Abstract
In mammals, there are four NOTCH receptors and five Delta-Jagged-type ligands regulating many aspects of embryonic development and adult tissue homeostasis. NOTCH proteins are type I transmembrane receptors that interact with ligands on adjacent cells and are activated by regulated intramembrane proteolysis (RIP). The activation mechanism of NOTCH1 receptors upon ligand binding is well understood and requires cleavage by ADAM10 metalloproteases prior to intramembranous cleavage by γ-secretase. How the other human NOTCH receptor homologues are activated upon ligand binding is not known. Here, we dissect the proteolytic activation mechanism of the NOTCH2 and NOTCH3 receptors. We show that NOTCH2 and NOTCH3 signaling can be triggered by both Delta-Jagged-type ligands and requires ADAM10 and presenilin-1 or -2. Importantly, we did not find any role for the highly related ADAM17/TACE (tumor necrosis factor alpha-converting enzyme) protease in ligand-induced NOTCH2 or NOTCH3 signaling. These results demonstrate that canonical ligand-induced proteolysis of the NOTCH1, -2, and -3 receptors strictly depends on consecutive cleavage of these receptors by ADAM10 and the presenilin-containing γ-secretase complex, leading to transcriptional activation.
INTRODUCTION
NOTCH signaling is a cell-cell communication pathway regulating cell fate decisions and cell renewal in developing embryos and adult animal tissues (1). Mammalian cells have four NOTCH receptors and five DSL (Delta and Serrate in Drosophila and LAG-2 in Caenorhabditis elegans) ligands. NOTCH receptors and ligands are type I transmembrane proteins that interact between neighboring cells, and the activation mechanism is governed by a highly regulated proteolytic cascade leading to transcriptional activation by cleaved NOTCH proteins in the nucleus (2, 3). During maturation in the trans-Golgi network, Notch polypeptides are cleaved by furin at site 1 (S1) and presented at the cell surface as noncovalently linked heterodimers. In the absence of ligand, NOTCH receptors are in an autoinhibited state maintained by the NOTCH negative regulatory region (NRR), which contains the three Lin12-Notch repeats (LNR) and the NOTCH heterodimerization domain (HD) (4). Ligand binding unfolds the NRR, allowing access to ADAM10 metalloprotease, which cleaves the Notch1 ectodomain at Val1711 or site 2 (S2) (2). Next, the membrane-tethered Notch intracellular domain is cleaved at site 3 (S3) by the intramembranous γ-secretase complex, leading to the release of the NOTCH intracellular domain (NICD), which translocates to the nucleus and binds to the nuclear DNA-bound repressor protein RBP-Jκ or CSL (named after CBF-1 in mammals, Suppressor of Hairless in Drosophila, and Lag-2 in C. elegans) to activate NOTCH target gene transcription (3). The γ-secretase complex consists of four main components: the aspartyl protease presenilin-1 (PS-1) or PS-2, nicastrin (Nct), presenilin enhancer 2 (PEN-2), and anterior pharynx-defective 1 (APH-1), which can be expressed as two isoforms depending on alternative splicing. Mice lacking Psen1 are embryonic lethal and resemble Notch1-deficient mice, and PS-1- and -2-deficient cells are defective in Notch1 cleavage and transcriptional activation (5, 6). Notch2-deficient mice show phenotypes similar to those reported in Notch1-deficient mice, but in a later time frame, and die around embryonic day 11.5 (E11.5) with massive cell death in the nervous system (7). In contrast, Notch3-deficient mice are viable and develop normally (8). PS-1 appears to be the main protease implicated in NICD formation for all four Notch proteins (9, 10), whereas PS-2 seems to play a less important and redundant role in Notch1 cleavage (11–13). While the activation mechanism and key players involved during ligand-dependent activation are well described for Notch1 (2, 14), the proteases implicated in the activation mechanism of Notch2 and Notch3 upon ligand binding are not well understood.
In adult tissues, Notch2 is expressed during development, is preferably expressed in mature B cells, and is required for the generation of splenic marginal zone B cells (15). Recently, Adam10 has been implicated in this process, although a direct effect of ADAM10 on Notch2 cleavage has not been shown (16). NOTCH3 is expressed in a wide variety of tissues during development, but in adult tissues it is mainly expressed in the smooth vascular muscle cells (17). The overall structure of NOTCH3 resembles that of NOTCH1 and NOTCH2 proteins. Important differences, however, are the absence of parts of epidermal growth factor (EGF) repeats 2 and 3, the lack of EGF repeat 21 in the extracellular domain, and a shorter C-terminal domain lacking the conserved transcriptional activation domain in NOTCH3, suggesting differences in ligand binding between receptors and target gene activation between NOTCH isoforms (18). Of particular interest is that amino acid sequences in the regions that are subjected to proteolytic cleavage are highly conserved between NOTCH receptors (19). All receptors and ligands have been implicated in sporadic and familial syndromes caused by deregulation of the canonical NOTCH pathway (20).
NOTCH2 is found to be deregulated in Alagille (21) and Hajdu-Cheney (22) syndromes, and mutated NOTCH3 receptors are found in CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy), a hereditary cerebrovascular disease characterized by tissue ischemia and stroke due to malfunctioning of smooth muscle cells (17). Mutations in NOTCH1 are associated with abnormalities of the aortic heart valve, and patients are more susceptible to thoracic aortic aneurysms (23, 24). More important are NOTCH1 receptor mutations found in the cancer leading directly to T-cell acute lymphoblastic leukemia (T-ALL) (25, 26). Genome sequencing has identified mutations in NOTCH2 and NOTCH3 in carcinomas of the ovary and in squamous head-and-neck and lung cancers, although the consequence of many of these mutations on NOTCH activity is not known (27). In solid breast tumors, expression of NOTCH2 is associated with a better overall survival and with differentiated tumors (28), whereas expression and amplification of NOTCH3 are prognostic for outcome in ovarian cancer and associated with resistance to alkylating chemotherapeutics (29).
In summary, NOTCH receptors are increasingly implicated in human disease processes, yet until now, little has been known about their mechanism of activation. Here, we investigated NOTCH2 and NOTCH3 receptor proteolysis and activation in ligand-dependent signaling using gain- and loss-of-function approaches. Our studies show that ADAM10 metalloproteases and the γ-secretase complex containing presenilins are required in the regulation of NOTCH2 and NOTCH3 proteolysis and transcriptional activation upon ligand binding in a manner similar to that of NOTCH1.
MATERIALS AND METHODS
Description of plasmids and expression vectors.
All constructs were generated using traditional cloning methods and were fully verified by DNA sequencing. The full-length human NOTCH3 vectors used are pLVX-puro-hNOTCH3-FLAG-HA and pcDNA5/FRT-hNOTCH3-FLAG-HA; these constructs, derived from the human NOTCH3 cDNA, a kind gift from T. Wang (30), contain the code for a 1- to 2,321-amino-acid (aa) transcript (identical to NCBI reference sequence NP_000426.2) of hNOTCH3 protein with, fused to the C terminus, a sequential FLAG and hemagglutinin (HA) tag followed by a stop. The construct pcDNA5/FRT-hN3ΔE-ΔPEST-FLAG-HA for hNOTCH3 aa 1501 to 1792 utilizes the signal peptide of hNOTCH1 at the N terminus and has, fused to the C terminus, a sequential FLAG and HA tag. The full-length human NOTCH2 vectors used are pLVX-puro-hNOTCH2-FLAG-HA and pcDNA5-FRT-hNOTCH2-FLAG-HA. These constructs were derived from human NOTCH2 cDNA, a kind gift from I. Prudovsky, and contain the code for a 1- to 2,471-aa transcript (identical to NCBI reference sequence AAA36377.2, with the exception of amino acids A2079V, delQ2263, and Y2264H) of the hNOTCH2 protein with, fused to the C terminus, a sequential FLAG and HA tag followed by a stop. Both untagged pQCXIH-mTACE/Adam17 (where TACE is TNF-α converting enzyme) and pQCXIH-mKUZ/Adam10 bear the 5′ untranslated region (UTR), transcription initiation site, and coding sequences (CDS) containing cDNAs of these genes until the original stop codon. Untagged human pLBCX-PSEN1wt was a kind gift of S. Weggen (31). MYC-tagged pBABE-puro-hJAGGED2 was a kind gift from J. P. Di Santo (32). pAP-hTNF-α-MYC-HIS was a gift of C. P. Blobel (33), and pAP-TGFα was a kind gift of S. Higashiyama (34). Lenti-ADAM short hairpin RNA (shRNA) interference vectors were a kind gift of F. M. Hess and A. Ludwig (35). The pGL4.24-12xCSL NOTCH luciferase reporter and pGL4.74 TK-hRL vector were described previously (36). pCMV-Gaussia was obtained from New England Biolabs (NEB).
Cell lines.
Adam and PS1/2−/− knockout (KO) cell lines were as described previously (37–40). U2OS, HEK293, 293FT, and NIH 3T3 cells were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and, for NIH 3T3 cells, with 10% newborn calf serum (NCS). OP9 and TSt-4 cells were maintained as described previously (2, 41). Pseudo- or lentiviral particles were produced in 293FT cells, and transgenes were packaged using the Lenti-X system (Clontech) according to the manufacturer's instructions or as described previously (35, 42). Stable cell lines were generated by repetitive viral transduction and selected as polyclonal lines, continuously maintained with medium supplemented with hygromycin, puromycin, or blasticidin prior to experiments. Transfections with plasmid DNA of cells were performed using linear polyethylenimine (P-PEI; Polysciences Inc.) or FuGENE (Promega), with the exception of AD10/17dKO cells or derivative lines. These cells were transected using an Amaxa Nucleofector protocol for mouse embryonic fibroblasts (MEFs) according to the manufacturer's instructions.
Western blotting.
In all experiments, equal amounts of cells, determined by using an automated Z2 cell counter (Beckman Coulter), were used prior to transfection, seeding, and starting monotypic and cocultures. For Western blot analysis, cultures of cells were quickly rinsed with phosphate-buffered saline (PBS) and then scraped in Laemmli loading buffer and immediately boiled, vortexed, and spun down. Proteins were separated on 6%, 7.5%, or 15% gradient or purchased 4 to 12% gradient (Bio-Rad) Tris-HCl SDS-PAGE gels, as needed, and transferred onto polyvinylidene difluoride (PVDF) membranes. Protein detection was performed with subsequent primary antibodies: mouse anti-Myc (9E10), rabbit anti-HA (Sigma), rabbit anti-cleaved Notch1-1744 (Cell Signaling), mouse anti-β-actin (MP Biomedicals), mouse anti-aryl hydrocarbon receptor nuclear translocator (anti-ARNT; BD Transduction Laboratories), rabbit anti-Adam10 (a kind gift of S. Weber or Genetex), rabbit anti-Adam17/TACE (GeneTex), rabbit anti-Jagged1 (Santa Cruz), rabbit anti-Jagged2 (Santa Cruz), mouse anti-PS-1 (Chemicon), rabbit anti-NOTCH2 antibody and rabbit anti-PS-2 (Cell Signaling), goat anti-Delta (Santa Cruz), and rabbit anti-lamin A/C (Sigma). Secondary antibodies used were anti-mouse and anti-rabbit IgG–horseradish peroxidase (HRP; Cell Signaling) and donkey anti-goat HRP-linked antibodies (Santa Cruz). ECL (Amersham Biosciences) was used for visualization as described by the manufacturer.
qRT-PCR.
Total RNA from monotypic cell lines was isolated using NucleoSpin RNA II (Macherey-Nagel). Real-time quantitative PCR (qRT-PCR) was performed as described previously (43). Most primer sets were published previously (42, 44), with the exception of ADAM10 and ADAM17 primer sets (see Table S1 in the supplemental material).
Chemicals and cell-based assays.
Dimethyl sulfoxide (DMSO; Sigma) or a γ-secretase inhibitor (GSI) (dibenzazepine [DBZ], 200 nM; Syncon, Groningen, Netherlands) was added to cell cultures 6 h after transfection and at the start of a monotypic culture or cocultures, and this was maintained no longer than 16 h, or else chemical-containing medium was replaced. Batimastat (BB94; Syncon, Groningen, Netherlands) was used at a concentration of 10 μM in coculture experiments and 5 μM in shedding experiments. Cells were coated overnight with Dll4-Fc (5 μg/ml; R&D Systems), 0.1% bovine serum albumin (BSA), and 0.2% gelatin in PBS at 4°C and rinsed once before they were plated. Alkaline phosphatase (AP) shedding of tumor necrosis factor alpha (TNF-α) and transforming growth factor α (TGF-α) by phorbol myristate acetate (PMA; 100 ng/ml; Sigma) was measured 1 h after stimulation in the medium according to the protocol described in reference 45 with the Phospha-Light system of Applied Biosystems, and Gaussia luciferase was measured with the BioLux Gaussia luciferase assay kit of NEB, according to the manufacturer's instructions, on a Fluostar Omega plate reader (BMG Labtech). Additionally, this plate reader was used to measure dual-luciferase activity, in Notch reporter gene assays; 16 h after initiated cocultures, cells were washed, lysed, and measured, as described by the manufacturer (dual-luciferase reporter [DLR] assay system; Promega).
RESULTS
Ligand-induced cleavage of NOTCH2 and NOTCH3.
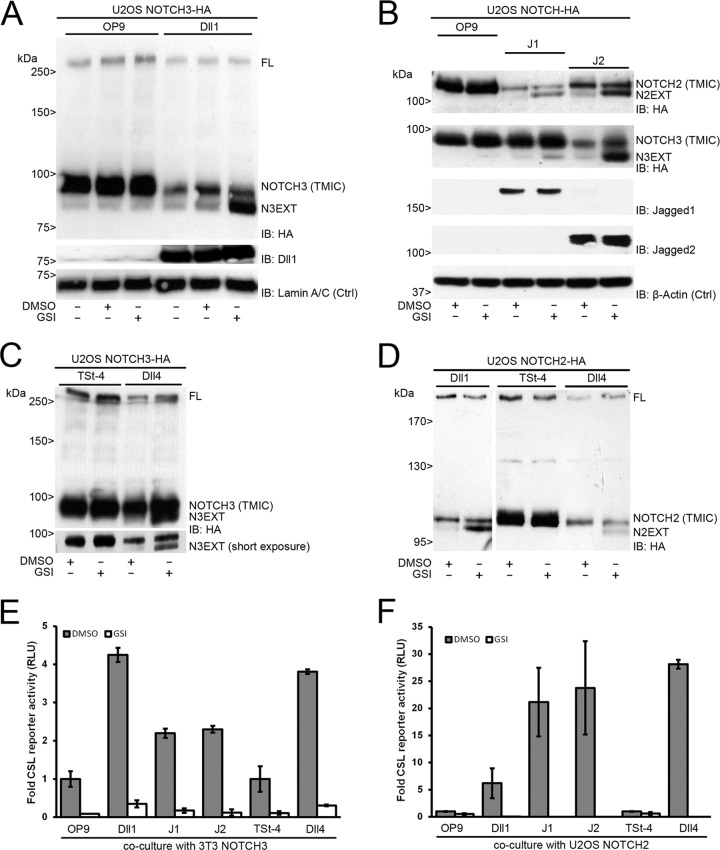
To study ligand-dependent signaling and processing of NOTCH3 receptors, we transduced U2OS cells with an HA-tagged human full-length NOTCH3 lentiviral expression vector and cocultured these with wild-type OP9 cells (control) or OP9 cells overexpressing Delta-like1 (Dll1). In the absence of ligand, an unprocessed full-length precursor product (FL) migrated at a molecular mass of ∼260 kDa, and a faster-migrating product at the ∼90-kDa fragment could be observed, which we designated trans-membrane and intracellular fragment (TMIC). Coculturing with OP9-Dll1 cells resulted in a reduction of the ∼90-kDa fragment and the appearance of a faster-migrating product that accumulated when γ-secretase activity was blocked using γ-secretase inhibitors, similar to the S2/NOTCH extracellular truncation (NEXT) cleavage fragment observed during ligand-induced Notch1 proteolysis (39). We therefore designated this fragment N3EXT (Fig. 1A).
FIG 1.
Human NOTCH2 and NOTCH3 proteolytic processing and transcriptional activation are triggered by DSL ligands. (A) Coculture experiment of U2OS NOTCH3-HA cells and OP9 parental or Dll1-expressing cells in the absence or presence of GSI. DMSO was used as a vehicle control. (Top panel) HA immunoblotting of cell lysates shows expression of both the full-length (FL) NOTCH3 precursor and mature, S1-processed, NOTCH3 TMIC. Dll1 stimulation leads to activation and diminished levels of NOTCH3 TMIC and accumulation of N3EXT in the presence of GSI. (Middle panel) Dll1 immunoblotting confirms expression of Dll1, which is absent in OP9 parental cells. (Bottom panel) Lamin A/C immunoblotting serves as a loading control (Ctrl). (B) Coculture experiment of U2OS NOTCH2-HA- and NOTCH3-HA-expressing cells with OP9 parental or Jagged1 (J1)- or Jagged2 (J2)-overexpressing cells in the absence or presence of GSI. (Top two panels) Stimulation by J1 and J2 leads to activation and diminished levels of NOTCH2 and NOTCH3 TMIC. N2EXT and N3EXT accumulate in the presence of GSI. (Middle two panels) Immunoblotting confirms expression of J1 and J2, which are absent in OP9 parental cells. (Bottom panel) β-Actin immunoblotting serves as a loading control (Ctrl). (C) Coculture experiment of U2OS NOTCH3-HA cells on top of TSt-4 parental or Dll4-overexpressing cells in the absence or presence of GSI. HA immunoblotting shows that Dll4 stimulation leads to NOTCH3 receptor proteolysis diminished TMIC expression and GSI-dependent accumulation of N3EXT (short exposure). (D) Coculture experiment of U2OS NOTCH2-HA-expressing cells with Dll1 or TSt-4 parental and TSt-4–Dll4-overexpressing cells in the absence or presence of GSI. HA immunoblotting of cell lysates shows expression of both the full-length (FL) NOTCH2 precursor and S1-processed NOTCH2 TMIC. Dll1 (left) and Dll4 (right) were able to activate NOTCH2 proteolytic processing, as accumulation of N2EXT fragments could be visualized by addition of GSI. Molecular mass marker proteins are indicated. (E and F) Dual NOTCH (CSL) luciferase reporter gene activity, corrected for Renilla luciferase expression, in either 3T3 cells expressing NOTCH3 (E) or U2OS cells expressing NOTCH2 (F). In response to coculture with ligand-expressing cells, an increased transcriptional activation was observed in comparison to coculture without ligand (OP9/TSt-4), which was completely blocked by GSI treatment. NOTCH-dependent signals were in parental cells arbitrarily set to 1. Measurements correspond to at least two experiments in triplicate and are displayed as relative light units (RLU). Error bars represent means ± standard deviations.
We next used the same coculture approach to study Jagged-induced proteolytic activation of human NOTCH2 and NOTCH3. Both Jagged1 (J1) and Jagged2 (J2) induced a ligand-dependent formation of N2EXT and N3EXT, which accumulated upon treatment with GSI. NOTCH2 TMIC migrates at ∼110 kDa, with N2EXT migrating at a slightly lower band (Fig. 1B). Furthermore, Delta-like4 (Dll4) ligand overexpression on murine thymic stromal TSt-4 cells also activated NOTCH3 and led to N3EXT (Fig. 1C). Dll4 mRNA expression in these cells is approximately 2,500-fold higher than in parental TSt-4 cells (see Fig. S1A in the supplemental material). Likewise, N2EXT was also readily formed when NOTCH2-HA-expressing cells were cocultured with Dll1- or Dll4-expressing cells (Fig. 1D). By means of the C-terminal tag, NOTCH2 proteins can be detected as precursor (FL) and mature NOTCH2 TMIC proteins, migrating at ∼300 and ∼110 kDa, respectively. A combination of coculture with ligand-expressing cells and GSI allowed detection of a faster-migrating additional band designated N2EXT (Fig. 1D). NOTCH2 and NOTCH3 show an identical patterning by proteolytic cleavage on Western blots, indicating that all four ligands (Dll1, Dll4, Jagged1, and Jagged2) activate NOTCH2 and NOTCH3 S2 cleavage to form N2EXT and N3EXT, albeit to a different extent (Fig. 1A to D). Coculture of NOTCH2- and NOTCH3-expressing cells with Dll4-Fc-coated plates also induced the formation of endogenous N2EXT and N3EXT in a manner similar to that of TSt-4–Dll4 cells (Fig. 2A; see also Fig. S1C in the supplemental material). Importantly, endogenous human N2EXT was also induced in a similar manner in untransfected HEK293 cells when stimulated with either Delta or Jagged ligand (see Fig. S1B in the supplemental material).
FIG 2.
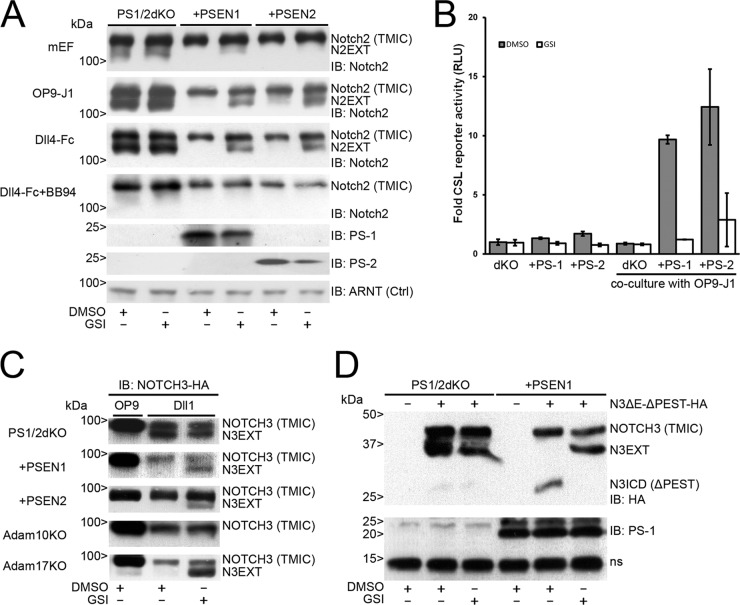
Notch2 and NOTCH3 proteolytic processing is regulated by presenilin-1 or -2. (A) Immunoblots showing expression of endogenous Notch2 in lysates from mouse embryonic fibroblasts (mEF) with double knockout for presenilin-1 and -2 (PS1/2dKO) or reconstituted with presenilin-1 (PSEN1) or 2 (PSEN2) (top panel), in coculture with OP9-J1 cells (second panel) or with coated Dll4-Fc molecules (third panel) in the absence or presence of GSI and GSI/BB94 (fourth panel). Immunoblots for presenilin-1 and -2 proteins (PS-1 and PS-2) show reconstitution of the respective presenilin, as indicated. ARNT (aryl hydrocarbon receptor nuclear translocator) served as a loading control (Ctrl). PS1/2dKO cells were unable to S3 process Notch2, indicated by accumulated N2EXT fragments after stimulation by either J1 or recombinant Dll4-Fc molecules. In the cells with PSEN1 or PSEN2, N2EXT fragments were observed only in the presence of GSI. Treatment with BB94 abrogates formation of N2EXT, indicating that the formation of N2EXT is dependent on metalloproteases. (B) Dual Notch (CSL) luciferase reporter gene activity, corrected for Renilla luciferase expression, in either PS1/2dKO cells or cells reconstituted with presenilin-1 (PS-1) or -2 (PS-2). In response to coculture with Jagged1-expressing cells, a γ-secretase-dependent increased transcriptional activation was observed only in cells reconstituted with either PS-1 or PS-2 and not in PS1/2dKO cells. All values were normalized to DMSO-treated PS1/2dKO cells and arbitrarily set to 1. Measurements correspond to at least two experiments in triplicate and are displayed as relative light units (RLU). Error bars represent means ± standard deviations. (C) Coculture experiment of NOTCH3-HA-expressing PS1/2dKO cells reconstituted with either PSEN1 or PSEN2, Adam10KO, and Adam17KO cells with Dll1-expressing cells compared to OP9 parental cells. HA immunoblots show diminished levels of TMIC after stimulation with Dll1. (Top three panels) N3EXT fragments accumulate in the absence of presenilin proteins, whereas in the presence of either PSEN1 or PSEN2, N3EXT accumulation was observed only when treated with GSI. (Bottom two panels) N3EXT fragments were absent in Adam10KO cells but not in Adam17KO cells after stimulation by Dll1 in the presence of GSI. (D) (Top) HA immunoblotting of cell lysates of nontransfected PS1/2dKO and PSEN1 reconstituted cells or transfected with an EGF repeat and PEST domain NOTCH3-HA deletion construct (hN3ΔE-ΔPEST) in the absence or presence of GSI. N3EXT accumulation was observed in hNOTCH3ΔE-ΔPEST-expressing PS1/2dKO cells without ligand stimulation. In PS1/2dKO cells reconstituted with PSEN1, N3EXT accumulation could be observed only in the presence of GSI. Furthermore, truncated N3ICD fragments were observed in PS1/2dKO cells reconstituted with PSEN1, which were absent in the presence of GSI. (Bottom) Immunoblot showing PS-1 expression with a nonspecific band (ns). Molecular masses are indicated.
Next, we measured ligand-induced NOTCH3 transcriptional activity in a coculture system, using a synthetic Notch reporter gene construct carrying 12 copies of the RBP-Jκ/CSL binding element driving luciferase and NOTCH3 in NIH 3T3 murine fibroblasts. Both Dll1 and Dll4 ligands strongly induced NOTCH3-dependent reporter gene activity. Similarly, J1 and J2 induced NOTCH3-dependent activity, albeit in a less pronounced manner than Dll1/4 ligands (Fig. 1E). For NOTCH2, we observed a consistent induction of luciferase reporter gene activity in U2OS cells upon stimulation with DSL ligands compared to parental cells, albeit to a different extent for the different ligands tested here (Fig. 1F). Overall, NOTCH2 activation was more robust and more responsive to Jagged1/2 ligands than was NOTCH3 activation. In contrast, Dll1 activated NOTCH3 receptors better than did Jagged1 and -2 ligands. In all experiments, reporter luciferase activity was blocked by GSI, indicating that this activity was dependent on cleavage of NOTCH receptors by γ-secretase (Fig. 1E and F). Taken together, all DSL ligands induce S2 cleavage of NOTCH2 and NOTCH3 receptors, resulting in transcriptional activation.
Both presenilin-1 and -2 cleave NOTCH2 and NOTCH3 proteins.
Vertebrates have two presenilin genes (PSEN1 and PSEN2), which encode the PS-1 and PS-2 proteins. Both proteins are aspartyl proteases and part of the γ-secretase complex. Since inhibition of γ-secretase by GSI led to the accumulation of N2EXT and N3EXT and blocked NOTCH transcriptional activity (Fig. 1), we studied the involvement of both presenilins in NEXT formation. In cells deficient for both PS1/2, stimulation with OP9-J1 cells or with soluble Dll4-Fc led to the accumulation of endogenous N2EXT. Accumulation of N2EXT was not influenced by pretreatment with GSI, demonstrating that this fragment is S2-cleaved Notch2 upstream of S3/γ-secretase cleavage. Reexpression of either PS-1 or PS-2 in PS1/2 double knockout (dKO) cells rescued S3/γ-secretase processing induced by OP9-J1 or Dll4-Fc, which could be inhibited by GSI, leading to N2EXT accumulation. Importantly, the formation of endogenous N2EXT was completely blocked when cells were incubated with the broad-spectrum metalloprotease inhibitor batimastat (BB94) (Fig. 2A). Next, we measured Notch-dependent transcriptional reporter activity in presenilin-deficient and -reconstituted cells. Only in cells reconstituted with either PSEN-1 or -2, ligand-dependent signaling that could be blocked by GSI was observed (Fig. 2B). Similarly, we studied the involvement of presenilins in NOTCH3 ligand-dependent activation. Similar to Notch2, NOTCH3 S3/γ-secretase activity required PS-1 and PS-2, and reconstitution with either PSEN1 or PSEN2 was sufficient to induce NOTCH3 cleavage upon Dll1 stimulation (compare Fig. 2C with Fig. 1A).
NOTCH3 receptors lacking EGF-LNR are constitutively active, and the PEST domain regulates N3ICD turnover.
Because we could not detect the S3/NICD-cleaved fragments of either NOTCH2 or NOTCH3 in our experiments, we hypothesized that because their NOTCH intracellular domains have a proline (P), glutamate (E), serine (S), and threonine (T) (PEST)-rich sequence, this led to their rapid degradation (9, 46, 47). To enable detection of S3 fragments of Notch1, constructs with deleted PEST domains or cleavage-specific antibodies have been used mostly (2). To study N3ICD formation from N3EXT, we generated NOTCH3 fusion proteins lacking the extracellular ligand binding epidermal growth factor-like (EGF) and Lin12-NOTCH repeats (LNR), leaving the heterodimerization domain (HD) intact and a deleted PEST domain. These hN3ΔE-ΔPEST proteins are highly transcriptionally active, independent of ligand, but remain dependent on S3/γ-secretase cleavage for their activity (our unpublished observations). hN3ΔE-ΔPEST proteins produced truncated N3EXT fragments in PS1/2dKO (Fig. 2D). Moreover, in PS1/2dKO cells reconstituted with PSEN1, N3EXT was processed to an ∼30-kDa product consistent with the calculated molecular mass of N3ICD-ΔPEST. The addition of GSI blocked N3ICD formation and led to the accumulation of N3EXT (Fig. 2D). Taken together, these data indicate that PS-1 induces N3EXT cleavage and produces N3ICD, which is very unstable, similar to what was seen for NOTCH1 and NOTCH2, and could be detected only when degradation was blocked by removal of the PEST domain (Fig. 2D).
N2EXT and N3EXT are generated by the metalloprotease Adam10 and not Adam17.
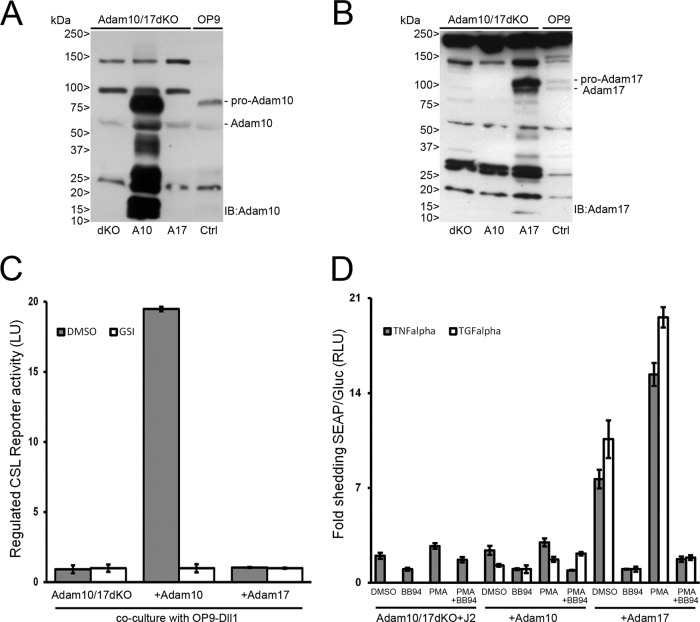
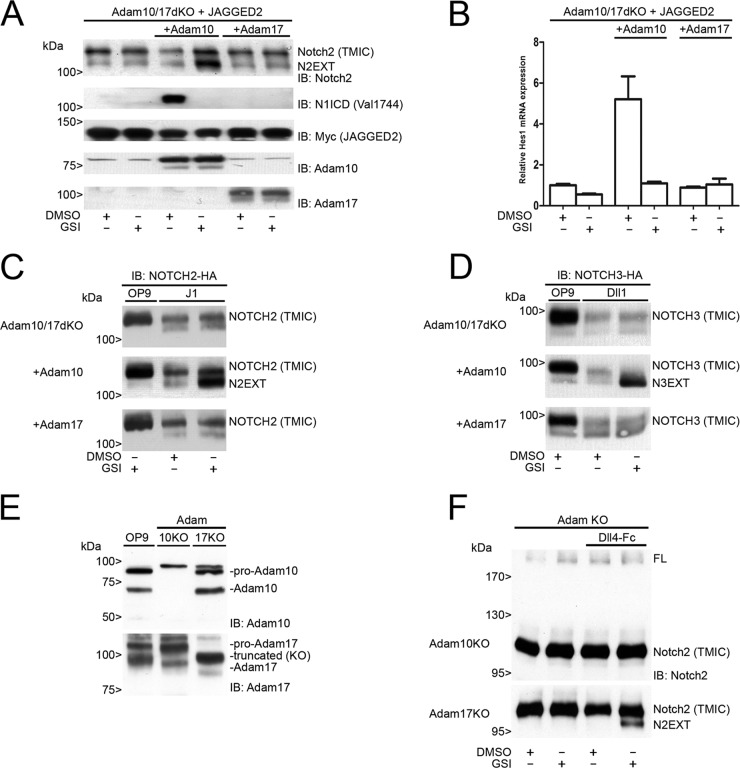
Upon ligand stimulation, murine Notch1 is sequentially cleaved by the Adam10 metalloprotease at S2-Val1711 and S3/γ-secretase at Val1744. There is no role for the highly related protease Adam17 in ligand-dependent Notch1 signaling (2). Here, we utilized fibroblasts lacking both Adam10 and Adam17 (Adam10/17dKO) (48) to study the requirements of these proteases in S2 processing for endogenous proteins and in cells transfected with NOTCH2 and NOTCH3 receptors. In parallel, Adam10/17dKO cells were generated with either murine ADAM10 or Adam17 expression by viral transduction, which resulted in stable mRNA (see Fig. S2A in the supplemental material) and protein expression of Adam10 and Adam17 (Fig. 3A and B). Since Adam10/17dKO fibroblasts express endogenous Notch1 and Notch2, we first determined transcriptional activation in the absence and presence of Adam10 or Adam17 proteases upon coculture with ligand. Only in cells where Adam10 was reintroduced was γ-secretase transcriptional activation of the Notch reporter gene observed in response to ligand (Fig. 3C). In Adam10/17dKO fibroblasts reconstituted with Adam17, no NOTCH-dependent RBP-Jκ/CSL reporter activity was observed. To confirm that Adam17 was catalytically active, we transfected cells with alkaline phosphatase (AP)-tagged TNF-α or TGF-α expression vectors that are selectively shed by Adam17 after stimulation with phorbol 12-myristate 13-acetate (PMA) (48). Only in Adam17-reconstituted cells was increased AP shedding, both constitutive and PMA induced, into the culture supernatant detected, confirming Adam17 catalytic activity in these cells. This activity was fully blocked by the addition of the metalloprotease inhibitor BB94 (Fig. 3D). Because Adam10/17dKO cells do not express detectable levels of ligand, they are deficient in endogenous Notch1 and -2 cleavage and transcriptional activation of the Notch pathway even when Adam10 or -17 is reconstituted (see Fig. S2C in the supplemental material). Only when stimulated with ligand in Adam10-expressing but not Adam17-expressing cells are Notch1 and Notch2 cleaved (Fig. 4A; see also Fig. S2C and D in the supplemental material), leading to RBP-Jκ/CSL transcriptional activation and restoration of the mRNA expression of NOTCH target genes Hes1 (Fig. 4B) and Hey1 (see Fig. S2B in the supplemental material).
FIG 3.
Characterization and functional analysis of Adam10/17dKO or Adam10/Adam17-reconstituted MEFs. (A and B) Immunoblotting with Adam10 and Adam17 antibodies on lysates from Adam10/17dKO MEF cells reconstituted with either Adam10 (A10) or Adam17 (A17). Pro- and mature Adam10 proteins could be detected only in Adam10-reconstituted Adam10/17dKO or OP9 cells (Ctrl), whereas Adam17 pro- and mature proteins were detected only in Adam17-reconstituted Adam10/17dKO or Ctrl cells. Molecular masses are indicated. (C) Firefly luciferase transcriptional Notch CSL reporter activity in Adam10/17dKO cells, reconstituted with either Adam10 or Adam17, cocultured with OP9-Dll1 cells. Regulated Notch reporter activity in light units (LU) could be measured only in cells expressing Adam10, which could be inhibited by GSI. Measurements were arbitrarily set to GSI values and correspond to at least two experiments in triplicate. Error bars represent means ± standard deviations. (D) AP-TNF-α- and AP-TGF-α-transfected Adam10/17dKO-J2 MEF cells reconstituted with either Adam10 or Adam17 were stimulated with DMSO (vehicle), BB94 (5 μM), PMA (100 ng/ml), or both BB94 and PMA. Gaussia luciferase excretion, unaffected by any of the stimuli, was used as an internal control. Medium was analyzed 1 h after stimulation for the presence of shed AP-TNF-α and AP-TGF-α molecules by enzymatic conversion of substrate into light units and normalized to Gaussia luciferase values. AP activity measurements of BB94-treated cells were arbitrarily set to 1. TGF-α shedding in dKO only cells could not be detected. Both constitutive and regulated PMA-induced TNF-α and TGF-α shedding could be observed only in cells expressing Adam17. Furthermore, shedding of both regulated and constitutive TNF-α and TGF-α is blocked by the addition of BB94. Measurements correspond to experiments performed in triplicate and are displayed in RLU. Error bars represent means ± standard deviations.
FIG 4.
Ligand-dependent NOTCH2 and NOTCH3 signaling requires Adam10. (A) Immunoblots on cell lysates of monotypic coculture experiments with Adam10/17dKO-JAGGED2 cells, reconstituted with either Adam10 or Adam17 treated with DMSO or GSI. Blots were probed with antibodies for Notch2, N1ICD (Val1744), Myc (JAGGED2), Adam10, or Adam17. Endogenous N2EXT can accumulate only in the presence of Adam10 and GSI. Endogenous activated Notch1 (Val1744) could be detected only in cultures of cells expressing Adam10 and is inhibited by GSI. Overexpression of Adam17 did not lead to proteolysis of either endogenous Notch1 or Notch2. Molecular masses are indicated. (B) Relative Hes1 mRNA expression in Adam10/17dKO-JAGGED2 cells, reconstituted with either Adam10 or Adam17, subjected to monotypic coculture experiments in the absence or presence of GSI. Hes1 mRNA expression was induced only in the presence of Adam10 and could be completely blocked by GSI treatment. Experiments were performed in triplicate. Error bars represent means and standard deviations. (C) Coculture experiment of NOTCH2-HA-expressing Adam10/17dKO cells, reconstituted with either Adam10 or Adam17 with J1-expressing cells compared to parental OP9 cells. Panels display HA immunoblots of cell lysates as indicated. Diminished levels of NOTCH2 TMIC indicative of J1-dependent NOTCH2 processing were found in all cell types in the absence or presence of GSI. Upon activation, N2EXT fragments accumulated in cells expressing Adam10 by the addition of GSI (middle) and not in the presence of Adam17 (bottom). (D) Coculture experiment of NOTCH3-HA-expressing Adam10/17dKO cells, reconstituted with either Adam10 or Adam17 with Dll1-expressing cells compared to parental OP9 cells. Diminished levels of NOTCH3 TMIC are shown by HA immunoblotting upon activation. Genuine N3EXT accumulation was observed only after GSI treatment in the presence of Adam10 (middle) and not in the presence of Adam17 (bottom). (E) Immunoblotting of Adam10 and Adam17 endogenous proteins in Adam10 or Adam17 single knockout cells with OP9 cells as a positive control. (F) Immunoblotting for endogenous Notch2 in Adam10 or Adam17 single knockout cells cultured on coated Dll4-Fc molecules shows the formation of N2EXT to be Adam10 dependent, as N2EXT formation could not be observed in Adam10 knockout cells. In Adam17 knockout cells, treatment with GSI leads to the formation and accumulation of an N2EXT fragment. Molecular masses are indicated.
Similarly, in Adam10/17dKO cells transduced with exogenous HA-tagged NOTCH2 (Fig. 4C) or NOTCH3 (Fig. 4D), S2 cleavage occurred only in the presence of Adam10 and ligand stimulation. In neither case did Adam17/TACE restore ligand-dependent S2 cleavage. In accordance, ligand-induced formation of N3EXT is completely abolished in independently derived Adam10KO cells but not in Adam17KO cells (Fig. 2C). Both mature forms and proforms of Adam10 or Adam17 are expressed in, respectively, Adam10 and Adam17 single-KO cells (Fig. 4E). Similar to NOTCH3, coculture of these cells with coated Dll4-Fc ligand revealed the accumulation of endogenous N2EXT in the presence of GSI only in Adam17- but not in Adam10-deficient cells (Fig. 4F). Interestingly, we observed that in cells lacking both Adam10 and Adam17, ligand stimulation still reduced the membrane-bound TMIC of Notch1, Notch2, and Notch3 in a manner similar to that of wild-type cells (Fig. 1 and 4C and D and data not shown). However, the appearance of NEXT-like cleavage fragments was not dependent on Adam17 or γ-secretase activity.
To further substantiate our findings, we used U2OS cells stably expressing shRNA against human ADAM10 and ADAM17 (U2OS double knockdown [dKD]); we obtained silencing up to ∼95% of ADAM10 and ADAM17 mRNA expression. No N3EXT in response to ligand compared to cells expressing a control scrambled shRNA (U2OS SCR) was observed (see Fig. S3A in the supplemental material). As expected, silencing of both Adam10 and Adam17 showed a strong reduction in NOTCH3-dependent RBP-Jκ/CSL transcriptional reporter gene activity (see Fig. S3B in the supplemental material). Taken together, these results lend further support to the key role of ADAM10 in ligand-induced NOTCH receptor proteolysis.
DISCUSSION
Previously, both our group and others have established that the Adam10 metalloprotease, but not Adam17, is essential in ligand-induced extracellular cleavage of Notch1 in cellular models and in vivo (2, 14, 49, 50). While much knowledge on Notch1 regulation in many different systems exists, relatively little is known about the activation mechanisms of the NOTCH2, NOTCH3, and NOTCH4 receptors in mammalian cells. Here, we disclose the proteolytic mechanism of ligand-induced human NOTCH2 and NOTCH3 receptor activation. We show that NOTCH2 and NOTCH3 receptor stimulation with Dll1, Dll4, J1, and J2 ligand-expressing cells or via recombinant Dll4-Fc induces a proteolytic cascade, which requires activity of the metalloprotease Adam10 and γ-secretase to lead to transcriptional activation of target genes. We found no role for the highly related TNF-α converting enzyme (TACE), also known as the Adam17 protease, in ligand-dependent signaling through the NOTCH1, NOTCH2, and NOTCH3 receptors.
Previously, it was demonstrated that murine NOTCH2 interacted with DSL ligands with different affinities in vitro and in cells, which resulted in receptor cleavage and target gene activation (51, 52). In these experiments, the S2/S3 cascade was not directly studied and the involvement of metalloproteases was not addressed. Here we show that DSL ligands can activate both endogenous murine and human NOTCH2, that the formation of N2EXT is a prerequisite in ligand-dependent downstream activation of NOTCH2 by ligands, and that this is blocked when cells are treated with a broad-spectrum metalloprotease inhibitor, batimastat. Similarly, the NOTCH3 extracellular domain has been shown to bind with different affinities to recombinant DSL ligands in vitro, suggesting that they could act as a ligand in cells (53); however, to the best of our knowledge, a comprehensive analysis of this is lacking. Here we show that all DSL ligands can trigger the NOTCH3 cleavage cascade, as we found for NOTCH2, leading to the formation of an extracellular truncated NOTCH3 receptor (N3EXT), which acts as a substrate for presenilin/γ-secretase-dependent cleavage producing N3ICD.
We therefore conclude that ligand-dependent NOTCH2 and NOTCH3 signaling follows the regulated intramembranous proteolysis cascade, similar to what is seen with NOTCH1 proteins (39). We show that human N2EXT and N3EXT are also subject to γ-secretase cleavage and that both presenilin-1 and -2 are sufficient to cleave NOTCH2 or NOTCH3. In our studies, we could not directly demonstrate the presence of human N2ICD and N3ICD, perhaps because these fragments comigrated with NOTCH2/3 TMIC, possibly due to phosphorylation (9, 10, 54). By using truncated NOTCH3 mutants lacking the PEST domain, we were able to demonstrate that N3ICD is formed in these and depends on the activity of PS-1.
PS-1 is best known for its involvement in the RIP of many substrates (55), and cells lacking PS-1 are deficient in proteolysis of both NOTCH and amyloid precursor protein (APP), the best-known physiological substrates (5). Mice lacking Psen1 only or both PSEN1 and -2 are embryonic lethal and exhibit Notch1-associated deficiencies in hematopoiesis, somitogenesis, and neurogenesis (6, 56). Mice lacking only Psen2, however, are viable, have no apparent NOTCH1 phenotype, and have no defects in APP cleavage (38). Mice lacking Notch3 develop normally, are viable and fertile, and cannot compensate for lack of NOTCH1 during embryogenesis (8).
Our study indicates that NOTCH2 and NOTCH3 are substrates for S3/γ-secretase complex cleavage containing either PS-1 or PS-2 proteins. Because NOTCH3 is nonessential for embryogenesis, its function may become apparent only in a temporal and spatial manner in adult tissues. Like the Notch receptors, PS-1 and -2 proteins have evolved differently. PS-1 appears to be crucial both during embryogenesis and in adult tissues, and its most important substrate seems to be NOTCH1. The relative importance of PS-1 versus PS-2 in Notch2 and Notch3 activation in adult tissues is not clear. Interestingly, we found that in the absence of Psen1, human NOTCH2 and -3 are also substrates for PS-2. Expression studies did not show any colocalization between NOTCH2 and NOTCH3 with PS-2 in human embryos, but these receptors did colocalize with PS-1 in the neuroepithelial layer, like NOTCH1 (57). Although previous studies, using coimmunoprecipitation, have also shown PS-2 to be able to bind with truncated Notch2 and Notch3 proteins (9), we provide the first functional evidence that PS-2 is also capable of producing N2ICD and N3ICD in mammalian cells. Conditional tissue-specific Psen1-deficient mice are needed to assess whether PS-2 can also function as a NOTCH2 and NOTCH3 intramembrane protease in vivo.
We observed that in the absence of Adam10, Notch receptors were being processed, as evidenced by the accumulation of proteolytic fragments with sizes approximately similar to that of NEXT, as well as a reduction in the cell surface S1/TMIC-cleaved forms. In contrast to wild-type signaling, these NEXT-like fragments formed in the absence of a γ-secretase inhibitor and did not lead to transcriptional activation. While we present no evidence to that effect here, we speculate that ligand-induced processing of NOTCH receptors in the absence of Adam10 could be a protection mechanism against the accumulation of “unused” NOTCH receptors at the cell surface, which may interfere with other signaling pathways or block other NOTCH ligand receptor interactions. This would ensure that when all components are present, ligand-receptor engagement would immediately lead to activation of the proteolytic cascade. While this is an interesting hypothesis, experimental proof for this is still lacking.
Using Adam10/17-deficient fibroblasts, we show for the first time that for ligand-dependent Notch2 and Notch3 signaling, Adam10 is absolutely required. TACE/Adam17 is neither necessary nor sufficient to rescue the endogenous Notch signaling defect in Adam10-deficient cells. Taken together, our data show that ligand-dependent human NOTCH2 and NOTCH3 signaling also follows the same paradigm upon ligand binding, i.e., cleavage of the receptors into the S2-cleaved NEXT fragment by the metalloprotease Adam10, followed by γ-secretase-dependent formation of NICD and transcriptional activation. Recently, it was shown that overexpression of membrane type I matrix metalloproteinase (MT1-MMP) could activate endogenous Notch1 in the absence of Adam10 and -17 (58). Using the same Adam10/17dKO cells, we show that these cells express no ligand and are deficient in Notch signaling. Only after ligand stimulation and reintroduction of Adam10 can Notch1 be cleaved and be transcriptionally active. Therefore, we infer that MT1-MMP overexpression may be involved in ligand-independent activation of Notch receptors, which we have not addressed here. This would be an interesting topic for further studies.
Adam17/TACE is crucial for the shedding of many signaling molecules, such as betacellulin, ICAM, TNF-α, L-Selectin, and TGF-α, as shown in experiments using Adam10-, Adam17-, or Adam10/17-deficient fibroblasts (40, 48). Recently, in vitro studies have demonstrated that recombinant human NOTCH2 peptides are a substrate for Adam17/TACE. In these experiments, ADAM10 was shown to be less efficient in NOTCH2 cleavage (59). Also, for NOTCH1, in vitro cleavage assays using Adam17/TACE protease have not been predictive for its role in ligand-dependent signaling in cultured cells or in vivo, therefore questioning the validity of recombinant (Adam17) protease and substrates in vitro. This is likely because purified proteins or peptides do not reflect the natural and complex environment in which the NRR must be folded and maintained. Indeed, only functional studies in mammalian cells directly proved that Adam10, and not Adam17, is responsible for ligand-induced shedding of NOTCH1 to -3 at S2 (2, 50), as well as this study. There is increasing evidence, however, that in ligand-independent Notch1 signaling, for example, in the case of mutated Notch1 receptors in T-ALL, other proteases like Adam17 may be involved in addition to Adam10 (14, 60). Whether this also holds true for the other Notch2, -3, and -4 receptors is currently not known. Such differential protease involvement may be exploited by specifically targeting Notch in diseased tissues only, while leaving Notch signaling in normal tissues less affected.
In summary, we demonstrate that the proteolytic cascade of canonical ligand-dependent signaling through Notch receptors is conserved and requires Adam10. Specific inhibitors targeting ADAM10 may be beneficial in diseases in which canonical ligand-dependent Notch signaling is affected. Caution against such approaches is warranted, however, because of the increasing evidence of loss-of-function mutations in diseases such as cancer.
Supplementary Material
ACKNOWLEDGMENTS
We kindly thank T. Wang (The University of Manchester, Manchester, United Kingdom) for providing the NOTCH3 cDNA and I. Prudovsky (Center for Molecular Medicine, Maine Medical Center Research Institute, ME) for providing the NOTCH2 cDNA. We thank H. Kawamoto (RIKEN Research Center for Allergy and Immunology, Yokohama, Japan) for providing TSt-4/Dll4 ligand-expressing cells.
This work is supported by the Deutsche Forschungsgemeinschaft (DFG grant SFB877) (to P.S.) and by the European Research Council under the European Community Seventh Framework Program (FP7/2007-2013)/ERC grant 208259 (to M.V.).
Footnotes
Published ahead of print 19 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00206-14.
REFERENCES
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770–776. 10.1126/science.284.5415.770 [DOI] [PubMed] [Google Scholar]
- 2.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. 2009. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284:31018–31027. 10.1074/jbc.M109.006775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. 2004. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 24:9265–9273. 10.1128/MCB.24.21.9265-9273.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518–522. 10.1038/19083 [DOI] [PubMed] [Google Scholar]
- 6.Donoviel DB, Hadjantonakis AK, Ikeda M, Zheng H, Hyslop PS, Bernstein A. 1999. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13:2801–2810. 10.1101/gad.13.21.2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. 1999. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 126:3415–3424 [DOI] [PubMed] [Google Scholar]
- 8.Krebs LT, Xue Y, Norton CR, Sundberg JP, Beatus P, Lendahl U, Joutel A, Gridley T. 2003. Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 37:139–143. 10.1002/gene.10241 [DOI] [PubMed] [Google Scholar]
- 9.Saxena MT, Schroeter EH, Mumm JS, Kopan R. 2001. Murine notch homologs (N1-4) undergo presenilin-dependent proteolysis. J. Biol. Chem. 276:40268–40273. 10.1074/jbc.M107234200 [DOI] [PubMed] [Google Scholar]
- 10.Mizutani T, Taniguchi Y, Aoki T, Hashimoto N, Honjo T. 2001. Conservation of the biochemical mechanisms of signal transduction among mammalian Notch family members. Proc. Natl. Acad. Sci. U. S. A. 98:9026–9031. 10.1073/pnas.161269998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steiner H, Capell A, Leimer U, Haass C. 1999. Genes and mechanisms involved in beta-amyloid generation and Alzheimer's disease. Eur. Arch. Psychiatry Clin. Neurosci. 249:266–270. 10.1007/s004060050098 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner BA. 2000. Presenilins are required for gamma-secretase cleavage of beta-APP and transmembrane cleavage of Notch-1. Nat. Cell Biol. 2:463–465. 10.1038/35017108 [DOI] [PubMed] [Google Scholar]
- 13.Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. 2000. Total inactivation of gamma-secretase activity in presenilin-deficient embryonic stem cells. Nat. Cell Biol. 2:461–462. 10.1038/35017105 [DOI] [PubMed] [Google Scholar]
- 14.Bozkulak EC, Weinmaster G. 2009. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29:5679–5695. 10.1128/MCB.00406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, Hirai H. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18:675–685. 10.1016/S1074-7613(03)00111-0 [DOI] [PubMed] [Google Scholar]
- 16.Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, Conrad DH. 2010. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J. Exp. Med. 207:623–635. 10.1084/jem.20091990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, Piga N, Chapon F, Godfrain C, Tournier-Lasserve E. 2000. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Invest. 105:597–605. 10.1172/JCI8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lardelli M, Dahlstrand J, Lendahl U. 1994. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech. Dev. 46:123–136. 10.1016/0925-4773(94)90081-7 [DOI] [PubMed] [Google Scholar]
- 19.van Tetering G, Vooijs M. 2011. Proteolytic cleavage of Notch: “HIT and RUN”. Curr. Mol. Med. 11:255–269. 10.2174/156652411795677972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penton AL, Leonard LD, Spinner NB. 2012. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 23:450–457. 10.1016/j.semcdb.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. 2006. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am. J. Hum. Genet. 79:169–173. 10.1086/505332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, Kim KH, Pauli RM, Aftimos S, Stewart H, Kim CA, Holder-Espinasse M, Robertson SP, Drake WM, Trembath RC. 2011. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 43:303–305. 10.1038/ng.779 [DOI] [PubMed] [Google Scholar]
- 23.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. 2005. Mutations in NOTCH1 cause aortic valve disease. Nature 437:270–274. 10.1038/nature03940 [DOI] [PubMed] [Google Scholar]
- 24.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM., III 2007. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J. Thorac. Cardiovasc. Surg. 134:290–296. 10.1016/j.jtcvs.2007.02.041 [DOI] [PubMed] [Google Scholar]
- 25.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269–271. 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- 26.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649–661. 10.1016/0092-8674(91)90111-B [DOI] [PubMed] [Google Scholar]
- 27.Egloff AM, Grandis JR. 2012. Molecular pathways: context-dependent approaches to Notch targeting as cancer therapy. Clin. Cancer Res. 18:5188–5195. 10.1158/1078-0432.CCR-11-2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parr C, Watkins G, Jiang WG. 2004. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int. J. Mol. Med. 14:779–786. 10.3892/ijmm.14.5.779 [DOI] [PubMed] [Google Scholar]
- 29.McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, Chen YS, Chin KT, Partridge JC, Poole BB, Cheng KH, Daggett J, Jr, Cullen K, Kantoff E, Hasselbatt K, Berkowitz J, Muto MG, Berkowitz RS, Aster JC, Matulonis UA, Dinulescu DM. 2012. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc. Natl. Acad. Sci. U. S. A. 109:E2939–E2948. 10.1073/pnas.1206400109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, Holt CM, Xu C, Ridley C, Jones RPO, Baron M, Trump D. 2007. Notch3 activation modulates cell growth behaviour and cross-talk to Wnt/TCF signalling pathway. Cell Signal. 19:2458–2467. 10.1016/j.cellsig.2007.07.019 [DOI] [PubMed] [Google Scholar]
- 31.Hahn S, Bruning T, Ness J, Czirr E, Baches S, Gijsen H, Korth C, Pietrzik CU, Bulic B, Weggen S. 2011. Presenilin-1 but not amyloid precursor protein mutations present in mouse models of Alzheimer's disease attenuate the response of cultured cells to gamma-secretase modulators regardless of their potency and structure. J. Neurochem. 116:385–395. 10.1111/j.1471-4159.2010.07118.x [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Ojeda ME, Klein Wolterink RG, Lemaitre F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, Di Santo JP. 2013. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood 121:1749–1759. 10.1182/blood-2012-06-440065 [DOI] [PubMed] [Google Scholar]
- 33.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. 2004. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164:769–779. 10.1083/jcb.200307137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokumaru S, Higashiyama S, Endo T, Nakagawa T, Miyagawa JI, Yamamori K, Hanakawa Y, Ohmoto H, Yoshino K, Shirakata Y, Matsuzawa Y, Hashimoto K, Taniguchi N. 2000. Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. J. Cell Biol. 151:209–220. 10.1083/jcb.151.2.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruessmeyer J, Martin C, Hess FM, Schwarz N, Schmidt S, Kogel T, Hoettecke N, Schmidt B, Sechi A, Uhlig S, Ludwig A. 2010. A disintegrin and metalloproteinase 17 (ADAM17) mediates inflammation-induced shedding of syndecan-1 and -4 by lung epithelial cells. J. Biol. Chem. 285:555–564. 10.1074/jbc.M109.059394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Tetering G, Bovenschen N, Meeldijk J, van Diest PJ, Vooijs M. 2011. Cleavage of Notch1 by granzyme B disables its transcriptional activity. Biochem. J. 437:313–322. 10.1042/BJ20110226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. 1997. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 385:729–733. 10.1038/385729a0 [DOI] [PubMed] [Google Scholar]
- 38.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, Baekelandt V, Dressel R, Cupers P, Huylebroeck D, Zwijsen A, Van Leuven F, De Strooper B. 1999. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. U. S. A. 96:11872–11877. 10.1073/pnas.96.21.11872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197–206. 10.1016/S1097-2765(00)80416-5 [DOI] [PubMed] [Google Scholar]
- 40.Le Gall SM, Maretzky T, Issuree PD, Niu XD, Reiss K, Saftig P, Khokha R, Lundell D, Blobel CP. 2010. ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J. Cell Sci. 123:3913–3922. 10.1242/jcs.069997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meek B, Cloosen S, Borsotti C, Van Elssen CH, Vanderlocht J, Schnijderberg MC, van der Poel MW, Leewis B, Hesselink R, Manz MG, Katsura Y, Kawamoto H, Germeraad WT, Bos GM. 2010. In vitro-differentiated T/natural killer-cell progenitors derived from human CD34+ cells mature in the thymus. Blood 115:261–264. 10.1182/blood-2009-05-223990 [DOI] [PubMed] [Google Scholar]
- 42.Theys J, Jutten B, Habets R, Paesmans K, Groot AJ, Lambin P, Wouters BG, Lammering G, Vooijs M. 2011. E-Cadherin loss associated with EMT promotes radioresistance in human tumor cells. Radiother. Oncol. 99:392–397. 10.1016/j.radonc.2011.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodin CM, Verdam FJ, Grootjans J, Rensen SS, Verheyen FK, Dejong CH, Buurman WA, Greve JW, Lenaerts K. 2011. Reduced Paneth cell antimicrobial protein levels correlate with activation of the unfolded protein response in the gut of obese individuals. J. Pathol. 225:276–284. 10.1002/path.2917 [DOI] [PubMed] [Google Scholar]
- 44.Watanabe K, Nagaoka T, Lee JM, Bianco C, Gonzales M, Castro NP, Rangel MC, Sakamoto K, Sun Y, Callahan R, Salomon DS. 2009. Enhancement of Notch receptor maturation and signaling sensitivity by Cripto-1. J. Cell Biol. 187:343–353. 10.1083/jcb.200905105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahin U, Weskamp G, Zheng Y, Chesneau V, Horiuchi K, Blobel CP. 2006. A sensitive method to monitor ectodomain shedding of ligands of the epidermal growth factor receptor. Methods Mol. Biol. 327:99–113. 10.1385/1-59745-012-X:99 [DOI] [PubMed] [Google Scholar]
- 46.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276:35847–35853. 10.1074/jbc.M103992200 [DOI] [PubMed] [Google Scholar]
- 47.Rechsteiner M, Rogers SW. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267–271. 10.1016/S0968-0004(96)10031-1 [DOI] [PubMed] [Google Scholar]
- 48.Le Gall SM, Bobe P, Reiss K, Horiuchi K, Niu XD, Lundell D, Gibb DR, Conrad D, Saftig P, Blobel CP. 2009. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, L-selectin, and tumor necrosis factor alpha. Mol. Biol. Cell 20:1785–1794. 10.1091/mbc.E08-11-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber S, Niessen MT, Prox J, Lullmann-Rauch R, Schmitz A, Schwanbeck R, Blobel CP, Jorissen E, de Strooper B, Niessen CM, Saftig P. 2011. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development 138:495–505. 10.1242/dev.055210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Groot AJ, Cobzaru C, Weber S, Saftig P, Blobel CP, Kopan R, Vooijs M, Franzke CW. 2013. Epidermal ADAM17 is dispensable for notch activation. J. Invest. Dermatol. 133:2286–2288. 10.1038/jid.2013.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. 2010. Therapeutic antibody targeting of individual Notch receptors. Nature 464:1052–1057. 10.1038/nature08878 [DOI] [PubMed] [Google Scholar]
- 52.Shimizu K, Chiba S, Hosoya N, Kumano K, Saito T, Kurokawa M, Kanda Y, Hamada Y, Hirai H. 2000. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol. Cell. Biol. 20:6913–6922. 10.1128/MCB.20.18.6913-6922.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu K, Chiba S, Saito T, Kumano K, Hirai H. 2000. Physical interaction of Delta1, Jagged1, and Jagged2 with Notch1 and Notch3 receptors. Biochem. Biophys. Res. Commun. 276:385–389. 10.1006/bbrc.2000.3469 [DOI] [PubMed] [Google Scholar]
- 54.Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. 2006. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281:5106–5119. 10.1074/jbc.M506108200 [DOI] [PubMed] [Google Scholar]
- 55.Haapasalo A, Kovacs DM. 2011. The many substrates of presenilin/gamma-secretase. J. Alzheimers Dis. 25:3–28. 10.3233/JAD-2011-101065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. 1997. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell 89:629–639. 10.1016/S0092-8674(00)80244-5 [DOI] [PubMed] [Google Scholar]
- 57.Kostyszyn B, Cowburn RF, Seiger A, Kjaeldgaard A, Sundstrom E. 2004. Distribution of presenilin 1 and 2 and their relation to Notch receptors and ligands in human embryonic/foetal central nervous system. Brain Res. Dev. Brain Res. 151:75–86. 10.1016/j.devbrainres.2004.04.005 [DOI] [PubMed] [Google Scholar]
- 58.Ma J, Tang X, Wong P, Jacobs B, Borden EC, Bedogni B. 2014. Noncanonical activation of notch1 protein by membrane type I matrix metalloproteinase (MT1-MMP) controls melanoma cell proliferation. J. Biol. Chem. 289:8442–8449. 10.1074/jbc.M113.516039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephenson NL, Avis JM. 2012. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc. Natl. Acad. Sci. U. S. A. 109:E2757–E2765. 10.1073/pnas.1205788109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sulis ML, Saftig P, Ferrando AA. 2011. Redundancy and specificity of the metalloprotease system mediating oncogenic NOTCH1 activation in T-ALL. Leukemia 25:1564–1569. 10.1038/leu.2011.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.