Abstract
While the E2F transcription factors (E2Fs) have a clearly defined role in cell cycle control, recent work has uncovered new functions. Using genomic signature methods, we predicted a role for the activator E2F transcription factors in the mouse mammary tumor virus (MMTV)-polyomavirus middle T oncoprotein (PyMT) mouse model of metastatic breast cancer. To genetically test the hypothesis that the E2Fs function to regulate tumor development and metastasis, we interbred MMTV-PyMT mice with E2F1, E2F2, or E2F3 knockout mice. With the ablation of individual E2Fs, we noted alterations of tumor latency, histology, and vasculature. Interestingly, we noted striking reductions in metastatic capacity and in the number of circulating tumor cells in both the E2F1 and E2F2 knockout backgrounds. Investigating E2F target genes that mediate metastasis, we found that E2F loss led to decreased levels of vascular endothelial growth factor (Vegfa), Bmp4, Cyr61, Nupr1, Plod 2, P4ha1, Adamts1, Lgals3, and Angpt2. These gene expression changes indicate that the E2Fs control the expression of genes critical to angiogenesis, the remodeling of the extracellular matrix, tumor cell survival, and tumor cell interactions with vascular endothelial cells that facilitate metastasis to the lungs. Taken together, these results reveal that the E2F transcription factors play key roles in mediating tumor development and metastasis in addition to their well-characterized roles in cell cycle control.
INTRODUCTION
Breast cancer remains a leading cause of death for women, with high mortality rates attributed to distant metastasis (1). To simplify the examination of signaling pathway requirements in metastatic breast cancer, research has turned to mouse model systems. Previous studies with mouse models of breast cancer have begun to reveal the mechanistic features of breast cancer metastasis, and in vivo selection has demonstrated the ability to select for tumors that metastasize to a specific location (2–5). Yet we lack a complete understanding of the pathways that govern the molecular circuitry of metastatic breast cancer. One model that has been integral in examining metastatic progression is the mouse mammary tumor virus (MMTV)-polyomavirus middle T oncoprotein (PyMT) model. Originally described as exhibiting rapid tumor onset and a high degree of pulmonary metastasis (6), this model has since been used to examine a number of facets of metastasis. For example, work using the MMTV-PyMT model led to the discovery of the prometastatic signaling exchange between tumors and macrophages (7). In addition, the metastatic contributions of individual signaling molecules, such as transforming growth factor β (TGF-β), AKT, and adiponectin, have also been uncovered using this model (8–10). Given that PyMT can activate multiple signaling pathways with relevance to human breast cancer (11), there is clear utility in this model for characterizing pathways that contribute to breast cancer metastasis.
The identification of signaling pathways contributing to tumor progression has been enhanced by recent progress in bioinformatic methods. One such method is the development of genomic signatures for determining signaling pathway activation status (12, 13). By generating gene expression training data for cell signaling pathways, a signature can be created and applied to subsequent gene expression data sets to predict whether the pathway in question is activated. This method has demonstrated heterogeneity in human breast cancer (14), moving beyond and refining the intrinsic classification of breast cancer (15, 16). In addition, this method has demonstrated tumor heterogeneity in mouse models of breast cancer (17, 18). As a predictive tool, genetic signatures allow the identification of signaling pathways that may contribute to tumor development. Indeed, the application of genomic signatures to Myc-induced tumors revealed that E2F transcription factors (E2Fs) were predicted to function in tumorigenesis, and a genetic test of this prediction demonstrated that E2Fs were involved in tumor onset and progression (19). Using a similar approach, the current study predicted a role for the activator E2F transcription factors in MMTV-PyMT-induced tumorigenesis.
The E2F transcription factor family is broadly classified into transcriptional activators (E2F1 to E2F3A) and repressors (E2F3B to E2F8), and the family members have been well characterized as regulators of the cell cycle (20–22). Prior data implicating the E2Fs in human cancer show that they are important regulators of apoptosis and proliferation (23). However, recent work has identified roles for E2Fs beyond simple cell cycle regulation (24). For example, a xenograft study utilizing short hairpin RNA (shRNA) knockdown of E2F1 in melanoma cell lines showed that E2F1 knockdown significantly reduced the sizes of pulmonary metastases (25). In esophageal squamous cell carcinoma, it was shown that patients with tumors that immunostained positive for E2F1 had a lower overall survival rate than patients with E2F1-negative tumors (26). Similarly, in prostate cancer, patients with detectable nuclear staining for E2F3 had worse prognoses than patients for whom E2F3 was undetectable (27). Furthermore, we demonstrated recently that E2Fs also play a role in relapse-free survival time in human breast cancer (19). Together, these prior studies demonstrate the clinical significance of the E2Fs in human cancer. Here we used genomic signatures to predict that E2F transcription factors are involved in a mouse model of breast cancer metastasis. We then genetically demonstrated that loss of E2F in MMTV-PyMT tumors alters tumor development, progression, and metastasis. Taken together, these data indicate that E2F transcription factors play a critical role in the regulation of metastasis.
MATERIALS AND METHODS
Bioinformatics.
Gene expression data for MMTV-PyMT tumor samples and cell lines were obtained from Gene Expression Omnibus (GEO) under accession numbers GSE13553, GSE13221, and GSE14457. These data were combined with data for other mouse mammary tumor models, with accession numbers GSE11259, GSE13230, GSE15904, GSE24594, GSE22406, GSE6246, GSE6453, GSE6581, GSE7595, GSE8516, GSE8828, GSE8863, GSE9343, GSE9355, GSE10450, GSE13259, GSE13916, GSE14226, GSE14457, GSE14753, GSE15119, GSE15263, GSE15632, GSE16110, and GSE17916. Batch effects between Affymetrix-derived data sets were removed using Bayesian factor regression modeling (BFRM) (28). COMBAT (29) was used to remove batch effects between Agilent and Affymetrix data sets. Pathways were predicted as described previously using a gene signature approach (12, 14, 30). Briefly, pathway predictions were made using gene expression training data for individual oncogenic pathways. By comparing oncogene overexpression to expression in control samples, the training data provided the transcriptional response of oncogenic pathway activation. For each training data set, metagene scores were calculated and signature genes were identified. Bayesian fitting of probit binary regression models was used to map MMTV-PyMT tumors to the metagene signature and to calculate the probability of activation of individual oncogenic pathways by utilizing BINREG software in Matlab. Metastasis gene sets were downloaded from the Molecular Signatures Database (MSigDB) (www.broadinstitute.org). Chromatin immunoprecipitation-DNA sequencing (ChIP-seq) data and chromatin immunoprecipitation with microarray technology (ChIP-chip) data for E2F1, E2F2, and E2F3 were obtained from previous publications (31–34). Unsupervised hierarchical clustering was performed with Cluster 3.0, and the results were visualized with Java TreeView. Figures were converted to a full-spectrum color scale using Matlab. To examine human breast cancer, we used the Kaplan-Meier plotter (KM-plot) (35) to examine distant-metastasis-free survival (DMFS).
Animal studies.
All animal work has been conducted according to national and institutional guidelines. All mice were in the FVB background. MMTV-PyMT634 mice were obtained from The Jackson Laboratory. PyMT transgenic mice were interbred with E2F1−/− (36), E2F2−/− (37), or E2F3+/− (38) mice. A very small number of mice of each genotype with runted growth were excluded from the experiment due to failure to thrive. Tumors were detected through weekly palpation, and tumor growth was measured twice per week. Kaplan-Meier curves for tumor latency were generated using GraphPad Prism. The tumor growth rate was measured by the amount of time it took for the primary tumor to reach a volume of 2,000 mm3 after palpation.
Mammary whole mounts were conducted as described previously (30). Once the primary tumor reached the approved endpoint, the number of lung metastases and the percentage of the area of the lungs occupied by metastases were quantified across both lobes of a single section of hematoxylin-and-eosin (H&E)-stained lungs. GraphPad Prism was used to conduct the Mann-Whitney statistical test.
Immunohistochemistry was performed on sections of mammary glands and representative end-stage tumors using the following antibodies: an anti-F4/80 rat monoclonal antibody (Q61549) from Serotec, anti-Ki67 (Ab15580) from Abcam, and anti-CD31 (DIA-310; Dianova) from HistoBioTec. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was done using the in situ cell death detection kit (catalog no. 11684817910) from Roche and a diaminobenzidine (DAB) substrate kit (SK-4100) from Vector Laboratories. For Western blotting to assess E2F3 protein levels, the primary antibody (sc-878) was purchased from Santa Cruz and was imaged using a goat anti-rabbit secondary antibody (product no. 326-32211) from Li-Cor.
A colony-forming assay was conducted based on previously published methods (39). Colonies were quantified using ImageJ software. GraphPad Prism was used to conduct the nonparametric t test using Mann-Whitney methods.
RNA was extracted from mammary glands, tumors, and blood cells by using the QIAamp RNA blood minikit (catalog no. 52304; Qiagen). Quantitative reverse transcription-PCR (RT-PCR) was performed using the QuantiTect SYBR green RT-PCR kit (catalog no. 204243; Qiagen). The primer sequences for the simian virus 40 (SV40) poly(A) transgene marker were GGAACCTTACTTCTGTGGTGT (SV40-F) and GGAAAGTCCTTGGGGTCTTCT (SV40-R). The other primers used were Actin-F (CATCATGCGTCTGGACCTG) and Actin-R (CTCACGTTCAGCTGTGGTCA), Adamts1-F (GATAATGGACACGGGGAATG) and Adamts1-R (GATAATGGACACGGGGAATG), Angpt2-F (GCCCAAGTACTAAACCAGACG) and Angpt2-R (CACTGGTCTGATCCAAAATCTG), Bmp4-F (CAATGGAGCCATTCCGTAGT) and Bmp4-R (CATGATTCTTGGGAGCCAAT), Gapdh-F (TCATGACCACAGTGGATGCC) and Gapdh-R (GGAGTTGCTGTTGAAGTCGC), Cyr61-F (ACGAGGACTGCAGCAAAACT) and Cyr61-R (TGAGCTCTGCAGATCCCTTT), Lgals3-F (CAACGCAAACAGGATTGTTC) and Lgals3-R (CGTGTTACACACAATGACTCTCC), Nupr1-F (CAATACCAACCGCCCTAGC) and Nupr1-R (CCTTATCTCCAGCTCCGTCTC), P4ha1-F (ACCTGTGAAGTTCCCCAAGA) and P4ha1-R (CAGTCATCTGACCAATTGACGTA), Plod2 Isoform 1-F (CCCCAAAGGGTGTGTTTATG) and Plod2 Isoform 1-R (TTCAAAAATCTGCCAGAAGTCA), Plod2 Isoform 2-F (TCAAGGAAAGACACTCCGATCT) and Plod2 Isoform 2-R (AACACACCCATATCTCTAGCATTG), and Vegfa-F (CAGGCTGCTGTAACGATGAA) and Vegfa-R (GCATTCACATCTGCTGTGCT).
Tumor cells were obtained from viably frozen tumor tissue (40) and were cultured for 2 passages prior to colonization assays. Adherent proliferating tumor cells were counted, and 5.0 × 105 cells were injected retro-orbitally into MMTV-Cre wild-type (WT) control mice. Mice were examined 35 days postinjection, and lungs were paraffin embedded prior to routine H&E staining.
Tumor transplant studies were conducted using viable frozen tumor samples generated during the study of tumor development and metastasis using the transgenic mice. Tumors (E2FWT/WT, E2F1−/−, or E2F2−/−) were implanted into the mammary fat pads of MMTV-Cre wild-type control mice. Once the primary tumor reached the endpoint, lung metastases and the percentage of the area of the lungs occupied by metastases were examined by routine histology and were quantified. GraphPad Prism was used to conduct the nonparametric t test using Mann-Whitney methods.
In vitro assays.
Tumor cells were obtained from viable frozen tumor tissue and were cultured for use in a wound-healing assay in the presence of 2 μg/ml mitomycin C using standard methods (41). Photomicrographs were taken at 0 h, 12 h, 24 h, 36 h, and 48 h.
Tumor cells were obtained from viable frozen tumor tissue, cultured, and serum starved for 24 h for use in a transwell invasion assay according to standard protocols (41). Serum-starved cells were resuspended in a serum-free medium with 2 μg/ml mitomycin C and were seeded at a density of 3.0 × 105 cells on the insert. Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 2 μg/ml mitomycin C was used as a chemoattractant. Cells were allowed to migrate for 6 h prior to fixation with 3% paraformaldehyde and staining with crystal violet.
RESULTS
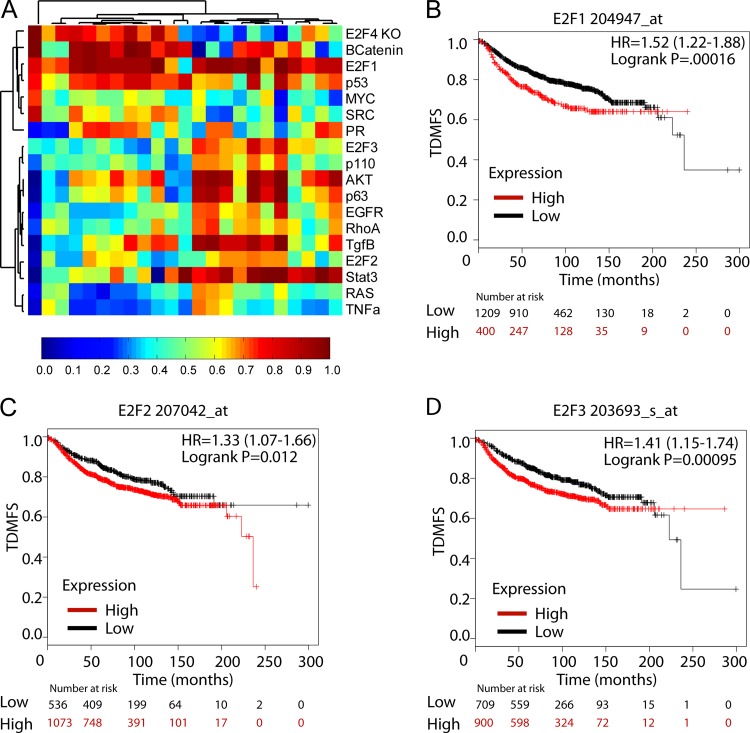
To identify signaling pathways associated with the metastatic progression of breast cancer, we applied a number of genomic signaling signatures to gene expression data from mouse models of breast cancer. Using previously described training data and methods (12–14, 17), we predicted signaling pathway activity across these models, which included the highly metastatic PyMT tumor model. When we examined the pathway activation predictions in the PyMT tumor model (Fig. 1A), we noted activation in a number of pathways known to be critical in PyMT tumors, such as AKT (8). We also observed a surprising degree of genomic heterogeneity in the tumor samples. Despite this heterogeneity, we found that virtually all samples had high levels of predicted activity for the E2F1 transcription factor. The two other activator E2Fs were also observed to have elevated probabilities of activation in a subset of tumor samples. The activation of E2F signatures in the majority of samples indicated that the E2F transcription factors may be involved in PyMT-mediated tumorigenesis.
FIG 1.
Pathway signatures predict E2F activation in metastatic breast tumors. (A) Heat map showing the probabilities of activation in transgenic MMTV-PyMT tumors for the signaling pathways listed on the right. Below the heat map, a scale bar depicts the range of probabilities from 0 to 1. The probabilities were used in unsupervised hierarchical clustering of pathways. Clusters were identified on the basis of pathway activation in the transgenic tumor samples. (B to D) Kaplan-Meier plots showing metastasis-free survival times for the indicated proportions of breast cancer patients (n, 1,610) stratified on the basis of expression of E2F1 (Affymetrix probe 204947_at) (P, 0.00016) (B), E2F2 (Affymetrix probe 207042_at) (P, 0.012) (C), or E2F3 (Affymetrix probe 203693_s_at) (P, 0.00095) (D). Patient stratification was conducted using the KM-plot autoselection option. HR, hazard ratio. TDMFS, distant-metastasis-free survival as a function of time.
PyMT-induced tumors are highly metastatic and have been used previously to examine the metastatic process. Accordingly, we used the Kaplan-Meier Plotter tool (KM-plot) to screen human breast cancer DMFS (distant-metastasis-free survival) clinical data for associations between individual E2F1, E2F2, and E2F3 probes or upregulated signature genes and DMFS times in human breast cancer. Importantly, elevated expression levels of either E2F1 (hazard ratio, 1.52 [95% confidence interval {95% CI}, 1.22 to 1.88]; P, 0.00016), E2F2 (hazard ratio, 1.33 [95% CI, 1.07 to 1.66]; P, 0.012), or E2F3 (hazard ratio, 1.41 [95% CI, 1.15 to 1.74]; P, 0.00095) in breast cancer patients were individually associated with shorter times to distant metastasis than those for patients whose tumors exhibited low levels of expression (Fig. 1B to D, respectively). Similarly, we assessed the upregulated genes in E2F1, E2F2, and E2F3 pathway signatures. Elevated expression of E2F1 (hazard ratio, 1.46 [95% CI, 1.19 to 1.79]; P, 0.00024) and E2F2 (hazard ratio, 1.52 [95% CI, 1.25 to 1.87]; P, 0.000037) signature genes also correlated with shorter times to distant metastasis in breast cancer patients than those for patients with low expression of these genes (see Fig. S1A and B in the supplemental material). In contrast, high expression of E2F3 signature genes (hazard ratio, 0.73 [95% CI, 0.59 to 0.91]; P, 0.0052) correlated with prolonged times to distant metastasis in breast cancer patients (see Fig. S1C). Taking into account intrinsic subtype status, we found that high levels of E2F1 and E2F1 signature genes were associated with a decreased time to distant metastasis in luminal A and luminal B breast cancers (see Fig. S2 in the supplemental material). High levels of E2F2 predicted a decreased time to metastasis in luminal A breast cancer, while E2F2 signature genes were similarly associated in both luminal subtypes (see Fig. S3 in the supplemental material). For E2F3, elevated probe levels and signature genes were not associated with decreased times to metastasis in a particular subtype (see Fig. S4 in the supplemental material). Considered with the PyMT mouse model data, these findings strongly suggested that activator E2F transcription factors have a functional role in breast cancer progression and metastasis.
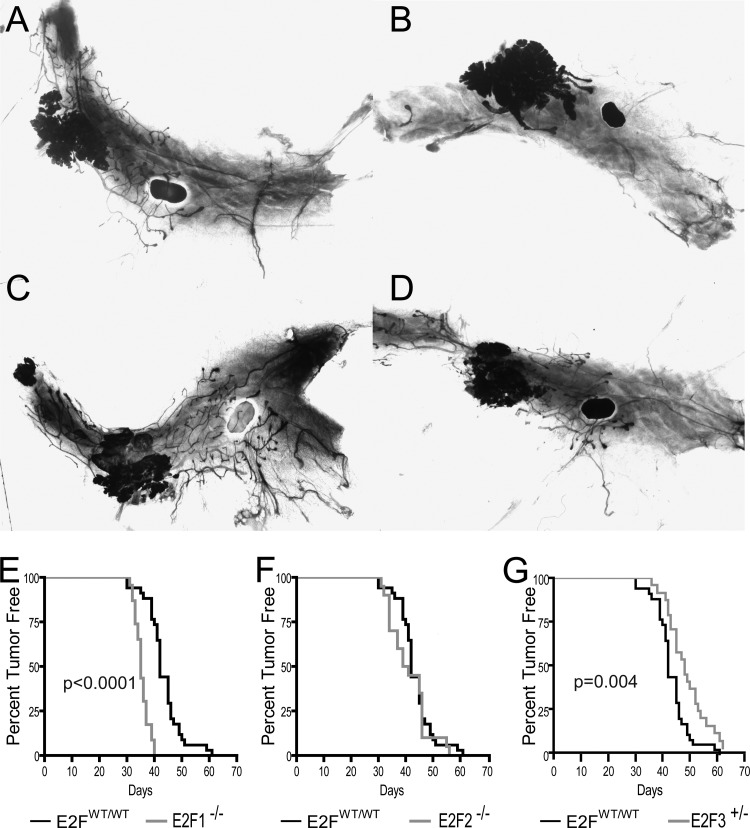
To test the hypothesis that E2Fs regulate metastatic breast cancer, we interbred MMTV-PyMT mice with mice in the E2F1, E2F2 and E2F3 knockout backgrounds. Due to embryonic lethality, E2F3 mice were maintained in the heterozygous state. To study E2F function in mammary tumor development, we examined mammary whole mounts of 35-day-old virgin MMTV-PyMT mice in the various E2F backgrounds (Fig. 2A to D). In all E2F backgrounds, transformation of the ductal tree was evident through whole-mount analysis. In comparison to the wild-type E2F control background (Fig. 2A), normal ductal epithelium was consistently absent in E2F1−/− mammary glands (Fig. 2B). Unlike the loss of E2F1, the loss of E2F2 or E2F3 did not result in mammary glands appreciably different from those of controls (Fig. 2C and D, respectively).
FIG 2.
Loss of E2Fs alters tumor onset. (A to D) Representative mammary whole mounts from 35-day-old virgin MMTV-PyMT E2FWT/WT (A), E2F1−/− (B), E2F2−/− (C), and E2F3+/− (D) mice. (E) The latencies of PyMT-induced tumors in wild-type (E2FWT/WT) (n, 34) and E2F1−/− (n, 22) backgrounds were compared in a Kaplan-Meier plot, revealing a significant acceleration of tumor onset with the loss of E2F1 (P, <0.0001). (F) Kaplan-Meier plot for tumor onset in the E2FWT/WT and E2F2−/− backgrounds (n, 34 and 20, respectively). (G) Kaplan-Meier plot for tumor onset in the E2FWT/WT and E2F3+/− backgrounds (n, 34 and 24, respectively), revealing a significant delay with the loss of E2F3 (P, 0.004).
Given that the loss of E2F1 resulted in transformation of the entire mammary epithelium, we postulated that this transformation should result in acceleration of tumor onset. Mammary glands were regularly palpated for the presence of mammary tumors. In agreement with the mammary whole-mount results, we noted a significant (P, <0.0001) acceleration of tumor onset in E2F1−/− mice relative to onset in the control wild-type E2F background (Fig. 2E). Indeed, 50% of control mice had developed tumors by 42 days (MMTV-PyMT median latency, 42 days), while the loss of E2F1 resulted in tumors in 50% of mice by 35 days (MMTV-PyMT E2F1−/− median latency, 35 days; hazard ratio, 0.2507 [95% CI, 0.02660 to 0.1458]). In contrast, no differences in latency were associated with the loss of E2F2 (median latency, 40 days; hazard ratio, 0.8195 [95% CI, 0.4109 to 1.461]) (Fig. 2F). However, E2F3 heterozygous (E2F3+/−) mice had a significant delay (P, 0.004) in tumor onset (median latency, 48 days; hazard ratio, 1.955 [95% CI, 1.297 to 4.124]) (Fig. 2G). Together these data demonstrate the differential roles of the E2Fs during the initiation of tumor development.
In order to examine the role of the E2F transcription factors in tumor proliferation, we compared the growth rates of PyMT-induced primary tumors in the various E2F backgrounds. Despite the differences in tumor onset, no significant alterations in the time from tumor palpation to the end stage were observed with loss of the E2Fs (see Fig. S5A in the supplemental material). In addition, we assessed the tumor burden at the endpoint, when the primary tumor reached 20 mm in the largest dimension. While no differences were observed for the E2F1−/− or E2F2−/− background, fewer tumors developed in the E2F3+/− background (P, 0.01) (see Fig. S5B). However, when total tumor volume was observed, the E2F3 mutants were indistinguishable from the wild-type E2F controls (see Fig. S5C). Moreover, Ki67 staining (see Fig. S6 in the supplemental material) and TUNEL staining (see Fig. S7 in the supplemental material) in early-stage (tumor diameter, 6 mm) and end-stage tumors indicated that E2F1 loss had no effect on tumor cell proliferation or apoptosis. Together, these data indicate that despite alterations to tumor latency, E2F loss had surprisingly few effects on growth rate, tumor burden, proliferation, or apoptosis in MMTV-PyMT tumors.
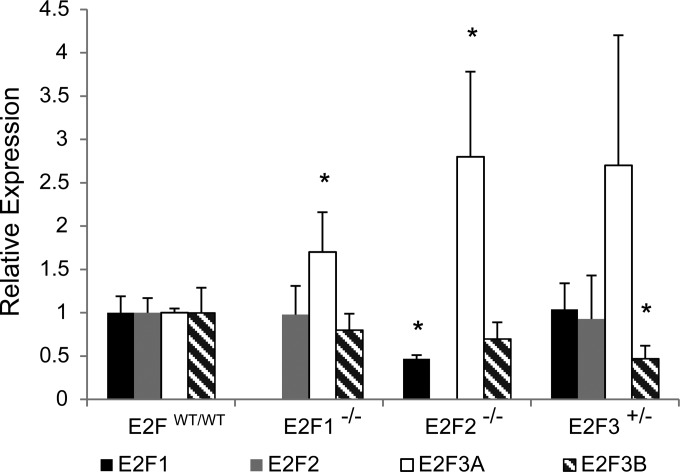
Since E2F loss had no impact on these features of tumor growth, we investigated whether compensatory upregulation of other E2F family members had occurred. To do this, we assayed levels of E2F1, E2F2, E2F3A, and E2F3B by RT-quantitative PCR (qRT-PCR) across tumor genotypes (Fig. 3). Compared to E2FWT/WT tumors (n, 4), E2F1−/− tumors (n, 4) showed similar levels of E2F2 and E2F3B but significant upregulation of E2F3A (P, 0.0232). In E2F2−/− tumors (n, 4), we detected a significant decrease in E2F1 levels (P, 0.0016) and significant upregulation of E2F3A (P, 0.0105). In E2F3+/− tumors, expression levels of E2F1 and E2F2 were similar to those in E2FWT/WT tumors. Interestingly, E2F3+/− mice had upregulation of E2F3A bordering on statistical significance (P, 0.0641) and significant downregulation of E2F3B (P, 0.0175). The mammary glands of 35-day-old E2F3+/− mice (n, 4) showed E2F3 protein levels slightly lower than those in the mammary glands of E2FWT/WT mice (n, 4) (see Fig. S8A in the supplemental material). In early-stage E2F3+/− tumors (diameter, 6 mm) (n, 3), we detected E2F3 levels similar to those in E2FWT/WT tumors (n, 3) (see Fig. S8B). Similar observations were made for end-stage tumors (diameter, 20 mm) (n, 4 for each genotype), although E2F3 protein levels were more variable (see Fig. S8C). As a whole, these results demonstrate that E2F loss in these tumors led to compensatory upregulation of the E2F3A isoform.
FIG 3.
E2F loss results in changes in the expression of the genes of other E2F family members. Shown are quantitative PCR results for the relative expression of E2F1, E2F2, E2F3A, and E2F3B in E2FWT/WT, E2F1−/−, E2F2−/−, and E2F3+/− tumors (n, 4 for each genotype). E2F1−/− tumors had upregulation of E2F3A (P, 0.0232). In E2F2−/− tumors, a decrease in E2F1 levels (P, 0.0016) and significant upregulation of E2F3A (P, 0.0105) were observed. In E2F3+/− tumors, significant downregulation of E2F3B (P, 0.0175) was observed.
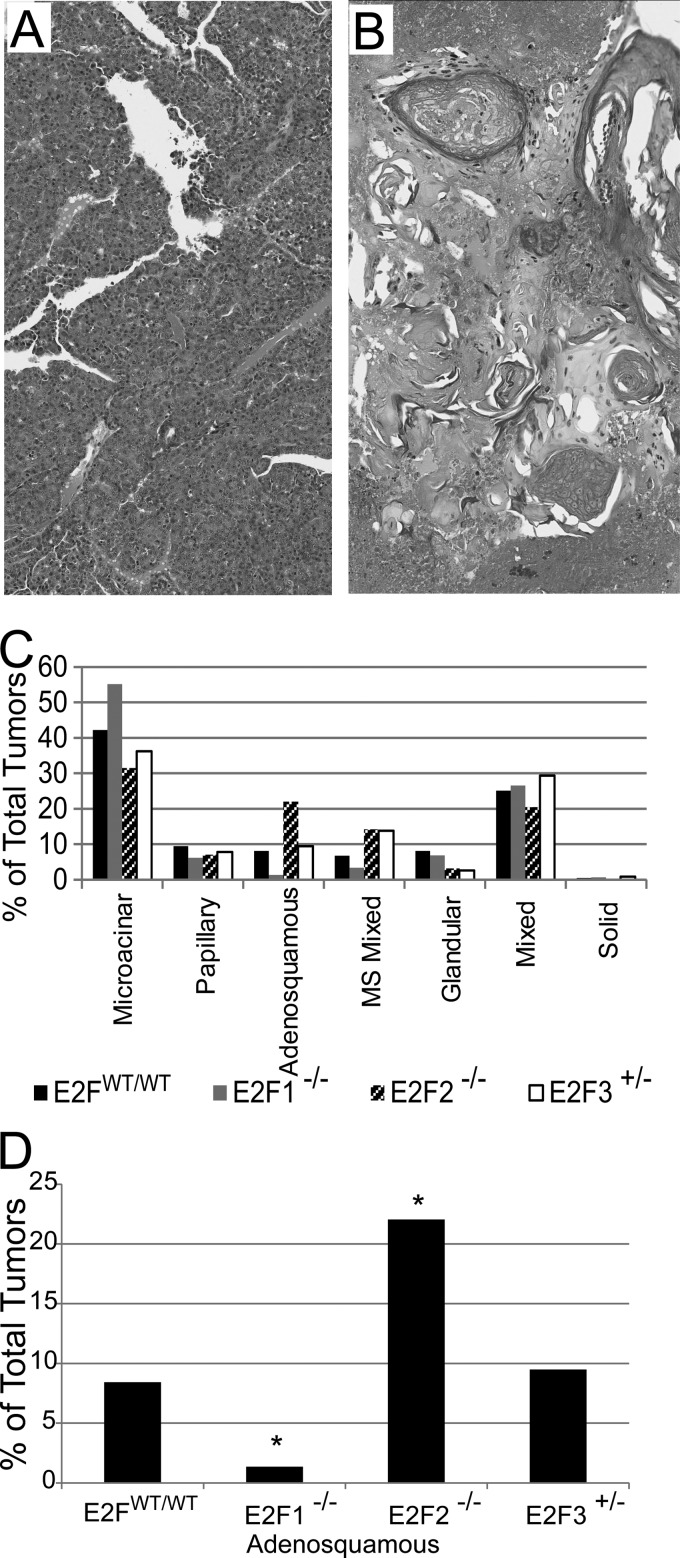
In addition to effects on latency, histological patterns observed in the tumors indicated that the E2F transcription factors play a role in tumor heterogeneity. In all backgrounds, the most common histological type of tumor was the microacinar subtype (Fig. 4A). In addition, we frequently noted adenosquamous tumors (Fig. 4B), as well as a number of other types at reduced frequencies across genetic backgrounds (Fig. 4C). The frequency of adenosquamous tumors was noticeably affected in E2F1−/− and E2F2−/− mice. Indeed, loss of E2F1 significantly reduced the frequency of adenosquamous tumors, from 8% to 1% (P, 0.001) (Fig. 4D). Conversely, loss of E2F2 significantly increased the proportion of adenosquamous tumors to 21% of all tumors (P, 0.0003). In contrast to the alterations in histology found with E2F1 and E2F2, no effects on tumor histology were noted for E2F3 mutants.
FIG 4.
Loss of E2Fs alters tumor histology. (A and B) H&E staining revealed microacinar tumors (A) and adenosquamous tumors (B) among other tumor types. (C) Proportions of different histological types of tumors in MMTV-PyMT mice of different genotypes. MS Mixed, mixture of microacinar and adenosquamous tumors. (D) Percentages of adenosquamous tumors within populations of E2F wild-type and mutant mice. Fisher's exact test was used to compare E2FWT/WT with E2F1−/− (P, 0.001) and E2F2−/− (P, 0.0003) populations.
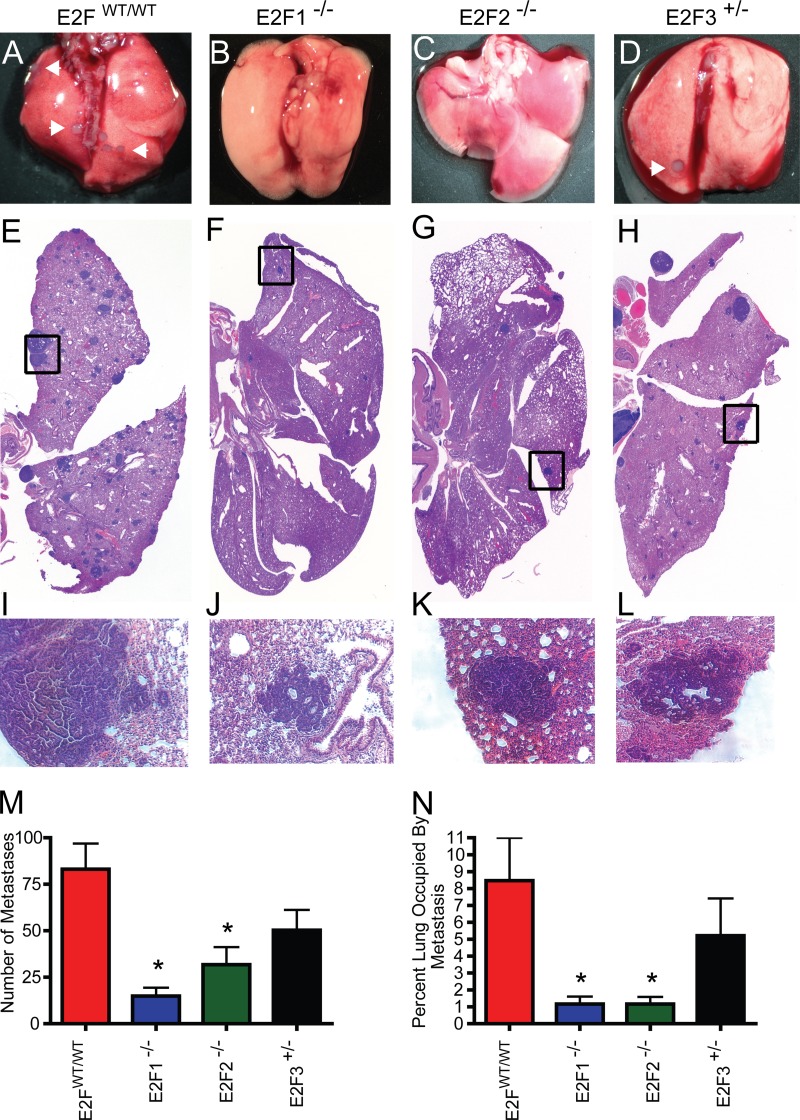
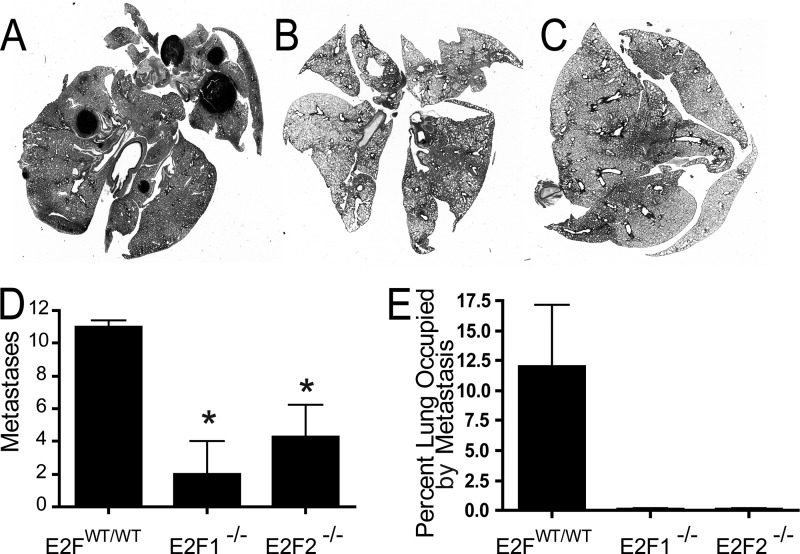
Since we predicted a high probability of E2F activation in the highly metastatic MMTV-PyMT mouse model and noted that high E2F levels were associated with decreased time to distant metastasis in human breast cancer, we hypothesized that the E2Fs were involved in breast cancer metastasis. To test this hypothesis, we examined the lungs of MMTV-PyMT mice in the various E2F backgrounds at the endpoint. Metastatic tumors were readily observed on the surfaces of the lungs of control MMTV-PyMT mice (Fig. 5A). Interestingly, we did not observe these metastases in the E2F1−/− or E2F2−/− background (Fig. 5B and C, respectively) but did note metastatic tumors on the surfaces of the lungs in the E2F3+/− background (Fig. 5D). Histological examination of matched sections of the lungs demonstrated widespread metastasis in the lungs of E2FWT/WT mice (Fig. 5E; histology for the median number of metastatic lesions is shown). In accordance with the gross observations, there were readily apparent decreases in the numbers of metastatic lesions in E2F1−/− and E2F2−/− mice (Fig. 5F and G) but not in E2F3 mutant mice (Fig. 5H). The metastases in the boxed areas of Fig. 5E to H are shown at a higher magnification in Fig. 5I to L. To quantitate the metastases, they were counted in a representative section of the lung for each of the tumor-bearing mice (37 E2FWT/WT mice, 21 E2F1−/− mice, 21 E2F2−/− mice, and 23 E2F3+/− mice). This revealed significant reductions in the numbers of metastases in the lungs of mice in both the E2F1−/− (P, <0.0001) and E2F2−/− (P, 0.002) backgrounds (Fig. 5M). Since many of the metastases noted were smaller in the E2F1−/− and E2F2−/− backgrounds, we also examined the area occupied by the metastases as a function of the total lung area. This demonstrated that loss of either E2F1 or E2F2 significantly reduced the metastatic burden (P, <0.0001 for both) (Fig. 5N).
FIG 5.
Loss of E2Fs decreases the level of pulmonary metastasis in MMTV-PyMT mice. (A to D) Representative wet-mount images of lungs from E2FWT/WT, E2F1−/−, E2F2−/−, and E2F3+/− mice. Arrowheads indicate surface metastases. (E to H) Representative images for H&E-stained sections of lungs. (I to L) Tenfold-enlarged images of the regions boxed in panels E to H show the histology of tumor metastases. (M) Comparison of the average numbers of lung metastases (with standard errors of the means) in pulmonary sections from E2FWT/WT mice with those for mice in E2F mutant backgrounds revealed significant differences in E2F1−/− (P, <0.0001) and E2F2−/− (P, 0.002) mice. (N) Comparison of the average percentage (with standard errors of the means) of the lung area occupied by metastases in pulmonary sections from E2FWT/WT mice with those for E2F1−/− mice (P, <0.0001), E2F2−/− mice (P, <0.0001), and E2F3+/− mice.
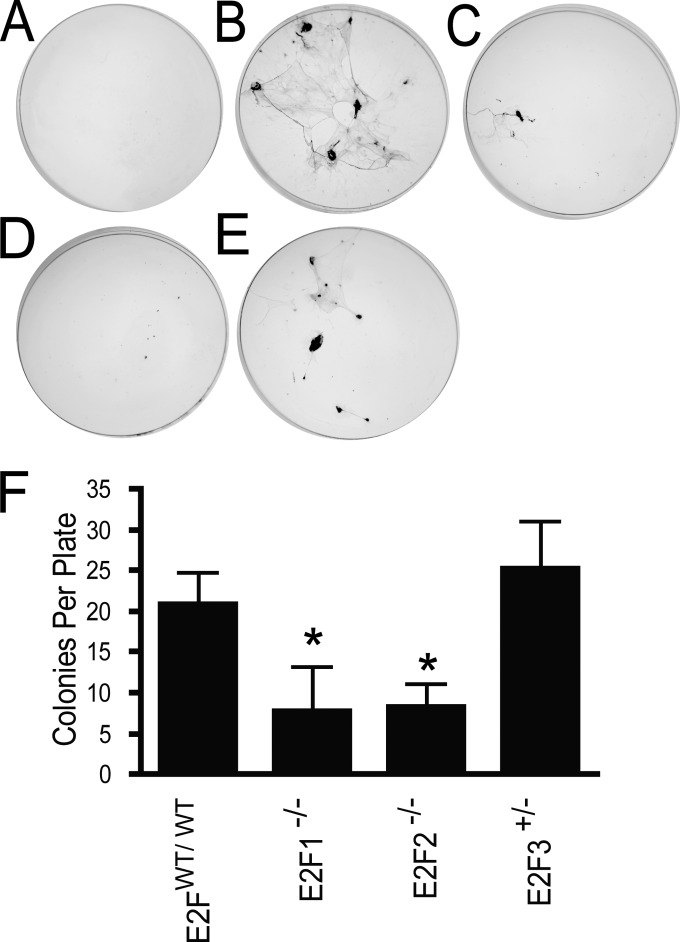
To determine the stage at which E2F1 or E2F2 loss blocked metastasis, we assayed the number of circulating tumor cells (CTCs) at the endpoint in tumor-bearing mice. To detect CTCs, we collected blood by cardiac puncture and cultured the CTCs in a colony-forming assay. Compared to age-matched wild-type tumor-free controls (n, 6), in which no colonies were detected (Fig. 6A), mice of the MMTV-PyMT strain (n, 14) were found to have a number of discrete colonies (Fig. 6B). The number of CTCs was visibly reduced in both E2F1−/− (n, 7) (Fig. 6C) and E2F2−/− (n, 10) (Fig. 6D) mice but not in E2F3+/− (n, 10) mice (Fig. 6E). Quantitation revealed that this decrease in the number of CTC colonies from that for the PyMT control was significant in the E2F1−/− (P, 0.02) and E2F2−/− (P, 0.006) mice (Fig. 6F). A reduction in the number of CTCs was confirmed by extracting RNA from blood at the tumor end stage and using qRT-PCR to assay transgene expression as an indicator of circulating tumor cells. This demonstrated that E2F1−/− mice had transgene levels in blood 4.8-fold-lower than those for controls (see Fig. S9 in the supplemental material), indicating a reduction in the number of circulating tumor cells. Further, in FVB negative controls, we observed no amplification of the transgene. Taken together, these results strongly suggest that loss of E2F1 or E2F2 inhibits metastasis by reducing the number of circulating tumor cells.
FIG 6.
Loss of E2Fs decreases the number of circulating tumor cells in MMTV-PyMT mice. (A to E) Representative colony-forming assay plates for negative-control, nontransgenic FVB mice (n, 6) (A), MMTV-PyMT E2FWT/WT mice (n, 14) (B), MMTV-PyMT E2F1−/− mice (n, 7) (C), MMTV-PyMT E2F2−/− mice (n, 10) (D), and MMTV-PyMT E2F3+/− mice (n, 10) (E). (F) Comparison of the average numbers of circulating tumor cells (with standard errors of the means) detected in MMTV-PyMT E2FWT/WT mice, MMTV-PyMT E2F1−/− mice (P, 0.02), MMTV-PyMT E2F2−/− mice (P, 0.006), and MMTV-PyMT E2F3+/− mice.
To test whether E2F1 or E2F2 also regulates colonization ability, we injected 5.0 × 105 cells derived from E2FWT/WT, E2F1−/−, or E2F2−/− PyMT tumors into the circulation of wild-type control mice. In mice injected with E2FWT/WT PyMT cells (n, 8), we observed robust lung colonization (Fig. 7A). In contrast, we observed a dramatic reduction in the level of colonization for mice injected with E2F1−/− (n, 9) or E2F2−/− (n, 7) PyMT tumor cells (Fig. 7B and C). Mice injected with E2FWT/WT PyMT tumor cells had an average of 8.5 metastases per section of lung, while a significant reduction was observed in mice injected with E2F1−/− (average number of metastases, 1.9 [P, 0.01]) or E2F2−/− (average number of metastases, 0.85 [P, 0.02]) tumor cells (Fig. 7D). Additionally, we measured the area of the lung occupied by metastases in these mice and observed an 18-fold reduction for mice injected with E2F1−/− tumor cells and a 50-fold reduction for mice injected with E2F2−/− tumor cells relative to the area for mice receiving E2FWT/WT PyMT tumor cells (Fig. 7E). Interestingly, the reduction in the number of circulating tumor cells (Fig. 6) or in tumor cell colonization ability (Fig. 7) was not related to defects in cell migration measured through scratch assays (see Fig. S10 in the supplemental material) and transwell migration assays (see Fig. S11 in the supplemental material); the latter experiments revealed no difference in motility between E2FWT/WT and E2F−/− tumor cells. Nonetheless, these data clearly demonstrate that E2F1 and E2F2 are necessary for tumor cell metastasis.
FIG 7.
Loss of E2Fs decreases the level of pulmonary tumor cell colonization. (A to C) Representative sections of lungs for mice injected with E2FWT/WT (n, 8) (A), E2F1−/− (n, 9) (B), or E2F2−/− (n, 7) (C) tumor cells. (D) Comparison of the average numbers (with standard errors of the means) of metastases detected in sections of lungs of mice injected with E2FWT/WT, E2F1−/− (P, 0.01), or E2F2−/− (P, 0.02) tumor cells. (E) Comparison of the average percentages (with the standard error of the mean) of the lung occupied by metastases in pulmonary sections from mice injected with E2FWT/WT tumor cells with those for mice injected with E2F1−/− or E2F2−/− tumor cells.
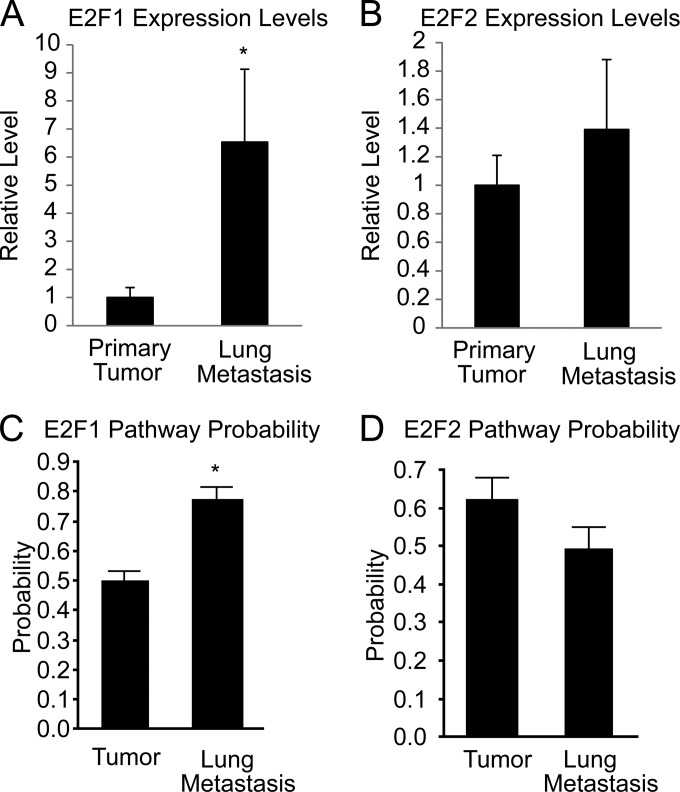
To further demonstrate the significance of the E2Fs for pulmonary colonization, we performed qRT-PCR for comparison of E2F1 and E2F2 levels in E2FWT/WT pulmonary metastases to those in E2FWT/WT primary tumors. Interestingly, E2F1 expression levels were nearly 7 times higher in lung metastases than in primary tumors (P, 0.0004) (Fig. 8A). E2F2 levels were similar in lung metastases and primary tumors (Fig. 8B). We also applied our gene signatures for E2F1 and E2F2 activity to previously published gene expression data for MMTV-PyMT tumors and lung metastases (42). In agreement with the qRT-PCR results, predicted E2F1 activity was significantly higher (P, 0.0007) in lung metastases than in primary tumors (Fig. 8C), while no differences in activity were observed for E2F2 (Fig. 8D).
FIG 8.
E2F1 expression levels and pathway activity are elevated in lung metastases. (A) Quantitative RT-PCR results showing the relative expression of E2F1 in primary tumors (n, 6) and in lung metastases (n, 6) (P, 0.0004). (B) Quantitative RT-PCR results showing the relative expression of E2F2 in primary tumors and in lung metastases. (C) The pathway signature for E2F1 shows predicted E2F1 activation in primary tumors (n, 6) and lung metastases (n, 4) within GEO data set GSE43566 (P, 0.0007). (D) The pathway signature for E2F2 shows predicted E2F2 activation in primary tumors and lung metastases within GEO data set GSE43566.
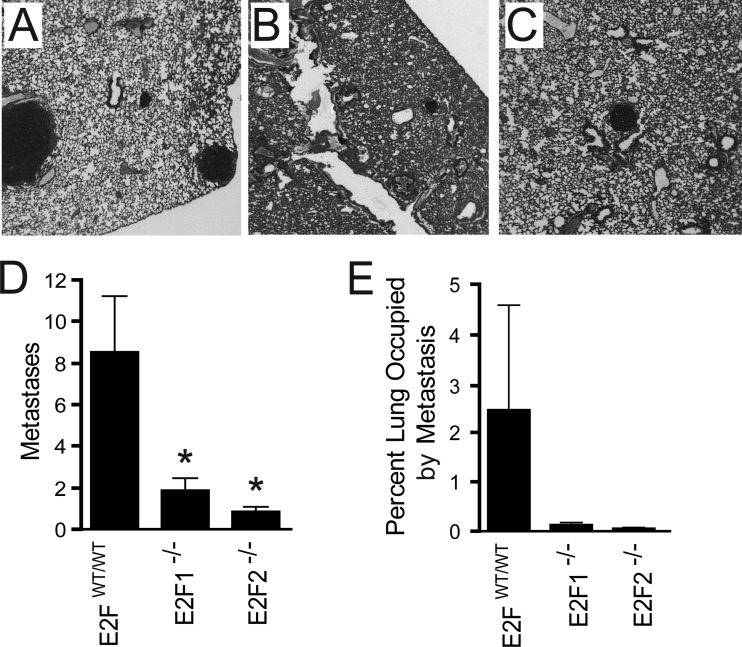
To test whether metastatic regulation by E2F1 or E2F2 is cell autonomous or is a result of E2F-associated tumor microenvironment defects, we assayed metastatic progression by utilizing a tumor transplant study. For this experiment, viable E2FWT/WT, E2F1−/−, or E2F2−/− frozen tumor samples (four samples of each type) from transgenic mice were transplanted into MMTV-Cre E2FWT/WT control mice. At the primary tumor endpoint, metastasis was analyzed. Histological sections of the lungs of mice implanted with a PyMT E2FWT/WT tumor demonstrated extensive metastasis (Fig. 9A). In contrast, metastatic lesions were rarely observed in the lungs of mice receiving a PyMT E2F1−/− (Fig. 9B) or PyMT E2F2−/− (Fig. 9C) tumor. These results revealed significantly fewer metastases in the lungs of mice implanted with a PyMT E2F1−/− (P, 0.003) or PyMT E2F2−/− (P, 0.01) tumor than in those of mice implanted with a PyMT E2FWT/WT tumor (Fig. 9D). Further, these metastatic defects also resulted in a dramatically lower proportion of the lungs occupied by metastases in mice implanted with an E2F1−/− or E2F2−/− tumor than in mice receiving an E2FWT/WT tumor (Fig. 9E).
FIG 9.
Transplantation of tumors into E2F wild-type mice shows that E2F regulation of metastasis is cell autonomous. Viable frozen tumor samples (4 samples each for E2FWT/WT, E2F1−/−, and E2F2−/− tumors) were used for transplantation into E2F wild-type MMTV-Cre control mice. (A to C) Representative histological sections of lungs of mice implanted with an E2FWT/WT (A), E2F1−/− (B), or E2F2−/− (C) tumor. (D) Quantification revealed significantly fewer metastases in the lungs of mice implanted with an E2F1−/− (P, 0.003) or E2F2−/− (P, 0.01) tumor than in mice implanted with an E2FWT/WT tumor. (E) Quantification of the percentages of lungs occupied by metastases shows reduced metastatic burdens in mice receiving an E2F1−/− or E2F2−/− tumor.
To determine if compensation by other E2F family members was occurring, E2F expression in transplanted tumors was analyzed (see Fig. S12 in the supplemental material). Significant increases in E2F2 (P, 0.0032) and E2F3A (P, 0.0254) expression and a significant decrease in E2F3B expression (P, 0.0358) were observed in PyMT E2F1−/− tumors (n, 4). Like the spontaneous tumors, transplanted E2F2−/− tumors showed a significant decrease in E2F1 expression (P, 0.0046). Interestingly, E2F3A upregulation in E2F2−/− tumors was not statistically significant. However, E2F3B was significantly downregulated (P, 0.0024) in these tumors.
To test for tumor microenvironment effects, we also performed F4/80 staining. In agreement with the observations from the tumor transplant study, we observed no differences in macrophage infiltration across E2F mutant backgrounds (see Fig. S13 in the supplemental material). Together, these data suggested that metastatic defects associated with E2F1 or E2F2 loss were intrinsic to the tumor cells.
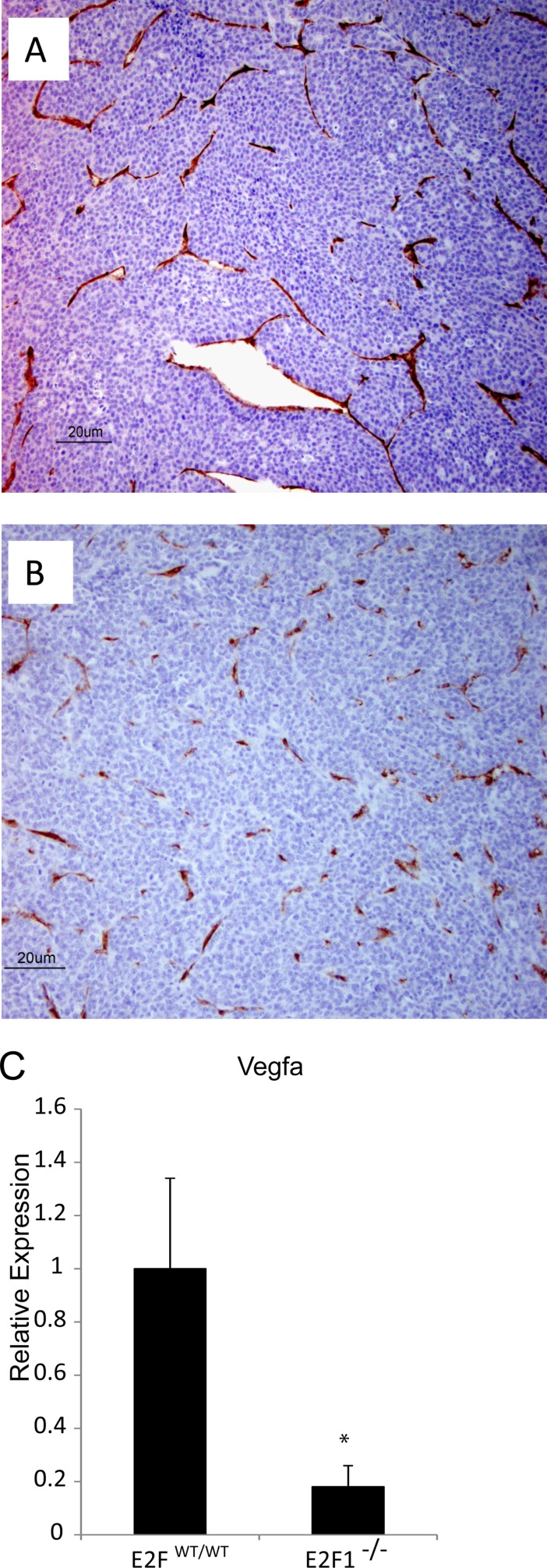
For initial investigation of the intrinsic mechanistic features of metastatic defects, we performed CD31 staining to assay tumor vasculature in end-stage tumors. In PyMT E2FWT/WT tumors (n, 5), we observed well-defined and continuous staining for CD31, indicating well-developed vasculature (Fig. 10A). In contrast, PyMT E2F1−/− tumors (n, 5) showed remarkably altered tumor vasculature (Fig. 10B). This was accompanied by significantly lower (P, 0.0002) gene expression of the proangiogenic signaling molecule vascular endothelial growth factor (Vegfa) in E2F1−/− tumors (n, 6) than in E2FWT/WT (n, 6) tumors (Fig. 10C). Jointly, these results indicate that loss of Vegfa expression in E2F1−/− tumors is associated with altered blood vessel development.
FIG 10.
Loss of E2F1 alters CD31 staining and reduces VEGFA expression in MMTV-PyMT tumors. (A and B) Representative sections of E2FWT/WT (A) and E2F1−/− (B) tumors stained for CD31 to reveal vascular structure. (C) Quantitative RT-PCR results showing the relative expression levels of VEGFA in E2FWT/WT and E2F1−/− tumors (P, 0.0002). Six tumors of each type were examined.
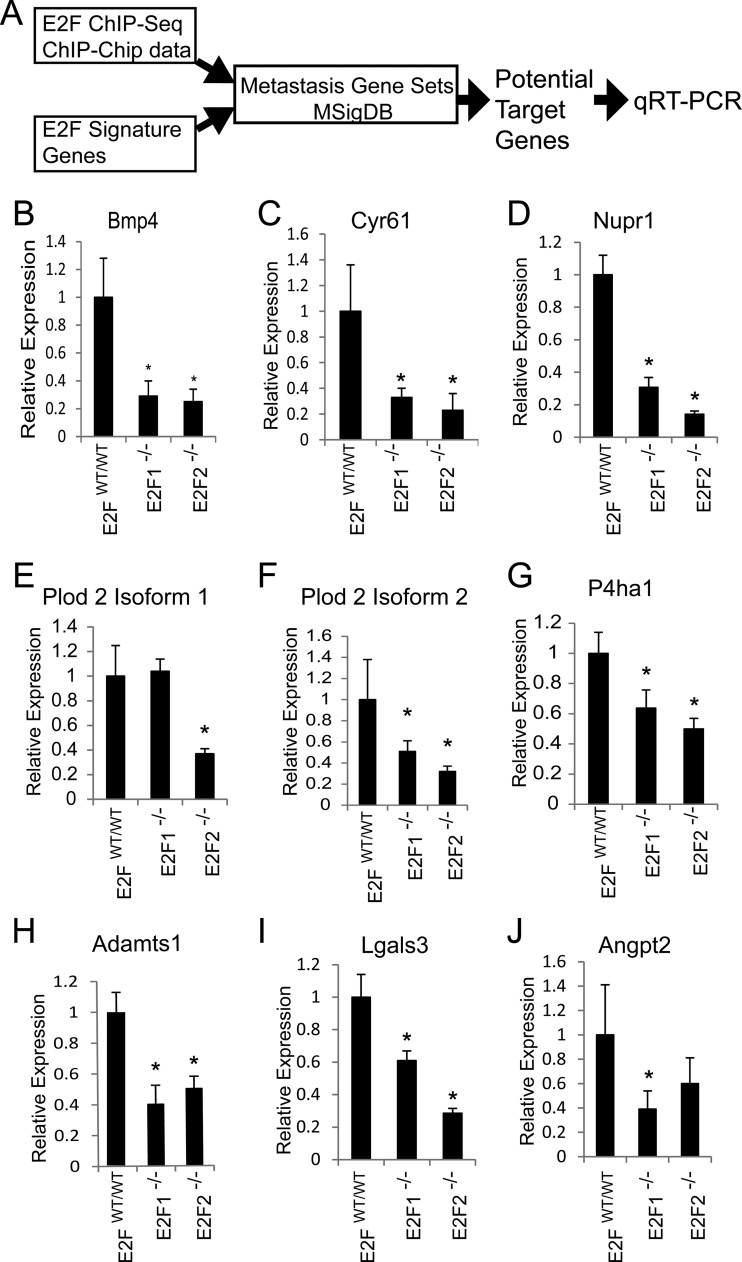
For further investigation of potential mechanisms of metastasis, we utilized an informatics approach to identify potential E2F targets mediating metastatic potential. As outlined in Fig. 11A, we utilized E2F signature gene expression data with published ChIP-seq and ChIP-chip data to identify direct E2F target genes. Next, we filtered our potential targets using gene sets for metastasis available on MSigDB. Testing potential target genes via qRT-PCR, we found that E2F1−/− and E2F2−/− tumors (six of each type were analyzed) had significantly lower levels of Bmp4 (P, 0.0002 and <0.0001, respectively), cysteine-rich angiogenic inducer 61 (Cyr61) (P, 0.0009 and 0.0006, respectively), Nupr1 (P, <0.0001 for both types), Plod 2 isoform 1 (P, <0.0001 for E2F2−/− tumors only), Plod 2 isoform 2 (P, 0.0122 and 0.0015, respectively), P4ha1 (P, 0.0006 and <0.0001, respectively), Adamts1 (P, <0.0001 for both types), Lgals3 (P, <0.0001 for both types), and Angpt2 (P, 0.0065 for E2F1−/− tumors only) (Fig. 11B to J). These results show which E2F target genes associated with metastatic function are downregulated in E2F1−/− and E2F2−/− tumors. In addition, these results provide important mechanistic insights into the molecular basis of E2F regulation of tumor metastasis.
FIG 11.
Analysis of E2F target genes reveals expression changes in prometastatic genes with E2F loss. (A) Informatics pipeline for filtering candidate genes for qRT-PCR testing. (B to J) Testing potential target genes via qRT-PCR, we found that E2F1−/− and E2F2−/− tumors (six of each type were analyzed) have significantly lower levels of Bmp4 (P, 0.0002 and <0.0001, respectively) (B), Cyr61 (P, 0.0009 and 0.0006, respectively) (C), Nupr1 (P, <0.0001 for both types) (D), Plod 2 isoform 1 (P, <0.0001 for E2F2−/− tumors only) (E), Plod 2 isoform 2 (P, 0.0122 and 0.0015, respectively) (F), P4ha1 (P, 0.0006 and <0.0001, respectively) (G), Adamts1 (P, <0.0001 for both types) (H), Lgals3 (P, <0.0001 for both types) (I), and Angpt2 (P, 0.0065 for E2F1−/− tumors only) (J).
DISCUSSION
In this study, we applied genomic signaling pathway signatures to mouse tumor model microarray data and predicted a role for the E2F transcription factors in PyMT-induced tumors. By genetically testing this prediction, we found that E2F1 loss enhanced ductal transformation and accelerated tumor onset. In contrast to this finding, we noted delayed tumor onset in E2F3+/− mice. Histologically, we observed that loss of E2F1 resulted in a significant decrease in the incidence of adenosquamous tumors, while loss of E2F2 led to an increase in the frequency of this tumor type. In some of the most striking findings, we identified a role for the E2F transcription factors in tumor metastasis. Indeed, loss of E2F1 or E2F2 dramatically reduced the metastatic capacity of MMTV-PyMT tumors. These metastatic defects were associated with a reduction in the number of circulating tumor cells and were cell autonomous. Together, these data demonstrate significant roles for individual E2Fs in tumor development and progression.
E2F transcription factors have previously been associated with defined roles in cell cycle control, proliferation, and apoptosis. While exploring the role of E2F1 in tumor development, we found that loss of E2F1 enhanced ductal transformation and accelerated tumor onset. In a previous study of Myc-mediated tumorigenesis, we noted a similar acceleration of tumor onset with loss of E2F1 (19). In agreement with the role of E2F1 in Myc-induced apoptosis (43), we noted that loss of E2F1 reduced the level of apoptosis and caused Myc tumors to grow more quickly. In contrast to the Myc model, we observed no effects of E2F1 loss on apoptosis (see Fig. S7 in the supplemental material). This suggests that E2Fs can respond to different pathway stimuli uniquely to differentially regulate specific genes and separate cellular processes.
In view of the delay in tumor onset in E2F3+/− mice and the previously defined roles for the E2Fs in cell cycle progression and apoptosis, alterations in tumor growth were expected in E2F mutant mice, but none were observed. However, the levels of the various E2F alleles indicated that there was significant compensation in the E2F3+/− strain. Given the previously noted potential for E2F compensation (44), the lack of defects associated with tumor growth in E2F mutant mice may not be surprising. In agreement with our data, it has been shown that with the loss of E2F1, E2F3A is upregulated (44). Further work has also demonstrated that E2F3A can compensate for the loss of E2F1 to sustain cell proliferation (45), while E2F3A is necessary for cellular proliferation (38, 45). Along with upregulation of E2F3A in each of the E2F mutant tumors, these previous data support our tumor growth and proliferation observations.
In addition to tumor development, we found that loss of E2F transcription factors altered tumor heterogeneity. As is apparent in Fig. 4C, PyMT induced tumors with wide histological heterogeneity. Interestingly, E2F1 and E2F2 had opposite effects on the incidence of adenosquamous tumors. Together, the latency, histological differences, and metastatic capacity findings clearly demonstrate that the E2Fs have unique and individual roles. The facts that the E2F DNA binding site is conserved in all E2Fs (46) and that ChIP-seq studies have demonstrated that various E2Fs bind the same targets (22) reinforce the idea that cooperating transcription factors, such as YY1, are required for specificity of function (47).
Perhaps the most notable of the experimental findings was the identification of E2Fs as regulators of breast cancer metastasis. Importantly, high levels of E2F1, E2F2, and E2F3 were predictive of accelerated onset of distant metastasis in human breast cancer. While our data for E2F1−/− and E2F2−/− mice corroborate these predictions, we did not observe metastatic impairment in E2F3+/− mice. However, E2F1, E2F2, and E2F3A levels were maintained in E2F3+/− mice (Fig. 3), potentially allowing for these E2Fs to compensate, resulting in normal regulation of metastatic progression. Importantly, our tumor transplant study provides evidence that the E2Fs regulate tumor metastasis in a cell-autonomous manner. Indeed, E2F loss had no effect on the tumor growth rate, tumor burden, proliferation, apoptosis, or macrophage staining but did reduce metastatic potential. This indicated that the metastatic defects are not a result of altered tumor development but are due to inherent properties of the tumor.
While both E2F1−/− and E2F2−/− mice present reductions in the level of tumor metastasis, our data suggest that E2F1 loss is responsible for the metastatic deficiency. Indeed, E2F1 levels and activity were higher in lung metastases than in primary tumors. In addition, PyMT E2F1−/− tumors transplanted into wild-type recipients had significantly higher levels of E2F2 yet still had significantly lower numbers of metastases. Meanwhile, both spontaneous and transplanted PyMT E2F2−/− tumors had significantly lower levels of E2F1 than PyMT E2FWT/WT tumors while failing to metastasize. Interestingly, E2F1−/− and E2F2−/− tumors exhibited shared expression patterns for most of the metastatic target genes we surveyed. These findings, taken together, may indicate that the mechanism behind the metastatic defects noted in both E2F1−/− and E2F2−/− tumors is mediated primarily by E2F1-regulated genes.
Considering both the reduction in the number of CTCs and the reduction in tumor cell colonization ability, our data suggest that the E2Fs regulate metastasis in early and late stages of metastatic progression. Through qRT-PCR of E2F target genes with known metastatic properties, we have identified molecular features of metastasis regulated by E2Fs. We found that in the early steps of metastasis, E2F1−/− tumors had lower levels of Vegfa than E2FWT/WT tumors and that, as a result, the tumor vasculature was altered. Indeed, previous reports have shown that E2F1 controls angiogenesis through the VEGF signaling axis (48). In addition, we detected other altered E2F target genes known to function in tumor angiogenesis. Previous studies have demonstrated that Angpt2 can promote angiogenesis (49, 50, 71) and tumor cell invasion (51, 72–74). In addition, E2F1 tumors have significantly low levels of Cyr61, and blocking of Cyr61 function decreased metastasis in a xenograft of the MDA-MB231 human breast cancer cell line (52). Further, given the finding that Cyr61 can regulate tumor angiogenesis independently of Vegfa expression (53), it seems that loss of Cyr61 together with loss of Vegfa and Angpt2 provides a molecular context for the pronounced tumor vasculature effects associated with E2F loss. Since blood vessel recruitment is a key rate-limiting step for metastasis (54), these data reveal the molecular alterations contributing to the angiogenesis defects associated with E2F loss and indicate one of the mechanisms by which E2Fs are involved in mediating the early steps of metastasis.
In addition to tumor angiogenesis, we detected gene expression changes that indicate that the loss of E2F1 or E2F2 may impact tumor cell remodeling of the extracellular matrix (ECM). Specifically, we noted a 2-fold reduction in the level of the extracellular metalloprotease Adamts1 in PyMT E2F1−/− and PyMT E2F2−/− tumors. Adamts1 has been ablated in the MMTV-PyMT mouse model previously, revealing the requirement for Adamts1 during tumor metastasis (55). Importantly, that work demonstrated that Adamts1 remodels the ECM to facilitate the transition from ductal carcinoma in situ to invasive and metastatic disease. Also critical to the remodeling of the ECM to facilitate a metastatic niche is collagen deposition (56). The PyMT tumors in the E2F1−/− and E2F2−/− backgrounds had significantly lower expression of P4ha1 and Plod 2, whose products both function as collagen hydroxylases. Recent work has demonstrated the necessity for these genes in tumor cell collagen deposition (57). Further, knockdown of these genes in metastatic human breast cancer cell lines reduced the number of CTCs in the blood, as well as the levels of pulmonary and lymphatic metastasis in a xenograft study (58). Taken together, these data indicate that the reduction in metastatic potential in the E2F1−/− and E2F2−/− backgrounds may be due to an inability to form extracellular fibrillar collagen, resulting from the loss of expression of E2F target collagen hydroxylase genes.
In addition to molecular signals that recruit blood vessels and remodel the extracellular matrix, we found that major cell-signaling pathways were impacted by E2F loss. Our qRT-PCR analysis suggests that pathways related to the TGF-β superfamily and Smad activation are impacted by the loss of E2F1 and E2F2. BMP4 expression was reduced >3-fold in E2F1−/− and E2F2−/− tumors. BMP4, a member of the TGF-β superfamily, leads to the activation of Smad1, Smad5, and Smad8 (59, 60), and studies have demonstrated a role for BMP4 in breast cancer cell invasion (61, 62). These data may indicate that loss of BMP4 signaling reduces the invasive potential of MMTV-PyMT tumor cells, thus contributing to the observed reduction in the number of CTCs. In addition, we also detected a >3-fold reduction in the level of Nupr1. Nupr1 expression has been shown to increase in response to TGF-β1 and to facilitate Smad transactivation (63). The prometastatic functions of Nupr1 were originally identified in work that showed that Nupr1 supports the growth of human breast cancer cells after seeding of a distant organ (64). In light of these findings, loss of NUPR1 expression may contribute to the observed defects in colonization of the lungs by tumor cells lacking E2F1 or E2F2.
We also detected significantly lower expression levels of Lgals3 in E2F1−/− and E2F2−/− tumors. A wide array of prometastatic functions, with relevance to early and late steps of the metastatic cascade, have been described for LGALS3 (65). For instance, it has been shown that the LGALS3 protein, galectin-3, mediates tumor cell adhesion to the ECM (66) and promotes the dissemination of tumor cells from the primary tumor (67). Galectin-3 has also been shown to be critical for the recognition and interaction of human breast cancer cells with endothelial cells of the vasculature (68, 69), and those tumor cell–endothelial cell interactions are necessary for tumor cell invasion and metastasis (70). In the later steps of metastasis, galectin-3 induces apoptosis in T cells and monocytes (65). This suggests that LGALS3 deficiency in E2F1−/− and E2F2−/− tumor cells may have allowed immune cells to reduce the number of CTCs and eliminate those cells before they could colonize the lungs in our study using retro-orbital injection of tumor cells into the bloodstream.
In conclusion, these data demonstrate that E2F1 and E2F2 play a critical role in MMTV-PyMT-mediated tumorigenesis, with E2F loss leading to alterations in tumor latency, histology, and metastasis. Importantly, E2F1 and E2F2 were shown to be critical regulators of intrinsic cell signaling, which allows tumor cells to progress through both the early and late steps of metastasis. Importantly, we identified the potential E2F target genes that are associated with these changes in metastatic ability. These gene expression changes suggest that the E2F transcription factors mediate the expression of genes critical to tumor angiogenesis, tumor cell remodeling of the extracellular matrix, tumor cell survival, and tumor cell interactions with vascular endothelial cells to facilitate metastasis to the lungs. Taken together, these findings indicate that E2Fs regulate key functions involved in metastasis in both mouse models and human breast cancer, and as such, our data extend the paradigm of E2F function.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Susan G. Komen Foundation (grant KG1110510) and by NIH R01CA160514 (to E.R.A.).
We thank the members of the Andrechek laboratory for helpful discussions.
We declare no conflict of interest.
Footnotes
Published ahead of print 16 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00737-14.
REFERENCES
- 1.Weigelt B, Peterse JL, van 't Veer LJ. 2005. Breast cancer metastasis: markers and models. Nat. Rev. Cancer 5:591–602. 10.1038/nrc1670 [DOI] [PubMed] [Google Scholar]
- 2.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. 2009. Genes that mediate breast cancer metastasis to the brain. Nature 459:1005–1009. 10.1038/nature08021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3:537–549. 10.1016/S1535-6108(03)00132-6 [DOI] [PubMed] [Google Scholar]
- 4.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. 2005. Genes that mediate breast cancer metastasis to lung. Nature 436:518–524. 10.1038/nature03799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen DX, Bos PD, Massague J. 2009. Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9:274–284. 10.1038/nrc2622 [DOI] [PubMed] [Google Scholar]
- 6.Guy CT, Cardiff RD, Muller WJ. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin EY, Nguyen AV, Russell RG, Pollard JW. 2001. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193:727–740. 10.1084/jem.193.6.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ. 2009. Akt1 and Akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 69:5057–5064. 10.1158/0008-5472.CAN-08-4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester E, Chytil A, Bierie B, Aakre M, Gorska AE, Sharif-Afshar AR, Muller WJ, Moses HL. 2005. Effect of conditional knockout of the type II TGF-β receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 65:2296–2302. 10.1158/0008-5472.CAN-04-3272 [DOI] [PubMed] [Google Scholar]
- 10.Landskroner-Eiger S, Qian B, Muise ES, Nawrocki AR, Berger JP, Fine EJ, Koba W, Deng Y, Pollard JW, Scherer PE. 2009. Proangiogenic contribution of adiponectin toward mammary tumor growth in vivo. Clin. Cancer Res. 15:3265–3276. 10.1158/1078-0432.CCR-08-2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluck MM, Schaffhausen BS. 2009. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev. 73:542–563. 10.1128/MMBR.00009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Jr, Marks JR, Dressman HK, West M, Nevins JR. 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439:353–357. 10.1038/nature04296 [DOI] [PubMed] [Google Scholar]
- 13.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. 2001. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. U. S. A. 98:11462–11467. 10.1073/pnas.201162998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, Datto MB, Kelley M, Mathey-Prevot B, Potti A, Nevins JR. 2010. A pathway-based classification of human breast cancer. Proc. Natl. Acad. Sci. U. S. A. 107:6994–6999. 10.1073/pnas.0912708107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. 2000. Molecular portraits of human breast tumours. Nature 406:747–752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 16.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98:10869–10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, Nevins JR. 2009. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc. Natl. Acad. Sci. U. S. A. 106:16387–16392. 10.1073/pnas.0901250106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollern DP, Yuwanita I, Andrechek ER. 2013. A mouse model with T58A mutations in Myc reduces the dependence on KRas mutations and has similarities to claudin-low human breast cancer. Oncogene 32:1296–1304. 10.1038/onc.2012.142 [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara K, Yuwanita I, Hollern DP, Andrechek ER. 2011. Prediction and genetic demonstration of a role for activator E2Fs in Myc-induced tumors. Cancer Res. 71:1924–1932. 10.1158/0008-5472.CAN-10-2386 [DOI] [PubMed] [Google Scholar]
- 20.Attwooll C, Lazzerini Denchi E, Helin K. 2004. The E2F family: specific functions and overlapping interests. EMBO J. 23:4709–4716. 10.1038/sj.emboj.7600481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevins JR. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699–703. 10.1093/hmg/10.7.699 [DOI] [PubMed] [Google Scholar]
- 22.Trimarchi JM, Lees JA. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11–20. 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
- 23.Hallstrom TC, Mori S, Nevins JR. 2008. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 13:11–22. 10.1016/j.ccr.2007.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HZ, Tsai SY, Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9:785–797. 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alla V, Engelmann D, Niemetz A, Pahnke J, Schmidt A, Kunz M, Emmrich S, Steder M, Koczan D, Putzer BM. 2010. E2F1 in melanoma progression and metastasis. J. Natl. Cancer Inst. 102:127–133. 10.1093/jnci/djp458 [DOI] [PubMed] [Google Scholar]
- 26.Ebihara Y, Miyamoto M, Shichinohe T, Kawarada Y, Cho Y, Fukunaga A, Murakami S, Uehara H, Kaneko H, Hashimoto H, Murakami Y, Itoh T, Okushiba S, Kondo S, Katoh H. 2004. Over-expression of E2F-1 in esophageal squamous cell carcinoma correlates with tumor progression. Dis. Esophagus 17:150–154. 10.1111/j.1442-2050.2004.00393.x [DOI] [PubMed] [Google Scholar]
- 27.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS. 2004. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene 23:5871–5879. 10.1038/sj.onc.1207800 [DOI] [PubMed] [Google Scholar]
- 28.Carvalho CM, Chang J, Lucas JE, Nevins JR, Wang Q, West M. 2008. High-dimensional sparse factor modeling: applications in gene expression genomics. J. Am. Stat Assoc. 103:1438–1456. 10.1198/016214508000000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 30.Andrechek ER, Mori S, Rempel RE, Chang JT, Nevins JR. 2008. Patterns of cell signaling pathway activation that characterize mammary development. Development 135:2403–2413. 10.1242/dev.019018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asp P, Acosta-Alvear D, Tsikitis M, van Oevelen C, Dynlacht BD. 2009. E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes Dev. 23:37–53. 10.1101/gad.1727309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst J, Plasterer HL, Simon I, Bar-Joseph Z. 2010. Integrating multiple evidence sources to predict transcription factor binding in the human genome. Genome Res. 20:526–536. 10.1101/gr.096305.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132:1049–1061. 10.1016/j.cell.2008.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. 2007. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 17:1550–1561. 10.1101/gr.6783507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123:725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 36.Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85:549–561. 10.1016/S0092-8674(00)81255-6 [DOI] [PubMed] [Google Scholar]
- 37.Murga M, Fernandez-Capetillo O, Field SJ, Moreno B, Borlado LR, Fujiwara Y, Balomenos D, Vicario A, Carrera AC, Orkin SH, Greenberg ME, Zubiaga AM. 2001. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity 15:959–970. 10.1016/S1074-7613(01)00254-0 [DOI] [PubMed] [Google Scholar]
- 38.Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690–703 [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Yuan Y, Liao L, Kuang SQ, Tien JC, O'Malley BW, Xu J. 2009. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc. Natl. Acad. Sci. U. S. A. 106:151–156. 10.1073/pnas.0808703105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borowsky AD, Namba R, Young LJ, Hunter KW, Hodgson JG, Tepper CG, McGoldrick ET, Muller WJ, Cardiff RD, Gregg JP. 2005. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin. Exp. Metastasis 22:47–59. 10.1007/s10585-005-2908-5 [DOI] [PubMed] [Google Scholar]
- 41.Selvaraj N, Budka JA, Ferris MW, Jerde TJ, Hollenhorst PC. 2014. Prostate cancer ETS rearrangements switch a cell migration gene expression program from RAS/ERK to PI3K/AKT regulation. Mol. Cancer 13:61. 10.1186/1476-4598-13-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franci C, Zhou J, Jiang Z, Modrusan Z, Good Z, Jackson E, Kouros-Mehr H. 2013. Biomarkers of residual disease, disseminated tumor cells, and metastases in the MMTV-PyMT breast cancer model. PLoS One 8:e58183. 10.1371/journal.pone.0058183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, Thompson MA, Yang H, Fujiwara Y, Greenberg ME, Orkin S, Smith C, Nevins JR. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105–113. 10.1016/S1097-2765(01)00275-1 [DOI] [PubMed] [Google Scholar]
- 44.Kong LJ, Chang JT, Bild AH, Nevins JR. 2007. Compensation and specificity of function within the E2F family. Oncogene 26:321–327. 10.1038/sj.onc.1209817 [DOI] [PubMed] [Google Scholar]
- 45.Danielian PS, Friesenhahn LB, Faust AM, West JC, Caron AM, Bronson RT, Lees JA. 2008. E2f3a and E2f3b make overlapping but different contributions to total E2f3 activity. Oncogene 27:6561–6570. 10.1038/onc.2008.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng N, Fraenkel E, Pabo CO, Pavletich NP. 1999. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13:666–674. 10.1101/gad.13.6.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freedman JA, Chang JT, Jakoi L, Nevins JR. 2009. A combinatorial mechanism for determining the specificity of E2F activation and repression. Oncogene 28:2873–2881. 10.1038/onc.2009.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merdzhanova G, Gout S, Keramidas M, Edmond V, Coll JL, Brambilla C, Brambilla E, Gazzeri S, Eymin B. 2010. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene 29:5392–5403. 10.1038/onc.2010.281 [DOI] [PubMed] [Google Scholar]
- 49.He T, Qi F, Jia L, Wang S, Song N, Guo L, Fu Y, Luo Y. 2014. MicroRNA-542-3p inhibits tumour angiogenesis by targeting angiopoietin-2. J. Pathol. 232:499–508. 10.1002/path.4324 [DOI] [PubMed] [Google Scholar]
- 50.Zhang ZL, Zhang JF, Yuan YF, He YM, Liu QY, Mao XW, Ai YB, Liu ZS. 2014. Suppression of angiogenesis and tumor growth in vitro and in vivo using an anti-angiopoietin-2 single-chain antibody. Exp. Ther. Med. 7:543–552. 10.3892/etm.2014.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imanishi Y, Hu B, Jarzynka MJ, Guo P, Elishaev E, Bar-Joseph I, Cheng SY. 2007. Angiopoietin-2 stimulates breast cancer metastasis through the α5β1 integrin-mediated pathway. Cancer Res. 67:4254–4263. 10.1158/0008-5472.CAN-06-4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J, Huo R, Wang L, Zhou Z, Sun Y, Shen B, Wang R, Li N. 2012. A novel anti-Cyr61 antibody inhibits breast cancer growth and metastasis in vivo. Cancer Immunol. Immunother. 61:677–687. 10.1007/s00262-011-1135-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris LG, Pannell LK, Singh S, Samant RS, Shevde LA. 2012. Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated upregulation of cyr61. Oncogene 31:3370–3380. 10.1038/onc.2011.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fidler IJ. 2003. The pathogenesis of cancer metastasis: the ‘seed and soil' hypothesis revisited. Nat. Rev. Cancer 3:453–458. 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- 55.Ricciardelli C, Frewin KM, Tan Ide A, Williams ED, Opeskin K, Pritchard MA, Ingman WV, Russell DL. 2011. The ADAMTS1 protease gene is required for mammary tumor growth and metastasis. Am. J. Pathol. 179:3075–3085. 10.1016/j.ajpath.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shintani Y, Hollingsworth MA, Wheelock MJ, Johnson KR. 2006. Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH2-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res. 66:11745–11753. 10.1158/0008-5472.CAN-06-2322 [DOI] [PubMed] [Google Scholar]
- 57.Gilkes DM, Chaturvedi P, Bajpai S, Wong CC, Wei H, Pitcairn S, Hubbi ME, Wirtz D, Semenza GL. 2013. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 73:3285–3296. 10.1158/0008-5472.CAN-12-3963 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL. 2013. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol. Cancer Res. 11:456–466. 10.1158/1541-7786.MCR-12-0629 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. 1997. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 11:984–995. 10.1101/gad.11.8.984 [DOI] [PubMed] [Google Scholar]
- 60.Park ES, Woods DC, Tilly JL. 2013. Bone morphogenetic protein 4 promotes mammalian oogonial stem cell differentiation via Smad1/5/8 signaling. Fertil. Steril. 100:1468–1475. 10.1016/j.fertnstert.2013.07.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo D, Huang J, Gong J. 2012. Bone morphogenetic protein 4 (BMP4) is required for migration and invasion of breast cancer. Mol. Cell. Biochem. 363:179–190. 10.1007/s11010-011-1170-1 [DOI] [PubMed] [Google Scholar]
- 62.Lai D, Yang X. 2013. BMP4 is a novel transcriptional target and mediator of mammary cell migration downstream of the Hippo pathway component TAZ. Cell. Signal. 25:1720–1728. 10.1016/j.cellsig.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 63.García-Montero AC, Vasseur S, Giono LE, Canepa E, Moreno S, Dagorn JC, Iovanna JL. 2001. Transforming growth factor β-1 enhances Smad transcriptional activity through activation of p8 gene expression. Biochem. J. 357:249–253. 10.1042/0264-6021:3570249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ree AH, Pacheco MM, Tvermyr M, Fodstad O, Brentani MM. 2000. Expression of a novel factor, com1, in early tumor progression of breast cancer. Clin. Cancer Res. 6:1778–1783 [PubMed] [Google Scholar]
- 65.Newlaczyl AU, Yu LG. 2011. Galectin-3—a jack-of-all-trades in cancer. Cancer Lett. 313:123–128. 10.1016/j.canlet.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 66.Warfield PR, Makker PN, Raz A, Ochieng J. 1997. Adhesion of human breast carcinoma to extracellular matrix proteins is modulated by galectin-3. Invasion Metastasis 17:101–112 [PubMed] [Google Scholar]
- 67.Ochieng J, Furtak V, Lukyanov P. 2004. Extracellular functions of galectin-3. Glycoconj. J. 19:527–535 [DOI] [PubMed] [Google Scholar]
- 68.Shekhar MP, Nangia-Makker P, Tait L, Miller F, Raz A. 2004. Alterations in galectin-3 expression and distribution correlate with breast cancer progression: functional analysis of galectin-3 in breast epithelial-endothelial interactions. Am. J. Pathol. 165:1931–1941. 10.1016/S0002-9440(10)63245-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou J, Glinsky VV, Landon LA, Matthews L, Deutscher SL. 2005. Peptides specific to the galectin-3 carbohydrate recognition domain inhibit metastasis-associated cancer cell adhesion. Carcinogenesis 26:309–318. 10.1093/carcin/bgh329 [DOI] [PubMed] [Google Scholar]
- 70.Newton-Northup JR, Dickerson MT, Ma L, Besch-Williford CL, Deutscher SL. 2013. Inhibition of metastatic tumor formation in vivo by a bacteriophage display-derived galectin-3 targeting peptide. Clin. Exp. Metastasis 30:119–132. 10.1007/s10585-012-9516-y [DOI] [PubMed] [Google Scholar]
- 71.Buchanan CF, Szot CS, Wilson TD, Akman S, Metheny-Barlow LJ, Robertson JL, Freeman JW, Rylander MN. 2012. Cross-talk between endothelial and breast cancer cells regulates reciprocal expression of angiogenic factors in vitro. J. Cell. Biochem. 113:1142–1151. 10.1002/jcb.23447 [DOI] [PubMed] [Google Scholar]
- 72.Holopainen T, Saharinen P, D'Amico G, Lampinen A, Eklund L, Sormunen R, Anisimov A, Zarkada G, Lohela M, Helotera H, Tammela T, Benjamin LE, Yla-Herttuala S, Leow CC, Koh GY, Alitalo K. 2012. Effects of angiopoietin-2-blocking antibody on endothelial cell-cell junctions and lung metastasis. J. Natl. Cancer Inst. 104:461–475. 10.1093/jnci/djs009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imanishi Y, Hu B, Xiao G, Yao X, Cheng SY. 2011. Angiopoietin-2, an angiogenic regulator, promotes initial growth and survival of breast cancer metastases to the lung through the integrin-linked kinase (ILK)-AKT-B cell lymphoma 2 (Bcl-2) pathway. J. Biol. Chem. 286:29249–29260. 10.1074/jbc.M111.235689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minami T, Jiang S, Schadler K, Suehiro J, Osawa T, Oike Y, Miura M, Naito M, Kodama T, Ryeom S. 2013. The calcineurin-NFAT-angiopoietin-2 signaling axis in lung endothelium is critical for the establishment of lung metastases. Cell Rep. 4:709–723. 10.1016/j.celrep.2013.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.