Abstract
Unlike other Rho GTPases, RhoB is rapidly induced by DNA damage, and its expression level decreases during cancer progression. Because inefficient repair of DNA double-strand breaks (DSBs) can lead to cancer, we investigated whether camptothecin, an anticancer drug that produces DSBs, induces RhoB expression and examined its role in the camptothecin-induced DNA damage response. We show that in camptothecin-treated cells, DSBs induce RhoB expression by a mechanism that depends notably on Chk2 and its substrate HuR, which binds to RhoB mRNA and protects it against degradation. RhoB-deficient cells fail to dephosphorylate γH2AX following camptothecin removal and show reduced efficiency of DSB repair by homologous recombination. These cells also show decreased activity of protein phosphatase 2A (PP2A), a phosphatase for γH2AX and other DNA damage and repair proteins. Thus, we propose that DSBs activate a Chk2-HuR-RhoB pathway that promotes PP2A-mediated dephosphorylation of γH2AX and DSB repair. Finally, we show that RhoB-deficient cells accumulate endogenous γH2AX and chromosomal abnormalities, suggesting that RhoB loss increases DSB-mediated genomic instability and tumor progression.
INTRODUCTION
RhoB is a small GTPase from the Rho family of proteins implicated in various intracellular functions, including actin cytoskeletal organization (1). Besides its well-established roles, RhoB emerged as an early DNA damage-inducible gene. RhoB is readily induced in response to various genotoxic agents, including UV and cisplatin (2, 3), although the molecular mechanisms of induction and functional relevance remain unclear. RhoB also differs from other Rho proteins, as it possesses tumor suppressor functions. The RhoB expression level decreases during the progression of various tumors, and loss of RhoB promotes cell proliferation, invasion, and metastasis (4–8).
DNA double-strand breaks (DSBs) are among the most severe lesions, and their inefficient repair can initiate genomic instability, ultimately leading to cancer (9–11). DSB repair requires the recruitment of DNA damage response (DDR) proteins in the vicinity of damaged chromatin (12). The serine/threonine kinases ATM, ATR, and DNA-dependent protein kinase (DNA-PK) are readily activated by DSBs and phosphorylate various DDR proteins, including histone H2AX and checkpoint kinase 2 (Chk2). Phosphorylation of these proteins is critical for efficient DDR and repair (10, 13). These phosphorylations are reversible and removed by specific serine/threonine phosphatases, including protein phosphatase 2A (PP2A), PP4, PP1, PP6, and Wip1 (14). Accumulating studies indicate that the timely dephosphorylation of DDR proteins is required for DSB repair (15–17).
Topoisomerase I (Top1) removes DNA torsional stress generated during replication and transcription. It relaxes DNA by producing transient Top1-DNA cleavage complexes (Top1cc), which are Top1-linked DNA single-strand breaks (18). The rapid resealing of Top1cc is inhibited by camptothecin (CPT) and its derivatives, which are used to treat cancers and which bind selectively at the Top1-DNA interface (18). Stabilized Top1cc interfere with the progression of replication and transcription complexes, which results in the production of DSBs (19–21). CPT is a sharp tool to dissect the cellular response to DSBs, as it has no other target besides Top1. CPT also has the advantage of trapping Top1cc reversibly. Indeed, Top1cc reverse fully within minutes after washing out CPT (18). Here we used CPT to determine whether DSBs induce RhoB and examined both the mechanisms of induction and its functional relevance.
MATERIALS AND METHODS
Drugs, chemical reagents, and cell culture.
CPT, okadaic acid, fostriecin, and the DNA-PK inhibitor NU7026 were obtained from Sigma-Aldrich. Human osteosarcoma (U2OS) and colon carcinoma (HCT116 and HCT15) cells were obtained from the American Type Culture Collection (ATCC). HCT15 cells stably expressing wild-type Chk2 (Chk2-WT) or a kinase-dead Chk2 D347A mutant (Chk2-KD) were obtained from Yves Pommier (NIH, Bethesda, MD) (22, 23). WT and RhoB−/− E6-immortalized mouse embryonic fibroblast (MEF) cells were established in the laboratory from SV129 mice obtained from G. C. Prendergast (Lankenau Institute for Medical Research) by using a protocol described previously (24). WT and RhoB−/− primary mouse dermal fibroblast (MDF) cells were isolated from SKH1 mice (established in the laboratory from SV129 mice), as described previously (25), and cultured for a maximum of 9 passages. The subline RG37, containing the homologous recombination substrate (pDR-GFP), was made as described previously (26). The subline GC92, containing the nonhomologous end joining (NHEJ) substrate (pCOH-CD4), was made as described previously (27). All of the above-described cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Western blotting.
Whole-cell extracts were obtained by lysing cells in buffer (1% SDS, 10 mM Tris-HCl [pH 7.4]) supplemented with protease (Complete; Roche Diagnostics) and phosphatase (Cocktail 3; Sigma-Aldrich) inhibitors. Viscosity of the samples was reduced by brief sonication, and proteins were separated by SDS-PAGE and immunoblotted with the following antibodies: anti-Chk2 (catalog number 2662; Cell Signaling), anti-Chk2-pS516 (catalog number 2669; Cell Signaling), anti-Chk2-pT68 (catalog number 2661; Cell Signaling), anti-H2AX (catalog number ab11175; Abcam), anti-γH2AX (catalog number 05-636; Millipore), anti-HuR (catalog number sc-5261; Santa Cruz), anti-PP2A(C) (catalog number 1512-1; Epitomics), anti-Rad51 (catalog number PC-130; Millipore), anti-RhoA (catalog number sc-418; Santa Cruz), anti-RhoB (catalog number sc-180; Santa Cruz), and anti-α-tubulin (catalog number T5168; Sigma-Aldrich). Immunoblotting was revealed by chemiluminescence using autoradiography or a ChemiDoc MP system (Bio-Rad). Quantification of protein levels was done by using ImageJ (version 1.40g) in Fig. 1E and 3F and with Image Lab software (version 4.1) in Fig. 3L and 5D.
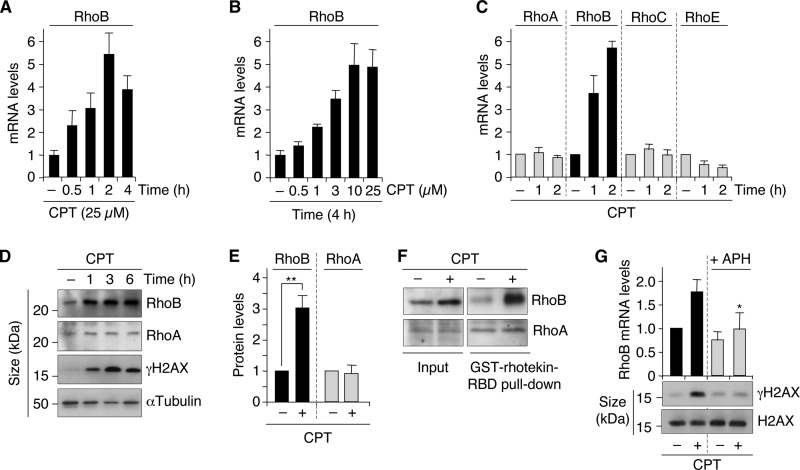
FIG 1.
Rapid and selective increase of RhoB by CPT-induced DSBs. (A and B) RhoB mRNA was analyzed by RT-qPCR in cells treated for the indicated times with 25 μM CPT (U2OS cells) (A) and with the indicated CPT concentrations for 4 h (HCT15/Chk2-WT cells) (B). Data shown are means ± SD for triplicate samples. (C) The indicated transcripts were analyzed by RT-qPCR in U2OS cells treated with 25 μM CPT (means ± SD for three independent experiments). (D) Western blotting of RhoB, RhoA, and γH2AX in U2OS cells treated with 25 μM CPT. α-Tubulin was the loading control. (E) Quantification of RhoB and RhoA protein levels by Western blotting in U2OS cells treated with CPT (25 μM for 4.5 h). Data shown are the means ± SD for three independent experiments. **, P < 0.01 by t test. (F) Active (GTP-bound) Rho proteins were pulled down with GST-rhotekin-RBD (Rho binding domain) beads in extracts from U2OS cells treated with 25 μM CPT for 4 h. The active forms of RhoB and RhoA were detected by Western blotting with antibodies against RhoB and RhoA, respectively. Total RhoB and RhoA protein levels were examined in the extracts before pulldown (input). (G) HCT15/Chk2-WT cells were treated with the replication inhibitor aphidicolin (APH) (1 μM for 15 min) before the addition of CPT (1 μM for 2 h). (Top) RT-qPCR analysis of RhoB mRNA. Data shown are means ± SD for three independent experiments. The asterisk denotes a significant difference from CPT-treated cells without APH (P < 0.05 by t test). (Bottom) Western blotting of γH2AX and H2AX.
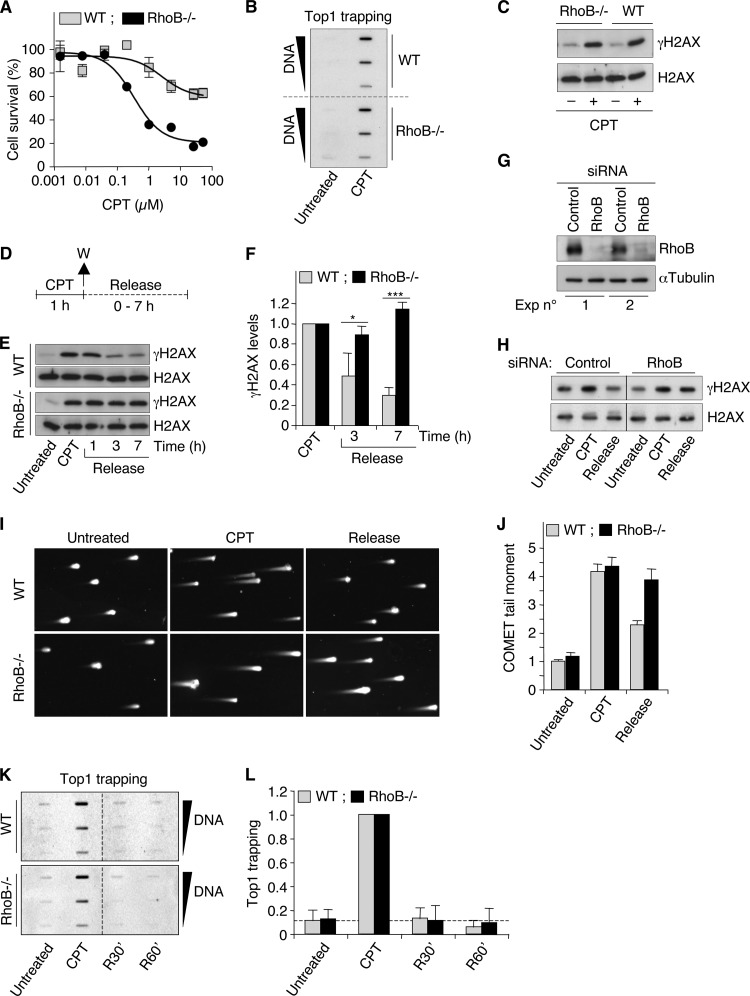
FIG 3.
RhoB-deficient cells are defective for γH2AX dephosphorylation after CPT removal. (A) WT and RhoB−/− MEF cells were treated with the indicated concentrations of CPT for 72 h, and cell survival was analyzed by a WST-1 assay (means ± SD for triplicate samples). (B) Detection of Top1-DNA cleavage complexes (Top1cc) in WT and RhoB−/− primary mouse dermal fibroblast (MDF) cells treated with CPT (25 μM for 1 h). Different amounts of genomic DNA (5, 2.5, and 1.25 μg) were probed with an anti-Top1 antibody. (C) Western blotting of γH2AX and H2AX in WT and RhoB−/− primary MDF cells treated with CPT (25 μM for 1 h). (D) Cell treatment protocol for the study of γH2AX dephosphorylation (E to H), DSB repair (I and J), and Top1cc reversal (K and L) in response to CPT. Cells were treated with CPT for 1 h and washed (W) and cultured in CPT-free medium (release) for the indicated times. (E and F) Western blotting of γH2AX and H2AX in WT and RhoB−/− primary MDF cells (CPT, 25 μM). (E) Representative experiment. (F) Quantification of γH2AX protein levels (means ± SD for three independent experiments). *, P < 0.05; ***, P < 0.001 (by t test). (G and H) HCT116 cells were transfected with RhoB-targeting or nontargeting (control) siRNAs. (G) Western blotting showing the efficiency of RhoB silencing in two independent experiments. α-Tubulin was the loading control. (H) Western blotting of γH2AX and H2AX (CPT, 1 μM; release, 6 h). Lines indicate that intervening wells have been spliced out. (I and J) Detection of DSBs by a neutral Comet assay in WT and RhoB−/− primary MDF cells (CPT, 25 μM; release, 1 h). (I) Representative pictures of nuclei. (J) Quantification of Comet tail moment (averages ± standard errors of the means). Sixty cells were examined per group. (K and L) Detection of Top1cc in MEF cells treated with 25 μM CPT for 1 h. R30′ and R60′ indicate cells harvested 30 and 60 min after CPT removal, respectively. Different amounts of genomic DNA (5, 2.5, and 1.25 μg) were probed with an anti-Top1 antibody. (K) Representative experiment. Dashed lines indicate where panels have been reorganized to facilitate reading. (L) Quantification of Top1cc normalized to values for CPT-treated cells (means ± SD for three independent experiments). The dashed line indicates Top1cc levels in WT untreated cells.
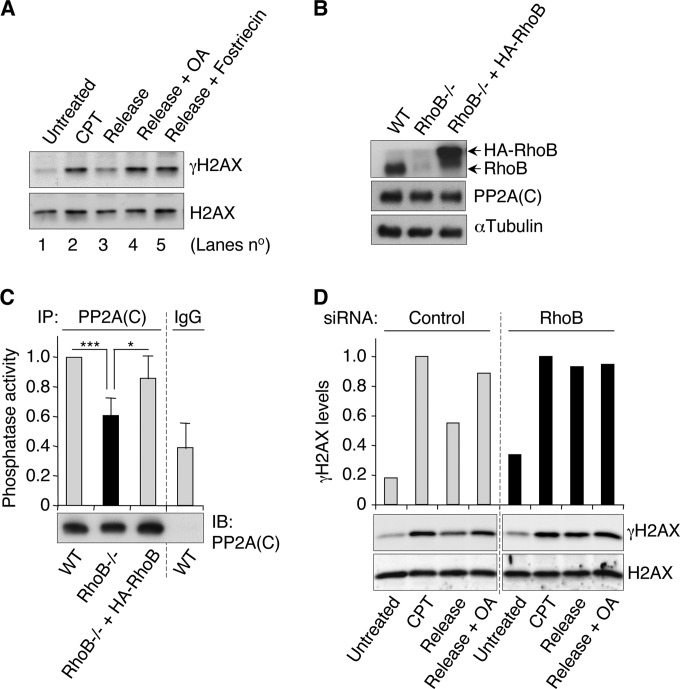
FIG 5.
RhoB-deficient cells are defective for PP2A activity. (A) Western blotting of γH2AX and H2AX in HCT15/Chk2-WT cells treated with 1 μM CPT for 1 h and washed and cultured in CPT-free medium for 5 h (release). Okadaic acid (OA) (100 nM) (lane 4) and fostriecin (100 nM) (lane 5) were added immediately after CPT removal (washes). See also the treatment protocol described in the legend of Fig. 3D. (B and C) Primary MDF cells of each genotype were analyzed for RhoB and PP2A(C) expression by Western blotting (B) and for PP2A activity after PP2A(C) immunoprecipitation (IP) using the Thr phosphopeptide K-R-pT-I-R-R as a substrate (C). Data shown are means ± SD for three independent experiments. *, P < 0.05; ***, P < 0.001 (by t test). IB, immunoblotting. (D) HCT15/Chk2-WT cells were transfected with RhoB-targeting or nontargeting (control) siRNAs before treatment, as described above for panel A. Western blotting of γH2AX and H2AX. The top panel shows quantification of γH2AX expression normalized to the expression level of H2AX, shown at the bottom.
RNA immunoprecipitation (RIP).
Immunoprecipitation of HuR-associated RNAs was done as described previously (28), with minor modifications. Forty million cells were lysed for 30 min at 4°C in 750 μl buffer containing 25 mM Tris-HCl (pH 7.4), 150 mM KCl, 0.5% NP-40, 2 mM EDTA, 1 mM NaF, and 0.5 mM dithiothreitol (DTT), supplemented with 0.2 U RNasin (Promega) and protease inhibitors (Complete; Roche Diagnostics). After centrifugation at 10,000 × g for 10 min, supernatants were precleared for 30 min at 4°C with 20 μl of protein A/G-agarose beads (Sigma-Aldrich) previously blocked for 5 h at 4°C in washing buffer (300 mM KCl, 50 mM Tris-HCl [pH 7.4], 1 mM MgCl2, 0.1% NP-40) containing 5 μg/μl yeast tRNA (Invitrogen), 1 μg/μl acetylated bovine serum albumin (BSA; Sigma-Aldrich), and protease inhibitors (Complete; Roche Diagnostics). Beads (20 μl) were coupled with 15 μg of mouse anti-HuR antibody (catalog number sc-5261; Santa Cruz) or 15 μg of mouse nonimmune antibody (control IgG) (catalog number 02-6502; Invitrogen) for 4 h at 4°C and incubated with 2 mg precleared cell lysate overnight at 4°C. After several washes in washing buffer and proteinase K (Roche Diagnostics) treatment, immunoprecipitated RNAs were extracted by using TRIzol LS reagent (Invitrogen) and treated with Turbo DNase (Ambion) before quantitative reverse transcription-PCR (RT-qPCR) experiments.
Quantitative reverse transcription-PCR.
Total RNAs (MasterPure RNA purification kit; Epicentre) were subjected to RT by using the iScript cDNA synthesis kit (Bio-Rad). qPCR analyses were performed on a CFX96 real-time system device (Bio-Rad) by using IQ SYBR green Supermix (Bio-Rad) according to the manufacturer's instructions. All samples were analyzed in triplicate, and β-actin mRNA was used as an endogenous control in the ΔΔCT analysis. The primer pairs used were RhoA-FW (5′-TGG AAG ATG GCA TAA CCT GTC-3′) and RhoA-RV (5′-AAC TGG TGG CTC CTC TGG-3′), RhoB-FW (5′-TTG TGC CTG TCC TAG AAG TG-3′) and RhoB-RV (5′-CAA GTG TGG TCA GAA TGC TAC-3′), RhoC-FW (5′-TGT CAT CCT CAT GTG CTT CTC-3′) and RhoC-RV (5′-GTG CTC GTC TTG CCT CAG-3′), RhoE-FW (5′-CCT GCT CCT CTC GCT CTC-3′) and RhoE-RV (5′-TCT GGC TGG CTC TTC TCT C-3′), and β-actin-FW (5′-TCC CTG GAG AGG AGC TAC GA-3′) and β-actin-RV (5′-AGG AAG GAA GGC TGG AAG AG-3′).
Cell transfection (siRNAs and plasmids).
For cell transfection with small interfering RNAs (siRNAs), cells were transfected with HuR-, Rad51-, or RhoB-targeting siRNAs or nontargeting siRNAs (Eurogentec) by using Oligofectamine transfection reagent (Invitrogen) according to the manufacturer's protocol. Target DNA sequences were 5′-GAG GCA ATT ACC AGT TTC A-3′ for HuR, 5′-GAA GCT ATG TTC GCC ATT A-3′ for Rad51, 5′-GGC ATT CTC TAA AGC TAT G-3′ for siRNA RhoB#1, 5′-GTC CAA GAA ACT GAT GTT A-3′ for siRNA RhoB#2, 5′-GCT AAG ATG GTG TTA TTT A-3′ for siRNA RhoB#3, and 5′-GAC GTG GGA CTG AAG GGG T-3′ for nontargeting siRNA. Experiments were performed 48 h after transfection. siRNA RhoB#1 was used in Fig. 4B and G, siRNA RhoB#2 was used in Fig. 4B to E and G and 5D, and siRNA RhoB#3 was used in Fig. 3G and H. For cell transfection with plasmids, cells were transfected with a plasmid encoding hemagglutinin (HA)-RhoB (29) by using jetPEI DNA transfection reagent (Polyplus transfection) according to the manufacturer's protocol. Experiments were performed 24 h after transfection.
FIG 4.
RhoB-deficient cells are defective for DSB repair by homologous recombination. (A) Substrate and strategy used to measure DSB-induced homologous recombination (26, 48). The pDR-GFP substrate contains two inactive genes coding for GFP under the control of a promoter (P). The 5′ gene is inactive because of the insertion of a cleavage site for I-SceI. The 3′ gene is inactive because it is deleted in both the 5′ and 3′ directions. When a DSB is produced by I-SceI, recombination between these two inactive genes (specifically gene conversion) restores a functional GFP coding sequence by either intrachromatid homologous recombination (represented) or unequal sister chromatid exchange (not represented). (B) RG37 cells stably expressing the pDR-GFP substrate were transfected with RhoB-targeting or nontargeting (control) siRNAs for 2 days and then transfected with an I-SceI plasmid for an additional 3 days. Cells transfected with Rad51-targeting siRNAs were used as a control for the pDR-GFP substrate. “No I-SceI” corresponds to cells transfected with an empty plasmid for 3 days. (Top) Percentages of GFP-positive recombinant cells determined by flow cytometry and normalized to the level of cells cotransfected with control siRNAs and I-SceI, which was set to a value of 1. Data shown are means ± standard errors of the means for five (with RhoB#1 siRNA) or three (with RhoB#2 and Rad51 siRNAs) independent experiments. ***, P < 0.001; **, P < 0.01 (by t test). (Bottom) Western blotting of RhoB and Rad51. α-Tubulin was the loading control. (C and D) HCT15/Chk2-WT cells were transfected with RhoB-targeting or nontargeting (control) siRNAs and left untreated or were treated with 1 μM CPT for 1 h. Rad51 foci were analyzed by immunofluorescence microscopy at 6 h post-CPT treatment (Release) (see protocol described in the legend to Fig. 3D). (C) Representative images. DAPI, 4′,6-diamidino-2-phenylindole. Bar, 10 μm. (D) Percentages of cells with at least five Rad51 foci. At least 200 cells were analyzed in each group (means ± standard errors of the means). (E) HCT15/Chk2-WT cells were transfected with RhoB-targeting or nontargeting (control) siRNAs, labeled with 30 μM BrdU for 30 min, and analyzed by flow cytometry. Numbers indicate percentages of BrdU-positive cells (means ± SD for three experiments). Unlabeled cells were used as negative controls for anti-BrdU staining. PI, propidium iodide. (F) Substrate and strategy used to measure DSB-induced end joining (49). The pCOH-CD4 substrate contains genes coding for the membrane antigens H2Kd, CD8, and CD4. The only expressed gene is H2Kd. CD8 is not expressed because it is in an inverted orientation, and CD4 is not expressed because it is too far from the promoter (P). Two cleavage sites for I-SceI are present in noncoding sequences, which are in direct orientation generating cohesive ends between the two sites. When two DSBs are produced by I-SceI, the internal fragment H2Kd/CD8 is excised, and rejoining of the DNA ends leads to the expression of the CD4 gene. (G) GC92 cells stably expressing the pCOH-CD4 substrate were transfected as described above for panel B, and percentages of CD4-positive cells were determined by flow cytometry using an anti-CD4 antibody. Nontargeting-siRNA-transfected cells treated with the DNA-PK inhibitor NU7026 (DNA-PKi) (10 μM) at the time of transfection with the I-SceI plasmid were used as a control for the pCOH-CD4 substrate. Data shown are means ± standard errors of the means for seven (with siRNA RhoB#1) or three (with siRNA RhoB#2 and the siRNA control plus the DNA-PK inhibitor) independent experiments. ns, nonsignificant (P = 0.15 [with siRNA RhoB#1] and P = 0.14 [with siRNA RhoB#2]); ***, P < 0.001 (by t test).
Neutral Comet assays.
Neutral Comet assays were performed according to the manufacturer's instructions (Trevigen), except that electrophoresis was performed at 4°C. Comet tail moments were measured by using ImageJ (version 1.47v) using a macro provided by Robert Bagnell (https://www.med.unc.edu/microscopy/resources/imagej-plugins-and-macros/comet-assay).
DSB repair assays.
RG37 and GC92 cells, for homologous recombination and end joining assays, respectively, were plated at 5 × 105 cells per well into six-well plates and transfected after 24 h with nontargeting or RhoB-targeting siRNAs. Forty-eight hours after siRNA transfection, cells were transfected with 1 μg of an I-SceI-encoding plasmid (pBASCe-ISceI) by using JetPEI reagent. Cells were collected 72 h later with phosphate-buffered saline (PBS)–50 mM EDTA and fixed in PBS–2% paraformaldehyde for 15 min at room temperature. Green fluorescent protein (GFP)-positive RG37 cells were detected by flow cytometry using a FACSCalibur instrument. For NHEJ assays, GC92 cells were incubated with PBS and 2% (wt/vol) BSA and then stained for 15 min with 1 μl of anti-CD4-phycoerythrin (PE) (Miltenyi Biotech) in PBS–1% (wt/vol) BSA. The cells were washed in PBS before fluorescence-activated cell sorter (FACS) analysis.
WST-1 cell viability assays.
WT and RhoB−/− MEF cells were seeded in triplicate into 96-well microplates at a density of 1,000 cells per well. Twenty-four hours after plating, cells were treated with various concentrations of CPT (from 1.6 nM to 50 μM) and cultured for 72 h. The WST-1 reagent (Roche Diagnostics) was then applied for 1 h at 37°C. The formazan dye was quantified at 450 nm by using a plate reader (LabSystems Multiskan). Data are expressed as the percentage of cell survival (mean ± standard deviation [SD] of treated cells normalized to the mean ± SD of untreated cells, which was set to 100%).
Protein phosphatase 2A activity assays.
Protein phosphatase 2A (PP2A) activity was assayed by using a PP2A immunoprecipitation phosphatase assay kit (Upstate) according to the manufacturer's protocol. Briefly, cells were lysed on ice in phosphatase extraction buffer (20 mM imidazole-HCl [pH 7.0], 2 mM EDTA, 2 mM EGTA) supplemented with a protease inhibitor cocktail (Sigma-Aldrich). The cell lysate (500 μg) was incubated for 2 h at 4°C with protein A-agarose beads coupled with 4 μg of mouse anti-PP2A(C) antibody (clone 1D6; Upstate) or mouse nonimmune antibody (control IgG) (catalog number 02-6502; Invitrogen). Phosphatase activity was assayed by incubating the immunoprecipitated proteins with the synthetic threonine phosphopeptide K-R-pT-I-R-R at 30°C for 10 min prior to detection with malachite green phosphate detection solution. After 15 min, free phosphate was quantified by measuring the absorbance at 620 nm in a microplate reader. Phosphatase activity was calculated by using a phosphate standard curve. All samples were analyzed in triplicate.
Detection of Top1-DNA cleavage complexes.
Cellular Top1-DNA cleavage complexes (Top1cc) were detected as previously described (30), except that immunoblotting was revealed with a rabbit monoclonal anti-Top1 antibody from Abcam (ab109374) and by chemiluminescence using autoradiography or a ChemiDoc MP system (Bio-Rad). Quantification for Fig. 3L was performed by using Image Lab software (version 4.1).
Immunofluorescence microscopy.
Immunofluorescence microscopy of γH2AX was performed, as described previously (20), with an anti-γH2AX antibody from Millipore (catalog number 05-636). Immunofluorescence microscopy of Rad51 was performed as described previously (31) by using an anti-Rad51 antibody from Millipore (catalog number PC-130), except that cells were fixed/permeabilized with ice-cold methanol for 15 min at 4°C and further permeabilized with 0.5% Triton X-100 for 5 min at room temperature. Slides were visualized by using a fluorescence microscope (Eclipse 90i; Nikon), and pictures were analyzed with Photoshop CS3 (Adobe).
DNA extraction and comparative genomic hybridization (CGH) arrays.
Genomic DNA was extracted from WT and RhoB−/− primary MDF cells by using the QIAamp DNA kit (Qiagen). DNA from WT and RhoB−/− cells was labeled with Cy3 and Cy5 (Dual color labeling kit; Roche-Nimblegen), respectively, before hybridization on 720K whole-genome tiling arrays (Roche-Nimblegen). Slides were scanned by using the MS200 scanner (Tecan), and images were analyzed by using DEVA 2.1 software (Roche-Nimblegen) with segmentation and background correction. Amplification and deletion were considered significant events when at least 5 consecutive probes had a log2 value of ≥0.3 (amplification) or a log2 value of ≤−0.4 (deletion). We determined the genomic instability index with the formula (number of deletions + number of amplifications)2/number of altered chromosomes, as previously described (32). Sexual chromosomes were excluded from the analysis.
RhoB and RhoA activity assays.
The Rho binding domain (RBD) of rhotekin, an effector of Rho proteins that selectively binds to the GTP-loaded form, was expressed as a recombinant fusion with glutathione S-transferase (GST) in Escherichia coli and purified through binding to glutathione (GSH)-Sepharose beads. Cells (8 × 106) were lysed on ice in 800 μl lysis buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 10 mM DTT) supplemented with a protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitors (Halt phosphatase inhibitor cocktail; Thermo Scientific). GST-RBD beads (30 μl) were incubated with the cell lysate for 30 min at 4°C. An aliquot from each lysate was removed as a control for equivalent input for the assay. After three washes in ice-cold washing buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10 mM MgCl2, 1% Triton X-100), bound Rho proteins were eluted from the beads with SDS-PAGE sample buffer at 95°C and analyzed by Western blotting with anti-RhoB (catalog number sc-180; Santa Cruz) or anti-RhoA (catalog number sc-418; Santa Cruz) antibodies.
RhoB promoter activity.
U2OS cells were cotransfected with a plasmid encoding the RhoB promoter linked to the firefly luciferase reporter gene and a plasmid encoding the cytomegalovirus (CMV) promoter linked to the Renilla luciferase reporter gene (internal control), as described previously (2). Luciferase activities were measured 48 h after transfection by using the Dual Luciferase assay system (Promega), and results were expressed as the ratio of the activity of the firefly luciferase to the activity of the Renilla luciferase.
BrdU incorporation assays.
Cells were incubated with 30 μM bromodeoxyuridine (BrdU; Sigma-Aldrich) for 30 min and labeled with anti-BrdU antibody according to the manufacturer's protocol (clone B44; BD Biosciences). Cells were analyzed on a Becton, Dickinson FACScan flow cytometer (BD Biosciences).
RESULTS
RhoB is rapidly and selectively induced in response to DSBs.
Exposure of human cancer cells to 25 μM CPT revealed a 2-fold increase of the RhoB mRNA level within 30 min (Fig. 1A). At this CPT concentration, the levels of RhoB mRNA reached a maximum 5-fold increase after 2 h (Fig. 1A). To investigate whether the induction of RhoB mRNA was dose dependent, cells were treated for 4 h with increasing CPT concentrations. RhoB induction was clearly detected at 1 μM and increased with increasing CPT concentrations (Fig. 1B). In contrast, the two RhoB homologs RhoA and RhoC were not induced after short exposures to CPT (Fig. 1C). RhoE, another member of the Rho family, has been identified as a p53-inducible gene in response to genotoxic agents (33). The authors of that study focused on upregulation of RhoE mRNA after long exposures to genotoxic agents, typically 12 h or longer. Figure 1C shows no increase in the RhoE mRNA level after short exposures to CPT in p53 wild-type U2OS cells under conditions where RhoB mRNA levels reached a maximum increase.
The increase in the RhoB mRNA level was associated with an increase in the RhoB protein level that was detectable 1 h after CPT treatment (Fig. 1D and E). Pulldown of active (GTP-bound) Rho proteins followed by immunoblotting with an anti-RhoB antibody indicated that CPT induced both total and active RhoB proteins (Fig. 1F), although it is still unclear whether these two events are connected. Under these conditions, the levels of total and active RhoA remained unchanged (Fig. 1D to F), which is consistent with the lack of induction of RhoA mRNA after CPT treatment (Fig. 1C).
Because DSBs are readily produced in CPT-treated cells (18), we tested whether they could be the initiating events for RhoB induction. Figure 1D shows that the increase in the level of RhoB protein coincided with the phosphorylation of H2AX at Ser139 (referred to as γH2AX), a marker for DSBs (9). To assess more directly the role of DSBs, we prevented their production in CPT-treated cells. DSBs are primarily produced during DNA replication at low concentrations (≤1 μM) of CPT (18). As expected (19), inhibition of replication with aphidicolin prevented the induction of γH2AX in response to CPT (Fig. 1G, bottom). Under these conditions, aphidicolin also prevented the induction of RhoB mRNA (Fig. 1G, top). These results suggest that DSBs promote RhoB upregulation.
Although RhoB can be induced in cells undergoing apoptosis (34), it is unlikely that the early increase in the level of RhoB induced by CPT resulted from the activation of apoptotic pathways. The increase of the RhoB level preceded the caspase-dependent cleavage of poly(ADP-ribose) polymerase (PARP) by several hours and was not prevented by the pancaspase inhibitor benzyloxycarbonyl-Val-Ala-dl-Asp(OMe)-fluoromethyl ketone (zVAD-fmk) (data not shown). Together, these results indicate that RhoB is induced rapidly and selectively in response to CPT-induced DSBs prior to and independently of apoptosis.
HuR-dependent stabilization of RhoB mRNA in CPT-treated cells.
Increases of both transcription and transcript stability have been involved in the upregulation of RhoB mRNA in UV-exposed cells (2, 35). To examine RhoB transcription, we linked its promoter to a luciferase reporter gene. Figure 2A shows that CPT did not increase luciferase activity in cells transfected with this construct, indicating that transcription is unlikely to account for the upregulation of RhoB mRNA. Next, we compared the stability of RhoB mRNA between untreated and CPT-treated cells. Experiments performed in the presence of the transcription inhibitor flavopiridol revealed that the half-life of RhoB mRNA was greatly prolonged in CPT-treated cells (Fig. 2B).
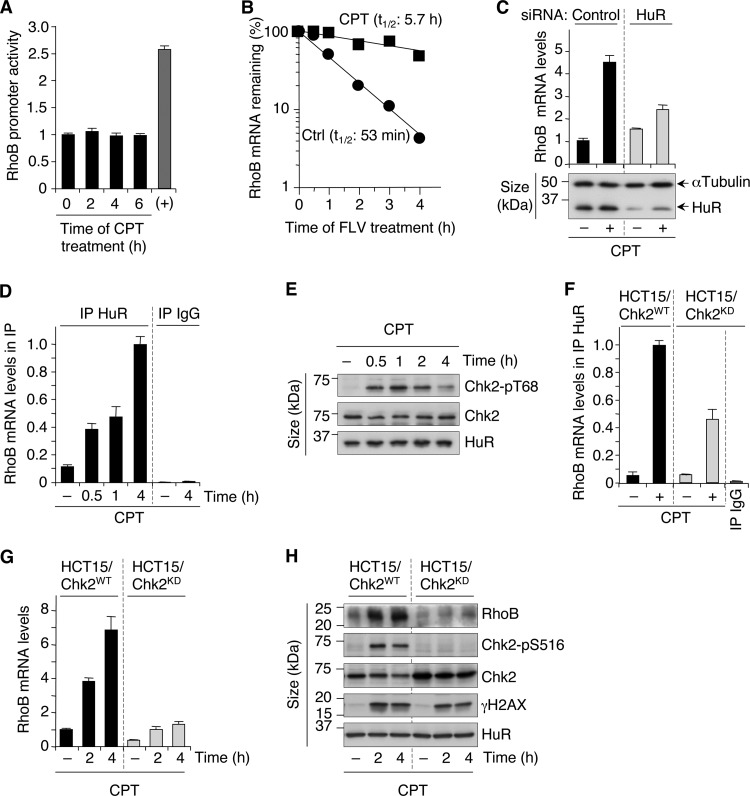
FIG 2.
Chk2- and HuR-dependent stabilization of RhoB mRNA in response to CPT. (A) RhoB promoter activity was determined after transfection of U2OS cells with a RhoB promoter-luciferase reporter gene construct before treatment with 25 μM CPT. Data shown are means ± SD for three independent experiments. The positive control (+) was cells treated with trichostatin A (1 μM for 15 h). (B) U2OS cells were left untreated (Ctrl) or were treated with CPT (25 μM for 2 h) before the addition of the transcription inhibitor flavopiridol (FLV) (1 μM). RhoB mRNA was analyzed by RT-qPCR and normalized to the level at the time of flavopiridol addition, which was set to 100% (averages of data from two independent experiments). The half-life (t1/2) of RhoB mRNA is indicated. (C) HCT116 cells were transfected with HuR-targeting or nontargeting (control) siRNAs before treatment with CPT (25 μM for 6 h). (Top) RT-qPCR analysis of RhoB mRNA (means ± SD for triplicate samples). (Bottom) Western blotting showing the efficiency of HuR silencing. α-Tubulin was the loading control. (D) Increased HuR-RhoB mRNA interaction upon CPT treatment. HuR was immunoprecipitated (IP) from HCT15/Chk2-WT cells treated with 25 μM CPT. The control was immunoprecipitation with nonimmune IgG. Coimmunoprecipitated RhoB mRNA was analyzed by RT-qPCR relative to β-actin mRNA levels in the input samples (means ± SD for triplicate samples). (E) Phosphorylation of Chk2 on Thr68 was examined by Western blotting in HCT15/Chk2-WT cells treated with 25 μM CPT. Chk2 and HuR were examined in parallel. (F) Chk2-dependent binding of HuR to RhoB mRNA. HCT15/Chk2-WT and HCT15/Chk2-KD cells were treated with 25 μM CPT for 4 h, and RhoB mRNA was analyzed in HuR immunoprecipitations, as described above for panel D. (G and H) HCT15/Chk2-WT and HCT15/Chk2-KD cells were treated with 25 μM CPT. (G) RhoB mRNA was analyzed by RT-qPCR (means ± SD for triplicate samples). (H) Western blotting of the indicated proteins. Phosphorylation of Chk2 on Ser516, which is an autophosphorylation site in response to DNA damage (64), was used to control Chk2 kinase activity in cells expressing WT or kinase-dead Chk2.
The RNA binding protein HuR is known to bind to and stabilize target mRNAs (36). RhoB mRNA contains a HuR binding site in its 3′ untranslated region (UTR) (28), and a constitutive HuR-RhoB mRNA interaction has been found by RNA immunoprecipitation coupled with microarray analyses (RIP-chip) (37, 38). To test the potential role of HuR in the enhanced stability of RhoB mRNA, we tested whether HuR inhibition affected CPT-induced RhoB mRNA. Figure 2C shows that siRNA-mediated depletion of HuR decreased the induction of RhoB mRNA. To further implicate HuR, we examined its binding to RhoB mRNA by RIP experiments. Endogenous HuR was immunoprecipitated, and the levels of coimmunoprecipitated RhoB mRNA were analyzed by RT-qPCR. In untreated cells, RhoB transcripts were enriched in HuR immunoprecipitates compared with those in control IgG immunoprecipitates (Fig. 2D), indicating that HuR binds to RhoB mRNA under normal conditions, as expected (37, 38). Cellular exposure to CPT resulted in a further enrichment of RhoB mRNA in HuR immunoprecipitates, which was detected within 30 min and increased with the time of CPT exposure (Fig. 2D). A similar increase in HuR-RhoB mRNA interactions has been observed after short exposures to UV (35), suggesting that HuR-dependent stabilization of RhoB mRNA is a common mechanism for the early induction of RhoB by genotoxic agents.
Chk2-dependent HuR-RhoB mRNA interaction in response to DSBs.
Chk2 is a serine/threonine kinase readily activated by DSBs (39). Active Chk2 phosphorylates HuR with RNA recognition motifs and modulates HuR binding to target mRNAs (37, 40–42). As expected (43), CPT induced rapid phosphorylation of Chk2 at Thr68 (Fig. 2E), which reflects its activation (44). Chk2 Thr68 phosphorylation was detected 30 min after CPT treatment (Fig. 2E) and coincided with the increased association of HuR with RhoB mRNA (Fig. 2D).
To assess directly the involvement of Chk2, we used HCT15 cells (Chk2 deficient) stably expressing wild-type Chk2 (Chk2-WT) or kinase-dead Chk2 (Chk2-KD). The levels of HuR protein were comparable in Chk2-WT and Chk2-KD cells and were unaffected by CPT treatment (Fig. 2H). Figure 2F shows that HuR binding to RhoB mRNA was reduced in Chk2-KD cells compared to Chk2-WT cells in response to CPT. These results led us to test whether the level of CPT-induced RhoB was also reduced in Chk2-KD cells. The induction of RhoB mRNA (Fig. 2G) and protein (Fig. 2H) was defective in Chk2-KD cells treated with CPT. As a control, parental HCT15 cells (Chk2 deficient) exhibited a similar induction defect in RhoB mRNA (data not shown) as that of Chk2-KD cells (Fig. 2G). Both WT and Chk2-KD cells showed identical induction of γH2AX in response to CPT (Fig. 2H), indicating that the defective response of Chk2-KD cells is not caused by a decreased amount of DSBs.
RhoB facilitates γH2AX dephosphorylation in CPT-treated cells.
To assess the potential role of RhoB in the cellular response to CPT, we compared survival of WT and RhoB-deficient (RhoB−/−) E6-immortalized MEF cells after CPT treatment. Cells were treated with increasing concentrations of CPT, and CPT sensitivity was assessed by WST-1 survival assays. Figure 3A shows that RhoB−/− cells are more sensitive to CPT than are WT cells, indicating that RhoB participates in the cellular response to CPT.
Because the cytotoxicity of CPT depends on Top1-linked DNA single-strand break (Top1cc)-induced DSBs (18), we analyzed the influence of RhoB on both the formation of Top1cc and the production of DSBs. Primary cells (WT and RhoB−/−) were used, as they normally have low background levels of γH2AX (45). Our results indicated that CPT-induced Top1cc (Fig. 3B) and γH2AX (Fig. 3C) levels were similar in WT and RhoB−/− cells.
We therefore hypothesized that the hypersensitivity of RhoB−/− cells could instead result from a defect in the repair of these DSBs. To determine the kinetics of DSB repair, we analyzed the kinetics of γH2AX dephosphorylation (45). WT and RhoB−/− cells were exposed to CPT for 1 h and washed, and γH2AX dephosphorylation was monitored post-CPT treatment (release) (Fig. 3D). Unlike continuous exposure to CPT, this protocol allows the study of DSB repair, as Top1cc reverse fully within minutes after washing out CPT (18), and DSBs are then no longer produced. After termination of the CPT treatment, γH2AX levels decreased by approximately 70% within 7 h in WT cells (Fig. 3E and F), which is consistent with the kinetics and magnitude of γH2AX focus loss after exposure to ionizing radiation (46). Under these conditions, γH2AX levels were not significantly reduced in RhoB−/− cells (Fig. 3E and F). Immunofluorescence microscopy confirmed the pronounced defect of γH2AX dephosphorylation in RhoB−/− cells after removal of CPT (data not shown). In addition, siRNA-mediated depletion of RhoB in human HCT116 cells (Fig. 3G) also resulted in a marked reduction of γH2AX dephosphorylation after removal of CPT (Fig. 3H). Neutral Comet assays confirmed that the persistence of γH2AX in RhoB-deficient cells post-CPT treatment corresponded to unrepaired DSBs (Fig. 3I and J). Analysis of endogenous Top1cc showed that they reversed efficiently in both WT and RhoB−/− cells after removal of CPT (Fig. 3K and L). Thus, it is unlikely that the persistent γH2AX signal in RhoB−/− cells resulted simply from the further production of DSBs after termination of the CPT treatment. These results indicate that RhoB promotes γH2AX dephosphorylation and DSB repair.
RhoB promotes DSB repair by homologous recombination.
Homologous recombination and nonhomologous end joining (NHEJ) are the prevalent pathways for the repair of DSBs (47). To assess directly the involvement of RhoB in DSB repair, we used human RG37 fibroblast cells that contain a single chromosomally integrated copy of the pDR-GFP substrate (26) (Fig. 4A). This substrate allows the monitoring of homologous recombination, specifically gene conversion, induced by a DSB produced by the nuclease I-SceI (48). Plasmid pDR-GFP consists of a tandem repeat of two inactive GFP genes, one of them containing a cleavage site for I-SceI. Transient expression of I-SceI produces a DSB in the chromosomal recombination substrate, which can induce homologous recombination and recreates a functional GFP (Fig. 4A). The recombinant cells become fluorescent and can be detected by flow cytometry. In RG37 cells, siRNA-mediated depletion of RhoB decreased the induction of GFP-positive recombinant cells in response to I-SceI expression (Fig. 4B). In agreement with these results, siRNA-mediated depletion of RhoB also prevented the formation of Rad51 foci post-CPT treatment (Fig. 4C and D), a key protein in homologous recombination that is recruited at DSB sites (47). Flow cytometry analysis of BrdU incorporation versus DNA content showed that the percentages of S-phase cells were similar in cells transfected with control and RhoB-targeting siRNAs (Fig. 4E) as well as in WT and RhoB−/− MEF cells (WT cells, 46.3% ± 2%; RhoB−/− cells 45.0% ± 0.7%) (data not shown). Thus, RhoB deficiency did not result in a lower proportion of S-phase cells, which excludes the possibility that fewer cells in S phase account for the reduced DSB repair by homologous recombination.
To assess the potential involvement of RhoB in NHEJ repair, we used human GC92 fibroblast cells stably expressing the pCOH-CD4 substrate (27, 49, 50) (Fig. 4F). This substrate contains genes encoding the membrane antigens H2Kd, CD8, and CD4. Before expression of I-SceI, neither CD8 nor CD4 is expressed. I-SceI expression produces the excision of the H2Kd/CD8 fragment, and rejoining of the DNA ends leads to the expression of the CD4 gene (Fig. 4F). Cells expressing CD4 at the plasma membrane can be detected by flow cytometry using an anti-CD4 antibody. Figure 4G shows that siRNA-mediated depletion of RhoB did not significantly affect the induction of CD4-positive cells in response to I-SceI expression. Together, these results indicate that RhoB is involved in DSB repair primarily by homologous recombination.
RhoB-deficient cells are defective for PP2A activity.
Inhibition of PP2A is known to impair DSB repair by homologous recombination (16). In addition, although several serine/threonine phosphatases (PP2A, PP4, PP1, PP6, and Wip1) can dephosphorylate γH2AX (14), PP2A seems to be the main phosphatase for γH2AX in response to CPT (15). PP2A inhibition induces persistent γH2AX and DSBs in CPT-treated cells and increases cellular sensitivity to CPT (15), effects similar to those observed in RhoB-deficient cells (Fig. 3). We therefore examined whether RhoB inhibition could affect PP2A activity.
Consistent with the prevalent role of PP2A in removing γH2AX in CPT-treated cells (15), inhibition of PP2A with fostriecin or okadaic acid completely prevented γH2AX dephosphorylation following removal of CPT (Fig. 5A). Next, we measured PP2A activity in WT, RhoB−/−, and RhoB−/− cells complemented with HA-RhoB. The levels of PP2A(C) protein were similar in all cell populations (Fig. 5B). Figure 5C shows that PP2A activity was reduced in RhoB−/− cells compared to WT and RhoB−/− cells complemented with HA-RhoB. To determine whether RhoB and PP2A are in the same pathway to dephosphorylate γH2AX, we compared the levels of γH2AX post-CPT treatment when RhoB is expressed or not under conditions where PP2A activity is inhibited. Figure 5D shows that RhoB suppression with siRNA did not further increase the level of γH2AX in cells exposed to okadaic acid post-CPT treatment. From these results, we propose that RhoB promotes PP2A activity and DSB repair.
RhoB-deficient cells reveal endogenous γH2AX foci and genomic instability.
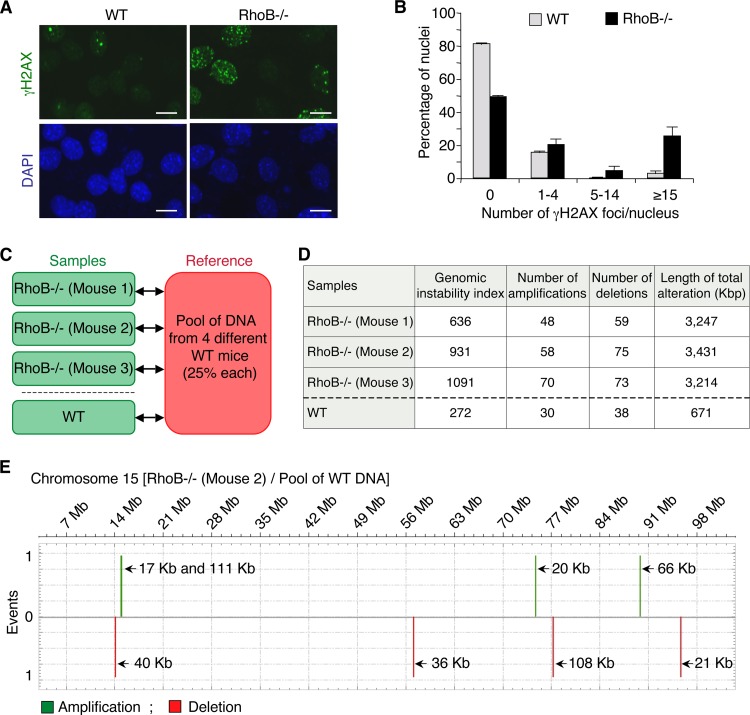
Because we found that RhoB promotes DSB repair, we examined whether RhoB-deficient cells would accumulate endogenous DSBs. We analyzed γH2AX nuclear foci by immunofluorescence microcopy in WT and RhoB−/− cells in the absence of treatment. This technique is much more sensitive than Western blotting, as it can detect a single DSB focus per cell (9). Figure 6A reveals increased numbers of γH2AX foci in RhoB−/− cells compared to WT cells. Quantitative analyses of the microscopy images showed that approximately 25% of RhoB−/− nuclei formed >15 γH2AX foci, compared to 3% in WT cells (Fig. 6B).
FIG 6.
RhoB loss is accompanied by increased γH2AX levels and chromosomal abnormalities. (A) Representative images of WT and RhoB−/− primary MDF cells after staining for γH2AX. DNA was counterstained with DAPI (blue). Bar, 10 μm. (B) Quantification of the number of γH2AX foci per nucleus. A minimum of 500 nuclei was analyzed per cell type (means ± SD for three independent experiments). (C) Design of the CGH array analysis. Genomic DNA of RhoB−/− MDF cells was compared to a pool of genomic DNA of MDF cells from four different WT syngeneic mice. As a control, to determine the number of genomic alterations due to interindividual variability, genomic DNA of WT MDF cells was compared to the pool of genomic DNA from WT mice. (D) Table showing whole-genome chromosomal amplification and deletion events as well as the total length (in kbp) of chromosomal alterations in RhoB−/− MDF cells compared to WT MDF cells analyzed by CGH arrays. The genomic instability index was calculated with the formula (number of deletions + number of amplifications)2/number of altered chromosomes, as previously described (32). (E) Chromosome plots for chromosome 15 of RhoB−/− MDF cells from mouse 2 compared to the pool of WT MDF cells, as determined by a CGH array. This chromosome is representative of the mean numbers of amplification and deletion events in RhoB−/− MDF cells.
Inefficient repair of DSBs can initiate genomic instability (9, 10). To evaluate genomic instability in RhoB−/− cells, we performed comparative genomic hybridization (CGH) whole-genome tiling arrays. Genomic DNA of primary mouse dermal fibroblast (MDF) cells from RhoB−/− mice was compared to a pool of genomic DNA of MDF cells from four WT syngeneic mice. As a control for genomic alterations due to interindividual variability, genomic DNA of MDF cells from a WT mouse was compared to the pool of WT genomic DNA (Fig. 6C). The three RhoB−/− mice analyzed revealed an increased number of chromosomal amplification and deletion events compared to the WT mouse (Fig. 6D). A representative chromosome is shown in Fig. 6E. In accordance with these results, the total length of chromosomal alterations and the genomic instability index, which reflects the number of alterations per chromosome (32), were also higher in RhoB−/− mice (Fig. 6D). Together, these experiments suggest that RhoB loss increases the number of endogenous DSBs and genomic instability.
DISCUSSION
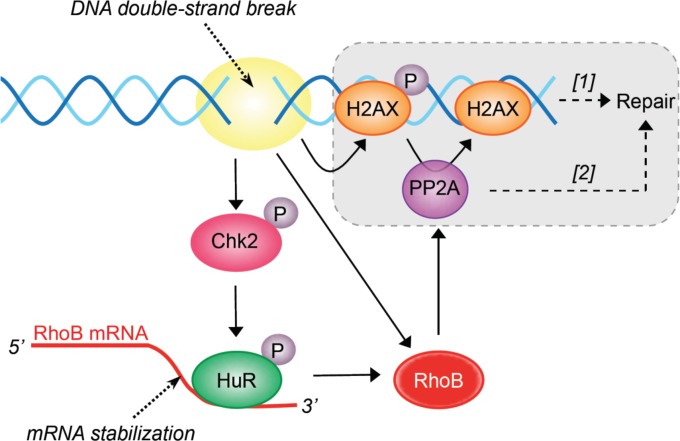
Here we identify RhoB as the first GTPase involved in the signaling and repair of DSBs. Our data support a model in which DSBs activate a Chk2-HuR-RhoB pathway that promotes PP2A-mediated dephosphorylation of γH2AX and repair (Fig. 7). DSBs are likely the initiating events for RhoB upregulation, as inhibition of CPT-induced DSBs by blocking of replication suppressed the induction of RhoB mRNA (Fig. 1G). Also, the induction of RhoB depends on Chk2 (Fig. 2), a checkpoint kinase readily activated by DSBs (39). Other parallel pathways besides Chk2 probably also contribute to RhoB induction, as Chk2-KD cells are not completely defective for HuR-RhoB mRNA interaction (Fig. 2F) and RhoB induction in response to CPT (Fig. 2G and H). The Chk2-independent induction of RhoB (Fig. 7) may be sufficient to promote γH2AX dephosphorylation, as Chk2-KD cells dephosphorylate γH2AX with an efficiency similar to that of Chk2-WT cells 5 h after CPT removal (data not shown). Moreover, because Chk2 is a major player in DSB repair (it phosphorylates/activates Brca1, p53, and PP2A) (39), these results also raise the possibility that Chk2-KD cells have evolved and compensate by increasing Chk2-independent pathways to repair DSBs. Such pathways may implicate Chk1. Indeed, CPT is known to activate the ATR-Chk1 pathway (18, 19), and Chk1 can promote HuR-dependent stabilization of mRNAs by inhibiting Cdk1-mediated phosphorylation of HuR (51, 52). Moreover, Chk1 can also promote DSB repair by phosphorylating the repair factor Rad51 (53). In addition, the effect of Chk2 loss seems to be less pronounced on HuR-RhoB mRNA interactions than on RhoB mRNA levels after CPT treatment, which raises the possibility that other pathways besides HuR also participate in the induction of RhoB mRNA by Chk2. Further support for the role of DSBs is provided by independent studies showing that RhoB is also upregulated early in response to UV and cisplatin (2, 3), which can produce DSBs in replicating cells (54–56).
FIG 7.
Proposed molecular pathways for the role of RhoB in the DDR and repair. The gray box indicates that PP2A promotes DSB repair by dephosphorylating γH2AX ([1]) and/or non-DDR proteins ([2]) (see Discussion).
RhoB appears to be implicated in the repair of DSBs from different origins. Indeed, RhoB-deficient cells maintain elevated γH2AX levels in response to CPT (Fig. 3), which produces DSBs indirectly during replication (19) and transcription (20) as well as in response to endogenous DSBs (Fig. 6). RhoB-deficient cells are also defective in the repair of DSBs that are produced directly by the endonuclease I-SceI (Fig. 4). DSBs are potentially lethal DNA lesions if not repaired (9). In agreement with the involvement of RhoB in DSB repair, RhoB-deficient cells are hypersensitive to CPT (Fig. 3A) and other genotoxic agents that can also induce DSBs, such as ionizing radiation (57) and UV (2).
It is now well documented that inhibition of PP2A activity impairs DSB repair (15–17). Hence, our finding that RhoB-deficient cells are defective for PP2A activity (Fig. 5) links RhoB expression to DSB repair. Although RhoB can bind PP2A(C) (58), it is not known whether this interaction is important for PP2A activity. PP2A may stimulate DSB repair by the timely dephosphorylation of γH2AX (15). Our analysis showing that RhoB-deficient cells are defective for DSB repair by homologous recombination but not for DSB repair by end joining (Fig. 4) further suggests that γH2AX dephosphorylation might be specifically required for homologous recombination. In line with this possibility, it has been reported that γH2AX dephosphorylation (removal) is required for efficient DNA end resection, which is a prerequisite for homology-mediated DSB repair (59). Besides RhoB promoting DSB repair by PP2A-mediated γH2AX dephosphorylation (Fig. 7), it is possible that RhoB also promotes DSB repair by PP2A-independent pathways, which may in turn further increase PP2A-mediated γH2AX dephosphorylation. PP2A may also stimulate DSB repair by homologous recombination by dephosphorylating non-DDR proteins such as Akt1 (60). Indeed, phosphorylated/active Akt1 has been reported to inhibit homologous recombination by inducing cytoplasmic retention of Brca1 and Rad51 (61). It is therefore possible that RhoB inhibition prevents DSB repair by inhibiting the PP2A-dependent dephosphorylation of Akt1. This hypothesis is concordant with our previous and current findings that the loss of RhoB expression promotes the activation of Akt1 (7) and prevents DSB repair by homologous recombination (Fig. 4). Besides γH2AX (and Akt), it is likely that RhoB loss affected other PP2A substrates, as RhoB loss decreased the global activity of PP2A, which was measured by using a nonspecific threonine phosphopeptide (Fig. 5C). Although PP2A can also promote DSB repair by NHEJ by dephosphorylating Ku70, Ku80, and DNA-PKcs (17), RhoB downregulation did not significantly affect end-joining-repair events (Fig. 4). Hence, the predominant role of RhoB-dependent PP2A activity is likely to promote DSB repair by homologous recombination. Besides PP2A, it is possible that RhoB loss also decrease a PP4 activity. Indeed, PP4 loss is primarily involved in the basal increase in the level of γH2AX (62), and we found that RhoB-deficient cells have elevated endogenous γH2AX levels (Fig. 6).
RhoB expression commonly decreases during tumor progression (4–8), and RhoB knockout (KO) mice are more susceptible to tumor formation and/or progression in response to UVB (63) and 7,12-dimethylbenz[a]anthracene (DMBA), whose metabolites induce DNA damage (24). However, very little is known about the molecular mechanisms by which the loss of RhoB promotes tumor progression. We show here that RhoB-deficient cells are defective for DSB repair (Fig. 3 and 4) and that, consistent with this, they have elevated endogenous γH2AX levels and chromosomal abnormalities (Fig. 6). We recently reported that RhoB-deficient human skin tumors also have elevated γH2AX levels compared to RhoB-proficient tumors (63). Hence, our findings suggest that loss of RhoB could promote oncogenesis by increasing DSB-mediated genomic instability.
ACKNOWLEDGMENTS
We thank Y. Pommier for Chk2-WT and Chk2-KD HCT15 cells and for stimulating discussions; G. C. Prendergast for WT and RhoB−/− mice; A. Peyret-Lacombe, J. Cherier, R. Gence, and I. Lajoie-Mazenc for preparing primary mouse dermal fibroblast cells from these mice; and L. Trouilh from the GenoToul Biochips platform for her kind help in the analyses of CGH arrays. We also thank S. Cabantous, E. Nicolas, and C. E. Redon for critically reading the manuscript.
This research was supported by grants from the Ligue Nationale Contre le Cancer and the Region Midi-Pyrénées.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 9 June 2014
REFERENCES
- 1.Etienne-Manneville S, Hall A. 2002. Rho GTPases in cell biology. Nature 420:629–635. 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- 2.Canguilhem B, Pradines A, Baudouin C, Boby C, Lajoie-Mazenc I, Charveron M, Favre G. 2005. RhoB protects human keratinocytes from UVB-induced apoptosis through epidermal growth factor receptor signaling. J. Biol. Chem. 280:43257–43263. 10.1074/jbc.M508650200 [DOI] [PubMed] [Google Scholar]
- 3.Fritz G, Kaina B, Aktories K. 1995. The ras-related small GTP-binding protein RhoB is immediate-early inducible by DNA damaging treatments. J. Biol. Chem. 270:25172–25177. 10.1074/jbc.270.42.25172 [DOI] [PubMed] [Google Scholar]
- 4.Adnane J, Muro-Cacho C, Mathews L, Sebti SM, Munoz-Antonia T. 2002. Suppression of rho B expression in invasive carcinoma from head and neck cancer patients. Clin. Cancer Res. 8:2225–2232 [PubMed] [Google Scholar]
- 5.Mazieres J, Tovar D, He B, Nieto-Acosta J, Marty-Detraves C, Clanet C, Pradines A, Jablons D, Favre G. 2007. Epigenetic regulation of RhoB loss of expression in lung cancer. BMC Cancer 7:220. 10.1186/1471-2407-7-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J, Zhu Y, Zhang G, Liu N, Sun L, Liu M, Qiu M, Luo D, Tang Q, Liao Z, Zheng Y, Bi F. 2011. A distinct role of RhoB in gastric cancer suppression. Int. J. Cancer 128:1057–1068. 10.1002/ijc.25445 [DOI] [PubMed] [Google Scholar]
- 7.Bousquet E, Mazieres J, Privat M, Rizzati V, Casanova A, Ledoux A, Mery E, Couderc B, Favre G, Pradines A. 2009. Loss of RhoB expression promotes migration and invasion of human bronchial cells via activation of AKT1. Cancer Res. 69:6092–6099. 10.1158/0008-5472.CAN-08-4147 [DOI] [PubMed] [Google Scholar]
- 8.Huang M, Prendergast GC. 2006. RhoB in cancer suppression. Histol. Histopathol. 21:213–218 [DOI] [PubMed] [Google Scholar]
- 9.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. 2008. GammaH2AX and cancer. Nat. Rev. Cancer 8:957–967. 10.1038/nrc2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper JW, Elledge SJ. 2007. The DNA damage response: ten years after. Mol. Cell 28:739–745. 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- 11.Jeggo PA, Lobrich M. 2007. DNA double-strand breaks: their cellular and clinical impact? Oncogene 26:7717–7719. 10.1038/sj.onc.1210868 [DOI] [PubMed] [Google Scholar]
- 12.Lukas J, Lukas C, Bartek J. 2011. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13:1161–1169. 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- 13.Shiloh Y. 2006. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 31:402–410. 10.1016/j.tibs.2006.05.004 [DOI] [PubMed] [Google Scholar]
- 14.Freeman AK, Monteiro AN. 2010. Phosphatases in the cellular response to DNA damage. Cell Commun. Signal. 8:27. 10.1186/1478-811X-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. 2005. Gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20:801–809. 10.1016/j.molcel.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 16.Kalev P, Simicek M, Vazquez I, Munck S, Chen L, Soin T, Danda N, Chen W, Sablina A. 2012. Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition. Cancer Res. 72:6414–6424. 10.1158/0008-5472.CAN-12-1667 [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Gao F, Wang T, Flagg T, Deng X. 2009. A nonhomologous end-joining pathway is required for protein phosphatase 2A promotion of DNA double-strand break repair. Neoplasia 11:1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pommier Y. 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6:789–802. 10.1038/nrc1977 [DOI] [PubMed] [Google Scholar]
- 19.Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, Pilch DR, Rogakou EP, Celeste A, Chen HT, Nussenzweig A, Aladjem MI, Bonner WM, Pommier Y. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303–20312. 10.1074/jbc.M300198200 [DOI] [PubMed] [Google Scholar]
- 20.Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E, Kohn KW, Bonner WM, Pommier Y. 2009. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 10:887–893. 10.1038/embor.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. 2011. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 39:3607–3620. 10.1093/nar/gkq1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aris SM, Pommier Y. 2012. Potentiation of the novel topoisomerase I inhibitor indenoisoquinoline LMP-400 by the cell checkpoint and Chk1-Chk2 inhibitor AZD7762. Cancer Res. 72:979–989. 10.1158/0008-5472.CAN-11-2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kass EM, Ahn J, Tanaka T, Freed-Pastor WA, Keezer S, Prives C. 2007. Stability of checkpoint kinase 2 is regulated via phosphorylation at serine 456. J. Biol. Chem. 282:30311–30321. 10.1074/jbc.M704642200 [DOI] [PubMed] [Google Scholar]
- 24.Liu AX, Rane N, Liu JP, Prendergast GC. 2001. RhoB is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Mol. Cell. Biol. 21:6906–6912. 10.1128/MCB.21.20.6906-6912.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aasen T, Izpisua Belmonte JC. 2010. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat. Protoc. 5:371–382. 10.1038/nprot.2009.241 [DOI] [PubMed] [Google Scholar]
- 26.Dumay A, Laulier C, Bertrand P, Saintigny Y, Lebrun F, Vayssiere JL, Lopez BS. 2006. Bax and Bid, two proapoptotic Bcl-2 family members, inhibit homologous recombination, independently of apoptosis regulation. Oncogene 25:3196–3205. 10.1038/sj.onc.1209344 [DOI] [PubMed] [Google Scholar]
- 27.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819–824. 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- 28.Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. 2011. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 18:1692–1701. 10.1038/cdd.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lajoie-Mazenc I, Tovar D, Penary M, Lortal B, Allart S, Favard C, Brihoum M, Pradines A, Favre G. 2008. MAP1A light chain-2 interacts with GTP-RhoB to control epidermal growth factor (EGF)-dependent EGF receptor signaling. J. Biol. Chem. 283:4155–4164. 10.1074/jbc.M709639200 [DOI] [PubMed] [Google Scholar]
- 30.Regairaz M, Zhang YW, Fu H, Agama KK, Tata N, Agrawal S, Aladjem MI, Pommier Y. 2011. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195:739–749. 10.1083/jcb.201104003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cron KR, Zhu K, Kushwaha DS, Hsieh G, Merzon D, Rameseder J, Chen CC, D'Andrea AD, Kozono D. 2013. Proteasome inhibitors block DNA repair and radiosensitize non-small cell lung cancer. PLoS One 8:e73710. 10.1371/journal.pone.0073710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagarde P, Perot G, Kauffmann A, Brulard C, Dapremont V, Hostein I, Neuville A, Wozniak A, Sciot R, Schoffski P, Aurias A, Coindre JM, Debiec-Rychter M, Chibon F. 2012. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin. Cancer Res. 18:826–838. 10.1158/1078-0432.CCR-11-1610 [DOI] [PubMed] [Google Scholar]
- 33.Ongusaha PP, Kim HG, Boswell SA, Ridley AJ, Der CJ, Dotto GP, Kim YB, Aaronson SA, Lee SW. 2006. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr. Biol. 16:2466–2472. 10.1016/j.cub.2006.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Kim CH, Won M, Choi CH, Ahn J, Kim BK, Song KB, Kang CM, Chung KS. 2010. Increase of RhoB in gamma-radiation-induced apoptosis is regulated by c-Jun N-terminal kinase in Jurkat T cells. Biochem. Biophys. Res. Commun. 391:1182–1186. 10.1016/j.bbrc.2009.12.012 [DOI] [PubMed] [Google Scholar]
- 35.Westmark CJ, Bartleson VB, Malter JS. 2005. RhoB mRNA is stabilized by HuR after UV light. Oncogene 24:502–511. 10.1038/sj.onc.1208224 [DOI] [PubMed] [Google Scholar]
- 36.Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266–277. 10.1007/PL00000854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda K, Abdelmohsen K, Kim MM, Srikantan S, Lee EK, Tominaga K, Selimyan R, Martindale JL, Yang X, Lehrmann E, Zhang Y, Becker KG, Wang JY, Kim HH, Gorospe M. 2011. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. EMBO J. 30:1040–1053. 10.1038/emboj.2011.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calaluce R, Gubin MM, Davis JW, Magee JD, Chen J, Kuwano Y, Gorospe M, Atasoy U. 2010. The RNA binding protein HuR differentially regulates unique subsets of mRNAs in estrogen receptor negative and estrogen receptor positive breast cancer. BMC Cancer 10:126. 10.1186/1471-2407-10-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pommier Y, Sordet O, Rao VA, Zhang H, Kohn KW. 2005. Targeting chk2 kinase: molecular interaction maps and therapeutic rationale. Curr. Pharm. Des. 11:2855–2872. 10.2174/1381612054546716 [DOI] [PubMed] [Google Scholar]
- 40.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25:543–557. 10.1016/j.molcel.2007.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. 2009. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell 20:4885–4898. 10.1091/mbc.E09-07-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY. 2011. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 39:8472–8487. 10.1093/nar/gkr567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takemura H, Rao VA, Sordet O, Furuta T, Miao ZH, Meng L, Zhang H, Pommier Y. 2006. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J. Biol. Chem. 281:30814–30823. 10.1074/jbc.M603747200 [DOI] [PubMed] [Google Scholar]
- 44.Melchionna R, Chen XB, Blasina A, McGowan CH. 2000. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2:762–765. 10.1038/35036406 [DOI] [PubMed] [Google Scholar]
- 45.Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. 2010. GammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9:662–669. 10.4161/cc.9.4.10764 [DOI] [PubMed] [Google Scholar]
- 46.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31:167–177. 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- 47.Khanna KK, Jackson SP. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247–254. 10.1038/85798 [DOI] [PubMed] [Google Scholar]
- 48.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633–2638. 10.1101/gad.13.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14:611–623. 10.1016/j.molcel.2004.05.008 [DOI] [PubMed] [Google Scholar]
- 50.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. 2007. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. U. S. A. 104:20902–20907. 10.1073/pnas.0708541104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Khalaf HH, Aboussekhra A. ATR controls the UV-related upregulation of the CDKN1A mRNA in a Cdk1/HuR-dependent manner. Mol. Carcinog., in press [DOI] [PubMed] [Google Scholar]
- 52.Kim HH, Abdelmohsen K, Gorospe M. 2010. Regulation of HuR by DNA damage response kinases. J. Nucleic Acids 2010:981487. 10.4061/2010/981487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7:195–201. 10.1038/ncb1212 [DOI] [PubMed] [Google Scholar]
- 54.Ward IM, Chen J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762 [DOI] [PubMed] [Google Scholar]
- 55.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. 2002. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma-H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. U. S. A. 99:233–238. 10.1073/pnas.231611798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frankenberg-Schwager M, Kirchermeier D, Greif G, Baer K, Becker M, Frankenberg D. 2005. Cisplatin-mediated DNA double-strand breaks in replicating but not in quiescent cells of the yeast Saccharomyces cerevisiae. Toxicology 212:175–184. 10.1016/j.tox.2005.04.015 [DOI] [PubMed] [Google Scholar]
- 57.Ader I, Delmas C, Bonnet J, Rochaix P, Favre G, Toulas C, Cohen-Jonathan-Moyal E. 2003. Inhibition of Rho pathways induces radiosensitization and oxygenation in human glioblastoma xenografts. Oncogene 22:8861–8869. 10.1038/sj.onc.1207095 [DOI] [PubMed] [Google Scholar]
- 58.Lee WJ, Kim DU, Lee MY, Choi KY. 2007. Identification of proteins interacting with the catalytic subunit of PP2A by proteomics. Proteomics 7:206–214. 10.1002/pmic.200600480 [DOI] [PubMed] [Google Scholar]
- 59.Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, Sleckman BP. 2011. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature 469:245–249. 10.1038/nature09585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. 2008. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J. Biol. Chem. 283:1882–1892. 10.1074/jbc.M709585200 [DOI] [PubMed] [Google Scholar]
- 61.Plo I, Laulier C, Gauthier L, Lebrun F, Calvo F, Lopez BS. 2008. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res. 68:9404–9412. 10.1158/0008-5472.CAN-08-0861 [DOI] [PubMed] [Google Scholar]
- 62.Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. 2008. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol. Cell 31:33–46. 10.1016/j.molcel.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyer N, Peyret-Lacombe A, Canguilhem B, Medale-Giamarchi C, Mamouni K, Cristini A, Monferran S, Lamant L, Filleron T, Pradines A, Sordet O, Favre G. 2014. RhoB promotes cancer initiation by protecting keratinocytes from UVB-induced apoptosis but limits tumor aggressiveness. J. Investig. Dermatol. 134:203–212. 10.1038/jid.2013.278 [DOI] [PubMed] [Google Scholar]
- 64.Wu X, Chen J. 2003. Autophosphorylation of checkpoint kinase 2 at serine 516 is required for radiation-induced apoptosis. J. Biol. Chem. 278:36163–36168. 10.1074/jbc.M303795200 [DOI] [PubMed] [Google Scholar]