Abstract
Osteoblasts and adipocytes arise from a common mesenchymal precursor cell. The cell fate decision of a mesenchymal precursor cell is under the influence of molecular cues and signaling pathways that lead to the activation or repression of lineage-specific transcription factors. The molecular mechanisms determining osteoblast versus adipocyte lineage specificity in response to bone morphogenic protein (BMP) remain unclear. In this study, we describe the mechanism through which Zfp521 (ZNF521), a regulator of lineage progression in multiple immature cell populations, regulates lineage specification of mesenchymal progenitor cells during BMP-induced differentiation events. In vivo deletion or in vitro knockdown of Zfp521 in mesenchymal precursors resulted in increased expression of the adipocyte determinant factor Zfp423 (ZNF423). This was concurrent with the loss of histone H3K9 methylation and an increase in histone H3K9 acetylation at the Zfp423 promoter, which together are indicative of decreased gene repression. Indeed, we found that Zfp521 occupies and represses the promoter and intronic enhancer regions of Zfp423. Accordingly, conditional deletion of Zfp521 inhibited heterotopic bone formation in response to local injection of BMP2. In contrast, marrow adiposity within BMP2-induced bone was markedly enhanced in Zfp521-deficient mice, suggesting that precursor cells lacking Zfp521 differentiate preferentially into adipocytes instead of osteoblasts in response to BMP2. Consistent with a cell-autonomous role of Zfp521 in mesenchymal precursors, knockdown of Zfp521 in stromal cells prevented BMP2-induced osteoblast marker expression and simultaneously enhanced lipid accumulation and expression of adipocyte-related genes. Taken together, the data suggest that Zfp521 is a cell fate switch critical for BMP-induced osteoblast commitment and identify Zfp521 as the intrinsic repressor of Zfp423 and hence of adipocyte commitment during BMP-induced mesenchymal precursor differentiation.
INTRODUCTION
Multipotent mesenchymal precursor cells can differentiate into several cell types, including osteoblasts and adipocytes. Commitment to a lineage appears to be mutually exclusive, so that factors that enhance osteoblastogenesis are often inhibitory to adipogenesis and vice versa. Imbalances between osteoblast and adipocyte commitment is evident in some human diseases, such as age-related osteoporosis (1), where decreased bone formation is accompanied by increased bone marrow fat, or in progressive osseous heteroplasia, where bone formation is initiated within subcutaneous fat (2). Thus, understanding the regulatory mechanisms that alter precursor cell fate is of potential clinical significance.
Mesenchymal progenitor cell fate is fine-tuned by transcription factors and signaling pathways, including the bone morphogenic protein (BMP) pathway. Although BMPs were first discovered based on their ability to induce the recruitment and differentiation of mesenchymal precursors into bone-forming osteoblasts (3), BMPs are now known to be important in multiple aspects of cell biology during early development and adult tissue homeostasis. The molecular mechanisms by which BMPs alter cell fate toward osteogenesis or adipogenesis are not fully understood. In particular, whereas considerable evidence indicates that BMP signaling enhances osteoblast and chondrocyte commitment (4) while repressing myoblastogenesis (5), the role of BMPs in adipocyte commitment remains unclear. Although initial reports suggested that BMP2 inhibits adipogenic differentiation (6), positive effects of BMPs on adipogenesis have also been reported in both genetic and cell culture studies (7–10). Thus, even though BMPs can induce osteoblast differentiation, they also appear capable of switching mesenchymal precursors toward adipogenesis under certain conditions. How precursor cells integrate a BMP signal into different fate decisions is not fully understood.
The transcriptional cascades downstream of BMP leading to a cell fate decision are mediated by the Smad-dependent activation or repression of lineage-specific transcription factors. During BMP2-induced osteoblast differentiation, BMP-activated Smads induce the expression of the osteoblast-specific cascade of transcription factors Dlx5 (11), Msx2 (12), Runx2 (13), and Osterix (also known as SP7 in humans) (14), which work in concert to drive the expression of genes defining the osteoblast phenotype (15, 16). In contrast, the adipogenic transcriptional cascade involves the induction of the peroxisome proliferator-activated receptor (PPAR) and C/EBP transcription factors by the Smad-associated cofactor Schnurri3 (9) and the recently identified preadipocyte commitment factor zinc finger protein 423 (Zfp423; ZNF423 in humans; also known as OAZ) (17). Zfp423 is particularly enriched in preadipocytes (18) and drives the expression of PPARγ (PPARG) in synergy with BMPs.
Zinc finger protein 521 (Zfp521 in mice or ZNF521 in humans; also known as EVI3 and EHZF), a transcriptional regulator containing 30 C2H2-type zinc fingers, is a paralog of Zfp423. Unlike Zfp423, which is specifically enriched in preadipocytes, Zfp521 expression is fairly ubiquitous but is particularly abundant in immature cell populations, such as hematopoietic progenitors (19), neuronal precursors (19), mesenchymal condensations (20), and embryonic stem (ES) cells (21). In all these cell types, Zfp521 functions as a regulator of lineage progression. In mesenchymal tissues, we have recently shown that Zfp521 modulates Ebf1 activity in osteoblasts and osteoclasts to maintain bone homeostasis in the postnatal skeleton (22). During skeletal development, we previously showed that Zfp521 suppresses Runx2 activity and prevents the hypertrophic conversion of prehypertrophic chondrocytes within the growth plate (23) and the maturation of osteoblasts during prenatal intramembranous bone formation (24). Interestingly, the balance between Zfp521 and Ebf1 is also important for adipose tissue formation in vivo (25): Ebf1 is a potent proadipogenic factor, and its repression by Zfp521 blocks adipogenesis, so that Zfp521−/− mice exhibit increased brown adipose tissue and skin adipocytes (25).
Importantly, Zfp521 expression can be regulated by BMPs (20, 21), but the functional relevance of this convergence of BMP signaling and Zfp521 expression in the context of mesenchymal lineage commitment has not been explored. Given that (i) Zfp521 is a key regulator of cell progression and lineage specification, (ii) Zfp521 is regulated by BMPs, and (iii) Zfp521 plays an important role in bone homeostasis and adipose tissue development, we hypothesized that Zfp521 might be a key factor in the BMP-mediated osteoblast versus adipocyte cell fate decision. We report here that Zfp521 regulates the fate of mesenchymal precursors during BMP-mediated differentiation events by repressing Zfp423, and thereby PPARγ, so that the osteogenic response to BMP is switched to an adipogenic response in vivo and in vitro in the absence of Zfp521. We present the following novel findings: (i) Zfp521 directly represses Zfp423 by binding to its promoter and intronic enhancer, (ii) Zfp521 is required for an osteoinductive response to BMP2, and (iii) loss of Zfp521 predisposes the fate of mesenchymal cells toward adipogenesis (versus osteogenesis) during BMP2-induced differentiation.
MATERIALS AND METHODS
Mice.
Zfp521+/− and Zfp521 floxed (Zfp521f/f) mice were provided by S. Warming (Genentech) and have been described previously (22). Dermo1-Cre (also known as Twist2-Cre) (26) were obtained from Jackson Laboratories. Littermate controls were used for all experiments. Genotyping primers were as follows: Zfp521-WT-F, 5′-GCTCCTTCACATGCTCACA-3′; Zfp521-WT-R, 5′-CTGGCATGGGTAAGGCAGT-3′; Zfp521-KO-R, 5′-CTCCGAGAGTTTGAGTGCAA-3′; Cre-F, 5′-CCAATTTACTGACCGTACACC-3′; Cre-R, 5′-CCCGGCAAAACAGGTAGTTA-3′. All animal protocols were approved by the Harvard Medical School Institutional Animal Care and Use Committee.
Calvarial injection.
In vivo analysis of BMP2 responses by calvarial injection was conducted as previously described (27). Briefly, 6-day-old mice were injected with recombinant BMP2 (Medtronic) 3 times a day for 5 days into the periosteal tissue overlying the right parietal bone. Saline vehicle control was injected onto the left parietal bone. Animals were sacrificed 6 days or 14 days after the last injection for analysis of bone formation or 24 h after the last injection for gene expression analysis.
μCT and light microscopy.
Dissected calvaria from BMP-injected animals were fixed immediately in 70% ethanol. Imaging was performed using μCT 35 (Scanco Medical) and reconstructed into three-dimensional (3D) volumes using μCT Ray V3.8 and μCT Evaluation Program V6.5-1 (Scanco Medical). The total volume of heterotopic new bone formation was quantified by delineating heights and widths from tangential micro-computed tomography (μCT) sections. For light microscopy, dissected calvaria were fixed in 70% ethanol or 10% neutral buffered formalin and embedded undecalcified in methyl methacrylate (Sigma-Aldrich). Sections (5 μm) were stained for bone by the von Kossa method (silver nitrate staining) and counterstained with toluidine blue. Histomorphometry was performed with Osteomeasure software (OsteoMetrics).
Cell culture.
W20-17 murine bone marrow stromal cells (28) (courtesy of V. Rosen) and HEK293 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). The Zfp521 expression plasmid (pCMV6-Kan/Neo-Zfp521), empty plasmid (pCMV6-Kan/Neo), Zfp521 short hairpin RNA (shRNA) (sequences, ACTTCAGTGTCACCTGATAGAGCACAGCT, TAGCCGCCTCAGCTACCTAAAGCACCATG, GAATCAGTTGAACTAACCTGTATGTTACT, and ACAGCGCAGTCCAAGACCTTGCAGGATGT), and scrambled shRNA control (TR30013) were obtained from Origene. Stable cell lines were produced by transfecting cells with X-tremeGene HP (Roche) and selecting for stable clones in growth medium supplemented with puromycin (Life Technologies) or G418 (Life Technologies) for shRNA and cDNA plasmids, respectively. Adipocyte differentiation was induced 24 h after plating by treating confluent cells with medium containing 3 μM troglitazone (Sigma). The medium was replaced every 48 h over a 6-day period. Lipid droplets were stained using 0.2% oil red O (Sigma) in 60% isopropanol after fixation with 10% neutral buffered formalin. Alkaline phosphatase activity was visualized by staining fixed cultures with 5-bromo-4-chloro-3-indolyl phosphate–Nitro Blue Tetrazolium (Sigma). Alkaline phosphatase activity was determined spectrophotometrically (wavelength, 405 nm) from cell lysates, using p-nitrophenylphosphate (Sigma) as a substrate. One unit of alkaline phosphatase hydrolyzes 1 μmol of p-nitrophenylphosphate/min at 37°C. Recombinant BMP2 was obtained from R&D Systems.
Quantitative real-time RT (qRT)-PCR.
Total RNA was isolated from cells or bone using a High Pure RNA isolation kit (Roche). For bone RNA extraction, tissue was first flash frozen with liquid nitrogen and then crushed into powder with a Bio-Pulverizer (Biospec). The RNA was reverse transcribed using a Transcriptor First Strand cDNA kit (Roche), and real-time PCR was carried out on an iCyclerIQ thermocycler (Bio-Rad) using FastStart Universal SYBR master mix (Roche). Relative quantification of gene expression was performed by the ΔCT method using Gapdh as the housekeeping gene for normalization. Primer (Integrated DNA Technologies) sequences were as follows: Gapdh, 5′-TGCACCACCAACTGCTTAG-3′ and 5′-GGATGCAGGGATGATGTTC-3′; Zfp521, 5′-ACAACGAGTGGGACATCCAGGT-3′ and 5′-GCTGTGCTCTATCAGGTGACAC-3′; Dlx5, 5′-AGGCTTATGCCGACTACGGCTA-3′ and 5′-CTCTGGCTCCGCCACTTCTTTC-3′; Sp7, 5′-GGCTTTTCTGCGGCAAGAGGTT-3′ and 5′-CGCTGATGTTTGCTCAAGTGGTC-3′; C/EBPα, 5′-GCAAAGCCAAGAAGTCGGTGGA-3′ and 5′-CCTTCTGTTGCGTCTCCACGTT-3′; Fabp4, 5′-TGAAATCACCGCAGACGACAGG-3′ and 5′-GCTTGTCACCATCTCGTTTTCTC-3′; Zfp423, 5′-GATCACTGTCAGCAGGACTT-3′ and 5′-TGCCTCTTCAAGTAGCTCA-3′; Pparγ2, 5′-GCATGGTGCCTTCGCTGA-3′ and 5′-TGGCATCTCTGTGTCAACCATG-3′; Alpl, 5′-CCAGAAAGACACCTTGACTGTGG-3′ and 5′-TCTTGTCCGTGTCGCTCACCAT-3′; Msx2, 5′-AAGACGGAGCACCGTGGATACA-3′ and 5′-CGGTTGGTCTTGTGTTTCCTCAG-3′; Pparγ1, 5′-TGAAAGAAGCGGTGAACCACTG-3′ and 5′-TGGCATCTCTGTGTCAACCATG-3′.
ChIP.
Genomic DNA extraction and chromatin immunoprecipitation (ChIP) analysis were performed using the SimpleChIP enzymatic chromatin IP kit (Cell Signaling) according to the manufacturer's instructions. Immunoprecipitation was performed using the following antibodies: acetyl-histone H3K9 (9649; Cell Signaling), anti-histone H3 trimethyl K9 (ab8898; Abcam), normal rabbit IgG (Cell Signaling), and Zfp521 (25). Primer sequences were as follows: Zfp423 promoter, 5′-AAAGTTTCCGAGAGGCAGGA-3′ and 5′-TGCTTCCGCCTGGACAT-3′; Zfp423 Intron 5, 5′-CAGCAGAAGGGGCTGTTCTA-3′ and 5′-GGGATTAGGCGGAATTTACC-3′; Zfp423 exon 3, 5′-GGAACAGCGTGACAAGTCAAG-3′ and 5′-TGACAGTGATCGCAGGTGTAA-3′.
Biotinylated oligonucleotide pulldown assay.
Nuclear protein extraction and oligonucleotide pulldown were performed based on a method described previously (29). Biotinylated oligonucleotides (IDT) were incubated for 1 h at 4°C with streptavidin-magnetic beads (Pierce). The beads were then washed three times with binding buffer (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 1.5 mM MgCl2, 150 mM KCl, 10% glycerol, 0.02% NP-40, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with a protease inhibitor cocktail (Sigma). The beads were then added to nuclear extracts from W20 cells supplemented with 20 μg/ml salmon sperm DNA (Sigma) and incubated at 4°C for 2 h under rotation. Bound proteins were eluted in 2× SDS sample buffer after washing the beads four times in binding buffer. The proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad), and immunoblotted with rabbit anti-Zfp521 (20) and anti-rabbit antibody conjugated to horseradish peroxidase (HRP) (Cell Signaling Technologies). The sense sequences to the biotinylated oligonucleotides were as follows: promoter, 5′-GAGGAGAAGGGGGAGAAGGGCTGCCCGGAGCGGCGGGGGGCGGTGCGGGGAGCCGACCGCGGACGGCGACGGAGCGCCCGGAGCCCCGGA-3′; intron, 5′-GCTGTTCTAATCTTGCCCACTGCACGCCGCCATCGGGAGAGCATTCCTAGGGCGCGAGCAGCTCCCTAGGGTGAGGCCGCGGTAAATTC-3′.
Reporter assays.
The BMP reporter plasmid (IDWTF4-luc) containing BMP response elements (BRE) from the ID1 gene and the mutant construct with mutated BMP response elements (IDMutB4-luc) (30) were kind gifts from T. Katagiri (Saitama Medical University). The previously described Zfp423 intron 5 enhancer reporter construct (pGL4-intron5-luc) (31) was kindly provided by B. Hamilton (University of California—San Diego). To construct the Zfp423 promoter luciferase reporter plasmid, a 1.4-kb fragment (−982/+405) of the 5′ region of the mouse Zfp521 gene was cloned by PCR using PrimeStar Max DNA polymerase (TaKaRa) with mouse genomic DNA as a template. The PCR product was then subcloned between the XhoI and HindIII (New England BioLabs) restriction sites of pGL4.14 Basic luciferase vector (Promega). The primer sequences containing spacer and restriction sites (lowercase) were as follows: 5′-agcctcgagCTACATTTCCAGCCAAGCCT-3′ and 5′-gccaagcttTCACCTTTCACCGATCGCGG-3′. Renilla luciferase (pRL-TK) (Promega) was used as an internal control for transfection efficiency. W20-17 or HEK293 cells were transfected using XtremeGene HP (Roche), and dual-luciferase activity was assessed using the Dual-Glo assay system (Promega). To examine BMP2 responses, cells were maintained in 0.5% fetal bovine serum and treated with 100 ng/ml BMP2 or vehicle 16 h prior to assay. Flag-Ebf1, Smad1, and Smad4 constructs were described previously (22, 32). Zfp521 deletion constructs (21) were kind gifts from D. Kamiya and Y. Sasai.
Coimmunoprecipitation.
W20 cells were transfected with Flag-Smad4 (32) and Zfp521 deletion constructs (21) using Lipofectamine 3000 (Invitrogen). Forty-eight hours after transfection, the cells were lysed with HKMG lysis buffer (10 mM HEPES, pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, and 0.5% NP-40) containing protease and phosphatase inhibitor cocktails (Cell Signaling). One thousand micrograms of protein was incubated with 20 μl of anti-Flag M2 (Sigma) or mouse IgG (Cell Signaling) magnetic beads overnight and then washed with lysis buffer five times. Bound proteins were eluted with 2× SDS sample buffer and analyzed by Western blotting. Flag epitope-tagged proteins were detected with anti-Flag–HRP conjugate antibody (Cell Signaling), and Zfp521 was detected with rabbit anti-Zfp521 (20) and anti-rabbit antibody conjugated to HRP (Cell Signaling).
RESULTS
Zfp521 directly represses Zfp423 transcription.
Despite the homology between Zfp521 and Zfp423, the expression levels of the two paralogs are inversely related within mesenchymal cells and regulated in an opposing manner during differentiation into osteoblasts or adipocytes (Fig. 1A). Knockdown of Zfp521 in W20-17 cells (28), a multipotent bone marrow stromal cell line capable of differentiating into osteoblasts or adipocytes, resulted in striking upregulation of Zfp423 expression (Fig. 1B), suggesting that Zfp423 may be a direct target of Zfp521. To determine whether Zfp423 is a direct target of Zfp521, we performedChIP analysis of endogenous Zfp521 in the promoter region and in a recently identified enhancer region of the Zfp423 locus (31). ChIP indicated that Zfp521 is recruited in both the promoter and intronic enhancer regions of Zfp423 (Fig. 1C) but is absent in a negative-control region within the coding sequence at exon 3. Direct association of Zfp521 with DNA sequences derived from the Zfp423 promoter and enhancer was confirmed by oligonucleotide pulldown using nuclear extracts from W20-17 cells (Fig. 1D).
FIG 1.
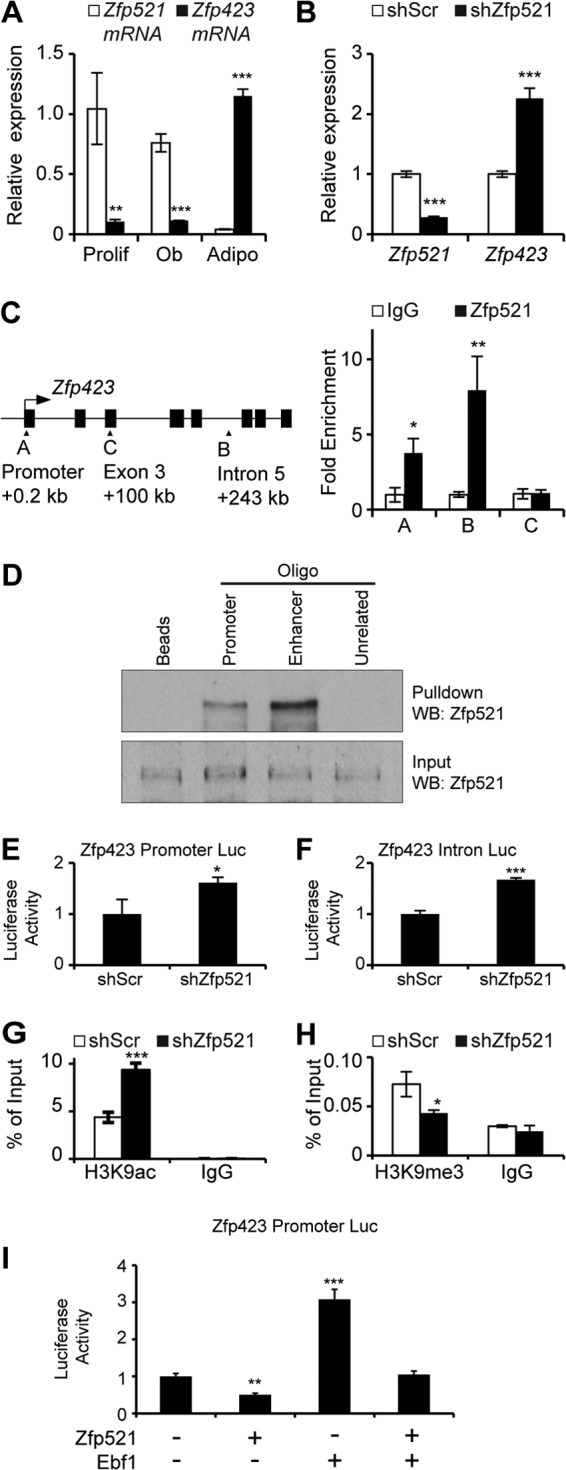
Zfp521 represses Zfp423 promoter activity. (A) qRT-PCR of Zfp521 and Zfp423 gene expression in undifferentiated proliferating W20 cells (Prolif) and in W20 cells 6 days following the induction of osteoblast (Ob) or adipocyte (Adipo) differentiation. (B) qRT-PCR for Zfp521 and Zfp423 gene expression in W20 cells stably expressing scrambled shRNA (shScr) or shRNA targeting Zfp521 (shZfp521). (C) ChIP analysis of Zfp521 on the Zfp423 promoter and intron 5. A coding region in Zfp423 exon 3 served as a negative control. The schematic shows the locations of the primers (A, B, and C) on the Zfp423 locus. (D) DNA affinity precipitation assay to determine binding of Zfp521 to the Zfp423 promoter or enhancer sequence. Nuclear extracts from W20 cells were incubated with biotinylated Zfp423 oligonucleotide probes and streptavidin-magnetic beads, after which Zfp521 binding was determined by Western blotting (WB). (E and F) Knockdown of Zfp521 enhanced the relative luciferase activity of a Zfp423 promoter luciferase construct (E) or a Zfp423 intron 5 luciferase construct (F) in HEK293 cells. (G and H) ChIP of acetylated histone H3 lysine 9 (H3K9ac) (G) or trimethylated histone H3 lysine 9 (H3K9me3) (H) in W20 cells stably expressing the indicated shRNA constructs using specific primers for the Zfp423 promoter region. (I) W20 cells were transfected with Zfp423 promoter luciferase plasmid and plasmids containing cDNA for the indicated transcription factors. Luciferase activity was measured 24 h after transfection. The data are presented as means ± standard deviations (SD); n ≥ 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student's t test.
To determine whether Zfp521 alters the transcriptional activity of the Zfp423 promoter or enhancer region, we analyzed the effect of Zfp521 knockdown on Zfp423 promoter- or enhancer-driven luciferase reporters. Zfp521 knockdown caused an increase in the luciferase activity of the Zfp423 gene promoter fragment (Fig. 1E), as well as the Zfp423 intronic enhancer fragment (Fig. 1F), suggesting that Zfp521 is an important endogenous regulator of Zfp423 transcription. We then determined whether Zfp521 association with the Zfp423 promoter is related to transcriptional silencing. For this purpose, we performed ChIP for histone H3K9 acetylation, an epigenetic mark of transcriptionally active chromatin, and for histone H3K9 trimethylation, a mark of transcriptional silencing. shRNA knockdown of Zfp521 resulted in increased H3K9 acetylation and decreased H3K9me3 in the Zfp423 promoter region, indicative of an increase in transcriptionally active chromatin following Zfp521 knockdown (Fig. 1G and H). We then assessed whether overexpression of Zfp521 could decrease Zfp423 promoter activity in the presence of Ebf1, a transcriptional activator known to induce Zfp423 expression (25). Overexpression of Zfp521 inhibited basal Zfp423 promoter activity, as well as Ebf1-induced Zfp423 promoter activity (Fig. 1I). Thus, these data indicate that Zfp521 directly binds to regulatory regions in the Zfp423 promoter and Zfp423 enhancer and represses Zfp423 gene transcription by facilitating transcriptional silencing of the Zfp423 locus.
Zfp521 is required for efficient BMP2 osteoinduction in vivo.
Both Zfp423 and Zfp521 are thought to act in response to BMP signaling. Subcutaneous injection of BMP2 onto the progenitor-rich periosteum lining the calvaria induces the local recruitment and differentiation of precursors into osteoblasts, resulting in the de novo formation of cancellous bone and bone marrow (27). To investigate the in vivo function of Zfp521 in BMP2-induced bone formation, BMP2 was injected onto the right parietal bone of 6-day-old wild-type and Zfp521−/− mice, whereas the left side was injected with vehicle (saline) as a control. Injections were performed 3 times/day for 5 days, and the mice were sacrificed 6 days after the last injection (Fig. 2A). BMP2 injection onto the calvaria of Zfp521−/− mice induced markedly less bone than with wild-type mice (Fig. 2B and C), with the volume of newly formed bone approximately 70% smaller in Zfp521−/− mice (Fig. 2D). Vehicle injected onto the left parietal bone did not induce any bone formation (Fig. 2B). Thus, BMP2-induced bone formation is impaired in the absence of Zfp521, suggesting that Zfp521 favors the osteogenic response downstream of BMP2.
FIG 2.
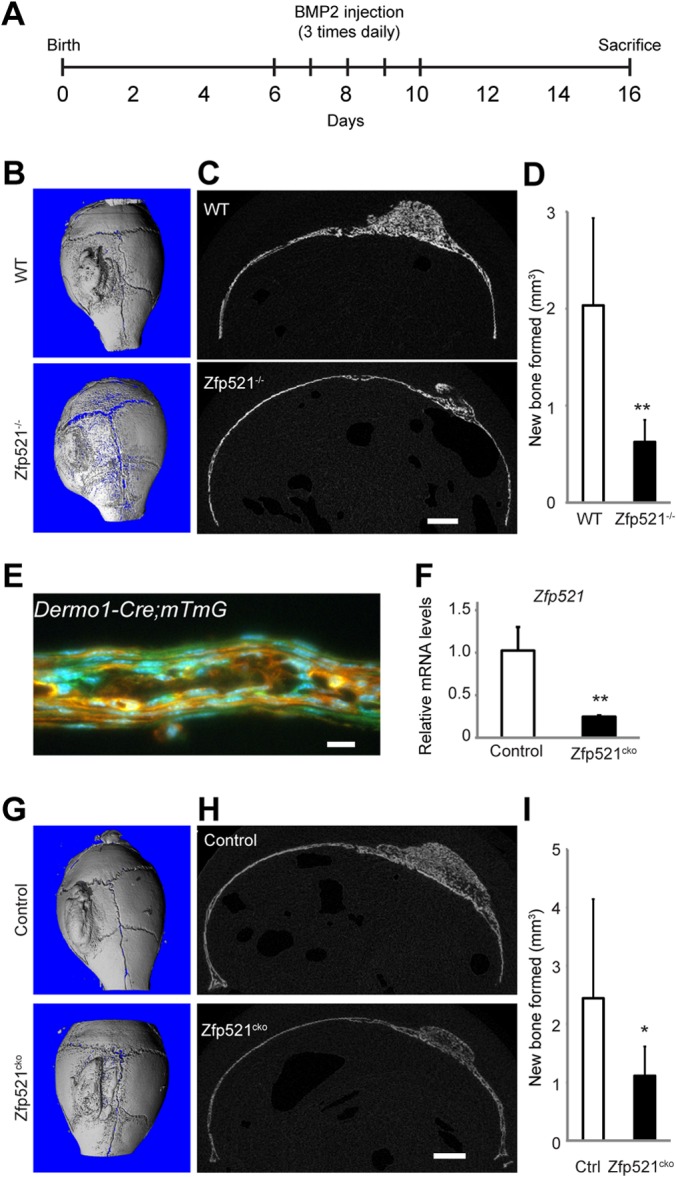
Reduced BMP2-induced bone formation in Zfp521−/− mice. (A) Schematic of a BMP2 calvarial injection model. (B and C) μCT 3D reconstruction (B) and cross-sectional image (C) of wild-type (WT) and Zfp521−/− calvarium following treatment with BMP2. (D) μCT quantification of newly formed bone in response to BMP2. (E) Fluorescence image of a 6-day-old Dermo1-Cre:mT/mG calvarium demonstrating Cre activity (green) in periosteal cells. (F) Expression of Zfp521 in calvaria of 6-day-old Dermo1-Cre:Zfp521f/f (Zfp521cko) mice. (G and H) μCT 3D (G) and cross-sectional (H) images of control (Dermo1-Cre; Zfp521+/+) and Zfp521cko mice following treatment with BMP2. (I) μCT quantification of newly formed bone in response to BMP2. The data are presented as means and SD; n ≥ 3. *, P < 0.05; **, P < 0.01; Student's t test. Scale bars, 1 mm (C and H); 10 μm (E).
To determine whether Zfp521 acts within the mesenchymal lineage to regulate BMP2-induced bone formation, we conditionally inactivated Zfp521 in mesenchymal progenitors by crossing Zfp521f/f mice with Dermo1-Cre (also called Twist2-Cre) mice (26). To confirm that Dermo1-Cre drives recombination within progenitor cells on the parietal bone periosteum, we used the mT/mG reporter strain (33) to detect Cre activity in vivo. Green fluorescent cells, indicative of Cre activity, were abundantly detected in Dermo1-Cre:mT/mG parietal bone periosteum (Fig. 2E). Quantitative PCR (qPCR) analysis of calvaria from control and Zfp521 conditional knockout (cKO) (Dermo1-Cre:Zfp521f/f) mice showed that Zfp521 expression was decreased by >60% (Fig. 2F). Similar to our observations in Zfp521−/− mice, BMP2 injection onto the calvaria of Zfp521cko mice induced significantly less bone than in control animals (Fig. 2G to I). These data demonstrate that Zfp521 deletion within mesenchymal cells impairs bone formation in response to BMP2.
Zfp521 deletion favors adipocyte differentiation in response to BMP2.
Histological analysis of the local response to BMP2 showed that, whereas osteoclast numbers were unaffected by Zfp521 deletion, and in contrast with the unchanged osteoblast numbers and decrease in bone formation, the number of adipocytes was markedly (10-fold) and significantly increased (90 ± 16 cells/mm2 versus 1,030 ± 100 cells/mm2; P < 0.001) within the marrow spaces of the newly formed bone in Zfp521−/− mice (Fig. 3A) relative to wild-type mice, leading to a significant increase in the adipocyte-to-osteoblast ratio (15 ± 6 to 110 ± 11; P < 0.001).
FIG 3.
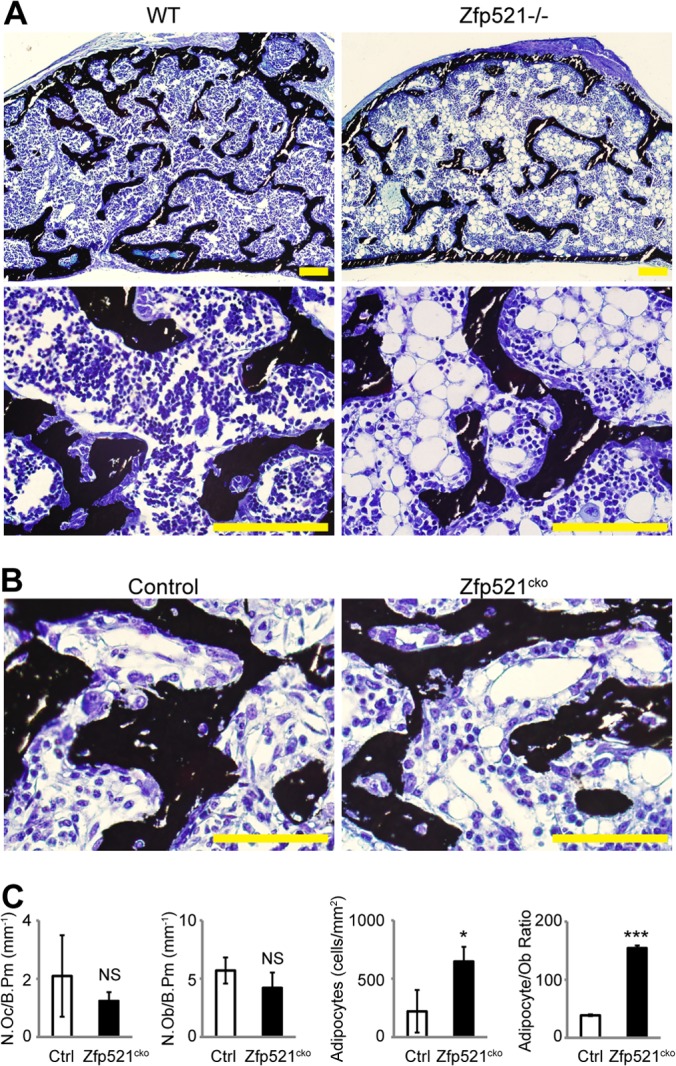
Increased adiposity in Zfp521−/− mice. (A) Toluidine blue and von Kossa staining of BMP-induced newly formed bone from WT and Zfp521−/− mice injected according to the protocol shown in Fig. 2A. (B and C) Toluidine blue and von Kossa staining (B) and histomorphometric quantification of osteoclast numbers (N. Oc) and osteoblast numbers (N. Ob) per bone perimeter (B. Pm) or adipocyte numbers (C) in BMP-induced newly formed bone from control and Zfp521cko mice. The data are presented as means ± SD; n ≥ 3. *, P < 0.05; ***, P < 0.001; NS, not significant; Student's t test. Scale bars, 500 μm (A); 270 μm (B).
Adipocytes were not observed at saline-injected sites in Zfp521−/− mouse calvaria (data not shown), indicating that adipocyte formation was BMP2 mediated and that loss of Zfp521 alone was insufficient to trigger ectopic adipocyte formation. Histomorphometry performed in the BMP2-injected cKO mice confirmed the increased adipocyte formation in the absence of Zfp521 (Fig. 3B and C). Despite the use of fluorescent labels to measure bone formation, it was not possible to measure bone formation rates due to the fact that the bone formed under these experimental conditions is mostly woven, leading to diffuse fluorescent labeling. Taken together, these data suggest that in the absence of Zfp521, mesenchymal progenitor cells preferably differentiate into adipocytes versus osteoblasts in response to BMP2. Thus, Zfp521 is an inhibitor of the adipocyte fate following BMP2 stimuli, possibly favoring their commitment to the bone-forming osteoblast lineage.
Zfp521 knockdown suppresses BMP-induced osteoblast commitment in vitro.
To explore possible mechanisms by which loss of Zfp521 results in diminished osteogenic and increased adipogenic responses to BMP2, we examined whether Zfp521 is also required for BMP-induced osteogenesis in vitro. Using W20-17 cells, we stably knocked down Zfp521 with shRNA (Fig. 4A). As expected, treatment of W20-17 cells with BMP2 induced osteoblast differentiation, as evidenced by an increase in alkaline phosphatase (Alpl, or AKP2) activity. Importantly, Zfp521 knockdown inhibited this response significantly both under basal conditions and after BMP2 stimulation (Fig. 4B and C). In response to BMP2, the early transcription factors Msx2 and Dlx5 are rapidly induced to initiate the osteogenic transcriptional cascade involving the master transcription factors Runx2 and Sp7 (Osterix). Zfp521 knockdown suppressed BMP2-induced expression of Msx2 and Dlx5 and also prevented expression of downstream markers of osteoblast differentiation, Sp7 and Alpl (Fig. 4D). Thus, Zfp521 is required for the initiation of the BMP2-stimulated osteoblast differentiation transcriptional program. To test this hypothesis, we examined the effect of Zfp521 on a BMP2-responsive reporter assay in which a luciferase gene is driven by BRE from the ID1 gene (30), a canonical BMP target. Zfp521 knockdown significantly inhibited the activation of BMP2-dependent luciferase activity (Fig. 4E), and overexpression of Zfp521 increased the activity of the BMP reporter under basal and BMP2-treated conditions (Fig. 4F). We next examined whether loss of Zfp521 altered Smad1/5 phosphorylation, a readout of BMP-induced transcription factor activation. As shown in Fig. 4G, Zfp521 knockdown in W20 cells reduced the basal level of phosphorylated Smad1/5 in untreated W20 cells. Smad1/5 phosphorylation following BMP2 treatment was reduced but not completely inhibited in Zfp521 knockdown cells. Taken together, these data demonstrate that Zfp521 is a critical component of the osteogenic response to BMP2 that favors the BMP2-mediated progression of undifferentiated cells to alkaline phosphatase-positive osteoblasts.
FIG 4.
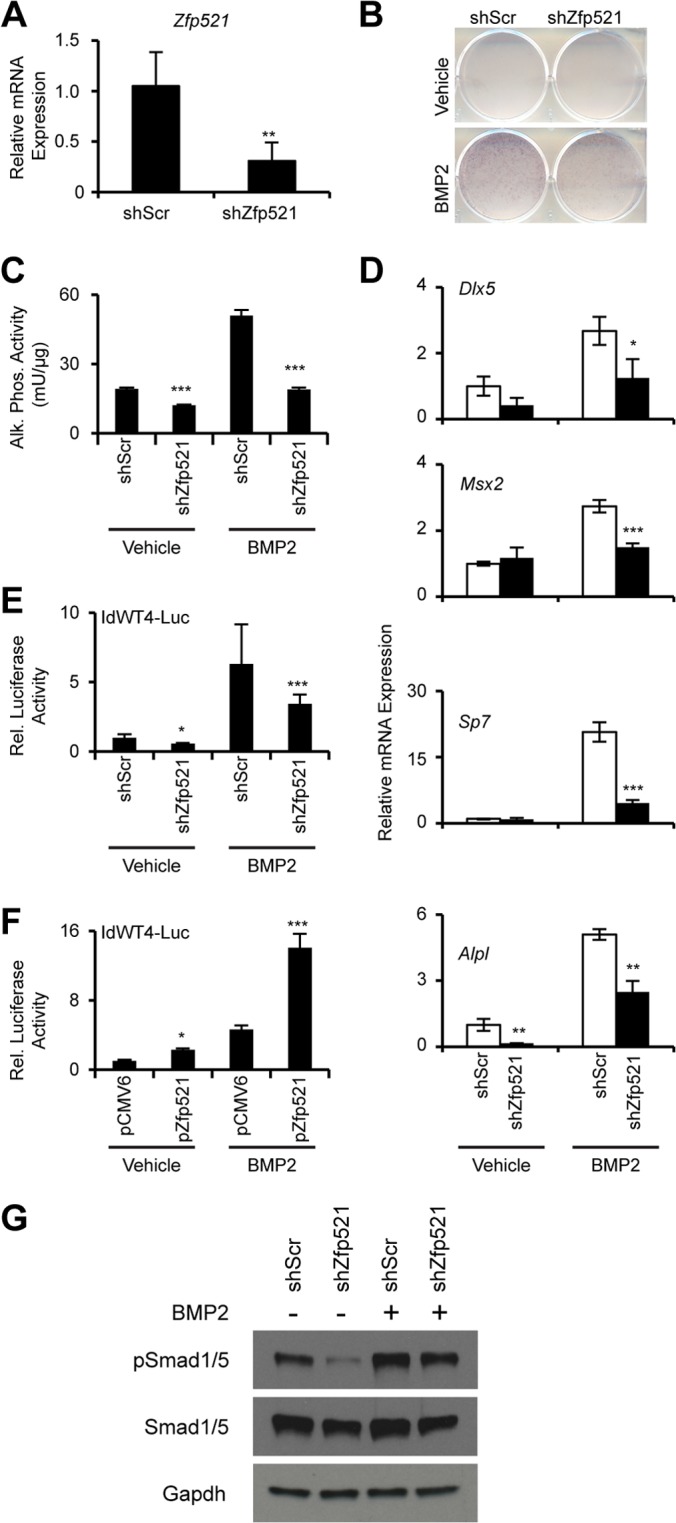
Zfp521 mediates BMP2-induced osteogenesis. (A) Confirmation of Zfp521 knockdown by qRT-PCR. (B and C) W20 cells expressing Zfp521 (shZfp521) or scrambled (shScr) shRNA were cultured with or without 100 ng/ml BMP2 for 48 h, after which alkaline-phosphatase activity was visualized by staining (B) or quantified enzymatically (C). (D) qRT-PCR of osteogenesis-related BMP target gene expression in W20 cells cultured with or without 100 ng/ml BMP2 for 24 h. (E and F) W20 cells were cotransfected with a BMP-responsive luciferase reporter together with vectors expressing shZfp521, shScr, Zfp521 cDNA (pZfp521), or empty-vector control (pCMV6) and then treated with 100 ng/ml BMP2 for 16 h. (G) shZfp521 or shScr stable W20 cells were treated with 100 ng/ml BMP2 for 1 h, after which phosphorylation of Smad1/5 was analyzed by Western blotting. The data are presented as means ± SD; n ≥ 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001 relative to shScr or pCMV6 cells within the same vehicle or BMP2 treatment group; Student's t test.
Zinc fingers 9 to 19 of Zfp521 share a high degree of homology to the well-characterized Smad-binding domain of Zfp423 (Fig. 5A). Thus, we next asked whether the Smad-interacting domain of Zfp521 is required for the increased BRE promoter activity following Zfp521 overexpression. We compared W20 cells expressing wild-type Zfp521 to cells expressing mutant deletion constructs of Zfp521 lacking the first eight zinc finger motifs (ΔZF1-8), the postulated Smad-binding domain (ΔZF9-19), or the last 28 zinc finger motifs (ΔZF9-30). We observed that full-length Zfp521 and ΔZF1-8 increased BRE promoter activity, but ΔZF9-19 and ΔZF9-30 failed to increase BRE promoter activity (Fig. 5B). Moreover, coimmunoprecipitation analysis showed that Zfp521 and ΔZF1-8 physically associated with Smad4, whereas ΔZF9-19 and ΔZF9-30 did not (Fig. 5C). These data suggest that the zinc finger motifs between 9 and 19 are required for the interaction of Zfp521 with Smad4 and the promotion of BRE promoter activity by Zfp521. To determine whether regulation of transcriptional activity is also dependent on the Smad-binding sites on the promoter, we asked whether mutation of the BRE on the promoter would affect the ability of Zfp521 to induce luciferase activity. As shown in Fig. 5D, Zfp521 failed to increase BMP-induced transcriptional activity following mutation of the BRE on the luciferase vector (IdMutB4-Luc). Finally, to determine whether Zfp521 can synergize with Smads to increase promoter activity, we transfected Zfp521, together with Smad1 and Smad4, and observed a synergistic increase in promoter activity following simultaneous transfection (Fig. 5E).
FIG 5.
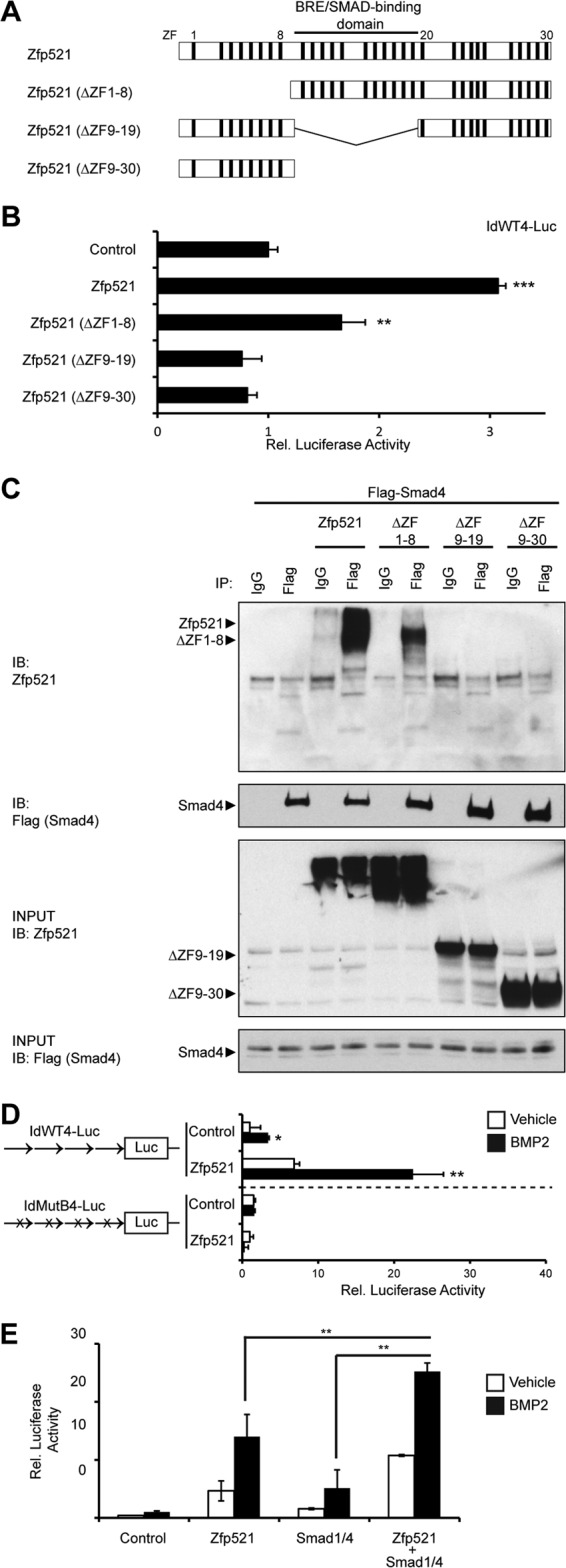
Modulation of BRE-dependent transcriptional activity by Zfp521. (A) Schematic demonstrating the zinc finger (ZF) motifs and Smad-binding and BRE binding domains of wild-type Zfp521 and Zfp521 deletion constructs. (B) BMP-responsive luciferase (IdWT4-luc) activity in W20 cells 24 h after transfection with wild-type Zfp521 or Zfp521 deletion constructs. (C) Coimmunoprecipitation analysis of Smad4 and deletion mutants of Zfp521. W20 cells were transfected with the indicated expression vectors, after which the cells were lysed and immunoprecipitated (IP) with anti-Flag antibody and blotted (IB) with anti-Zfp521 or anti-Flag antibody. Flag-Smad4 bound wild-type Zfp521 and the ΔZF1-8 mutant but did not bind ΔZF9-19 or Δ9-30. (D) Luciferase reporter activities of the BRE-dependent IDWT4-luc and a mutant reporter in which the four BMP response elements had been mutated (IDMutB-luc) to prevent Smad binding. Reporters were cotransfected into W20 cells together with Zfp521 or empty-vector control and then treated with 100 ng/ml BMP2 for 16 h. (E) Zfp521 works in concert with Smad1/4. W20 cells were transfected with IDWT4F-luc and the indicated expression vectors. Luciferase activity was measured 16 h after treatment with 100 ng/ml BMP2. The data are presented as means and SD; n ≥ 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001 relative to the empty-vector control unless otherwise indicated.
Zfp521 suppresses adipocyte commitment.
To further understand how Zfp521 deletion results in increased adipocyte formation following BMP2 injection in vivo, we examined the effect of Zfp521 loss of function on adipocyte commitment in vitro using the W20-17 cell line. shRNA knockdown of Zfp521 strongly enhanced the adipogenic differentiation of W20-17 cells when cultured in adipogenic medium, whether in the presence or absence of BMP2 (Fig. 6A). In concordance with these observations, gene expression analysis revealed enrichment of adipocyte-related genes, including Zfp423, a recently identified early determinant of preadipocyte commitment that acts upstream of Pparγ and other preadipocyte-enriched genes (17), in Zfp521 knockdown cells relative to control cells (Fig. 6B) and a concurrent decrease in osteoblast-related genes (Fig. 6C). Zfp521 is thus an intrinsic suppressor of adipocyte commitment, and loss of Zfp521 is sufficient to initiate the adipogenic transcriptional program. To determine whether this also occurred in vivo, we isolated RNA from the newly formed bone tissue of Zfp521cko calvaria after injection of BMP2 (Fig. 6D). qRT-PCR analysis of adipocyte-related genes revealed that, similar to our in vitro observations, Zfp423 was increased in Zfp521cko calvaria relative to those of wild-type mice. Later markers of adipocyte differentiation were only slightly (Pparγ1) or insignificantly (Fabp4 and Pparγ2) changed. Thus, Zfp521 appears to be a repressor of the early stages of preadipocyte commitment upstream of Zfp423 and PPARγ.
FIG 6.
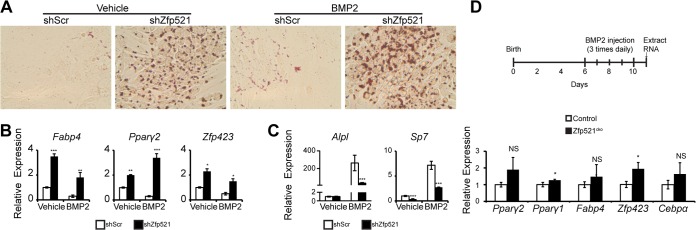
Zfp521 suppresses adipocyte commitment. (A) Oil red O staining of W20 cells expressing scrambled or Zfp521 shRNA differentiated in adipogenic medium supplemented or not with BMP2 for 6 days. (B and C) qRT-PCR of adipocyte selective genes (B) or osteogenic selective genes (C) in the differentiated cultures shown in panel A. (D) In vivo gene expression of key adipocyte commitment factors within the BMP2-induced newly formed bone of wild-type and Zfp521cko mice. Mice were injected with BMP2 as indicated, and qRT-PCR was performed on mRNA isolated directly from newly formed bone tissue 24 h after the final injection. The data are presented as means ± SD; n ≥ 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant relative to shScr within the same vehicle or BMP2 treatment group; Student's t test.
DISCUSSION
The in vivo data presented here using global and conditional Zfp521 knockout mice, combined with our in vitro observations, demonstrate that Zfp521 is a core component of the osteoinductive response to BMP2. Ablation of Zfp521 impairs BMP-induced bone formation and induces a shift of mesenchymal precursors toward bone marrow adipose tissue formation (Fig. 7). Thus, Zfp521 acts downstream of BMP as a switch to prevent adipocytic commitment and to favor the osteoblastic lineage. This study establishes further that Zfp521 is a strong repressor of adipogenesis. We show that Zfp521 exerts this strong repression by binding directly to the promoter and enhancer regions of Zfp423, a key proadipocytic transcription factor regulating the expression of PPARγ (21, 25, 33). Zfp423 is an early determinant of preadipocyte commitment and initiates the transcriptional cascades driving adipocyte differentiation. The mechanisms by which Zfp423 expression is regulated are poorly understood. Recently, Zfp423 promoter activity in fetal tissues has been linked to maternal obesity (34), and in silico analysis of transcription factor motifs identified a classical enhancer region within Zfp423 intron 5 (31). Our ChIP analysis provides the first data demonstrating that Zfp521 regulates the expression of Zfp423 by associating with chromatin and promoting chromatin modifications, such as H3K9 acetylation and H3K9me3, at the Zfp423 promoter, pointing to a role for Zfp521 in epigenetic regulation of the transcription of a subset of genes involved in cell lineage determination. Our findings therefore support the growing evidence that lineage determination of mesenchymal precursor cells is epigenetically regulated through histone modifications (35, 36). DNA methylation in the Zfp423 promoter was also observed in adipose tissue from mice subjected to an obesogenic diet (34). Additional studies will be required to further elucidate the epigenetic mechanism by which Zfp521 alters Zfp423 expression. Since Zfp521 acts upstream of Zfp423 expression, it is likely that Zfp521 expression would need to be downregulated for adipocyte differentiation to take place. Indeed, we observed that Zfp521 is highly expressed in undifferentiated cells and osteoblasts but absent in adipocytes. Since we show that Zfp521 expression is suppressed by BMP signaling (20), this suggests that during BMP-induced adipocyte differentiation, BMP downregulates Zfp521 to allow Zfp423 expression, thus enhancing PPARγ expression and adipogenesis. In another study, we showed that Zfp521 is also a strong repressor of Ebf1, which induces PPARγ through enhancement of Zfp423 expression (33) and represses bone formation (15). We show here that Zfp521 prevents the commitment of mesenchymal precursors to the adipocyte lineage and favors the osteoblast lineage, not only via the repression of Ebf1 but also through the direct epigenetic repression of Zfp423, imposing a two-pronged repressive mechanism to prevent adipocytic differentiation.
FIG 7.
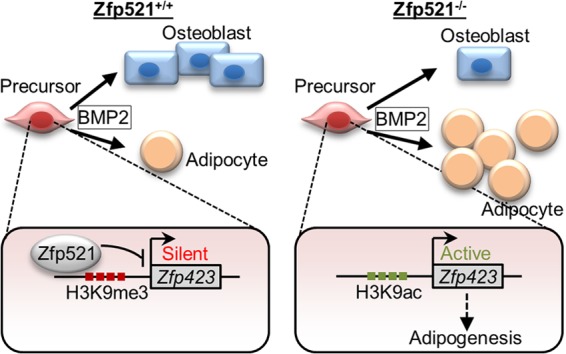
Model for the coordinated regulation of adipocyte and osteoblast commitment by Zfp521. BMP signals promote the differentiation of mesenchymal progenitor cells. In uncommitted precursors, Zfp521 acts as an intrinsic block to Zfp423 expression by maintaining the Zfp423 promoter and enhancer in a transcriptionally silent state. In the absence of Zfp521, Zfp423 expression is triggered, allowing BMP or proadipogenic stimuli to further stimulate Zfp423 expression, leading to exaggerated adipogenic lineage commitment.
Zfp521 has been shown to be particularly abundant in immature cell populations, such as neural progenitors, hematopoietic progenitors, ES cells, and mesenchymal condensations (20, 21, 37). It is generally accepted that in these populations, Zfp521 acts as a regulator of cell commitment, lineage determination, and differentiation. These effects of Zfp521 have been attributed to its ability to strongly repress the transcriptional activity of target genes, preventing the progression of cells in their differentiation or, through repression of repressors, favoring their progression in specific lineages. We have previously shown that Zfp521 is involved in osteoblast, chondrocyte, and adipocyte differentiation via its interactions with and repression of the transcription factors Ebf1 and Runx2 (20, 22, 23, 25), and we add here a key adipogenic factor, Zfp423, to the target genes that are strongly repressed by Zfp521. Intriguingly, like Zfp423, Zfp521 is a 30-zinc-finger nuclear protein, and it is highly homologous to Zfp423. The two proteins are therefore generally considered members of the same family. Our results show that although structurally highly homologous, these two 30-zinc-finger proteins fulfill exactly opposite functions in mesenchymal stem cell commitment, with Zfp423 favoring and Zfp521 repressing adipocyte differentiation. In the present study, we have extended our understanding of the role of Zfp521 in mesenchymal progenitors by showing that it is required downstream of BMP signaling to ensure BMP-induced osteogenesis through the repression it exerts on adipogenesis. It is interesting that adipocyte accumulation was specifically a BMP-induced response, since Zfp521−/− calvaria injected with saline did not display increased adiposity at the injected sites. This suggests that during the initiation of mesenchymal cell recruitment and differentiation by BMP2, the adipose lineage is intrinsically suppressed in wild-type mice. Zfp521 is thus an endogenous brake on BMP2-induced adipogenesis in vivo, perhaps explaining why BMP2 responses are predominantly osteogenic, even though BMP2 is known to induce adipogenesis in vitro.
Zfp423/OAZ was originally identified as a cofactor of BMP-activated Smads in Xenopus embryos (38). Although Zfp521 has also been implicated in BMP signaling in hematopoietic cells (37), the mechanistic details have yet to be clarified. In this study, we used Zfp521 mutant constructs to demonstrate that zinc finger motifs 9 to 19 are required for Smad4 binding and subsequent mediation of BMP-induced transcriptional activity. This raises the intriguing possibility that Zfp521 and Zfp423 target distinct or overlapping Smad target genes in a context-dependent manner.
Thus, Zfp521 acts downstream of BMP signaling to regulate the balance of osteoblasts and adipocytes. The reduced BMP-induced bone formation observed in Zfp521−/− mice is consistent with our recent observations that Zfp521 increases bone mass (20) and that its deletion leads to osteopenia (22). The “switch” effect we observe being exerted here by Zfp521 between osteoblasts and adipocytes may be specific to BMP2 responses and/or to the bone microenvironment, since decisions on the osteoblast lineage are normally restricted to the skeletal tissues. These findings may therefore apply to marrow adipose tissue and to the overall regulation of the bone-bone marrow niche and its regulation. It appears, however, that if it does not act as a switch between osteoblasts and adipocytes in other tissues, Zfp521 still conserves its biological function as a strong repressor of adipogenesis. Indeed, Zfp521−/− mouse embryos exhibit increased brown adipose and subcutaneous white adipose tissue (25), clearly indicating that Zfp521 represses adipogenesis independently of its positive role in osteogenesis.
Alltogether, we have shown that Zfp521 mediates BMP responses in vivo and in vitro and identify Zfp521 as an endogenous suppressor of adipocyte fate, both during BMP-induced osteogenesis here and under basal conditions in other tissues (15, 33). Our results suggest that this antiadipogenic function of Zfp521 is mostly exerted through repression of Zfp423 and the downstream target PPARγ gene.
ACKNOWLEDGMENTS
These studies were funded by grants to R.B. from the National Institutes of Health (AR048218 and AR57769) and by a fellowship to W.N.A. from the Canadian Institutes of Health Research.
We thank L. Neff, V. Rodriguez, K. Takeyama, H. Nistala, S. Kota, S. Kokabu, and all the members of the Baron laboratory for insightful comments over the course of this work. We are also grateful to D. Kamiya and Y. Sasai for Zfp521 deletion constructs, T. Katagiri for Id1 reporter constructs, and B. Hamilton for Zfp423 intron reporter constructs.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Verma S, Rajaratnam JH, Denton J, Hoyland JA, Byers RJ. 2002. Adipocytic proportion of bone marrow is inversely related to bone formation in osteoporosis. J. Clin. Pathol. 55:693–698. 10.1136/jcp.55.9.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan FS, Shore EM. 2000. Progressive osseous heteroplasia. J. Bone Miner. Res. 15:2084–2094. 10.1359/jbmr.2000.15.11.2084 [DOI] [PubMed] [Google Scholar]
- 3.Urist MR. 1965. Bone: formation by autoinduction. Science 150:893–899. 10.1126/science.150.3698.893 [DOI] [PubMed] [Google Scholar]
- 4.Lowery JW, Pazin D, Intini G, Kokabu S, Chappuis V, Capelo LP, Rosen V. 2011. The role of BMP2 signaling in the skeleton. Crit. Rev. Eukaryot. Gene Expr. 21:177–185. 10.1615/CritRevEukarGeneExpr.v21.i2.60 [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi A, Katagiri T, Ikeda T, Wozney JM, Rosen V, Wang EA, Kahn AJ, Suda T, Yoshiki S. 1991. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J. Cell Biol. 113:681–687. 10.1083/jcb.113.3.681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gimble JM, Morgan C, Kelly K, Wu X, Dandapani V, Wang CS, Rosen V. 1995. Bone morphogenetic proteins inhibit adipocyte differentiation by bone marrow stromal cells. J. Cell. Biochem. 58:393–402. 10.1002/jcb.240580312 [DOI] [PubMed] [Google Scholar]
- 7.Hata K, Nishimura R, Ikeda F, Yamashita K, Matsubara T, Nokubi T, Yoneda T. 2003. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol. Biol. Cell 14:545–555. 10.1091/mbc.E02-06-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Song TJ, Li X, Hu L, He Q, Liu M, Lane MD, Tang QQ. 2009. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. U. S. A. 106:12670–12675. 10.1073/pnas.0906266106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin W, Takagi T, Kanesashi SN, Kurahashi T, Nomura T, Harada J, Ishii S. 2006. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev. Cell 10:461–471. 10.1016/j.devcel.2006.02.016 [DOI] [PubMed] [Google Scholar]
- 10.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. 2008. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454:1000–1004. 10.1038/nature07221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. 1999. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126:3795–3809 [DOI] [PubMed] [Google Scholar]
- 12.Ichida F, Nishimura R, Hata K, Matsubara T, Ikeda F, Hisada K, Yatani H, Cao X, Komori T, Yamaguchi A, Yoneda T. 2004. Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation. J. Biol. Chem. 279:34015–34022. 10.1074/jbc.M403621200 [DOI] [PubMed] [Google Scholar]
- 13.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754. 10.1016/S0092-8674(00)80257-3 [DOI] [PubMed] [Google Scholar]
- 14.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17–29. 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- 15.Lee MH, Kim YJ, Kim HJ, Park HD, Kang AR, Kyung HM, Sung JH, Wozney JM, Ryoo HM. 2003. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 278:34387–34394. 10.1074/jbc.M211386200 [DOI] [PubMed] [Google Scholar]
- 16.Ryoo HM, Lee MH, Kim YJ. 2006. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene 366:51–57. 10.1016/j.gene.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 17.Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. 2010. Transcriptional control of preadipocyte determination by Zfp423. Nature 464:619–623. 10.1038/nature08816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, Spiegelman BM. 2012. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 15:230–239. 10.1016/j.cmet.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bond HM, Mesuraca M, Amodio N, Mega T, Agosti V, Fanello D, Pelaggi D, Bullinger L, Grieco M, Moore MA, Venuta S, Morrone G. 2008. Early hematopoietic zinc finger protein-zinc finger protein 521: a candidate regulator of diverse immature cells. Int. J. Biochem. Cell Biol. 40:848–854. 10.1016/j.biocel.2007.04.006 [DOI] [PubMed] [Google Scholar]
- 20.Wu M, Hesse E, Morvan F, Zhang JP, Correa D, Rowe GC, Kiviranta R, Neff L, Philbrick WM, Horne WC, Baron R. 2009. Zfp521 antagonizes Runx2, delays osteoblast differentiation in vitro, and promotes bone formation in vivo. Bone 44:528–536. 10.1016/j.bone.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamiya D, Banno S, Sasai N, Ohgushi M, Inomata H, Watanabe K, Kawada M, Yakura R, Kiyonari H, Nakao K, Jakt LM, Nishikawa S, Sasai Y. 2011. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470:503–509. 10.1038/nature09726 [DOI] [PubMed] [Google Scholar]
- 22.Kiviranta R, Yamana K, Saito H, Ho DK, Laine J, Tarkkonen K, Nieminen-Pihala V, Hesse E, Correa D, Maatta J, Tessarollo L, Rosen ED, Horne WC, Jenkins NA, Copeland NG, Warming S, Baron R. 2013. Coordinated transcriptional regulation of bone homeostasis by Ebf1 and Zfp521 in both mesenchymal and hematopoietic lineages. J. Exp. Med. 210:969–985. 10.1084/jem.20121187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Correa D, Hesse E, Seriwatanachai D, Kiviranta R, Saito H, Yamana K, Neff L, Atfi A, Coillard L, Sitara D, Maeda Y, Warming S, Jenkins NA, Copeland NG, Horne WC, Lanske B, Baron R. 2010. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev. Cell 19:533–546. 10.1016/j.devcel.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesse E, Saito H, Kiviranta R, Correa D, Yamana K, Neff L, Toben D, Duda G, Atfi A, Geoffroy V, Horne WC, Baron R. 2010. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J. Cell Biol. 191:1271–1283. 10.1083/jcb.201009107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang S, Akerblad P, Kiviranta R, Gupta RK, Kajimura S, Griffin MJ, Min J, Baron R, Rosen ED. 2012. Regulation of early adipose commitment by Zfp521. PLoS Biol. 10:e1001433. 10.1371/journal.pbio.1001433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. 2003. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130:3063–3074. 10.1242/dev.00491 [DOI] [PubMed] [Google Scholar]
- 27.Mundy G, Garrett R, Harris S, Chan J, Chen D, Rossini G, Boyce B, Zhao M, Gutierrez G. 1999. Stimulation of bone formation in vitro and in rodents by statins. Science 286:1946–1949. 10.1126/science.286.5446.1946 [DOI] [PubMed] [Google Scholar]
- 28.Thies RS, Bauduy M, Ashton BA, Kurtzberg L, Wozney JM, Rosen V. 1992. Recombinant human bone morphogenetic protein-2 induces osteoblastic differentiation in W-20-17 stromal cells. Endocrinology 130:1318–1324 [DOI] [PubMed] [Google Scholar]
- 29.Deng W-G, Zhu Y, Montero A, Wu KK. 2003. Quantitative analysis of binding of transcription factor complex to biotinylated DNA probe by a streptavidin-agarose pulldown assay. Anal. Biochem. 323:12–18. 10.1016/j.ab.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 30.Katagiri T, Imada M, Yanai T, Suda T, Takahashi N, Kamijo R. 2002. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells 7:949–960. 10.1046/j.1365-2443.2002.00573.x [DOI] [PubMed] [Google Scholar]
- 31.Cho YW, Hong CJ, Hou A, Gent PM, Zhang K, Won KJ, Hamilton BA. 2013. Zfp423 binds autoregulatory sites in p19 cell culture model. PLoS One 8:e66514. 10.1371/journal.pone.0066514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatakos G, Rowe GC, Kveiborg M, Wu M, Neff L, Chiusaroli R, Philbrick WM, Baron R. 2008. Doubly truncated FosB isoform (Δ2ΔFosB) induces osteosclerosis in transgenic mice and modulates expression and phosphorylation of Smads in osteoblasts independent of intrinsic AP-1 activity. J. Bone Miner. Res. 23:584–595. 10.1359/jbmr.080110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 34.Yang Q-Y, Liang J-F, Rogers CJ, Zhao J-X, Zhu M-J, Du M. 2013. Maternal obesity induces epigenetic modifications to facilitate Zfp423 expression and enhance adipogenic differentiation in fetal mice. Diabetes 62:3727–3735. 10.2337/db13-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, Ge K. 2013. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. EMBO J. 32:45–59. 10.1038/emboj.2012.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park NH, Wang CY. 2012. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11:50–61. 10.1016/j.stem.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bond HM, Mesuraca M, Carbone E, Bonelli P, Agosti V, Amodio N, De Rosa G, Di Nicola M, Gianni AM, Moore MA, Hata A, Grieco M, Morrone G, Venuta S. 2004. Early hematopoietic zinc finger protein (EHZF), the human homolog to mouse Evi3, is highly expressed in primitive human hematopoietic cells. Blood 103:2062–2070. 10.1182/blood-2003-07-2388 [DOI] [PubMed] [Google Scholar]
- 38.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. 2000. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100:229–240. 10.1016/S0092-8674(00)81561-5 [DOI] [PubMed] [Google Scholar]


