Abstract
Histidine protein methylation is an unusual posttranslational modification. In the yeast Saccharomyces cerevisiae, the large ribosomal subunit protein Rpl3p is methylated at histidine 243, a residue that contacts the 25S rRNA near the P site. Rpl3p methylation is dependent upon the presence of Hpm1p, a candidate seven-beta-strand methyltransferase. In this study, we elucidated the biological activities of Hpm1p in vitro and in vivo. Amino acid analyses reveal that Hpm1p is responsible for all of the detectable protein histidine methylation in yeast. The modification is found on a polypeptide corresponding to the size of Rpl3p in ribosomes and in a nucleus-containing organelle fraction but was not detected in proteins of the ribosome-free cytosol fraction. In vitro assays demonstrate that Hpm1p has methyltransferase activity on ribosome-associated but not free Rpl3p, suggesting that its activity depends on interactions with ribosomal components. hpm1 null cells are defective in early rRNA processing, resulting in a deficiency of 60S subunits and translation initiation defects that are exacerbated in minimal medium. Cells lacking Hpm1p are resistant to cycloheximide and verrucarin A and have decreased translational fidelity. We propose that Hpm1p plays a role in the orchestration of the early assembly of the large ribosomal subunit and in faithful protein production.
INTRODUCTION
Components of the translational apparatus are extensively posttranscriptionally and posttranslationally modified in all three domains of life. One of the most common modifications is the addition of methyl groups to rRNA, tRNA, mRNA, snRNA, translation factors, and ribosomal proteins (1–4). These reactions are catalyzed by the members of several families of structurally related methyltransferases that use S-adenosylmethionine as a methyl donor; some 1% of the genes in a variety of organisms encode such enzymes (5). In the yeast Saccharomyces cerevisiae, more than half of the 64 known methyltransferases are devoted to the modification of the RNA and protein components of the translational machinery (6–8). The majority of rRNA modifications are found clustered in functional centers of the ribosome and increase in complexity in higher organisms. They are found in the peptidyl transferase center (PTC); the A, P, and E sites; the polypeptide exit channel; and the interface between the large and small ribosomal subunits, suggesting that these modifications help maintain the proper structures of these functional centers to ensure accurate and efficient translation (9). Methylation of tRNA is conserved in most organisms and can affect structural stability and translational fidelity (10, 11). Methylation of translation release factors has been implicated in translation termination and fidelity (12, 13). To date, relatively little is known about the roles of ribosomal protein methylation.
Previous mass spectrometric analyses of intact small and large ribosomal subunit proteins in S. cerevisiae have revealed eight stoichiometrically methylated ribosomal proteins (Rps3p, Rps25p, Rps27p, Rpl1p, Rpl3p, Rpl12p, Rpl23p, and Rpl42p) and one substoichiometrically methylated protein (Rps2p) (14–22). Unlike rRNA methylation, the sites of protein methylation are dispersed throughout the ribosome. The methylation sites in Rpl12p, Rps25p, and Rps27p are exposed to the cytoplasm, whereas the methylated sites in Rpl3p, Rpl23p, Rpl42p, and Rps3p are in close proximity to rRNA. Although it is unknown what the precise roles of these modifications are, it is likely that protein modifications in close contact with rRNA play structural or assembly roles, whereas protein modifications exposed to the cytoplasm may act as binding platforms for translation factors or be involved in signaling.
We previously reported that the large-subunit protein Rpl3p is methylated in an unusual reaction that results in the formation of 3-methylhistidine at position 243 (16). Although a few animal proteins have been found with 3-methyl- and 1-methylhistidine, including actin and myosin, this modification was novel to yeast and no enzymes catalyzing the modification in any organism had previously been described (16). Methylation of His-243 is dependent upon the presence of a putative methyltransferase that we designated Hpm1p (histidine protein methyltransferase 1) (16). His-243 lies on the tryptophan finger domain of Rpl3p that extends deeply into the large ribosomal subunit core, making extensive contacts with the 25S rRNA (23). The tryptophan finger is involved in the accommodation of aminoacyl-tRNAs into the ribosomal A site and in the activation of the PTC (24). Interestingly, the Escherichia coli ortholog of yeast Rpl3p is also methylated (25). Ribosomal protein L3 is methylated at a glutamine residue that aligns 12 residues from His-243 of S. cerevisiae Rpl3p. E. coli cells lacking the PrmB methyltransferase, responsible for L3 methylation, demonstrated a defect in the biogenesis of the small and large ribosomal subunits, indicating that L3 methylation might play a role in the early steps of ribosome biogenesis (25). It is not known whether mammalian Rpl3 is methylated, although the human C1orf156 protein (homolog of Hpm1p) has been found in a complex with human Rpl3 (26).
In this paper, we have elucidated the biological roles of Hpm1p by analyzing its activities in vivo and in vitro. Hpm1p is responsible for all of the detectable protein histidine methylation in S. cerevisiae, and its substrates are present in the ribosome and nucleus-containing organelles. We demonstrate that loss of Hpm1p leads to a large-subunit biogenesis defect that stems from a defect in early rRNA processing. Hpm1p-deficient cells are significantly more resistant to the ribosome-targeting drugs cycloheximide and verrucarin A, indicating an altered ribosomal structure. Additionally, Hpm1p-deficient cells exhibit defects in translational fidelity shown by an increase in amino acid misincorporation and stop codon readthrough. We propose that Hpm1p acts as an assembly factor during early ribosome biogenesis and methylation of Rpl3p is needed for proper ribosome assembly and faithful protein production.
MATERIALS AND METHODS
Strains and growth media.
All of the strains in this study are of the BY4742 background (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and were obtained from the Open Biosystems yeast knockout collection (Thermo Scientific), containing a kanamycin resistance marker that replaces the open reading frame (ORF) of the gene. Yeast strains were grown in yeast peptone dextrose medium containing 1% yeast extract, 2% peptone, and 2% (wt/vol) d-glucose (YPD; Difco) or minimal synthetic defined medium lacking uracil and methionine (SD-Ura-Met). This medium contains 0.67% yeast nitrogen base with ammonium sulfate without amino acids (BD Biosciences), 0.2% amino acid mixture without uracil or methionine (Fisher), and 2% synthetic anhydrous dextrose (EMD Millipore).
Plasmids and expression of recombinant proteins.
The HPM1 rescue plasmid was made by amplifying its ORF from genomic DNA of the wild-type (WT) BY4742 strain. The inclusion of terminal SpeI and ClaI sites allowed its insertion into the pUG35 expression vector. Similarly, the human C1orf156 rescue plasmid was prepared by amplifying DNA from its ORF from pCMVSPORT6 (Open Biosystems) and cloning it into the pUG35 expression vector at the SpeI and ClaI restriction sites. The VEIG:AAAA and EIGCG:KIVCE substitutions in S-adenosylmethionine binding motif 1 of Hpm1p were constructed with the Stratagene QuikChange Lightning kit. All sequences were confirmed by Sanger dideoxy sequencing (Laragen, Inc.). The following primers were used: HPM1 F SpeI, GACTGACTACTAGTATGTCATTTTCCTTCGGCTTTAC; HPM1 R ClaI, GACTGACTATCGATCTATCGGATAGCTTTATTTGTTTC; HPM1 (VEIG:AAAA) F, AACGATATCGACGCGGTTGCCGCAGCAGCCTGTGGTACGGCACTACCC; HPM1 (VEIG:AAAA) R, GGGTAGTGCCGTACCACAGGCTGCTGCGGCAACCGCGTCGATATCGTT; HPM1 (EIGCG:KIVCE) F, GATATCGACGCGGTTGTCAAAATAGTCTGTGAGACGGCACTACCCTCAG; HPM1 (EIGCG:KIVCE) R, CTCTGAGGGTAGTGCCGTCTCACAGACTATTTTGACAACCGCGTCGATATC; C1orf156 F SpeI, GACTGACTACTAGTATGACCTTTCAGTTTAATTTCAC; and C1orf156 R ClaI, GACTGACTATCGATTTAACCAGGAAACTTAAAAGTTATTTC.
To prepare recombinant His-tagged proteins, the ORFs of HPM1, RPL3, and C1orf156 were cloned into the pET100/D-TOPO Escherichia coli expression vector as instructed (Invitrogen). The HPM1 ORF was amplified from a BG1805 plasmid containing the gene (Open Biosystems) by PCR with the forward primer CACCATGTCATTTTCCTTCG and the reverse primer CTATCGGATAGCTTTATTTG. The RPL3 ORF was amplified from BY4742 WT genomic DNA with the forward primer CACCATGTCTCACAGAAAG and the reverse primer TTACAAGTCCTTCTTCAAAGTA. The C1orf156 ORF was amplified from a pCMVSPORT6 plasmid containing the gene (Open Biosystems) with the forward primer CACCATGACCTTTCAGTTTA and the reverse primer TTAACCAGGAAACTTAAAAG. Proper insertion into the pET100/D-TOPO vector was verified by DNA sequencing with T7 forward and reverse primers (GENEWIZ). The vector was transformed into E. coli BL21(DE3) cells (Invitrogen). The recombinant N-terminally His-tagged protein was overexpressed by growing 2 liters of the cells at 37°C in LB medium (Difco) with 100 μg/ml carbenicillin to an optical density at 600 nm (OD600) of 0.6 and then adding isopropyl-β-d-thiogalactopyranoside (catalog no. I1003; Anatrace, Maumee, OH) to a final concentration of 1 mM and growing the cells for 6 h before harvesting them. Cells were then washed, resuspended in lysis buffer (50 mM sodium phosphate, 500 mM NaCl, 5% glycerol, protease inhibitor cocktail [11836145001; Roche Applied Science], 5 mM β-mercaptoethanol, pH 8.0), and lysed with an Avestin EmulsiFlex-C3 emulsifier (C315270) set to 18,000 lb/in2. Lysates were centrifuged at 20,000 × g for 20 min, and the resulting supernatant was loaded onto a 5-ml His-Trap HP nickel affinity column (part no. 17-5248-1; GE Healthcare), and the recombinant protein was eluted with a gradient of 20 to 500 mM imidazole. Recombinant proteins were then buffer exchanged (50 mM sodium phosphate, 300 mM NaCl, 5 mM imidazole, 5% glycerol, 5 mM β-mercaptoethanol, pH 8.0) and stored at −80°C. Expression of the recombinant proteins was verified by Western blot analysis with a mouse anti-6×His antibody (Abcam) and by bottom-up electrospray ionization Fourier-transform ion cyclotron resonance tandem mass spectrometry (Thermo Scientific).
In vivo radiolabeling with [3H]AdoMet and subcellular fractionation.
Overnight cultures of WT (BY4742) and hpm1Δ mutant cells were diluted in 200 ml of YPD medium to an OD600 of 0.0001 and incubated on a rotary shaker at 30°C until the OD reached 0.6. At that time, cells were harvested from 23 ml of the culture (14 OD units) by centrifugation at 5,000 × g for 5 min at room temperature; the remainder of the culture was placed at 4°C. The pelleted cells were washed with 1 ml of water three times and then resuspended in 1,696 μl of YPD and 304 μl of S-adenosyl-l-[methyl-3H]methionine ([3H]AdoMet; 83.3 Ci/mmol; 0.55 mCi/ml in 10 mM H2SO4–ethanol at 9:1; PerkinElmer). Cells were labeled for 30 min at 30°C on a rotatory shaker and then harvested at 5,000 × g for 5 min at 4°C. Radiolabeled cells were washed twice with water, resuspended in 1 ml of YPD, and then remixed with the remaining 177 ml of the original culture. These cells were harvested at 5,000 × g for 5 min, washed twice with water, and stored at −80°C. Cells in the thawed pellet were then lysed by resuspension in 5 ml of buffer A (20 mM Tris base, 15 mM magnesium acetate, 60 mM KCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, Roche protease inhibitor cocktail, adjusted to pH 7.5 with HCl) and 10 cycles of vortexing for 1 min with 1.5 g of baked glass beads (Biospec Products, Bartlesville, OK), followed by cooling on ice for 1 min. The crude lysates were centrifuged at 800 × g for 5 min at 4°C to pellet unlysed cells, and the supernatant volume was measured (this fraction is referred to as lysate). The lysates were then centrifuged at 12,000 × g for 5 min at 4°C in a JA17 rotor (Beckman). The pellet (subcellular organelles) was resuspended in 500 μl of buffer A, the volume was measured again, and the suspension was frozen at −80°C. The supernatant was centrifuged at 20,000 × g for 15 min at 4°C, and the pellet was discarded. The resulting supernatant was transferred into a fresh tube and centrifuged at 159,000 × g for 2 h at 4°C with a Ti65 rotor (Beckman). The pellet (ribosome fraction) was resuspended in 500 μl of buffer A at 0°C, and the final volume was measured and the supernatant (cytosol fraction) volume was measured as well. Samples were stored at −80°C. The protein concentration of each fraction was determined by the Lowry method (27) after protein precipitation with 10% trichloroacetic acid.
Amino acid analysis by high-resolution cation-exchange chromatography.
WT and hpm1Δ mutant subcellular fractions (lysate, subcellular organelles, ribosome, and cytosol) were acid hydrolyzed as follows. Four hundred micrograms of protein from each sample was transferred to a glass vial (6 by 50 mm), precipitated with equal volume of 25% trichloroacetic acid for 30 min at room temperature, and centrifuged at 4,000 × g for 30 min at room temperature. The pellets were washed with 100 μl of cold acetone and centrifuged again for 30 min. A 50-μl volume of 6 M HCl was added to each glass vial, and it was placed in a reaction chamber (catalog no. 1163; Eldex Labs) containing 200 μl of 6 M HCl. The vials were heated for 20 h in vacuo at 109°C with a Pico-Tag vapor phase apparatus (Waters). Residual HCl was removed by vacuum centrifugation. Samples were resuspended in 50 μl of water and 100 μl of 0.2 M sodium citrate, pH 2.2. Each sample was added and spiked with 2 μmol of a 3-methyl-l-histidine standard (Fisher no. 50-750-2805). Samples were loaded onto a cation-exchange column (0.9-cm inner diameter by 12-cm height; Beckman AA-15 sulfonated polystyrene beads) and equilibrated with sodium citrate buffer (0.2 M Na+, pH 5.84, at 28°C). Amino acids were eluted in the same buffer at 1 ml/min. One-minute fractions were collected from 50 to 105 min, corresponding to the approximate elution time of 3-methyl-l-histidine standard. A 500-μl volume of each fraction was added to 5 ml of scintillation fluor (Safety Solve; Research Products International) and counted. The 3-methyl-l-histidine standard was detected by the ninhydrin method (28), with the following modifications. A 50-μl volume of each fraction was added to a well of a flat-bottom 96-well plate (Fisher no. 12565501) and mixed with 100 μl of ninhydrin reagent (2% [wt/vol] ninhydrin and 3 mg/ml hydrindantin in a solvent of 75% [vol/vol] dimethyl sulfoxide and 25% [vol/vol] 4 M lithium acetate at pH 4.2). The plate was heated at 100°C for 15 min, and the absorbance at 570 nm was measured with a SpectraMax M5 microplate reader.
Isolation of ribosomes.
Overnight cultures of WT (BY4742) and hpm1Δ mutant cells were diluted to an OD600 of 0.1 in 500 to 1,000 ml of YPD medium and grown at 30°C until they reached late log phase at ODs of 1 to 2. The cells were harvested by centrifugation at 5,000 × g for 5 min, washed twice with water, and stored at −80°C. Cells were resuspended in 5 ml of buffer A and lysed by vortexing in glass beads as described above. The crude lysate was centrifuged for 5 min at 12,000 × g at 4°C. The supernatant was transferred to a new tube and centrifuged at 20,000 × g for 15 min at 4°C. The supernatant was again transferred into a fresh tube and centrifuged at 159,000 × g for 2 h at 4°C with a Ti65 Beckman rotor. The resulting ribosomal pellet was resuspended in buffer A and stored at −80°C. The protein concentration was determined by the Lowry method (27) after precipitation with trichloroacetic acid.
SDS-PAGE and fluorography.
Polypeptides from cell lysates and subcellular fractions were fractionated by SDS-PAGE. Samples were loaded onto a Tris-glycine polyacrylamide gel (15 by 17 by 0.2 cm) and resolved by applying 80 V until the bromphenol blue dye reached the interface of the stacking (4%) and resolving (12%) gels, followed by 180 V until the dye reached the bottom of the resolving gel. Gels were Coomassie stained and destained overnight (10% acetic acid, 5% methanol). After imaging, gels were washed with water and then treated with En3Hance (PerkinElmer Life Sciences) solution for 1 h, followed by a 30-min wash with water. The gel was then dried on Whatman 3MM filter paper under a vacuum for 2 h at 80°C, followed by 1 h without heat. The gel was placed on Kodak BioMax XAR film at −80°C.
Polysome profile analysis and ribosomal subunit quantification.
For polysome profile analysis, overnight cultures of yeast cells were diluted in 100 to 200 ml of the appropriate medium to an OD600 of 0.1 (0.01 for cells growing in SD-Ura-Met). Growth was monitored until the cells reached an OD of 0.8 to 1.0. Cycloheximide was added to a final concentration of 100 μg/ml, and cells were immediately placed in an ice bath for 10 min. Cells were then pelleted by centrifugation at 3,000 × g at 4°C for 5 min and washed once with 20 ml of ice-cold lysis buffer (10 mM Tris-Cl [pH 7.4], 100 mM NaCl, 30 mM MgCl2, 100 μg/ml cycloheximide, 200 μg/ml heparin, 0.1% diethylpyrocarbonate) and transferred to 15-ml conical polypropylene tubes. Cell pellets were stored at −80°C. For ribosomal subunit dissociation, yeast cells were grown and harvested similarly, except that the cells were not treated with cycloheximide and were washed with buffer C (50 mM Tris-Cl [pH 7.4], 50 mM NaCl, 1 mM DTT). For lysis, the pellet was resuspended in 1 ml of lysis buffer (buffer C for subunit dissociation), a quarter volume of glass beads was added, and the cells were vortexed for 30 s, followed by 30 s on ice, for a total of 12 cycles. The crude lysate was centrifuged at 5,000 × g for 5 min at 4°C. The supernatant was then transferred to prechilled microcentrifuge tubes and centrifuged again at 12,000 × g for 8 min at 4°C. The supernatant was transferred again to fresh prechilled tubes and absorbance at 260 nm was measured with a NanoDrop 2000c spectrophotometer (Thermo Scientific). Ten A260 units of lysate was layered onto 11 ml of a 8 to 48% (wt/vol) sucrose gradient buffered with 50 mM Tris-Cl–(pH 7.0)–50 mM NH4Cl–12 mM MgCl2–1 mM DTT (freshly prepared from solid) with Beckman Ultra-Clear centrifuge tubes (344059). For subunit dissociation, 2 A260 units of lysate was layered onto 11 ml of an 8 to 48% (wt/vol) sucrose gradient buffered with buffer C. Tubes were centrifuged at 37,000 rpm (234,116.4 × g) at 4°C for 3.5 h with a Beckman SW41 Ti rotor. Continuous monitoring of the absorbance at 280 nm of the sucrose gradient was done with an ISCO gradient fractionator (model 185). The flow rate was set to 1.5 ml/min by using Fluorinert FC-40 as a displacing agent. The absorbance detector used was an ISCO model UA-5 absorbance monitor; the sensitivity was set to 0.5 absorbance U/V. For digitization of the absorbance readings, an RS-232 interface-equipped voltmeter (TekPower model TP4000ZC) connected to the external chart recorder outputs recorded the DC voltage at 1-s intervals with the included software (DMM version 2.0) and the results were then imported into Microsoft Excel for further data processing.
Northern blot analysis.
Cells were grown in YPD to mid-log phase and an OD600 of 0.4. Cells were harvested by centrifugation at 4,000 × g for 2 min at room temperature. Cell pellets were washed with 1 ml of ice-cold water, transferred to 1.5-ml tubes, and centrifuged, and the resulting pellets were frozen in dry ice. Total RNA was extracted according to the hot-phenol–acid-washed glass bead protocol (29). Five micrograms of RNA was denatured by glyoxal (30) and loaded onto 1.2% agarose–1× BPTE [piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)–Bis–Tris–EDTA] gels. The separated RNAs were passively transferred to Hybond N+ nylon membranes (GE Healthcare) in 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]). Membranes were cross-linked with 254-nm light with a UV Stratalinker 2400 as instructed by the manufacturer. Membranes were hybridized in standard Church's buffer (1% bovine serum albumin, 1 mM EDTA, 0.5 M sodium phosphate [pH 7.2], 7% SDS) with oligonucleotide probes labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. The oligonucleotides used were based on 18S rRNA (5′-CATGGCTTAATCTTTGAGAC-3′), 25S rRNA (5′-CTCCGCTTATTGATATGC-3′), the E-C2 region of pre-rRNA for the detection of 7S pre-rRNA and total 27S pre-rRNA (5′-GGCCAGCAATTTCAAGTTA-3′), the 5′ external transcribed spacer (ETS) region upstream of A0 for the detection of 35S and 23S pre-rRNAs (5′-CGCTGCTCACCAATGG-3′), and the D-A2 region for the detection of 20S pre-rRNA (5′-CGGTTTTAATTGTCCTA-3′). PDR5 mRNA was detected with a riboprobe antisense to the PDR5 ORF. The 25S and 18S Northern blot assay signals were quantified with the Quantity One software from the Bio-Rad FX Plus Phosphorimaging System.
Pulse-chase labeling of rRNA.
The pulse-chase assay protocol used was a modified form of that described by Tollervey et al. (31). Cells were first transformed with an empty pUG35 plasmid (URA+) to allow growth in the absence of uracil, according to the lithium acetate (LiOAc)–single-stranded DNA (ssDNA)–polyethylene glycol (PEG) method (32). The transformed cells were then grown at 25°C in 10 ml synthetic dextrose medium lacking uracil (SD-Ura) to mid-log phase (OD600 of 0.4). For the pulse, 400 μCi of [5,6-3H]uracil (Amersham) was added directly to the 10-ml culture. After a 2-min pulse, an excess of unlabeled uracil (1 ml of 2.5 mg/ml uracil dissolved in SD-Ura) was added directly to the labeled cultures. At the designated time points, 1 ml of culture was added to 10 ml of 100% ethanol prechilled on dry ice in 15-ml tubes. The tubes were then warmed to room temperature and centrifuged at 3,000 × g for 3 min. The pellets were washed with 1 ml of ice-cold water to remove precipitate from the pellet and then frozen on dry ice in 1.5-ml tubes. The total RNA was extracted according to the hot-phenol–acid-washed glass bead protocol (29). The resulting RNA pellet was resuspended in 15 μl of nuclease-free water (Ambion), and 1 μl was denatured in glyoxal buffer and loaded onto 1.2% agarose–1× BPTE gels (30). The separated RNAs were then transferred to Hybond N+ nylon membrane (GE Healthcare), and the membrane was dried and exposed to BioMax MS film with a BioMax TranScreen LE (Kodak) at −80°C for 5 days.
Dual-luciferase assay for measurement of translational fidelity.
The dual-luciferase systems described in references 33 and 34 were used here. Luciferase reporters and control vectors were generously provided by David Bedwell and Ming Du at the University of Alabama, Birmingham, AL. For amino acid misincorporation, vectors used were CTY775/luc CAAA FF K529 (AAA to AAT) and CTY775/luc CAAA (control vector). For stop codon readthrough, vectors used were CTY775/luc UAAC, CTY775/luc UAGC, and CTY775/luc UGAC, with the respective control vectors CTY775/luc CAAC, CTY775/luc CAGC, and CTY775/luc CGAC. The control vectors were used to adjust for differences in protein expression between WT and hpm1Δ mutant cells. All eight vectors were transformed into WT and hpm1Δ mutant cells by the LiOAc-ssDNA-PEG method (32). The dual-luciferase assay was done as described in references 33 and 34 with the Dual-Luciferase Reporter Assay system (Promega), with a few modifications. Lysates from each strain were diluted 10-fold with 1× Passive Lysis Buffer (Promega) to ensure that measurements were in the linear range and spotted into each well of a white 96-well plate (Costar 3912). Firefly and Renilla luciferase luminescence levels were measured at room temperature with a SpectraMax M5 microplate reader set to the following parameters: readtype = endpoint, readmode = luminescence with a 1,000-ms integration time, and wavelength = all. The remaining parameters were set to the default settings.
RESULTS
Hpm1p is the major histidine methyltransferase in S. cerevisiae with a primary target corresponding to ribosomal protein Rpl3p.
We previously reported that S. cerevisiae ribosomal protein Rpl3p contains a 3-methylhistidine modification on residue 243 by mass spectrometry and high-resolution cation-exchange chromatography (16). This modification is dependent on the presence of the seven-beta-strand putative methyltransferase product of the HPM1 gene (16). To determine other potential substrates of Hpm1p, we radioactively labeled all of the methylated products in both WT and hpm1 null yeast cells by incubating log-phase cells with [3H]AdoMet. Cell lysates were acid hydrolyzed, and amino acids were resolved by high-resolution cation-exchange chromatography. Fractions containing 3-methylhistidine were collected, and radioactivity was counted. WT lysate showed a radioactive peak coeluting with a 3-methylhistidine standard (Fig. 1A). The radioactive species eluted 1 min before the nonisotopically labeled 3-methylhistidine standard because of a known isotope effect (16, 35). Strikingly, the lysate from Hpm1p-deficient cells had complete loss of radioactivity in the 3-methylhistidine fractions. This result indicates that Hpm1p is responsible for all of the detectable histidine methylation in S. cerevisiae under these growth conditions.
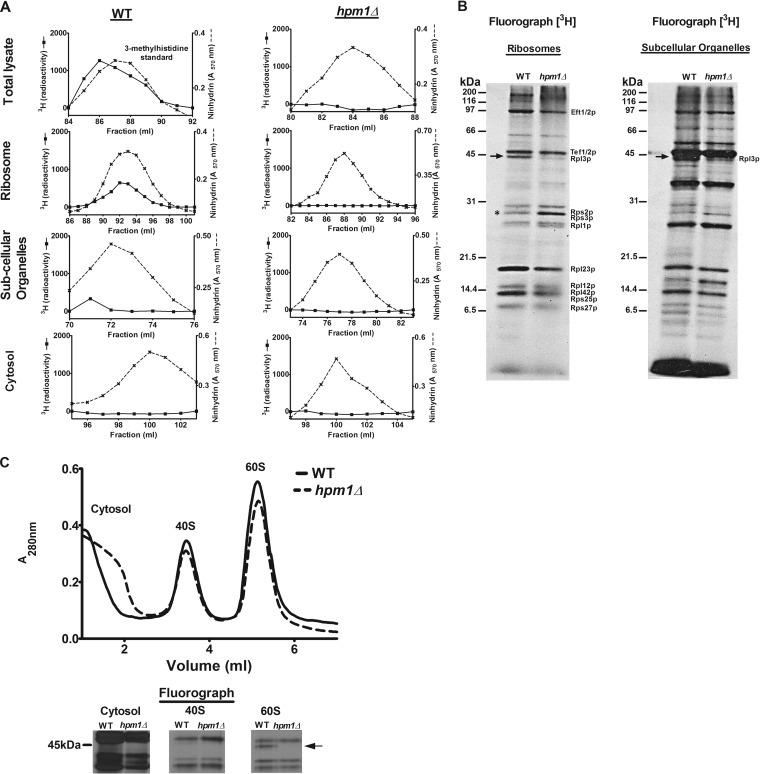
FIG 1.
Yeast protein 3-methylhistidine is found primarily on a ribosomal protein corresponding to Rpl3p and is dependent on HPM1. (A) Total lysate, ribosomes, subcellular organelles, and cytosol were prepared from WT (BY4742) and hpm1Δ mutant cells (BY4742) for amino acid analysis as described in Materials and Methods. The radioactivity shown (in counts per minute) has been normalized by background subtraction. Ninhydrin absorbance at 570 nm was measured to detect the 3-methylhistidine standard. (B) In vivo 3H-radiolabeled ribosomes and subcellular organelles from WT and hpm1Δ mutant cells (40 μg of protein) were pretreated with 2 U of Benzonase (Novagen 70746-4) for 30 min at 37°C and resolved by SDS-PAGE. 3H-methylated proteins were detected by fluorography for 13 weeks at −80°C as described in Materials and Methods. The arrow shows the methylated polypeptide corresponding to the molecular weight of Rpl3p, and the asterisk shows the positions of Rps2 and Rps3, whose levels of methylation are altered in the absence of HPM1. Radiolabeled bands corresponding to known methylated ribosomal proteins are identified on the right (14–22). (C) Seven OD units of WT and hpm1Δ mutant cells was radiolabeled with [3H]AdoMet, and the small 40S and large 60S ribosomal subunits were dissociated as described in Materials and Methods. One-hundred-microliter volumes of the cytosol, 40S, and 60S peak fractions were resolved by SDS-PAGE as described in Materials and Methods, except that a 12% Bis-Tris gel was prepared and run with MOPS running buffer. 3H-methylated proteins were detected by fluorography for 10 weeks at −80°C as described in Materials and Methods. At the bottom are three portions of lanes from the same gel.
To determine the subcellular distribution of Hpm1p-dependent methylation, ribosomes, nucleus-containing organelles, and cytosolic fractions were isolated from the lysates of [3H]AdoMet-labeled WT and hpm1 null cells and acid hydrolyzed for amino acid analysis as described above. Most of the radioactive 3-methylhistidine was present in WT ribosomes and absent from the hpm1 mutant (Fig. 1A). A small amount of the modification was present in WT organelles and again absent from hpm1Δ mutant cells. Importantly, no radiolabeled 3-methylhistidine was found in the ribosome-depleted cytosol fraction of WT and hpm1Δ mutant cells. These results suggest that ribosomes are the major target of Hpm1p-dependent methylation.
To obtain a profile of methylated ribosomal proteins in the WT and hpm1Δ mutant strains, radiolabeled ribosomes prepared from [3H]AdoMet-labeled cells as described above were analyzed by SDS-PAGE and fluorography. The absence of Hpm1p resulted in the loss of only one radiolabeled polypeptide that corresponds to the size of Rpl3p, suggesting that it is the only ribosomal protein substrate of Hpm1p (Fig. 1B, arrow). To unambiguously confirm that this polypeptide is present on the large ribosomal subunit, radiolabeled lysates were resolved by sucrose density ultracentrifugation to separate the small and large ribosomal subunits. Peak fractions were analyzed by SDS-PAGE and fluorography and showed one methylated band beneath the 45-kDa marker only in the 60S fraction and not in the 40S or cytosol fraction (Fig. 1C). This band completely disappeared from hpm1Δ mutant cells.
Interestingly, a band corresponding to the size of the small ribosomal subunit proteins Rps2 and Rps3 showed hypermethylation in the absence of Hpm1p (Fig. 1B, asterisk). Mass spectrometry analysis of intact ribosomal proteins confirmed that Rps2 is hypermethylated and that the content of fully methylated Rps3 is increased in the absence of Hpm1p (data not shown). It appears that the methylation of other proteins can be affected in the absence of Hpm1p, indicating a possible compensatory mechanism. This was also seen in the parallel analysis of the in vivo [3H]AdoMet-labeled organelle fraction. This analysis also revealed a radiolabeled methylated polypeptide corresponding to the size of Rpl3p from WT cells but not from hpm1Δ mutant cells, as well as alterations in the methylation patterns of other proteins (Fig. 1B). It is unclear whether this represents methylation of the protein in membrane-bound ribosomes of the endoplasmic reticulum or in an assembly intermediate of the nucleus. Altogether, these data suggest that the bulk of 3-methylhistidine modification resides in the ribosome on Rpl3p and that a small portion of this modification may be found in subcellular organelles.
Hpm1p is a bona fide methyltransferase that modifies ribosome-associated but not free Rpl3p.
Since Rpl3p methylation is dependent on Hpm1p, we next investigated whether purified recombinant Hpm1p is, in fact, a methyltransferase that can directly methylate Rpl3p in vitro. Ribosomes from the WT and hpm1Δ mutant strains were incubated with recombinant Hpm1p in the presence of [3H]AdoMet. Proteins were then separated by SDS-PAGE and visualized by fluorography. A radioactive 43-kDa polypeptide band corresponding to Rpl3p was present in hpm1Δ mutant ribosomes but not in WT ribosomes (Fig. 2A). This result is consistent with ribosomal proteins in WT cells being stoichiometrically methylated and therefore unable to be further methylated in vitro. However, hpm1Δ mutant cells would contain unmodified Rpl3p and any other potential Hpm1p substrates, which would be competent for methylation in vitro. To verify that this radiolabeled protein is the substrate of Hpm1p, amino acid analyses of these radiolabeled ribosomes were performed. A radioactive species comigrating with the 3-methylhistidine standard was found only when recombinant Hpm1p was incubated with ribosomes from the hpm1Δ mutant strain and not when it was incubated with ribosomes from the WT strain, confirming that Hpm1p catalyzes a 3-methylhistidine modification on a ribosomal protein corresponding to the size of Rpl3p (Fig. 2B). We also tested the ability of recombinant Hpm1p to methylate Rpl3p in the organelle fraction. This experiment showed that Hpm1p can also methylate a 43-kDa protein only in the organelle fraction from hpm1Δ mutant cells (data not shown).
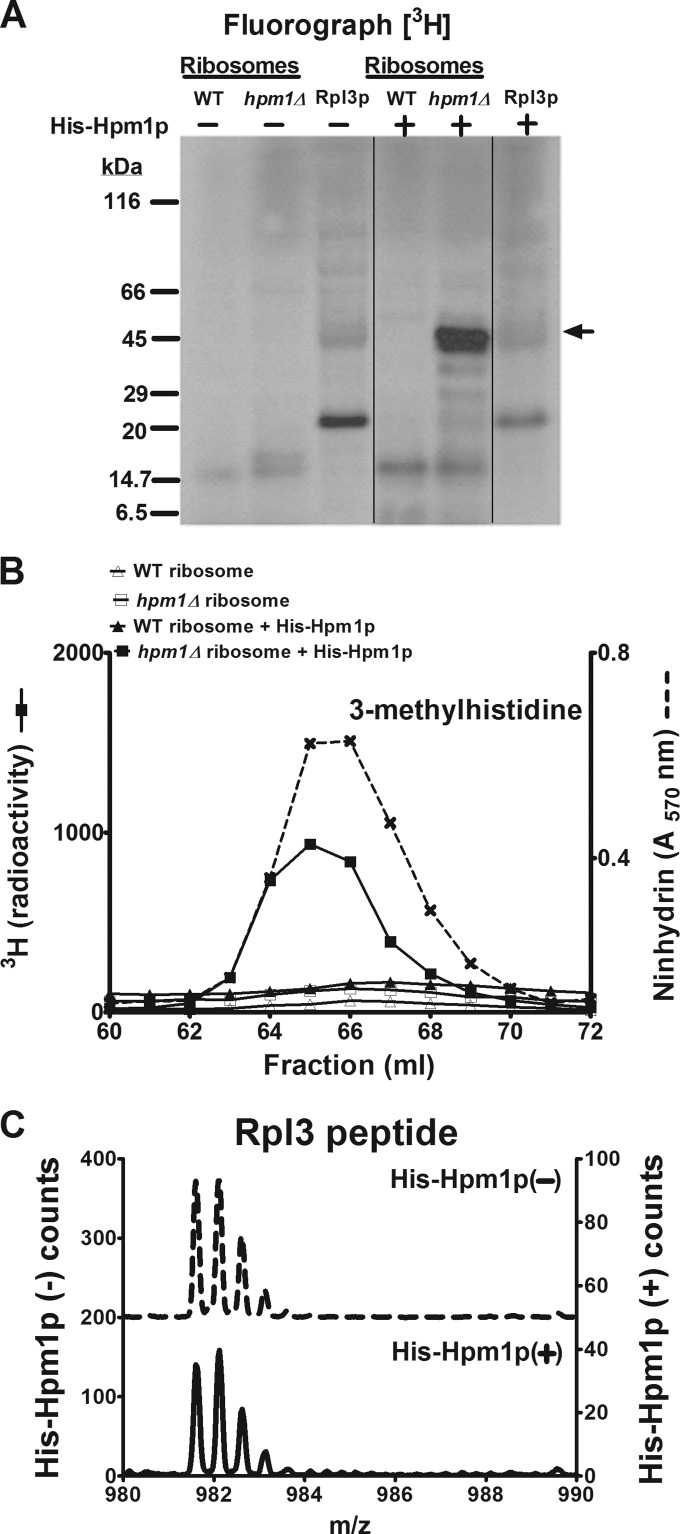
FIG 2.
Hpm1p can methylate Rpl3p on intact ribosomes but not free Rpl3p. (A) A 46-μg sample of crude ribosome protein isolated as described in Materials and Methods from WT and hpm1Δ mutant cells or 46 μg of recombinant Rpl3p was incubated with or without recombinant, histidine-tagged Hpm1p (His-Hpm1p; 12 μg) in the presence of 1 μM [3H]AdoMet and 100 mM sodium chloride–100 mM sodium phosphate (pH 7.5) (methylation buffer) for 5 h at 30°C. Reactions were terminated by the addition of equal volume of 2× Laemmli sample buffer, and proteins were analyzed by SDS-PAGE and fluorography as described in Materials and Methods. Film was incubated with the dried gel for 7 weeks at −80°C. The arrow indicates the approximate position of recombinant His-Rpl3p. The radiolabeled band at about 20 kDa appears to reflect a bacterial contaminant in the His-Rpl3p preparation. Vertical lines show where nonrelevant lanes were removed from the single gel. (B) Thirty-seven-microgram samples of crude ribosome protein from WT and hpm1Δ mutant cells were incubated with or without His-Hpm1p as described above. Proteins were trichloroacetic acid precipitated and acid hydrolyzed for amino acid analysis as described in Materials and Methods, except that fractions eluting at 60 to 72 ml were collected and the 3H radioactivity of 900 μl of each fraction was counted (reported in counts per minute). Ninhydrin, absorbance of the 3-methylhistidine standard. (C) The synthetic peptide WGTKKLPRKTHRGLRK (Biosynthesis, Lewisville, TX), corresponding to the methylated region of Rpl3p, was incubated with or without His-Hpm1p (30 μg) in the presence of 200 μM S-adenosyl-l-methionine p-toluenesulfonate (Sigma) and methylation buffer for 16 h at 30°C. Reactions were terminated with trifluoroacetic acid to a final concentration of 1%, and the products were fractionated by high-performance liquid chromatography with a PLRP-S reverse-phase column (pore size, 300 Å; bead size, 5 μm; 120 by 2 mm; Polymer Laboratories, Amherst, MA). The column was maintained at 50°C and initially equilibrated in 95% solvent A (0.1% trifluoroacetic acid in water) and 5% solvent B (0.1% trifluoroacetic acid in acetonitrile) at a flow rate of 0.5 ml/min. The following program was used: 10 min of 5% B, 25 min of a gradient to 60% B, 1 min of a gradient to 100% B, 5 min of 100% B, 1 min of a gradient to 5% B, and 8 min of 5% B. The column effluent was directed to the electrospray ion source of a QSTAR Elite (Applied Biosystems) mass spectrometer running in MS-only mode and was calibrated with external peptide standards. The left y axis represents the counts in the absence of His-Hpm1p, and the right y axis represents the counts in the presence of His-Hpm1p.
We next investigated the ability of recombinant Hpm1p to recognize and methylate a synthetic 16-amino-acid-long peptide corresponding to the methylated region of Rpl3p. This peptide was incubated with recombinant Hpm1p and nonradioactive AdoMet and analyzed by liquid chromatography-MS. Methylation of the peptide should result in a mass increase of 14 Da. However, addition of Hpm1p to the peptide resulted in no mass change, indicating that Hpm1p cannot detectably methylate the peptide (Fig. 2C). This result suggests that Hpm1p may require secondary or tertiary structures present in full-length Rpl3p needed for recognition and subsequent methylation. We therefore tested the ability of recombinant Hpm1p to methylate full-length recombinant Rpl3p when incubated with [3H]AdoMet. No radiolabeled polypeptide band corresponding to the size of recombinant Rpl3p was detected (Fig. 2A). These data suggest that Hpm1p may require other factors associated with the ribosome to efficiently methylate Rpl3p.
Loss of Hpm1p results in reduced levels of 60S ribosomal subunits.
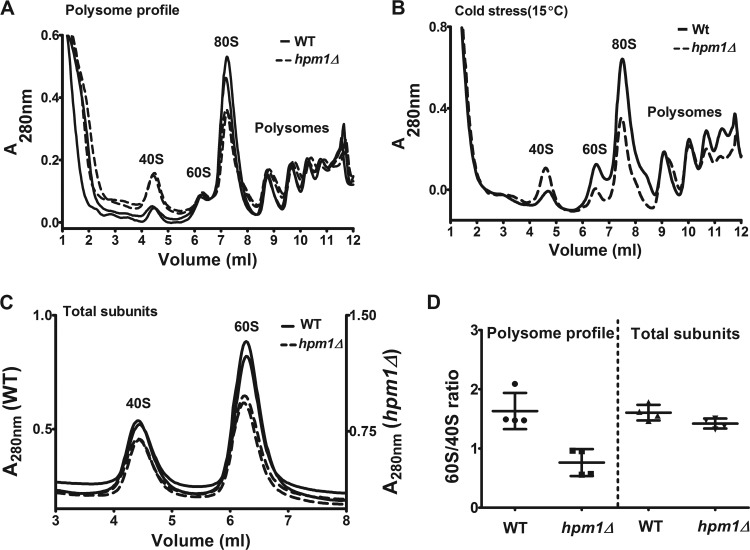
Since Rpl3p is the only known substrate of Hpm1p and many of the ribosome modifications occur during ribosome assembly, we next investigated the potential role of this enzyme in ribosome biogenesis and/or translation. Studies of WT and hpm1Δ mutant cell growth in rich medium (YPD) at 15, 30, or 38°C or in the presence of 250 mM NaCl showed no difference in the growth rates (data not shown). To dissect any abnormalities in ribosome biogenesis and translation, we performed sucrose density centrifugation analysis to separate the small (40S) and large (60S) ribosomal subunits from intact monosome ribosomes (80S) and polysomes. Loss of Hpm1p resulted in abnormal polysome profiles, most strikingly seen with increased levels of 40S subunits and decreased levels of 60S subunits and 80S monosomes when the background is taken into account (Fig. 3A). Additionally, hpm1Δ mutant cells exhibited increased ribosome runoff, as noted by decreased levels of polysomes at the end of the profile. This phenotype was accentuated under cold stress conditions (Fig. 3B). These changes are reflected in the 60S/40S ratio, which decreased by >50% in hpm1Δ mutant cells (Fig. 3D, left). To confirm this, lysates from WT and hpm1Δ mutant yeast cells were prepared with a buffer that results in complete dissociation of the subunits (no cycloheximide or Mg2+). Dissociation of the monosomes and polysomes revealed a clear reduction of the large ribosomal subunit and no observable increase in the small subunit (Fig. 3C), which is also reflected in an ∼10% decrease in the 60S/40S ratio (Fig. 3D, right). Since monosomes and polysomes contain stoichiometric amounts of large and small subunits, a decrease in the number of free 60S subunits would result in increased levels of free 40S subunits.
FIG 3.
Lack of HPM1 results in deficiencies in large-subunit biogenesis and translation initiation. (A) Polysome profile analyses of WT and hpm1Δ mutant cells were done as described in Materials and Methods. Shown are two independent profiles each of the WT and of the hpm1Δ mutant. (B) Polysome profile analysis of cold-stressed (15°C) cells was done as described for panel A. (C) Total subunit analysis of WT and hpm1Δ mutant cells was done as described for panel A, except that cells were not pretreated with cycloheximide, the lysis buffer was replaced with buffer C, and 2 A260 units of extract was loaded. (D) Quantification of ratios of large to small ribosomal subunits for both the polysome profiles (A) and total subunits (C). Peak areas were determined with Graphical Analysis 3.8.4 software. Error bars represent standard deviations of four independent experiments. Unpaired t test two-tailed P values for the differences in the subunit ratios were 0.004 for the polysome ratio and 0.057 for the dissociated subunit ratio.
Loss of Hpm1p results in early processing defects and delayed kinetics of pre-rRNA maturation.
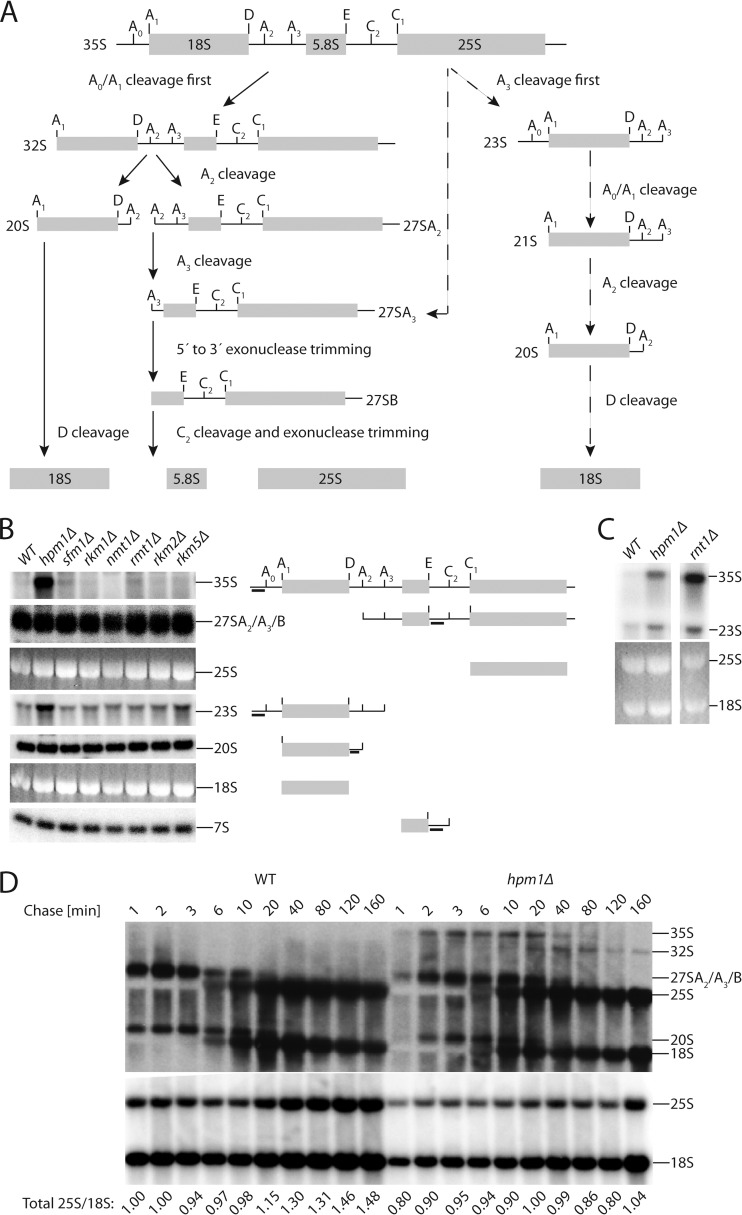
To examine the basis for the large-subunit biogenesis defect, we probed for alterations in the rRNA processing pathway. The 35S pre-rRNA transcript, which encodes both the small- and large-subunit rRNAs, undergoes a series of sequential modifications and endo- and exonucleolytic cleavages to produce the mature 18S rRNA of the small subunit and 5.8S and 25S rRNAs of the large subunit (Fig. 4A) (36, 37). The predominant processing pathway of the 35S rRNA transcript involves initial cleavages at sites A0, A1, and A2. Defects in various aspects of ribosome biogenesis can result in impaired cleavage at sites A0, A1, and A2, causing a shift toward more precursors undergoing the first cleavage at site A3 and leading to a greater steady-state accumulation of 23S and 21S intermediate transcripts (Fig. 4A, right side) (38). We performed Northern blot analyses with radiolabeled probes that bind to different regions of the pre-rRNA transcript to determine steady-state levels of pre-rRNAs (Fig. 4B). A probe hybridizing upstream of the A0 site revealed a substantial accumulation of the 35S precursor in the hpm1Δ mutant strain relative to that in the WT (Fig. 4B, top). There was also an accumulation of the 23S pre-RNA species in the hpm1Δ mutant strain, suggesting defects in A0, A1, and A2 cleavage in cells lacking Hpm1p. However, the levels of the large-subunit pre-rRNAs, 7S and 27S, and the small subunit pre-rRNA, 20S, were relatively unchanged. This suggests that loss of Hpm1p results in an rRNA processing defect early on in the processing pathway. In contrast, other ribosomal protein methyltransferase mutant strains did not exhibit this defect, highlighting the specificity of the phenotype detected in the hpm1Δ mutant strain. Comparison of the rRNA processing defect of the hpm1Δ mutant with a mutant with a deletion of RNT1, which processes the 3′ ETS of pre-rRNAs, showed comparable accumulations of 35S and 23S species (Fig. 4C).
FIG 4.
Loss of HPM1 results in early rRNA processing defects and delayed kinetics of pre-rRNA maturation. (A) rRNA precursor processing pathway in S. cerevisiae. The 35S pre-rRNA encodes the 18S rRNA of the small ribosomal subunit and the 5.8S and 25S rRNAs of the large subunit. The predominant pathway for the processing of the 35S pre-rRNA transcript is shown on the left, and the abnormal pathway is shown on the right (dashed arrow). First, the 5′ end of the 35S pre-rRNA is cleaved at sites A0 and A1, generating the mature 5′ end of the 18S rRNA. Cleavage at the A2 site separates the 20S and 27SA2 precursors. The 20S precursor is cleaved at site D in the cytoplasm to yield the mature 18S transcript. The 5′ end of the 27SA2 precursor is cleaved at site A3; this is followed by 5′-to-3′ exonuclease trimming to generate the mature 5′ end of the 5.8S rRNA. C2 cleavage separates the 5.8S and 25S precursors, which are then trimmed by exonucleases to yield the mature 5.8S and 25S rRNAs. (B) Northern blot analysis of rRNA precursor steady-state levels in the WT and the hpm1Δ mutant (lanes 1 and 2). Lanes 3 to 8 represent deletions of various ribosomal protein methyltransferases. The locations of the probes within each rRNA species are indicated on the right. The 25S and 18S blots are from the ethidium bromide fluorescence of the gel prior to transfer. (C) Top, Northern blot analysis of rRNA precursor steady-state levels with the 35S probe in WT and hpm1Δ and rnt1Δ mutant cells; bottom, 25S and 18S rRNAs detected by ethidium bromide fluorescence of the gel prior to transfer. (D) Top, pulse-chase assay with [3H]uracil for analysis of pre-rRNA processing kinetics in WT and hpm1Δ mutant cells. Cultures in the early exponential growth phase were pulsed with tritiated uracil for 2 min and then chased with an excess of nonisotopically labeled uracil. Bottom, analysis of the membrane from the pulse-chase assay by Northern blotting of 25S and 18S rRNAs. The ratios of the 25S to 18S rRNAs are normalized to the 1-min chase lane of the WT.
To further dissect the processing kinetics of rRNA precursors, a pulse-chase assay was performed with [3H]uracil. Radiolabeled rRNA precursors, intermediates, and mature products were fractionated on an agarose gel, transferred to a nylon membrane, and analyzed by autoradiography. hpm1Δ mutant cells exhibited a considerable delay in the processing of pre-rRNAs (Fig. 4D). A fraction of the unprocessed 35S pre-rRNA particle accumulated for over 20 min in the hpm1Δ mutant strain and was slowly converted to 32S over the course of the chase, whereas in the WT, these precursors did not accumulate (Fig. 4D). In addition, hpm1Δ mutant cells showed a delayed appearance of 20S and 27SA pre-rRNAs. To determine steady-state levels of mature rRNAs, the same membrane was probed for 18S and 25S rRNAs. It showed a reduced ratio of 25S to 18S rRNA, for cells lacking Hpm1p, throughout the chase (Fig. 4D). These data indicate that in addition to an overall delay in kinetics, a detectable fraction of the 35S precursor undergoes very slow processing, indicating that this fraction has not assembled properly. The delay in processing kinetics and increased steady-state levels of 35S and 23S pre-rRNAs are hallmarks of defects in early ribosome biogenesis (39).
Hpm1p methyltransferase activity is required for proper large-subunit biogenesis and translation.
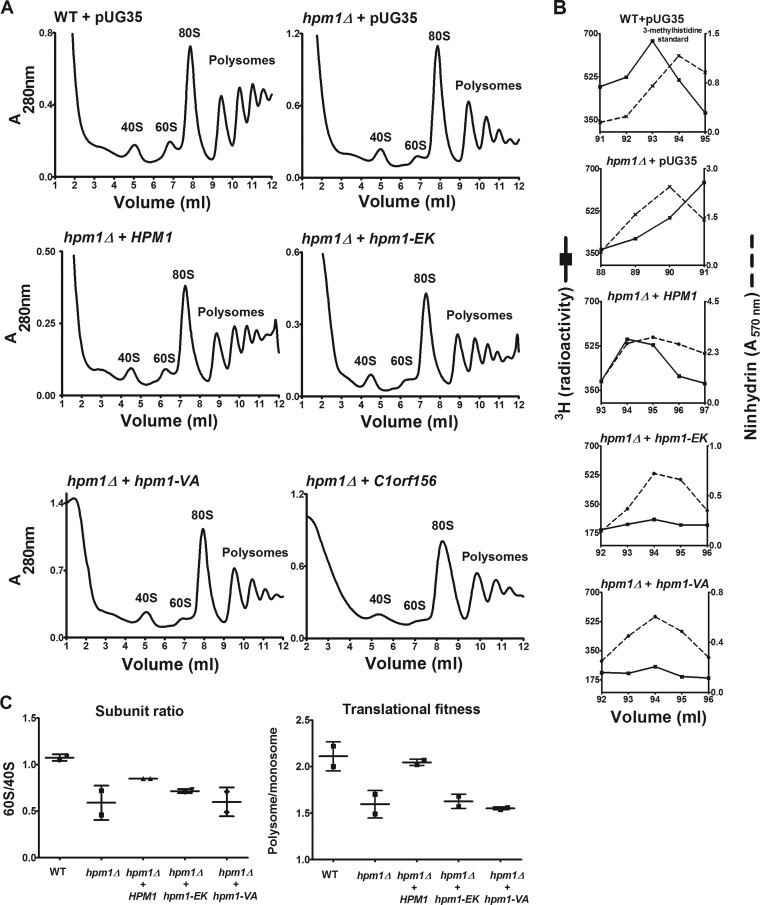
To see if ectopic Hpm1p could rescue the defects in large-subunit biogenesis and translational initiation associated with the hpm1Δ mutant, WT and hpm1Δ mutant cells were transformed with an inducible pUG35 vector containing the WT HPM1 ORF. Expression of the genes was induced by omitting methionine from the minimal growth medium. Cell lysates were prepared and layered onto sucrose gradients, and ribosomal particles were separated by ultracentrifugation. Consistent with the results shown in Fig. 3, hpm1Δ mutant cells containing an empty pUG35 vector exhibited a 60S biogenesis defect with an accumulation of the small 40S subunit and decreased amounts of the large 60S subunit (Fig. 5A). Significantly fewer ribosomes were associated with translation-active polysomes in the hpm1Δ mutant than in the WT, which is indicative of translation initiation defects. Reintroduction of Hpm1p into hpm1Δ mutant cells eliminated the 60S biogenesis defect phenotype and resulted in significantly more ribosomes involved in active translation, showing that Hpm1p can rescue the defects in large ribosomal subunit biogenesis and translation associated with Hpm1p-deficient cells (Fig. 5A). These rescued cells had levels of 3-methylhistidine similar to those in WT cells (Fig. 5B).
FIG 5.
Plasmids containing WT HPM1 but not the human homolog C1orf156 can rescue hpm1Δ mutant defects in large-subunit biogenesis and translational initiation. (A) hpm1Δ mutant cells were transformed with the pUG35 plasmid containing the HPM1 gene, its human homolog C1orf156, or HPM1 active-site mutants under the control of the Met25 promoter. As controls, WT and hpm1Δ mutant cells were transformed with empty pUG35 expressing only GFP. The hpm1(EK) and hpm1(VA) genes have mutations in motif 1 of the MT domain, which is involved in AdoMet binding, i.e., EIGCG to KIVCE (EK) and VEIG to AAAA (VA). Cells were grown overnight in SD-Ura-Met at 30°C in a rotary shaker. Cells were then diluted in 100 ml of fresh SD-Ura-Met to an OD600 of 0.01 and grown overnight at 30°C until they reached an OD600 of 0.8 to 1.0. Polysome profile analysis was done as described in Materials and Methods. The HPM1, C1orf156, hpm1(EK), and hpm1(VA) genes were cloned into pUG35 with their stop codons to express the genes without the GFP tag. (B) WT and hpm1Δ mutant cells containing the plasmids described in panel A were labeled in vivo with [3H]AdoMet as described in Materials and Methods, except that 3.5 OD600 units of cells was labeled for 1 h. Total lysates were prepared in buffer C and acid hydrolyzed for amino acid analysis as described in Materials and Methods, except that the column was kept at 35°C and that 900 μl of each fraction was taken for radioactivity determination. 3H radioactivity is shown in counts per minute, and ninhydrin absorbance of the 3-methylhistidine standard was measured. (C) Quantification of polysome profiles. Ratios were calculated by determining the areas of the 40S, 60S, monosome (80S), and polysome peaks with Graphical Analysis 3.8.4 software. Error bars represent standard deviations of two independent experiments. The subunit ratio and translational fitness differences were not statistically significant at the P < 0.05 level.
To determine if the loss of Hpm1p methyltransferase activity is sufficient to cause the subunit imbalance, two mutant sequences that alter the AdoMet-binding motif were expressed in hpm1Δ mutant cells. The mutation of EIGCG in motif I to KIVCE (hpm1-EK) reduced the presence of 3-methylhistidine by at least ∼80%, and mutation of VEIG in motif I to AAAA (hpm1-VA) resulted in the near absence of the modification, as determined by amino acid analysis (Fig. 5B). Neither of these hpm1 mutant plasmids was able to rescue hpm1Δ mutant cells, as shown by the retention of the subunit reversal phenotype and diminished amounts of ribosomes engaged in active translation (Fig. 5A). The human homolog of Hpm1p, C1orf156, was also unable to rescue the diminished large-subunit levels and translation defects. There is no evidence that human Rpl3 is methylated. Hence, C1orf156 may be unable to rescue Hpm1p-deficient cells because C1orf156 is not an ortholog of Hpm1p, because it is not able to recognize the yeast protein, or because it is nonfunctional when expressed in yeast cells. Subunit ratios (60S/40S) were quantified and showed that HPM1 partially restored the levels of 60S subunits to that of the WT, whereas hpm1-EK and hpm1-VA were unable to do so (Fig. 5C, left). The translational fitness of the various strains was quantified by using the polysome/monosome ratio; a higher ratio indicates that more ribosomes are engaged in active translation. Translational fitness is reduced in hpm1Δ mutant cells and was rescued only by the introduction of WT Hpm1p (Fig. 5C, right).
Cells lacking Hpm1p exhibit hyperresistance to the ribosome-targeting drugs cycloheximide and verrucarin A.
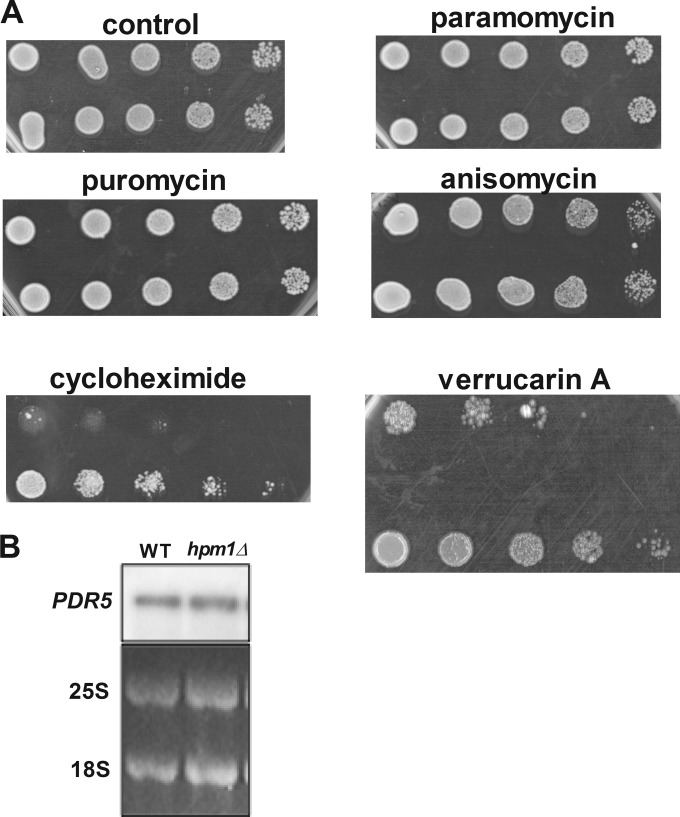
We next examined the sensitivities of WT and hpm1Δ mutant cells to various ribosome-binding drugs to determine any potential consequences of a lack of Hpm1p on ribosome structure and function. The growth rates of log-phase WT and hpm1Δ mutant cells on YPD agar plates containing puromycin, anisomycin, paromomycin, cycloheximide, and verrucarin A were compared. These drugs interfere with different aspects of translation by binding to different regions of the ribosome. No changes in the sensitivity of the hpm1Δ mutant to puromycin, anisomycin, or paromomycin were seen (Fig. 6A). However, hpm1Δ mutant cells were significantly more resistant than the WT to cycloheximide and verrucarin A (Fig. 6A). The dramatic increase in resistance to these drugs indicates structural alterations of their binding sites (see Discussion).
FIG 6.
Lack of HPM1 results in greatly increased resistance to the ribosome-targeting drugs cycloheximide and verrucarin A. (A) Dilution spot assays were done with mid-log-phase yeast cells (OD600 of 0.5) grown in YPD medium at 30°C in a rotary shaker for 2 generations from an overnight culture grown in YPD at 30°C. Cells were serially diluted 5-fold on YPD agar plates in the presence or absence of the ribosome-targeting antibiotics puromycin (50 μg/ml), cycloheximide (500 ng/ml), paromomycin (50 μg/ml), anisomycin (5 μg/ml), and verrucarin A (2 μg/ml) and incubated at 30°C for 2 days (control, puromycin, anisomycin, and paromomycin), 3 days (verrucarin A), or 5 days (cycloheximide). WT and hpm1Δ mutant cells containing the empty pUG35 vector were also spotted onto SD-Ura-Met and incubated for 3 days. (B) Northern blot analysis of mRNA expressed from the PDR5 gene encoding the multidrug exporter PDR5. The 25S and 18S blots are from the ethidium bromide fluorescence of the gel prior to transfer.
Northern blot analyses detected similar levels of the multidrug transporter PDR5, suggesting that the resistance phenotypes are not a consequence of different levels of the PDR5 exporter in WT and hpm1Δ mutant cells (Fig. 6B). Altogether, these results suggest that hpm1Δ mutant cells have an altered ribosome structure that is different from that of WT ribosomes, causing changes in ribosome-targeting drug sensitivity.
Cells deficient in Hpm1p have reduced translational fidelity.
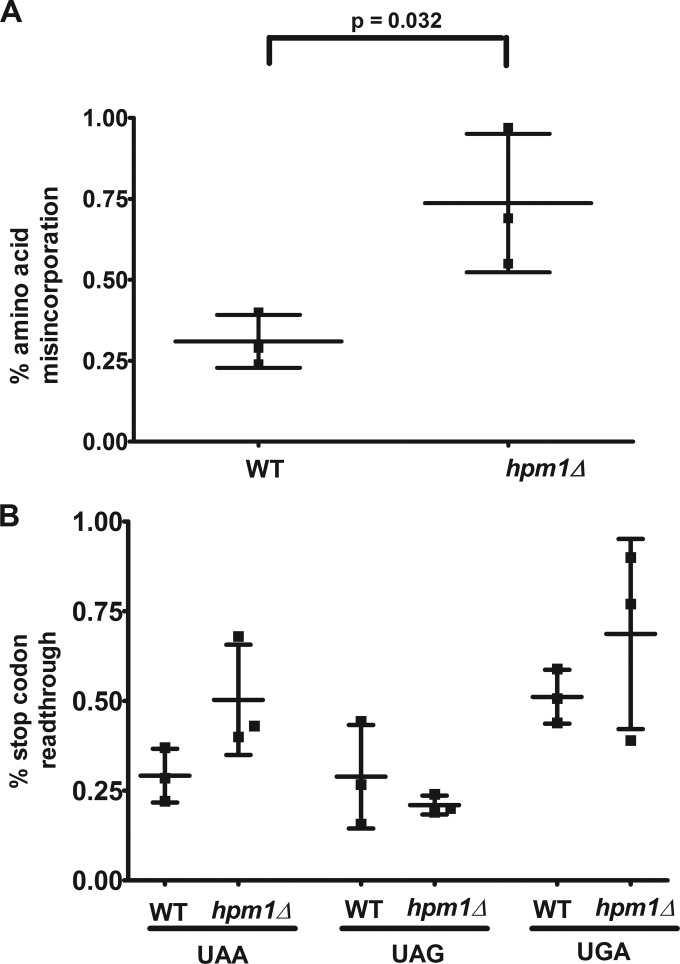
We next investigated the effect of Hpm1 depletion on translational fidelity by using a dual-luciferase reporter assay to measure amino acid misincorporation and stop codon suppression. This system uses a plasmid with a Renilla luciferase gene upstream of a firefly luciferase gene (33, 34). The amount of Renilla enzyme luminescence is used to normalize differences in mRNA abundance and translation initiation efficiency. To measure amino acid misincorporation, the firefly luciferase gene contains a near-cognate K529N mutation that renders firefly luciferase catalytically inactive (34). Misincorporation of the WT lysine residue through the recognition of the near-cognate lysyl-tRNALys would restore the enzymatic activity of the firefly enzyme and result in increased amounts of firefly luciferase luminescence. Stop codon readthrough reporter plasmids contain each of the three stop codons between the Renilla and firefly luciferase genes (34). Suppression of these stop codons would again result in increased firefly luciferase activity. Hpm1p-deficient cells displayed a significant increase in amino acid misincorporation that was more than 2-fold higher than that in WT cells (Fig. 7A). Additionally, there was increased readthrough of the UAA and UGA stop codons but not the UAG stop codon. These findings indicate that hpm1Δ mutant ribosomes have a reduced ability to discriminate between cognate and near-cognate aminoacyl-tRNAs in the A site and in the recognition of translation-terminating stop codons.
FIG 7.
Cells deficient in Hpm1p have increased amino acid misincorporation and stop codon readthrough. The dual-luciferase assay was done as described in Materials and Methods. Percentages of amino acid misincorporation (A) and stop codon readthrough (B) were calculated by dividing the firefly/Renilla luciferase luminescence ratio by the same ratio of the respective control vector. Error bars represent the standard deviations of three independent experiments. The unpaired t test two-tailed P value is shown for panel A. Differences in stop codon readthrough between the WT and the hpm1Δ mutant were not statistically significant at the P < 0.05 level.
DISCUSSION
The large ribosomal subunit protein Rpl3p of S. cerevisiae contains an unusual 3-methylhistidine modification that makes extensive contacts with the 25S rRNA near the PTC (16, 23, 24). In this work, we show that the loss of Rpl3p methylation in hpm1 null cells correlates with a deficit of large ribosomal subunits, decreased translational fitness, ribosomal structure alterations, and decreased fidelity of protein synthesis. These defects in large-subunit biogenesis and translation likely stem from delayed and aberrant processing of pre-rRNA transcripts, as demonstrated by the accumulation of the 23S precursor, delayed appearance of the 27S and 20S precursors of the 25S and 18S mature rRNAs, respectively, and the detection of a fraction of a 35S species that undergoes very slow A0/A1 cleavage to the 32S form in hpm1Δ mutant cells. Similar results have been reported for cells depleted of Rpl3p, which also demonstrated the subunit reversal phenotype and abnormal rRNA processing, as well as an accumulation of half-mer polysomes, indicative of initiation defects (40). Although we did not observe half-mers in hpm1Δ mutant cells, there was increased ribosome runoff, another manifestation of initiation defects. Furthermore, it has been reported that the loss of yeast Rrb1p, which physically interacts with Rpl3p, also results in a 60S biogenesis defect (41, 42). Rrb1p is proposed to target Rpl3p to the 35S pre-rRNA transcript. Notably, the human homolog of Hpm1p, C1orf156, copurifies with both human RPL3 and GRWD1 (human homolog of Rrb1p) (26). These reports are consistent with our hypothesis that Hpm1p is involved in early 60S subunit assembly.
Our ability to rescue the phenotypes of hpm1 null cells with a plasmid containing WT but not mutant HPM1 suggests that Hpm1p's role in early large-subunit assembly is dependent on its methyltransferase activity. This is unlike what has been reported for the Bud23p 18S rRNA methyltransferase in S. cerevisiae or the Rmt3p protein arginine methyltransferase in Schizosaccharomyces pombe; in both of these cases, the methyltransferase activities are dispensable for their roles in ribosome biogenesis (43, 44). While Rmt3p is implicated in 40S small subunit biogenesis (45), to our knowledge, Hpm1p is the only ribosomal protein methyltransferase shown to be involved in large-subunit biogenesis.
Using in vitro methylation assays, we now show that Rpl3p can be directly methylated by Hpm1p, confirming that this protein is an active methyltransferase and not simply a required accessory factor in the modification of Rpl3p at His-243. Hpm1p is capable of catalyzing the methylation of Rpl3p when it is in a complex with mature ribosomes but not as a purified, recombinant protein. Additionally, we find no evidence that Hpm1p can methylate a synthetic peptide containing the His-243 site. These results suggest that Hpm1p requires specific interactions with ribosomal components to efficiently methylate Rpl3p. This situation is similar to that observed with the E. coli PrmB L3 glutamine methyltransferase, where unfolded 70S ribosomes or a reconstituted mixture of extracted ribosomal proteins and rRNA is required for efficient methylation (25). The ability of Hpm1p to methylate ribosome-associated Rpl3p in vitro seems counterintuitive, since the methylation site on mature ribosomes is buried deep within the core of the 25S rRNA. This location makes it unlikely that Hpm1p can locate its target site on mature ribosomes in vivo. Green fluorescent protein (GFP) localization studies have shown that Hpm1p is present in both the nucleus and the cytoplasm, with higher levels in the nucleus (46). Hpm1p has also been reported to copurify with the Mlp2p nuclear protein (47) and the Rpa135p subunit of nucleolar RNA polymerase I (48). Coupled with the rRNA processing defects that we observed and the appearance of the 3-methylhistidine modification inside nucleus-containing subcellular organelle fractions, it is more plausible that methylation occurs on partially assembled preribosomes in the nucleus.
To examine the possibility that the alterations in ribosome biogenesis affect the function of the mature ribosome, Hpm1p-deficient cells were probed for changes in sensitivity to several ribosome-binding drugs. Altered sensitivities may indicate structural abnormalities in the functional centers that bind these drugs. Puromycin and anisomycin act as competitive inhibitors of aminoacyl-tRNAs by binding to a hydrophobic crevice in the A site of the PTC (49) that is approximately 20 Å away from the methylated H243 site. Paromomycin binds to the decoding center of the small 40S subunit greater than 80 Å away from the H243 site; changes in sensitivity to this drug have been linked to alterations in the decoding center (50). Our findings that hpm1 null cells had responses to sublethal levels of puromycin, anisomycin, and paromomycin similar to those of WT cells suggest that the structural integrity of the A site and the decoding center is largely unchanged. Cycloheximide binds to the E site of the eukaryotic ribosome and inhibits the peptidyl transferase activity of the large subunit (51). hpm1 null cells displayed significantly increased resistance to cycloheximide. Considering that the cycloheximide-binding site is more than 80 Å away from the methylated H243 site, loss of Hpm1p appears to have a long-ranging effect on the structural integrity of the ribosome. The precise binding site of verrucarin A is unclear; however, it is known to inhibit peptide chain initiation, resulting in ribosome runoff (52). It is therefore possible that this drug binds near the A site of the PTC or in the aa-tRNA accommodation corridor. The resistance of hpm1Δ mutant cells to verrucarin A might reflect an alteration of these two functional centers that could compromise the ability of hpm1Δ mutant ribosomes to discriminate between cognate and near-cognate aa-tRNAs. Chemical probing for rRNA structural changes is needed to confirm these hypothesis.
To test the possibility that ribosome function is altered in Hpm1p-null cells, we analyzed translational fidelity in vivo by measuring the extent of amino acid misincorporation and stop codon readthrough. There was >2-fold more misincorporation of near-cognate aminoacyl-tRNAs in hpm1 null cells than in WT cells. Hpm1p-deficient cells also displayed increased stop codon suppression of the UAA and UGA codons. This suggests that ribosomes with unmodified Rpl3p have difficulty in discerning between cognate and near-cognate tRNAs, as well as a decrease in translation termination efficiency.
In yeast, histidine 243 is located in close proximity (<20 Å) to three 2′-O-methylated nucleotides (U1888, C2337, U2980) and one pseudouridine (U2880) (23, 53); the clustering of these modifications may be functionally relevant. H243 lies in the tryptophan finger domain of Rpl3p at the core of the 25S rRNA, more specifically, in a positively charged “basic thumb” that protrudes perpendicularly to the finger (54). The basic thumb has recently been shown to play a role in coordinating the processes occurring in the PTC, the aminoacyl-tRNA binding region, and the elongation factor binding site to promote unidirectional translation (54). Mutations of various amino acids in the basic thumb result in growth defects, decreased translation fidelity and peptidyl transfer rates, and decreased binding of aminoacyl-tRNAs and the elongation factor eEF2 (54). Interestingly, H243 flanking residues, R240 and R247, displayed more severe phenotypes than the other basic thumb mutants examined (54). It is thus possible that methylation of H243 plays a role in the functionality of this basic thumb.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM026020 (to S.G.C.) and GM061518 (to G.F.C.). K.R. and M.C.D. were supported by NIH training program T32GM007185.
We thank James Wohlschlegel and Joseph Loo for their helpful advice. We also thank David Bedwell and Ming Du for providing the translational fidelity plasmids and help with the dual-luciferase assay.
Footnotes
Published ahead of print 27 May 2014
REFERENCES
- 1.Lapeyre B. 2005. Conserved ribosomal RNA modification and their putative roles in ribosome biogenesis and translation, p 263–284 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany [Google Scholar]
- 2.Johansson MO, Byström A. 2005. Transfer RNA modifications and modifying enzymes in Saccharomyces cerevisiae, p 87–120 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany [Google Scholar]
- 3.Bokar J. 2005. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA, p 141–177 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany [Google Scholar]
- 4.Polevoda B, Sherman F. 2007. Methylation of proteins involved in translation. Mol. Microbiol. 65:590–606. 10.1111/j.1365-2958.2007.05831.x [DOI] [PubMed] [Google Scholar]
- 5.Katz JE, Dlakic M, Clarke SG. 2003. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2:525–540 [DOI] [PubMed] [Google Scholar]
- 6.Petrossian TC, Clarke SG. 2009. Multiple motif scanning to identify methyltransferases from the yeast proteome. Mol. Cell. Proteomics 8:1516–1526. 10.1074/mcp.M900025-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wlodarski T, Kutner J, Towpik J, Knizewski L, Rychlewski L, Kudlicki A, Rowicka M, Dziembowski A, Ginalski K. 2011. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS One 6:e23168. 10.1371/journal.pone.0023168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke SG. 2013. Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38:243–252. 10.1016/j.tibs.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27:344–351. 10.1016/S0968-0004(02)02109-6 [DOI] [PubMed] [Google Scholar]
- 10.Motorin Y, Helm M. 2010. tRNA stabilization by modified nucleotides. Biochemistry 49:4934–4944. 10.1021/bi100408z [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T. 2005. Biosynthesis and function of tRNA wobble modifications, p 23–69 In Grosjean H. (ed), Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany [Google Scholar]
- 12.Dinçbas-Renqvist V, Engstrom A, Mora L, Heurgué-Harnard V, Buckingham R, Ehrenberg M. 2000. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19:6900–6907. 10.1093/emboj/19.24.6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heurgué-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. 2002. The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21:769–778. 10.1093/emboj/21.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. 2002. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl. Acad. Sci. U. S. A. 99:5942–5947. 10.1073/pnas.082119899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb KJ, Al-Hadid Q, Zurita-Lopez CI, Young BD, Lipson RS, Clarke SG. 2011. The ribosomal l1 protuberance in yeast is methylated on a lysine residue catalyzed by a seven-beta-strand methyltransferase. J. Biol. Chem. 286:18405–18413. 10.1074/jbc.M110.200410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb KJ, Zurita-Lopez CI, Al-Hadid Q, Laganowsky A, Young BD, Lipson RS, Souda P, Faull KF, Whitelegge JP, Clarke SG. 2010. A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J. Biol. Chem. 285:37598–37606. 10.1074/jbc.M110.170787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porras-Yakushi TR, Whitelegge JP, Clarke S. 2006. A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at lysine 10. J. Biol. Chem. 281:35835–35845. 10.1074/jbc.M606578200 [DOI] [PubMed] [Google Scholar]
- 18.Chern MK, Chang KN, Liu LF, Tam TC, Liu YC, Liang YL, Tam MF. 2002. Yeast ribosomal protein L12 is a substrate of protein-arginine methyltransferase 2. J. Biol. Chem. 277:15345–15353. 10.1074/jbc.M111379200 [DOI] [PubMed] [Google Scholar]
- 19.Porras-Yakushi TR, Whitelegge JP, Miranda TB, Clarke S. 2005. A novel SET domain methyltransferase modifies ribosomal protein Rpl23ab in yeast. J. Biol. Chem. 280:34590–34598. 10.1074/jbc.M507672200 [DOI] [PubMed] [Google Scholar]
- 20.Webb KJ, Laganowsky A, Whitelegge JP, Clarke SG. 2008. Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. J. Biol. Chem. 283:35561–35568. 10.1074/jbc.M806006200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG. 2010. Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry 49:5225–5235. 10.1021/bi100428x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young BD, Weiss DI, Zurita-Lopez CI, Webb KJ, Clarke SG, McBride AE. 2012. Identification of methylated proteins in the yeast small ribosomal subunit: a role for SPOUT methyltransferases in protein arginine methylation. Biochemistry 51:5091–5104. 10.1021/bi300186g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334:1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 24.Meskauskas A, Dinman JD. 2008. Ribosomal protein L3 functions as a ‘rocker switch' to aid in coordinating of large subunit-associated functions in eukaryotes and Archaea. Nucleic Acids Res. 36:6175–6186. 10.1093/nar/gkn642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lhoest J, Colson C. 1981. Cold-sensitive ribosome assembly in an Escherichia coli mutant lacking a single methyl group in ribosomal protein L3. Eur. J. Biochem. 121:33–37. 10.1111/j.1432-1033.1981.tb06425.x [DOI] [PubMed] [Google Scholar]
- 26.Cloutier P, Lavallee-Adam M, Faubert D, Blanchette M, Coulombe B. 2013. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 9:e1003210. 10.1371/journal.pgen.1003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowry OH, Rosebrough NJ, Farr NL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 28.Gary JD, Clarke S. 1995. Purification and characterization of an isoaspartyl dipeptidase from Escherichia coli. J. Biol. Chem. 270:4076–4087 [DOI] [PubMed] [Google Scholar]
- 29.Chanfreau G, Rotondo G, Legrain P, Jacquier A. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726–3737. 10.1093/emboj/17.13.3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10:573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Woods RA. 2002. Transformation of yeast by the LiOAc/SS carrier DNA/PEG method. Methods Enzymol. 350:87–96. 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- 33.Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10:691–703. 10.1261/rna.5147804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salas-Marco J, Bedwell DM. 2005. Discrimination between defect in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 348:801–815. 10.1016/j.jmb.2005.03.025 [DOI] [PubMed] [Google Scholar]
- 35.Gottschling H, Freese E. 1962. A tritium isotope effect on ion exchange chromatography. Nature 196:829–831. 10.1038/196829a0 [DOI] [PubMed] [Google Scholar]
- 36.Venema J, Tollervey D. 1995. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11:1629–1650 [DOI] [PubMed] [Google Scholar]
- 37.Granneman S, Baserga SJ. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43–50. 10.1016/j.yexcr.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 38.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. 2004. RNA polymerase 1 transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506–2517. 10.1101/gad.1226604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. 2009. Rrp17p is a eukaryotic exonuclease required for 5′end processing of pre-60S ribosomal RNA. Mol. Cell 36:768–781. 10.1016/j.molcel.2009.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosado IV, Kressler D, de la Cruz J. 2007. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome biogenesis. Nucleic Acids Res. 35:4203–4213. 10.1093/nar/gkm388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaper S, Fromont-Racine M, Linder P, de la Cruz J, Namane A, Yaniv M. 2001. A yeast homolog of chromatin assembly factor 1 is involved in early ribosome assembly. Curr. Biol. 11:1885–1890. 10.1016/S0960-9822(01)00584-X [DOI] [PubMed] [Google Scholar]
- 42.Iouk TL, Aitchison JD, Maquire S, Wozniak RW. 2001. Rrb1, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol. Cell. Biol. 21:1260–1271. 10.1128/MCB.21.4.1260-1271.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. 2008. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 28:3151–3161. 10.1128/MCB.01674-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perreault A, Gascon S, D'Amours A, Aletta JM, Bachand F. 2009. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 284:15026–15037. 10.1074/jbc.M109.004812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachand F, Silver PA. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23:2641–2650. 10.1038/sj.emboj.7600265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 47.Keck JM, Jones MH, Wong CC, Binkley J, Chen D, Jaspersen SL, Holinger EP, Xu T, Niepel M, Rout MP, Vogel J, Sidow A, Yates JR, III, Winey M. 2011. A cell cycle phosphoproteome of the yeast centrosome. Science 332:1557–1561. 10.1126/science.1205193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. 2006. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. U. S. A. 103:12707–12712. 10.1073/pnas.0605686103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen JL, Moore PB, Steitz TA. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061–1075. 10.1016/S0022-2836(03)00668-5 [DOI] [PubMed] [Google Scholar]
- 50.Ogle JM, Carter AP, Ramakrishnan V. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28:259–266. 10.1016/S0968-0004(03)00066-5 [DOI] [PubMed] [Google Scholar]
- 51.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6:209–217. 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernández F, Cannon M. 1982. Inhibition of protein synthesis in Saccharomyces cerevisiae by the 12,13-epoxytrichothecenes trichodermol, diacetoxyscirpenol and verrucarin A. J. Antibiot. (Tokyo) 35:875–881. 10.7164/antibiotics.35.875 [DOI] [PubMed] [Google Scholar]
- 53.Piekna-Przybylska D, Decatur WA, Fournier MJ. 2007. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA 13:305–312. 10.1261/rna.373107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meskauskas A, Dinman JD. 2010. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A site. Nucleic Acids Res. 38:7800–7813. 10.1093/nar/gkq641 [DOI] [PMC free article] [PubMed] [Google Scholar]