a PVDF membrane (ISEQ00010; Millipore). The membrane was blocked with 5% skim milk in PBS containing 0.1% Tween 20 for 1 h, and then antibodies detected the target proteins. To determine the HNF4A expression level upon DSS-induced inflammation by using immunohistochemistry, mice given 3% DSS in drinking water for 2, 4, or 6 days were euthanized and dissected, and the entire colon was fixed at 4°C overnight in 4% paraformaldehyde. Fixed tissues were gradually dehydrated in ethanol, embedded in paraffin, and cut into 5-μm sections. Ten millimolar sodium citrate (pH 6.0) was used to retrieve antigens, and endogenous peroxidase activity was inhibited in methanol containing 3% H2O2. Tissue sections were blocked with 5% fetal bovine serum (FBS; Gibco) for 4 h at ambient temperature and incubated overnight at 4°C with anti-HNF4A antibody (Ab; 1:1,000; sc-6556; Santa Cruz Biotechnology). Sections were washed in PBS containing 0.1% Tween 20 (Amresco, OH) for 2 h and treated with biotinylated anti-goat secondary IgG antibody (1:300; Vector Laboratories) for 1 h. Color reactions were developed using diaminobenzidine substrate (DAB; Sigma-Aldrich) and Vectastain avidin-biotin complex (ABC kit; Vector Laboratories). For quantitative immunofluorescence measurement of HNF4A levels during the DSS time course, mice and tissue sections were treated as described above but secondary antibody was replaced with Cy3-conjugated mouse anti-goat antibody (115-166-003; Jackson ImmunoResearch). Images were acquired using a Zeiss LSM510 META confocal microscope. Three-dimensional reconstructions of the z-stacks were analyzed using Imaris software (Bitplane AG, Zurich, Switzerland). Mean fluorescence intensity of HNF4A (Cy3) was measured in individual nuclei segmented by DAPI staining (n ≥ 100 nuclei per condition; P value < 0.005 calculated by analysis of variance [ANOVA] single factor).
ChIP, ChIP-seq, and data analysis.
Colon epithelial cells were prepared from wild-type (WT) control mice and DSS-treated mice (3% [wt/vol] for 6 days in drinking water), as described above. Chromatin immunoprecipitation was performed as described previously with little modification (13). For HNF4A ChIP, colon epithelial cells were isolated as described above and then cross-linked in 1% formaldehyde for 10 min at 4°C and then for 30 min at ambient temperature. Cells were washed 2 times with ice-cold PBS and collected by brief centrifugation (1 min, 300 × g, 4°C). Cells were resuspended in 1 ml of buffer 1 (10 mM Tris [pH 8.0], 0.25% Triton X-100, 100 mM EDTA) for 10 min on ice and collected by brief centrifugation (3,000 rpm, 1 min, 4°C). Cells were further treated twice with 1 ml of buffer 2 (10 mM Tris [pH 8.0], 200 mM Nacl, 10 mM EDTA, and 1× protease inhibitor cocktail mix) for 5 min on ice, and nuclei were collected by brief centrifugation. Collected nuclei were quickly (5 s) rinsed with 1 ml of buffer 3 (10 mM Tris-HCl [pH 8.0], 100 mM Nacl, 1 mM EDTA, 1% SDS, and 1× protease inhibitor cocktail mix), centrifuged, dissolved in buffer 3, and sonicated using a Diagenode Bioruptor to generate 200- to 1,000-bp fragments, as determined by agarose gel electrophoresis. Cell lysates were diluted 8.4-fold in binding buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.0]) and incubated with 7 μl of HNF4A antibody (lot number GR4841-13; ab41898; Abcam) coupled to magnetic beads for 16 h at 4°C. The immunoprecipitates were washed 5 times with RIPA buffer (50 mM HEPES [pH 7.6], 1 mM EDTA, 0.7% Na-deoxycholate, 1% NP-40, 0.5M LiCl) and 2 times with TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA). The DNA was recovered by reversing the cross-links in 1% SDS, 0.1 M NaHCO3 for 6 h at 65°C, purified, and quantified by using Picogreen (Life Technologies). Seven microliters of Ab was used per column; three columns were combined to get the desired amount of DNA for library preparation (30 ng).
H3K27ac histone ChIPs were done as described previously (9, 14). Isolated colon epithelial cells were first permeabilized with digestion buffer (50 mM Tris-HCl [pH7.6], 1 mM CaCl2, 0.2% Triton X-100, 5 mM Na-butyrate, 1× protease inhibitor cocktail, 0.5 mM PMSF) for 5 min on ice and then incubated with 0.2 to 0.4 unit of micrococcal nuclease (MNase; Sigma) until most of the genome was reduced to mononucleosome length (∼146 bp). MNase activity was stopped by using an equal volume of buffer (10 mM Tris [pH 7.6], 5 mM EDTA). Samples were dialyzed in 1,000 volumes of RIPA buffer (10 mM Tris [pH 7.6], 1 mM EDTA, 0.1% SDS, 0.1% Na-deoxycholate, 1% Triton X-100) for 2 h at 4°C. One milliter of mononucleosomal chromatin was supplemented with 10 μl of 10% SDS and incubated with H3K27ac Ab (lot GR28147-1; Ab4729) coupled to magnetic beads for 16 h at 4°C. The immunoprecipitates were washed 5 times with RIPA buffer (50 mM HEPES [pH 7.6], 1 mM EDTA, 0.7% Na-deoxycholate, 1% NP-40, 0.5M LiCl) and 2 times with TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA). The DNA was recovered by resuspending the beads in 100 μl of TE (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) supplemented with 3 μl of 10% SDS and 5 μl of a 20-mg/ml solution of proteinase K, incubated for 6 h at 55°C, purified, and quantified by using Picogreen (Life Technologies). Immunoprecipitated DNA was either used for qPCR or amplified to generate libraries using the TruSeq DNA sample prep kit (Illumina) and sequenced using an Illumina Hi-seq 2000 apparatus. Though Na-butyrate was used in the H3K27ac ChIP assays, a comparison ChIP-qPCR of 9 target regions with and without Na-butyrate showed no significant difference in the ChIP enrichment between samples with butyrate versus those without butyrate (1.17-fold ± 0.25 [mean ± standard deviation]; not significantly different from a ratio of 1, based on a paired t test). Prior to sequencing, qPCR was used to verify that positive and negative control ChIP regions were amplified in the linear range. Sequences were mapped to the Mus musculus reference genome mm9 by using ELAND. Sequencing depth was near saturation as determined by binding peak recovery and tag sampling using model-based analysis of ChIP-seq data (MACS) (15): HNF4A control, 17,886,946 unique tags, 92.08% saturation of binding peak recovery; HNF4A DSS, 15,034,270 unique tags, 90.42% recovery; H3K27ac control, 12,995,761 tags, 95.68% recovery; H3K27ac DSS, 15,479,369 tags, 94.73% recovery.
Binding peaks for HNF4A in WT and DSS inflamed colons were identified using the MACSV2 program (15) with a P value cutoff of 10−4 and default values for other parameters. Nucleosomes containing H3K27ac were called using nucleosome positioning from sequencing software (16), and nucleosome stability-destability (NSD) scores were calculated by using the BINOCh software package (17) and scoring scheme, as previously outlined (8). The NSD score assigned to a genomic region is calculated by measuring the differential ChIP-seq signal strength between nucleosome pairs and how this differential signal changes upon DSS treatment. Regions with large NSD scores represent regions gaining open, active enhancer chromatin upon DSS treatment; regions with little change score close to 0; regions losing active, open chromatin have negative scores. The most dynamic chromatin regions falling outside 2 kb from the nearest transcription start site (TSS) of RefSeq genes were used for subsequent analysis, to allow for a focus on putative enhancers rather than promoters.
GREAT analysis (version 2.0.2) was used to bin binding sites (MACSV2 P < 104) by distance to the nearest TSS (mm9 reference genome) with a maximum allowable distance of 1 Mb (18). Gene set enrichment analysis (GSEA) (19) was performed using public gene expression data from DSS-treated mice (GEO series GSE22307; expression data reported for C57BL/6J male mice, 12 to 14 weeks old, treated with 3% DSS for 6 days, and controls, with whole-colon tissue RNA measured using Affymetrix 430_2.0 arrays [20]). These GSE22307 expression data were compared to genes within 10 kb of the 1,000 most positive or 1,000 most negative NSD-scoring chromatin regions. GSEAV2.0 settings were set to defaults, except 5,000 permutations of the gene set and the weighted p2 scoring scheme with a signal-to-noise metric were used.
To compare changes in gene transcript levels in HNF4A knockout mice with HNF4A ChIP-seq binding frequencies near these genes, heat maps were created, with “bound genes” defined as those containing an HNF4A ChIP-seq site within 10 kb of the transcriptional start site of the dysregulated gene. Yellow heat corresponds to the number of genes with at least 1 HNF4A binding site in a sliding 10-gene window along the expression data rankings. The corresponding gene expression heat map and color scale were generated using dChip (21) to analyze publicly available HNF4A knockout colon gene expression data (GEO series GSE11759, which includes Affymetrix 430_2.0 expression array data from control and Hnf4a; Villin-Cre knockout mice at 1 year of age [22]), and genes with significant changes in transcript levels (>1.25 fold change; P < 0.05, t test) were arranged from most decreasing to most increasing upon HNF4A knockout. The supplemental material includes the results of this gene expression analysis and indicates which regulated genes harbored an HNF4A ChIP-seq site within 20 kb of their transcriptional start site.
Composite plots of ChIP-seq signal traces, evolutionary conservation scores, and DNA motif finding (SeqPos version 1.0.0) were generated and analyzed using cistrome tools (23). For motif finding, the SeqPos motif tool and Cistrome motif database were used to identify factor motifs in the top 5,000 regions in HNF4A ChIP-Seq (MACSV2 P < 104) or the 1,000 regions with the highest and lowest NSD scores in the H3K27ac ChIP-Seq results.
The integrated genome viewer (IGV) was used to visualize representative ChIP-seq data traces of normalized WIG files (24). WIG files were produced using MACSV1.4 (15), and WIG files were normalized to promoter signals at all TSSs (UCSC; mm9 reference genome) for H3K27ac or to the relative number of sequence tags for HNF4A. Variant set enrichment (VSE) analysis was done using HNF4A binding sites defined in CaCo2 cells (GEO data set GSM575229) at a MACS P value of >10−10 (9) as reported previously (25, 26), with GWAS loci for IBD, ulcerative colitis, and Crohn's disease (downloaded on 10 December 2013 from the GWAS catalog at www.genome.gov). Briefly, this analysis calculates a score and P value for enrichment of a set of disease-associated variants (variants are defined as single nucleotide polymorphisms [SNPs] identified in GWAS studies associated with disease risk, along with SNPs within a linkage disequilibrium with the risk-associated SNP) and whether these variants are enriched within a genomic annotation (in this case, at HNF4A binding sites, defined by ChIP-seq). The intersectBed program from the BEDTools suite was used to compute the overlap between chromosomal coordinates of the SNPs and the HNF4A binding sites. A null distribution for the mapping tally was based upon random permutation of the variant sets. Analysis was not done to determine whether disease SNPs were enriched specifically within HNF4A binding motifs at the HNF4A binding loci.
Statistics.
The R program was used for statistical analysis, including analysis of variance (ANOVA). The statistical calculations are embedded in the MACS, DAVID, GSEA, NPS, VSE, and Cistrome tools.
ChIP-seq data accession number.
All genome-scale data have been deposited in GEO under accession number GSE52426.
RESULTS
Inflammation-induced epigenomic changes in the intestinal epithelium reveal a shift in transcription factor regulatory networks.
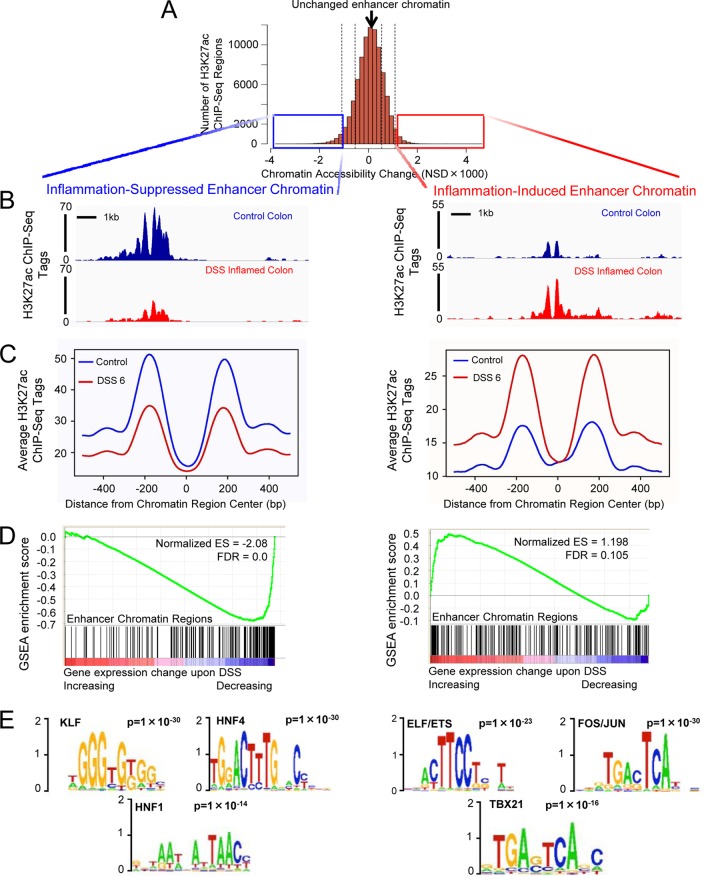
The intestinal epithelium is an integral component of inflammatory bowel disease, but the impact of acute inflammation on the transcriptional regulatory networks of the colonic epithelium is uncharacterized. The NSD methodology score was developed and has been applied to detect changes in enhancer chromatin structure and to discover new transcriptional regulatory networks (8, 9, 14). The NSD scoring algorithm identifies changes in regulatory chromatin by accounting for two properties of transcriptional enhancers: DNA accessibility where transcription factors bind and flanking nucleosomes with elevated levels of posttranslational modifications that are associated with active transcriptional enhancers (H3K27ac, H3K4me2, and H3K4me1) (8, 9, 17). Transcription factors occupying dynamic chromatin regions can be discovered by using DNA sequence motif enrichment analysis centered on these DNA-accessible regions flanked by modified nucleosomes. To discover transcription factor regulatory networks operating in the normal and inflamed colon, we first identified chromatin regulatory regions that responded dynamically during acute inflammation in mice. The inflammatory state was induced by DSS treatment of adult mice (27). DSS was chosen to induce inflammation because the acute and reproducible nature of the inflammatory response is best suited to detect short-term epigenomic changes. Colonic epithelial cells were scraped from the distal colon of control or DSS-treated mice (3% DSS in drinking water, 6 days) and purified from nonepithelial cells to ensure that measurements of enhancer chromatin changes were specific to the colonic epithelium (epithelial purity was verified by qRT-PCR [data not shown]). To determine genomic locations and levels of each H3K27ac-containing nucleosome in the genome, we generated nucleosome maps by using H3K27ac ChIP-seq on MNase-treated chromatin from each condition. Similar ChIP-seq levels and distributions of H3K27ac-containing nucleosomes were observed in the inflamed and control colons (209,118 and 195,024 nucleosomes containing H3K27ac were identified, respectively; P < 10−5). To identify dynamic chromatin regions, we applied the NSD methodology. While the majority of genomic regions are refractory to inflammation (indicated by NSD scores clustering around 0) (Fig. 1A), thousands of H3K27ac-containing genomic regions exhibited inflammation-induced changes (1,639 activated and 2,565 reduced genomic regions fell outside 2 standard deviations of the distribution of all NSD scores). H3K27ac modification dynamics were evident in individual ChIP-seq traces (Fig. 1B) and composite ChIP-seq data from the 1,000 regions with the highest (Fig. 1C, right) and 1,000 regions with the lowest (Fig. 1C, left) NSD scores. These genomic regions thus provided a window to view the impact of inflammation on the epigenome of the colonic epithelium.
FIG 1.
Epigenomic profiling of colonic epithelium before and during DSS treatment suggests an inflammation-induced shift in transcription factor regulatory networks. (A) Histogram of genomic regions in the colonic epithelium containing H3K27ac ChIP-seq at adjacent nucleosomes, binned by NSD score (8). More-negative scoring regions are less accessible and less acetylated on H3K27 at 6 days of DSS treatment than in controls (left), and more-positive scoring regions gain an active enhancer configuration during inflammation (right). Boxes indicate the 1,000 regions showing the most dynamic changes to a closed (left, blue) or active (right, red) conformation upon DSS treatment. (B) Raw sequence traces, showing a representative dynamic chromatin region that was either less (left) or more (right) active, based upon loss or gain of H3K27ac signal at nucleosomes flanking a more accessible, nucleosome-free region (gap in the sequence data). Transcription factors typically occupy this gap in enhancer chromatin. (C) Composite ChIP-seq plot of the 1,000 regions shown in the boxes of panel A. Note that flanking nucleosomes (approximately 200 bp on either side of the region center) either lose (left) or gain (right) H3K27ac signal upon DSS-induced inflammation. (D) GSEA revealed enhancer chromatin regions corresponded to DSS-induced changes in gene expression. Genes within 10 kb of the 1,000 chromatin regions that showed the greatest decrease in enhancer chromatin structure (black bars) were likely to exhibit decreased transcript levels upon DSS treatment (blue-red heat map). Genes within 10 kb of the 1,000 chromatin regions showing the greatest NSD score increases were likely to exhibit increased transcript levels upon DSS treatment (right). (E) The transcription factor DNA binding motifs most enriched at chromatin regions that become less (left) or more (right) active during DSS treatment. The supplemental material includes complete data outputs.
Consistent with a functional regulatory role, genomic regions that exhibit the most dynamic decrease in enhancer chromatin structure were located near genes with reduced transcript levels upon inflammation (20). Conversely, regions that gained enhancer chromatin signal were near genes with increased expression levels upon inflammation (Fig. 1D, GSEA data) (19). The strength of the correlations between dynamic enhancer chromatin and dynamic gene expression was especially strong considering that gene expression analysis (20) was performed on whole-colon tissues while ChIP-seq was epithelium-specific and data were generated in different laboratories.
Of potentially greater interest, the NSD analysis enabled discovery of transcription factors operating at regions of the genome undergoing inflammation-induced changes to enhancer chromatin structure. DNA sequence analysis of dynamic chromatin regions revealed stark differences in transcription factor binding motifs present at regions active in normal versus inflamed tissue (Fig. 1E; see also the supplemental material). At chromatin regions selectively activated upon DSS treatment, motifs recognized by members of the AP-1 and ETS transcription factor families were the top three most enriched (Fig. 1E, right). This is consistent with AP-1 and ETS roles in mediating the inflammatory response, as both families have been implicated in responding to tumor necrosis factor alpha signaling and have been reported to induce cytokine expression in endothelial and epidermal tissues (28–32). At genomic regions that lose enhancer chromatin structure upon inflammation, colonic developmental transcription factor motifs predominated (KLF, HNF4, and HNF1), suggesting that these regions lose their transcription factor occupancy during colitis (33–37). Indeed, previous studies demonstrated that KLF5 and HNF4A can protect against DSS treatment (36, 38, 39). In total, we identified a subset of nucleosomes comprising approximately 2% of all of the H3K27ac epigenome in the colonic epithelium that responds dynamically to DSS-induced inflammation (defined as regions with NSD scores greater than 2 standard deviations), and we found that the genomic regions harboring these dynamic chromatin structures correlated with the changing transcriptome. Motif analysis suggested a shift in the transcriptional regulatory network away from a stabilizing colon-specific transcription factor network of HNF4A, KLF, and HNF1A/B to an inflammation-specific network involving ETS and AP-1 factors.
The expression level and chromatin binding activity of HNF4A are compromised during inflammation.
Previous implications for HNF4A in inflammatory pathology prompted further investigation of the transcriptional regulatory mechanisms of HNF4A in the normal versus inflamed colon. HNF4A belongs to the nuclear receptor family of transcription factors and is one of the more prominent epithelial genes implicated in IBD. Genome-wide association studies have identified HNF4A as susceptibility locus for ulcerative colitis (40–43), and knockout mouse models have corroborated these genetic implications of HNF4A in IBD by demonstrating a role for Hnf4a in differentiation of gut epithelial cells (44), in the modulation of gut homeostasis (33, 45), and in a protective role against colitis (38).
Importantly, the temporal window of HNF4A function during the inflammatory process is not known; it is not clear from these models whether HNF4A plays a protective role prior to the inflammatory state and/or an active role during an inflammatory bout. Previous reports indicated that whole-colon HNF4A levels decrease during inflammation (22, 38). However, epithelium-specific expression has not been examined, and reduced HNF4A levels could be a secondary effect of the loss of epithelial cells that occurs during an inflammatory episode.
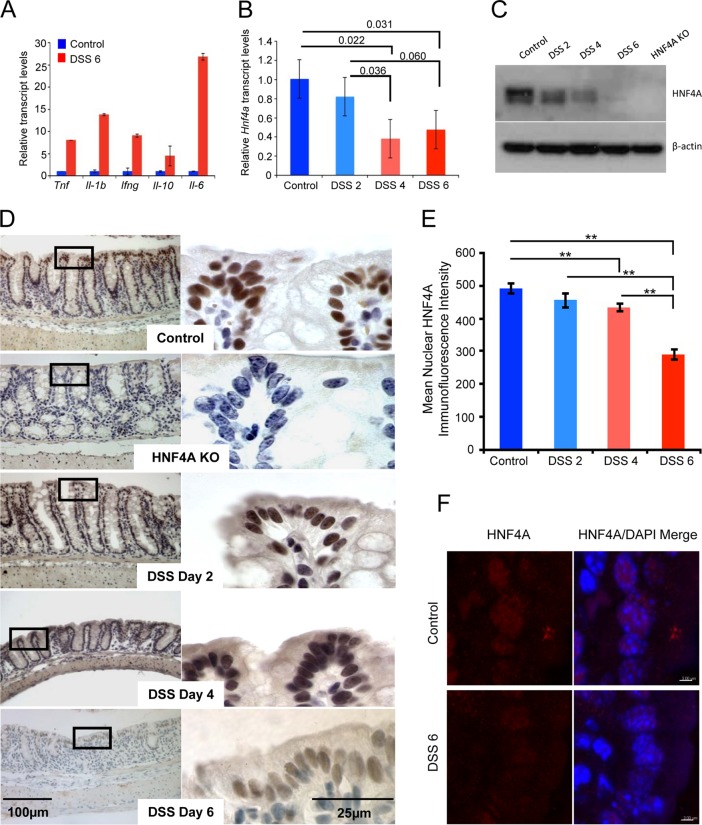
To determine if HNF4A is regulated at an epithelial cell-autonomous level, we monitored HNF4A mRNA and protein levels in isolated colonic epithelium during a time course of inflammation (Fig. 2). RNA transcripts from purified epithelium confirmed an inflammatory phenotype with elevated inflammatory cytokine levels in mice treated with DSS for 6 consecutive days (Fig. 2A). Hnf4a transcripts were significantly depleted by 4 days of DSS treatment (Fig. 2B), whereas markers specific to the epithelium (Krt18 and Vil1) were not affected (data not shown). Immunoblotting also demonstrated reduced HNF4A protein levels in isolated epithelial cells (Fig. 2C), and nuclear HNF4A protein levels were also reduced in the DSS-treated mouse epithelium in situ, as revealed by immunohistochemistry and quantitative immunofluorescence (Fig. 2D to F). Thus, based upon expression levels, protective mechanisms of HNF4A function may be diminished, but not necessarily eliminated, during inflammation, as HNF4A is downregulated in the colonic epithelium but still readily detectible during colitis.
FIG 2.
HNF4A levels are reduced, but detectable, in the colonic epithelium after DSS-induced inflammation. (A) Results of qRT-PCR to measure inflammatory markers in the colon of mice treated with 3% DSS for 6 days. (B) Results of qRT-PCR to measure relative Hnf4a transcript levels on mRNA isolated from the colonic epithelium during a time course of DSS treatment (2-tailed t test; n = 3). Bars show the standard errors of the means. (C) Immunoblot measuring HNF4A levels during a DSS treatment course. The immunoblot was underdeveloped to allow evaluation of the relative levels of HNF4A over time. Protein levels were detectable, though reduced, at day 6 of DSS treatment. (D) HNF4A immunoreactivity diminishes, but is still present, over the DSS treatment course. HNF4A was not detected in HNF4A KO epithelium. The colonic regions selected for this analysis were based upon the presence of an intact epithelium and do not fully represent the inflammatory pathology. (E) Quantitative immunofluorescence was also applied to measure HNF4A protein levels specifically within the nucleus. Mean fluorescence intensity of HNF4A (Cy3) was measured in individual nuclei segmented by 4′,6-diamidino-2-phenylindole staining (n ≥ 100 nuclei per condition). **, P < 0.005 by ANOVA single factor. Bars, standard errors of the means. (F) Representative confocal images used in the quantitative analysis in panel E are shown for control and DSS6 samples. Bar, 3 μm.
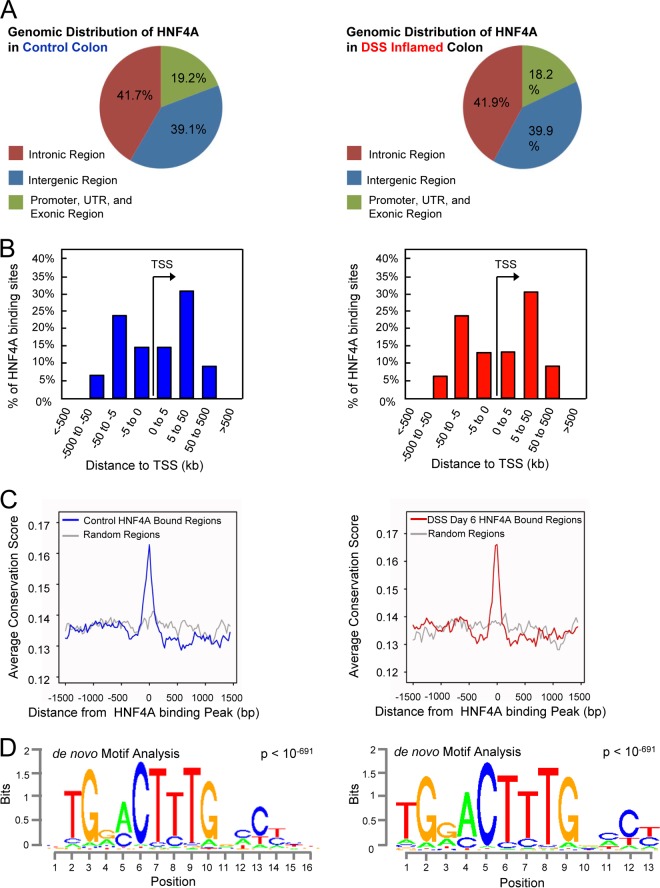
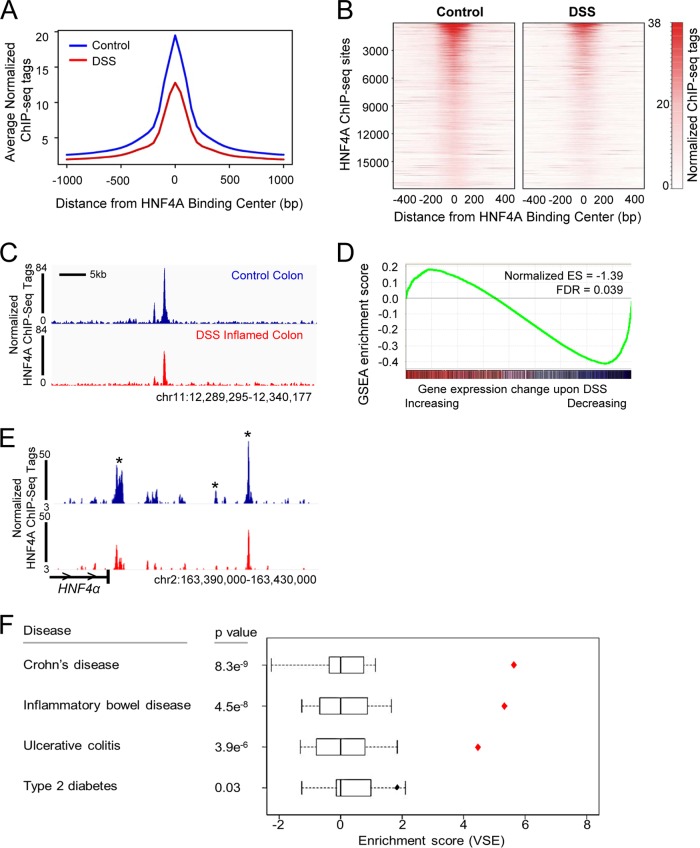
The direct regulatory targets of HNF4A in the colonic epithelium have not been determined in vivo or under inflammatory stress. To discern between possible HNF4A functions before and during the inflammatory state, we measured HNF4A binding to the genome of the colonic epithelium under each condition by using ChIP-seq analysis. ChIP of the healthy colonic epithelium revealed 17,886 HNF4A binding sites (MACS2 P, <10−4) scattered across the genome (Fig. 3A, left), with the majority of binding sites between 5 and 50 kb up or downstream from the nearest TSS (Fig. 3B, left). As expected for functional HNF4A binding sites, HNF4A-bound regions exhibited increased evolutionary conservation compared to genomic background levels (Fig. 3C, left). Moreover, the most prominent de novo DNA motif found to be enriched at HNF4A binding regions corresponded to previously defined HNF4A recognition sequences (Fig. 3D, left). Together, these data indicated robust identification of physical interactions between HNF4A and the colonic epithelial genome.
FIG 3.
The relative genomic binding distribution and target sequence preference of HNF4A do not change in the inflammatory state. (A) The genomic distribution of HNF4A in control (left) and DSS-inflamed colon (right; 6 days of 3% DSS treatment) indicates that HNF4A predominantly binds far from classical promoters. (B) The majority of binding sites occur 5 to 50 kb from the nearest TSS. (C) As expected for functional regulatory elements, HNF4A binding sites are enriched in conserved nucleotides across multiple vertebrate species. (D) The most frequently occurring DNA sequence motif enriched at HNF4A ChIP-seq sites matches the expected HNF4A binding sequence (14).
We next determined whether the chromatin binding profile of HNF4A was changed upon inflammation. HNF4A ChIP-seq for the inflamed colonic epithelium showed a binding profile similar to that observed in the healthy epithelium, with a similar distribution of genomic binding regions (Fig. 3, right), distance from transcriptional start sites (Fig. 3B, right), evolutionary conservation (Fig. 3C, right), and preferred DNA binding motif (Fig. 3D, right). However, the binding events observed in the inflamed colon were fewer (10,688 regions). A reduced magnitude of HNF4A ChIP-seq events was evident in the reduced average signal at all HNF4A binding sites (Fig. 4A) and in direct comparison on a site-by-site basis (Fig. 4B to D). As HNF4A binding is reduced during inflammation, we predicted genes near HNF4A binding sites would be compromised upon inflammation. Indeed, integration of ChIP-seq data and gene expression analysis demonstrated that transcripts that were decreased in the inflamed colonic epithelium were more likely to harbor HNF4A binding sites within 10 kb of their transcriptional start sites (Fig. 4D). Taken together, HNF4A binding to chromatin is compromised in the inflamed colon, and reduced binding appears to have a direct consequence on colonic gene expression. Notably, multiple HNF4A binding sites were observed at the HNF4A locus (Fig. 4E), suggesting that loss of an autoregulatory loop could destabilize HNF4A expression in the inflammatory condition and contribute to reduced HNF4A protein levels (Fig. 2C to F).
FIG 4.
The number and magnitude of HNF4A binding sites are compromised during inflammation. (A and B) The average, normalized ChIP-seq signal at HNF4A binding sites is diminished upon DSS treatment, as shown in a composite plot (A) or on a site-by-site basis (B). The heat map scale indicates normalized ChIP-seq tag counts. The heat map depicts each genomic region called an HNF4A binding site in the control condition across a 1-kb genomic window centered at the binding peak summit. (C) Example of reduced HNF4A binding upon DSS treatment at the Hnf4a locus. (D) GSEA comparing genes with HNF4A binding sites (MACS2 P < 10−10) with gene expression changes that occur upon DSS-induced inflammation. Genes that decrease upon DSS treatment are enriched in HNF4A binding sites, suggesting reduced HNF4A binding during inflammation may lead to their decreased levels of target gene expression. (E) Sample of reduced HNF4A binding upon DSS treatment at the Hnf4a locus, suggesting the possibility of HNF4A autoregulation. Asterisks denote HNF4A ChIP-seq peaks downstream of Hnf4a; horizontal carets indicate the direction of transcription at the Hnf4a locus. (F) VSE analysis showed that HNF4A binding regions in the human colon cell line Caco2 (GSM575229) are enriched with IBD risk-associated loci. Box plots show the null distributions based on 1,000 matched random variant sets for each disease against HNF4A binding sites. Diamonds show mapping tallies for the disease-associated clusters at HNF4A binding sites. Red diamonds highlight mapping tallies for genomic annotations that fall outside of the null distribution (P < 0.001). The overlap between HNF4A binding and disease-associated genetic variants suggests that altered HNF4A function at these loci may contribute to the genetic component of IBD. The enrichment analysis did not specifically test whether HNF4A binding motifs were disrupted by the genetic variants, only that the variants were enriched at HNF4A binding regions.
HNF4A binding regions are enriched in IBD risk-associated genetic variants.
Upon finding reduced HNF4A binding in the inflammatory condition, we wondered if HNF4A binding sites could also be targets of genetic predisposition to IBD. This was addressed by determining the relative enrichment of risk loci associated with IBD, Crohn's disease, or ulcerative colitis (GWAS catalog as of 10 December 2013) at HNF4A genomic binding regions previously reported in the human colorectal cell line CaCo2 (46). The VSE analysis (25, 26) revealed that IBD-associated risk loci map to HNF4A binding sites more than expected by chance (P = 4.5e−8) (Fig. 4F). Similar observations were shown between risk loci associated with Crohn's disease (P = 8.3e−9) and ulcerative colitis (P = 3.9e−6) (Fig. 4F). By contrast, HNF4A binding sites in CaCo2 cells were not overrepresented in SNPs associated with type 2 diabetes risk (Fig. 4F), even though HNF4A has a prominent role in this disease. Thus, colon-specific binding patterns of HNF4A are significantly and specifically associated with the genetic predispositions of human colon inflammatory pathologies.
Direct targets of HNF4A include immune regulatory genes.
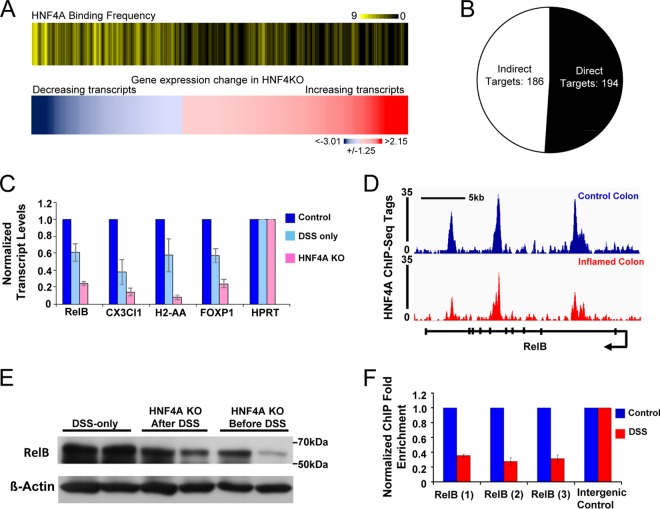
Despite being powerfully implicated in IBD, the direct transcriptional targets of HNF4A in the colon have not been identified. Microarray expression in HNF4A knockouts has revealed gene transcripts that increase and decrease upon HNF4A loss and are implicated in roles such as preventing oxidative stress and regulating lipid metabolism (22, 47). To define which transcripts are likely direct versus indirect regulatory targets of HNF4A, we integrated our ChIP-seq data with gene expression analysis results. We defined direct targets of HNF4A as genes with both an HNF4A binding site within its locus and dysregulated upon HNF4A knockout based on microarray analysis (22). HNF4A binding was far more prevalent at genes downregulated upon Hnf4a knockout than genes upregulated upon Hnf4a loss, implicating HNF4A as primarily a transcriptional activator (Fig. 5A). No significant association was observed between HNF4A binding at genes increased upon Hnf4a knockout compared to nonregulated genes, indicating that HNF4A does not have a prominent role in gene repression.
FIG 5.
HNF4A predominantly activates colon epithelial genes, and its direct targets include genes involved in the immune response. (A) Corresponding heat maps depict HNF4A binding frequency (top) at genes with significant expression changes upon HNF4A KO (GSE11759; bottom). Genes that significantly decrease upon HNF4A knockout are more likely to harbor an HNF4A site within 10 kb of their TSS than all other genes. (B) Pie chart depicting downregulated genes upon HNF4A knockout of panel A that included HNF4A binding sites within 20 kb of their transcriptional start sites (direct targets) and downregulated genes lacking HNF4A binding sites (indirect targets). The entire list of target genes appears in the supplemental material. (C) qRT-PCR confirmed that HNF4A is required in the inflamed condition to activate presumed immune regulatory genes. (D) The RelB locus harbors three HNF4A binding sites (the arrow indicates the transcriptional direction from the promoter), as detected by using ChIP-seq and independently confirmed by ChIP-qPCR. ChIP enrichment was reduced in the inflammatory condition (F). (E) RelB protein levels are reduced in isolated colonic epithelia from DSS-treated animals lacking HNF4A compared to DSS-only controls. Bars in the graphs of panels C and F indicate standard errors.
Of 380 transcripts significantly reduced in Hnf4a KO colon, 194 were bound by HNF4A within 20 kb of their transcriptional start sites (defined as direct HNF4A targets), whereas 186 were not bound (indirect HNF4A targets) (Fig. 5B). Identification of putative direct and indirect HNF4A regulatory targets in the colon will provide a useful resource for dissecting the downstream mechanisms of the protective function of HNF4A; these targets are presented in the supplemental material. For example, we were intrigued to find immune regulatory genes among direct targets of HNF4A, as this is an underappreciated role of HNF4A (48). Immune regulatory targets were tested for HNF4A-dependent regulation in the inflamed condition and exhibited reduced transcript levels in HNF4A KO mice (Fig. 5C). The corresponding loci of these genes also showed reduced HNF4A ChIP-seq binding levels during colitis (Fig. 5D and data not shown). Identification of HNF4A regulatory targets in the inflamed colon will open new avenues to explore HNF4A function. For example, RelB was of interest, as members of the NF-κB signaling pathway function in the epithelium to protect against inflammation and DSS colitis (49–53). Consistent with our epigenomic analysis (Fig. 5D), HNF4A was confirmed to bind 3 genomic regions at the RelB locus based on replicate ChIP-qPCR, and binding was reduced, but detectable, in the inflammatory condition (Fig. 5F). RelB protein levels were also reduced in isolated colonic epithelia from DSS-treated HNF4A knockout versus DSS-treated control animals (Fig. 5E). Taken together, genomic profiling of HNF4A binding regions in the normal and inflamed colon suggests mechanisms underlying the protective role of HNF4A against colonic inflammation, including a capacity to control immune regulatory genes both before and during an inflammatory episode.
HNF4A is protective during an active inflammatory bout.
When HNF4A is ablated from the fetal colonic epithelium, knockout animals are clearly more susceptible to DSS-induced colitis when they are adults (38), but it is unclear whether HNF4A is required to prevent the onset of colitis or actively suppress an ongoing inflammatory episode. As epithelial cells retain detectable levels of HNF4A protein and chromatin binding during inflammation (Fig. 2 and 4), we tested whether the protective effects of HNF4A include suppression of an active inflammatory bout. To refine the temporal window in which HNF4A protects against experimental colitis, we took advantage of the tamoxifen-inducible Villin-CreERT2 driver (54) and conditionally deleted floxed Hnf4a (55) in the epithelium either before or during acute inflammation in the colonic epithelium (Fig. 6A). HNF4A knockout by tamoxifen injection requires approximately 2 days to eliminate HNF4A protein expression and was initiated either 5 days before or concurrent with DSS-induced inflammation (Fig. 6A). We refer to these experimental conditions as HNF4A KO “before” and “concurrent with” HNF4A KO in DSS-induced colitis. Each group was treated with DSS for 5 days, and the degree of inflammation was inferred by changes in body weight, colon length, and histopathology of the distal colon. As reported previously (38), we observed that knockout of HNF4A before the inflammatory stimulus led to a severe and rapid onset of experimental colitis (Fig. 6B to E). However, despite reduced protein levels upon inflammation (Fig. 2C to F), HNF4A still performs a protective role in the colonic epithelium during inflammation, as loss of HNF4A after the onset of the inflammatory stimulus resulted in a more severe inflammatory phenotype than in control-treated mice. Representative histopathology results were consistent with these phenotypes (Fig. 6E). Taken together, these results indicate that HNF4A not only prevents the onset of inflammation but also plays an active role in the suppression of an inflammatory state, and thus expands the temporal window within which HNF4A functions to suppress colitis.
DISCUSSION
Disease-associated genetic variants are increasingly found to overlap with regulatory regions (26, 56–59), including in IBD (60), highlighting the importance of regulatory elements in human disease and necessitating their functional characterization. To identify inflammation-sensitive regulatory elements in the colonic epithelium, we employed an epigenomic approach. Analysis of these regions revealed that transcription factor regulatory networks change in the inflamed state, with HNF4A DNA binding motifs among the top transcription factor motifs found at genomic regions that lose enhancer chromatin modifications upon colonic inflammation. By integrating mouse genetic models with ChIP-seq mapping of HNF4A binding sites during an inflammatory bout, we found that gene regulatory activity of HNF4A is reduced but still essential to mollify inflammatory symptoms.
Genome-scale HNF4A binding studies in other tissues and species provide interesting insights on tissue-specific binding patterns, evolution of regulatory elements, and gene regulation in the liver, pancreas, and cancer (61–65). To our knowledge, no study has investigated HNF4A binding changes in altered or noncancer disease conditions. It will be interesting to see whether compromised HNF4A binding represents a common mechanism in inflammatory diseases of other HNF4A-expressing tissues. While many interesting single-gene studies have identified regulatory roles for HNF4A in barrier protection and lipid metabolism, these studies have been reported for Hnf4a mutant mice under normal conditions (22, 33, 38, 45, 47, 66). Our analysis of HNF4A binding in the inflamed colonic epithelium yielded primary HNF4A regulatory targets that will be useful in dissecting the protective mechanisms of HNF4A during colitis, including immune regulatory genes, a role not frequently attributed to direct HNF4A regulation, although it was recently observed in the liver (48, 67). Thus, HNF4A appears to support pleiotropic protective roles in the colonic epithelium, and our study opens new avenues for exploration and regulation of HNF4A in other inflamed tissues. While we observed exacerbated colonic phenotypes in DSS-treated mice upon HNF4A loss, we did not observe a significant increase in inflammatory cytokines compared to DSS-treated control mice (data not shown), suggesting that the phenotypic severity caused by HNF4A loss in these models may function independently of these cytokines (Fig. 2A). Functional characterization of HNF4A direct target genes should help elucidate the modalities by which HNF4A protects the colon.
In this work, we have revealed a protective but compromised role for HNF4A in the inflamed colon, and we observed the cooccurrence of HNF4A binding and IBD risk-associated genetic variants. Together, these findings justify exploring HNF4A as a therapeutic target in IBD, as restoration of HNF4A expression or binding activity would be predicted to ameliorate inflammation-induced changes in the epithelium. Along these lines, identifying the initial trigger that decreases HNF4A levels will also be important, and KLF factors, which have been implicated in inflammatory bowel models (36) and whose motifs are enriched at HNF4A binding sites, are good candidates to mediate HNF4A regulation. It is also important to consider the duration of HNF4A inactivation in both genetic and inflammatory models, as our experiments do not discern how the difference in the duration of HNF4A loss affects gene regulation. Such differences could also impact epithelial regeneration following the inflammatory bout.
As HNF4A is a nuclear receptor, the development of an activating agonist makes this an exciting and plausible option (68, 69). Indeed, HNF4A reversibly binds linoleic acid in endogenous contexts (70), and linoleic-acid derivatives have recently been identified as candidate therapeutics in a drug repositioning screen for IBD (71). Our work provides a context (active inflammation) and a mechanism (reduced HNF4A chromatin binding) with which to explore the function of these and other potential HNF4A ligands.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sylvie Robine for sharing Vill-CreERT2 mice, Jay Tishfield for support in quantitative image analysis, and Kiron Das for helpful discussions.
The study was supported by grants from the Human Genetics Institute of New Jersey (M.P.V.), National Institutes of Health grants K01DK088868 (M.P.V.) and R01CA155004 (M.L.), and the Princess Margaret Cancer Foundation (M.L.). M.L. holds a young investigator award from the Ontario Institute for Cancer Research and a new investigator salary award from the Canadian Institute of Health Research.
We declare no conflicts of interest.
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00349-14.
REFERENCES
- 1.Henderson P, van Limbergen JE, Schwarze J, Wilson DC. 2011. Function of the intestinal epithelium and its dysregulation in inflammatory bowel disease. Inflamm. Bowel Dis. 17:382–395. 10.1002/ibd.21379 [DOI] [PubMed] [Google Scholar]
- 2.Pastorelli L, De Salvo C, Mercado JR, Vecchi M, Pizarro TT. 2013. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front. Immunol. 4:280. 10.3389/fimmu.2013.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmerman NP, Vongsa RA, Wendt MK, Dwinell MB. 2008. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm. Bowel Dis. 14:1000–1011. 10.1002/ibd.20480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 107:21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zentner GE, Tesar PJ, Scacheri PC. 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21:1273–1283. 10.1101/gr.122382.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. 2010. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 42:343–347. 10.1038/ng.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. 2010. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19:713–726. 10.1016/j.devcel.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdelbaqi M, Chidlow JH, Matthews KM, Pavlick KP, Barlow SC, Linscott AJ, Grisham MB, Fowler MR, Kevil CG. 2006. Regulation of dextran sodium sulfate induced colitis by leukocyte beta 2 integrins. Lab. Invest. 86:380–390. 10.1038/labinvest.3700398 [DOI] [PubMed] [Google Scholar]
- 11.Laroui H, Ingersoll SA, Liu HC, Baker MT, Ayyadurai S, Charania MA, Laroui F, Yan Y, Sitaraman SV, Merlin D. 2012. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLoS One 7:e32084. 10.1371/journal.pone.0032084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega-Cava CF, Ishihara S, Rumi MA, Kawashima K, Ishimura N, Kazumori H, Udagawa J, Kadowaki Y, Kinoshita Y. 2003. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J. Immunol. 170:3977–3985. 10.4049/jimmunol.170.8.3977 [DOI] [PubMed] [Google Scholar]
- 13.Verzi MP, Shin H, Ho LL, Liu XS, Shivdasani RA. 2011. Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31:2026–2039. 10.1128/MCB.01250-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verzi MP, Shin H, San Roman AK, Liu XS, Shivdasani RA. 2013. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 33:281–292. 10.1128/MCB.01185-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. 2008. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics 9:537. 10.1186/1471-2164-9-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer CA, He HH, Brown M, Liu XS. 2011. BINOCh: binding inference from nucleosome occupancy changes. Bioinformatics 27:1867–1868. 10.1093/bioinformatics/btr279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28:495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang K, Bruce M, Pattillo CB, Zhang S, Stone R, II, Clifford J, Kevil CG. 2011. Temporal genomewide expression profiling of DSS colitis reveals novel inflammatory and angiogenesis genes similar to ulcerative colitis. Physiol. Genomics 43:43–56. 10.1152/physiolgenomics.00138.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Wong WH. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U. S. A. 98:31–36. 10.1073/pnas.98.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darsigny M, Babeu JP, Dupuis AA, Furth EE, Seidman EG, Levy E, Verdu EF, Gendron FP, Boudreau F. 2009. Loss of hepatocyte-nuclear-factor-4α affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One 4:e7609. 10.1371/journal.pone.0007609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, Pape UJ, Poidinger M, Chen Y, Yeung K, Brown M, Turpaz Y, Liu XS. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12:R83. 10.1186/gb-2011-12-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akhtar-Zaidi B, Cowper-Sal-lari, Corradin RO, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, Willis J, Moore JH, Tesar PJ, Laframboise T, Markowitz S, Lupien M, Scacheri PC. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336:736–739. 10.1126/science.1217277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M. 2012. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 44:1191–1198. 10.1038/ng.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. 1990. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 98:694–702 [DOI] [PubMed] [Google Scholar]
- 28.Oettgen P. 2006. Regulation of vascular inflammation and remodeling by ETS factors. Circ. Res. 99:1159–1166. 10.1161/01.RES.0000251056.85990.db [DOI] [PubMed] [Google Scholar]
- 29.Oliver JR, Kushwah R, Hu J. 2012. Multiple roles of the epithelium-specific ETS transcription factor, ESE-1, in development and disease. Lab. Invest. 92:320–330. 10.1038/labinvest.2011.186 [DOI] [PubMed] [Google Scholar]
- 30.Schonthaler HB, Guinea-Viniegra J, Wagner EF. 2011. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 70(Suppl 1):i109–i112. 10.1136/ard.2010.140533 [DOI] [PubMed] [Google Scholar]
- 31.Wang A, Al-Kuhlani M, Johnston SC, Ojcius DM, Chou J, Dean D. 2013. Transcription factor complex AP-1 mediates inflammation initiated by Chlamydia pneumoniae infection. Cell. Microbiol. 15:779–794. 10.1111/cmi.12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zingarelli B, Hake PW, Burroughs TJ, Piraino G, O'Connor M, Denenberg A. 2004. Activator protein-1 signalling pathway and apoptosis are modulated by poly(ADP-ribose) polymerase-1 in experimental colitis. Immunology 113:509–517. 10.1111/j.1365-2567.2004.01991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babeu JP, Darsigny M, Lussier CR, Boudreau F. 2009. Hepatocyte nuclear factor 4α contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 297:G124–G134. 10.1152/ajpgi.90690.2008 [DOI] [PubMed] [Google Scholar]
- 34.D'Angelo A, Bluteau O, Garcia-Gonzalez MA, Gresh L, Doyen A, Garbay S, Robine S, Pontoglio M. 2010. Hepatocyte nuclear factor 1α and β control terminal differentiation and cell fate commitment in the gut epithelium. Development 137:1573–1582. 10.1242/dev.044420 [DOI] [PubMed] [Google Scholar]
- 35.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. 2005. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology 128:935–945. 10.1053/j.gastro.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 36.McConnell BB, Kim SS, Bialkowska AB, Yu K, Sitaraman SV, Yang VW. 2011. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology 140:540–549. 10.1053/j.gastro.2010.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu T, Chen X, Zhang W, Li J, Xu R, Wang TC, Ai W, Liu C. 2012. Kruppel-like factor 4 regulates intestinal epithelial cell morphology and polarity. PLoS One 7:e32492. 10.1371/journal.pone.0032492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, Inoue Y. 2008. Hepatocyte nuclear factor 4α in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm. Bowel Dis. 14:908–920. 10.1002/ibd.20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tetreault MP, Alrabaa R, McGeehan M, Katz JP. 2012. Kruppel-like factor 5 protects against murine colitis and activates JAK-STAT signaling in vivo. PLoS One 7:e38338. 10.1371/journal.pone.0038338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, Nimmo ER, Massey D, Blaszczyk K, Elliott T, Cotterill L, Dallal H, Lobo AJ, Mowat C, Sanderson JD, Jewell DP, Newman WG, Edwards C, Ahmad T, Mansfield JC, Satsangi J, Parkes M, Mathew CG, Donnelly P, Peltonen L, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Duncanson A, Jankowski J, Markus HS, McCarthy MI, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Samani N, Trembath RC, Viswanathan AC, Wood N, Spencer CC, Bellenguez C, Davison D, Freeman C, Strange A, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Perez ML, Potter SC, Ravindrarajah R, Ricketts M, Waller M, Weston P, Widaa S, Whittaker P, Attwood AP, Stephens J, Sambrook J, Ouwehand WH, McArdle WL, Ring SM, Strachan DP. 2009. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 41:1330–1334. 10.1038/ng.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Sommeren S, Visschedijk MC, Festen EA, de Jong DJ, Ponsioen CY, Wijmenga C, Weersma RK. 2011. HNF4α and CDH1 are associated with ulcerative colitis in a Dutch cohort. Inflamm. Bowel Dis. 17:1714–1718. 10.1002/ibd.21541 [DOI] [PubMed] [Google Scholar]
- 43.Yang SK, Jung Y, Kim H, Hong M, Ye BD, Song K. 2011. Association of FCGR2A, JAK2 or HNF4A variants with ulcerative colitis in Koreans. Dig. Liver Dis. 43:856–861. 10.1016/j.dld.2011.07.006 [DOI] [PubMed] [Google Scholar]
- 44.Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. 2006. Hepatocyte nuclear factor 4α is essential for embryonic development of the mouse colon. Gastroenterology 130:1207–1220. 10.1053/j.gastro.2006.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cattin AL, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, Robine S, Pincon-Raymond M, Cardot P, Lacasa M, Ribeiro A. 2009. Hepatocyte nuclear factor 4α, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Biol. 29:6294–6308. 10.1128/MCB.00939-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, Clevers H, Liu XS, Shivdasani RA. 2010. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc. Natl. Acad. Sci. U. S. A. 107:15157–15162. 10.1073/pnas.1003822107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcil V, Seidman E, Sinnett D, Boudreau F, Gendron FP, Beaulieu JF, Menard D, Precourt LP, Amre D, Levy E. 2010. Modification in oxidative stress, inflammation, and lipoprotein assembly in response to hepatocyte nuclear factor 4α knockdown in intestinal epithelial cells. J. Biol. Chem. 285:40448–40460. 10.1074/jbc.M110.155358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. 2010. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology 51:642–653. 10.1002/hep.23357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckmann L, Nebelsiek T, Fingerle AA, Dann SM, Mages J, Lang R, Robine S, Kagnoff MF, Schmid RM, Karin M, Arkan MC, Greten FR. 2008. Opposing functions of IKKβ during acute and chronic intestinal inflammation. Proc. Natl. Acad. Sci. U. S. A. 105:15058–15063. 10.1073/pnas.0808216105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. 2004. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118:285–296. 10.1016/j.cell.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 51.Kajino-Sakamoto R, Inagaki M, Lippert E, Akira S, Robine S, Matsumoto K, Jobin C, Ninomiya-Tsuji J. 2008. Enterocyte-derived TAK1 signaling prevents epithelium apoptosis and the development of ileitis and colitis. J. Immunol. 181:1143–1152. 10.4049/jimmunol.181.2.1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, Gumucio D, Neurath MF, Pasparakis M. 2007. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 446:557–561. 10.1038/nature05698 [DOI] [PubMed] [Google Scholar]
- 53.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. 2008. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol. 180:2588–2599. 10.4049/jimmunol.180.4.2588 [DOI] [PubMed] [Google Scholar]
- 54.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39:186–193. 10.1002/gene.20042 [DOI] [PubMed] [Google Scholar]
- 55.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. 2001. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21:1393–1403. 10.1128/MCB.21.4.1393-1403.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karczewski KJ, Dudley JT, Kukurba KR, Chen R, Butte AJ, Montgomery SB, Snyder M. 2013. Systematic functional regulatory assessment of disease-associated variants. Proc. Natl. Acad. Sci. U. S. A. 110:9607–9612. 10.1073/pnas.1219099110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, Yao K, Kehoe SM, Lenz HJ, Haiman CA, Yan C, Henderson BE, Frenkel B, Barretina J, Bass A, Tabernero J, Baselga J, Regan MM, Manak JR, Shivdasani R, Coetzee GA, Freedman ML. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 41:882–884. 10.1038/ng.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. 2012. Linking disease associations with regulatory information in the human genome. Genome Res. 22:1748–1759. 10.1101/gr.136127.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mokry M, Middendorp S, Wiegerinck CL, Witte M, Teunissen H, Meddens CA, Cuppen E, Clevers H, Nieuwenhuis EE. 2013. Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology 146:1040–1047. 10.1053/j.gastro.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 61.Mizutani A, Koinuma D, Tsutsumi S, Kamimura N, Morikawa M, Suzuki HI, Imamura T, Miyazono K, Aburatani H. 2011. Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4α in HepG2 cells. J. Biol. Chem. 286:29848–29860. 10.1074/jbc.M110.217745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378–1381. 10.1126/science.1089769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328:1036–1040. 10.1126/science.1186176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, Crowley D, Whittaker CA, Meyerson M, Kimura S, Jacks T. 2013. Nkx2–1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol. Cell 50:185–199. 10.1016/j.molcel.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MS, Komorowski J, Wadelius C. 2009. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 37:7498–7508. 10.1093/nar/gkp823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lussier CR, Babeu JP, Auclair BA, Perreault N, Boudreau F. 2008. Hepatocyte nuclear factor-4α promotes differentiation of intestinal epithelial cells in a coculture system. Am. J. Physiol. Gastrointest. Liver Physiol. 294:G418–G428. 10.1152/ajpgi.00418.2007 [DOI] [PubMed] [Google Scholar]
- 67.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. 2011. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147:1233–1247. 10.1016/j.cell.2011.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Azmi AS, Bao GW, Gao J, Mohammad RM, Sarkar FH. 2013. Network insights into the genes regulated by hepatocyte nuclear factor 4 in response to drug induced perturbations: a review. Curr. Drug Discov. Technol. 10:147–154. 10.2174/1570163811310020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hwang-Verslues WW, Sladek FM. 2010. HNF4α—role in drug metabolism and potential drug target? Curr. Opin. Pharmacol. 10:698–705. 10.1016/j.coph.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. 2009. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One 4:e5609. 10.1371/journal.pone.0005609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. 2011. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 3:96ra76. 10.1126/scitranslmed.3002648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.