Abstract
More than 70% of mitochondrial proteins utilize N-terminal presequences as targeting signals. Presequence interactions with redundant cytosolic receptor domains of the translocase of the outer mitochondrial membrane (TOM) are well established. However, after the presequence enters the protein-conducting Tom40 channel, the recognition events that occur at the trans side leading up to the engagement of the presequence with inner membrane-bound receptors are less well defined. Using a photoaffinity-labeling approach with modified presequence peptides, we identified Tom40 as a presequence interactor of the TOM complex. Utilizing mass spectrometry, we mapped Tom40's presequence-interacting regions to both sides of the β-barrel. Analysis of a phosphorylation site within one of the presequence-interacting regions revealed altered translocation kinetics along the presequence pathway. Our analyses assess the relation between the identified presequence-binding region of Tom40 and the intermembrane space domain of Tom22. The identified presequence-interacting region of Tom40 is capable of functioning independently of the established trans-acting TOM presequence-binding domain during matrix import.
INTRODUCTION
Mitochondria receive the majority of their resident proteins from the cytoplasm. Therefore, mitochondria must mediate the influx of proteins, which have been translated on cytosolic ribosomes, in a spatially constrained manner, i.e., all substrates are required to be unfolded during membrane passage. This constraint is in part due to the static dimensions of the pore within the translocase of the outer mitochondrial membrane (TOM). The TOM complex is involved in the majority of mitochondrial import traffic and therefore serves as the main gateway into the mitochondrion (for reviews, see references 1 to 4).
About two-thirds of mitochondrial proteins are targeted into mitochondria by N-terminal targeting signals (presequences) (5). These signals consist of amphipathic α-helices with a net positive charge and direct the precursors across the outer and inner mitochondrial membranes. Presequence recognition at the TOM complex is thought to commence with the interaction of the hydrophobic surface of the amphipathic presequence-helix with the Tom20 binding groove (6–8). Subsequently, the hydrophilic side is recognized by the cytosolic domain of the central receptor Tom22 that interacts with the presequence in a salt-sensitive manner (6). The exact series of initial presequence-binding events is still debated; however, there is growing support for a tertiary complex in which both Tom20 and Tom22 interact with the presequence simultaneously (9, 10). Following the positioning of the presequence via Tom22, the substrate is passed to the pore-forming subunit of the TOM complex, Tom40. This process involves Tom5, which serves as a TOM assembly and presequence-interacting subunit (11). As the presequence emerges from the Tom40 pore on the trans side, the intermembrane space (IMS) domain of Tom22 engages with the precursor (10, 12–14). From the TOM complex, the precursor is passed to the presequence translocase of the inner mitochondrial membrane (TIM23 complex). The primary presequence receptor of TIM23 is Tim50, which is dedicated to presequence recognition and precursor transport to Tim23 (15–18). Tim23 exposes a presequence-binding domain into the IMS and is the pore-forming subunit of the presequence translocase (13, 15, 19, 20). The interaction of the presequence with Tim23 activates the channel and initiates the Δψ-dependent transfer of the presequence across the inner membrane (19, 20).
Tom40, a β-barrel protein of the outer mitochondrial membrane, represents the protein-conducting channel of the TOM complex. It was first identified via photo-cross-linking to an arrested artificial mitochondrial substrate (21). Shortly after, it was observed that the deletion of the gene resulted in the cytosolic accumulation of unprocessed mitochondrial precursors and subsequent lethality (22). Ever since Tom40's discovery, it has been postulated to play a more active role in the translocation of presequence-containing substrates across the outer membrane, rather than simply acting as a passive channel. Rapaport et al. observed that Tom40 interacts with presequences on both the cis and especially the trans side of the TOM complex (23). Similar findings on Tom40's presequence interaction have been reported by several studies (24–31), yet the identification of a dedicated presequence-interacting site within Tom40 has only been hinted at (32–37).
Here, we used a photoaffinity-labeling approach to identify presequence receptors and their binding sites in the TOM complex. Our investigation revealed Tom40 as a predominant presequence interactor of the TOM complex and uncovered two regions of presequence interaction on either side of the barrel. Tom40 has been recently shown to be phosphorylated at several locations (38). Importantly, one of these sites, the residue T220, is positioned between two identified presequence-interacting peptides. The corresponding phosphomimetic mutant exhibited altered Tom40 photoadduct formation. Import analysis revealed a selective presequence pathway import phenotype. Our analyses establish Tom40 as an active participant in the presequence pathway and examine its functional relationship with the IMS domain of Tom22.
MATERIALS AND METHODS
Yeast genetic manipulation, culture, and mitochondrial isolation.
Saccharomyces cerevisiae YPH499 was used as the parent strain for the creation of all mutants (39). Yeast cells were cultivated in YPG medium (1% yeast extract, 2% peptone, and 3% glycerol) at 30°C, unless specified otherwise. tom40E mut (T220E) and tom40V mut (T220V) alleles were created by site-directed mutagenesis (QuikChange Lightning; Stratagene) using wild-type TOM40 in pFL39 containing the endogenous promoter and terminator as a template (40). The resulting plasmids were transformed into a tom40Δ strain containing a YEp352 plasmid coding for wild-type TOM40 with a URA3 marker (41) using the lithium acetate method. Selection for pYEp352-TOM40 loss was achieved through growth on 5-fluoroorotic acid (5-FOA) medium.
Creation of tom22-2 mutants was done as previously described (14). In brief, a PCR-generated integration cassette containing HIS3MX6 from pYM10 (42) and flanking TOM22 regions was transformed into corresponding strains. Following homologous recombination, selection was imparted by growth on synthetic glucose medium (0.67% yeast nitrogen base, 0.07% complete supplement mixture [CSM] lacking histidine [MP Biomedicals], and 2% glucose), giving rise to the established tom22-2 allele encoding a Tom22 protein lacking 31 C-terminal resides (12). The TOM22His strain was created in part as described previously (43), in which the pFA-His10-HIS3MX5 vector was used to create an integration cassette via PCR containing HIS3MX5- and TOM22-flanking regions (44).
Growth tests were performed through normalization of optical density at 600 nm (OD600), spotting of cells at 1:10 serial dilutions on plates containing YPG solid medium, and growth for 3 days at the corresponding temperature. Mitochondria were isolated through Zymolyase treatment (7 mg/g [wet weight] of cells), Dounce homogenization, and differential centrifugation as previously described (45).
Presequence probe synthesis.
Presequence probes were synthesized as previously described (15) through the use of a peptide synthesizer (Applied Biosystems) with standard fluorenylmethoxycarbonyl chemistry for introducing the amino acid derivatives para-benzoylphenylalanine (BPA) and biotinylated lysine (Novabiochem).
Photo-cross-linking.
In organello photo-cross-linking was performed essentially as previously described (15). In short, mitochondria were suspended in import buffer (250 mM sucrose, 10 mM morpholinepropanesulfonic acid [MOPS]-KOH, pH 7.2, 80 mM KCl, 2 mM KH2PO4, 5 mM MgCl2, 5 mM methionine) at 1 μg/μl, supplemented at 2 μM with presequence probe, and incubated for 10 min on ice. For photo-cross-linking in TOM40 mutants, 75 mM NaCl was added to the reaction mixture during incubation on ice. UV irradiation was carried out for 30 min on ice in a custom-made device consisting of a halogen metal vapor lamp, below which a glass screen was placed to filter out wavelengths below 300 nm (46). After UV treatment, mitochondria were washed with SEM buffer (250 mM sucrose, 20 mM MOPS-KOH, pH 7.2, and 1 mM EDTA) and analyzed via SDS-PAGE and Western blotting. Photo-cross-linking using isolated TOM complex was performed similarly to that under in organello conditions in import buffer containing digitonin as specified below, with the samples being immediately resolved via SDS-PAGE after UV irradiation. Presequence probe at 2 μM and 13 μM was used for small- and large-scale photo-cross-linking utilizing isolated TOM complex, respectively.
TOM complex isolation.
The TOM complex was immunoisolated through the use of antiserum created against the cytosolic domain construct of Tom22 (6). Antiserum was incubated with protein A-Sepharose (4 μl serum/μl resin) according to the manufacturer's instructions (GE Healthcare) and cross-linked with dimethyl pimelimidate before use. Mitochondria were solubilized in solubilization buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 0.1 mM EDTA, 1% digitonin, 2 mM Pefabloc, 2 μg/ml leupeptin, and 2 mM phenylmethanesulfonyl fluoride [PMSF]) at 1 μg/μl for 30 min at 4°C. After the removal of insoluble particles via centrifugation (15 min, 20,000 × g, 4°C), mitochondrial lysates were incubated with resin at 4°C under gentle agitation. Column material was washed with solubilization buffer containing 0.3% digitonin and lacking Pefabloc and leupeptin. Bound proteins were subjected to acidic elution (100 mM glycine-HCl, pH 2.5), and all samples were analyzed via SDS-PAGE and Western blotting.
Small-scale TOM isolation via Tom22His was carried out by the solubilization of mitochondria in His solubilization buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10% glycerol, 0.1 mM EDTA, 30 mM imidazole, 1% digitonin, and 1 mM PMSF) at 1 μg/μl for 20 min on ice. Following a clarifying spin, mitochondrial lysates were added to nickel-nitrilotriacetic acid (Ni-NTA)–agarose (Qiagen) at 25 μg/μl resin. Resin was washed with His solubilization buffer containing 80 mM imidazole and 0.2% digitonin, and bound proteins were eluted with His solubilization buffer containing 0.2% digitonin and 200 mM imidazole. Prior to photo-cross-linking, eluates were concentrated and buffer was exchanged to import buffer with 0.2% digitonin using 3-kDa-cutoff centrifugal filters (Millipore).
Large-scale TOM isolation via Tom22His was performed in partial accordance with the previously established method (47). In brief, mitochondria were solubilized in TOM solubilization buffer (50 mM potassium acetate, 10 mM MOPS-KCl, pH 7.0, 20% glycerol, 1% digitonin, and 1 mM PMSF) at 1 μg/μl and mixed with Ni-NTA–agarose (Qiagen) at 25 μg/μl resin. Resin was washed with TOM solubilization buffer with 0.5% digitonin and 80 mM imidazole, followed by elution with solubilization buffer containing 0.5% digitonin and 500 mM imidazole. Eluates were concentrated in 50-kDa-cutoff centrifugal filters (Millipore), and then the buffer was exchanged to import buffer with 0.5% digitonin using PD G10 desalting columns according to the manufacturer's instructions (GE Healthcare). Following an additional centrifugal filter concentration step, the isolated complex was subjected to photo-cross-linking.
Import of precursor proteins.
Radiolabeled precursor proteins were generated by translation in a reticulocyte lysate system (Promega) in the presence of [35S]methionine. Recombinant precursor was isolated as previously described (48). Isolated mitochondria were suspended in import buffer containing 1% fatty acid-free bovine serum albumin (BSA), and import was performed at 25°C in the presence of 2 mM NADH and 2 mM ATP. AVO (1 μM valinomycin, 8 μM antimycin A, and 20 μM oligomycin) was used to dissipate the membrane potential where indicated. Radiolabeled Tim9 was urea denatured, imported, and subjected to reducing SDS-PAGE as described previously (49). Following import, mitochondria were proteinase K treated (with the exception of the OxaIGIP TOM translocation assay), washed with SEM buffer, and either solubilized for blue native PAGE (BN-PAGE) or analyzed via SDS-PAGE in conjunction with digital autoradiography or Western blotting.
Mass spectrometry.
Mass spectrometric analysis of photoadducts was performed as previously published (15). In brief, TOM-Holo photo-cross-linking was visualized on SDS-PAGE with colloidal Coomassie blue staining (50). Photoadduct bands were excised from the gel and subjected to in-gel digestion with trypsin prior to extraction with 1% trifluoroacetic acid and reversed-phase chromatography (Easy-nLC; Bruker Daltonics). Following robot-based target spotting (Proteineer fc II; Bruker Daltonics), samples were analyzed with a matrix-assisted laser deionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometer (Ultraflextreme; Bruker Daltonics). Tandem mass spectrometry (MS/MS) spectra containing precursor masses corresponding to possible cross-linked peptides were analyzed using a combination of Biotools and Flex-Analysis software (Bruker Daltonics), followed by manual validation.
Homology modeling.
Homology modeling was performed essentially as previously reported (40). In brief, the S. cerevisiae Tom40 sequence was submitted to the HHpred server (51), where the voltage-dependent anion-selective channel protein (VDAC1) from mouse (Protein Data Bank [PDB] ID 3EMN) (52) was identified as the best template exhibiting 15% sequence identity. The homology model was obtained using Modeler (53). Images were generated with Pymol. Residues 1 to 48 and 363 to 387 were removed from the models since they were located outside the barrel.
Miscellaneous.
BN-PAGE was performed as previously published (48). SDS-PAGE and Western blotting were performed according to standard protocols using polyvinylidene fluoride membrane. Membrane-bound proteins were detected using fluorescent dye-coupled secondary antibodies (Li-Cor) in conjunction with a fluorescent scanner (FLA-9000; Fujifilm) or via standard enhanced chemiluminescence (Thermo Scientific). Quantifications were performed using ImageQuant TL (GE Healthcare).
RESULTS
Tom40 represents a prominent presequence cross-link site within the TOM complex.
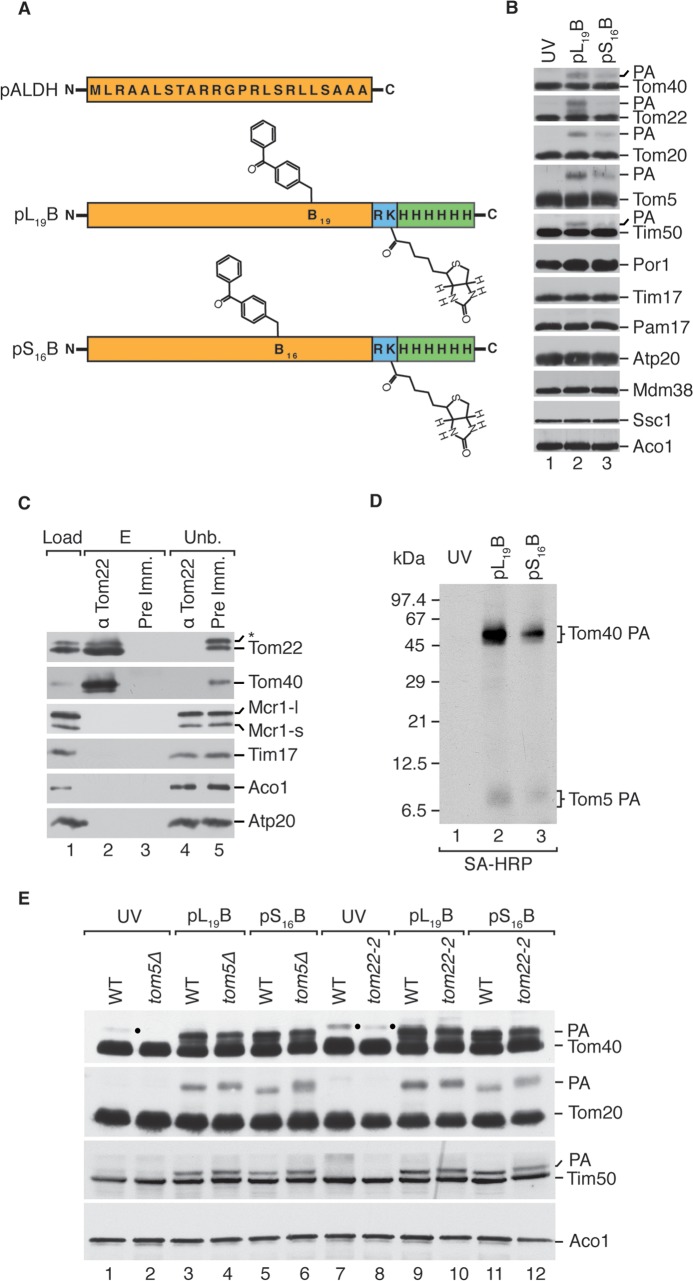
To investigate presequence interaction sites within the TOM complex, we employed the use of photoactivatable presequence probes derived from the rat aldehyde dehydrogenase presequence (pALDH) (15) (Fig. 1A). For photoaffinity labeling, the peptide probes contained the photoreactive amino acid derivative para-benzoylphenylalanine (BPA) either at the hydrophilic surface of the presequence helix (replacing serine 16; pS16B) or at the hydrophobic surface of the helix (replacing leucine 19; pL19B). These peptides are efficiently imported into isolated mitochondria, thus allowing for photoaffinity labeling during the transport process (15). Western blot analysis of in organello photo-cross-linking revealed Tom40 as a presequence interactor among all other established TOM complex presequence receptors (Fig. 1B). As the interaction of Tom40 with presequences has been repeatedly reported (21–24, 28–31), we set out to assess its relative contribution to presequence interaction within the TOM complex. Therefore, we isolated the TOM complex utilizing Tom22 antiserum. As expected, Tom22 immunoisolation of the TOM complex substantially enriched the core TOM components (Fig. 1C). Subsequently, we performed TOM immunoisolation via Tom22 after in organello presequence probe photo-cross-linking (Fig. 1D). The detection of the photoadducts, via the biotin tag on the peptide probe, allowed for the direct comparison of the relative presequence probe cross-linking levels between receptors. This analysis revealed Tom40 as a predominant presequence interactor of the TOM complex.
FIG 1.
Tom40 is a predominant presequence interactor of the TOM complex. (A) Schematic representation of presequence probes based upon rat ALDH presequence (pALDH) to which a BPA residue, trypsin site, biotinylated K residue, and His6 tag have been added. (B) In organello photo-cross-linking of presequence probes in isolated wild-type mitochondria visualized via Western blotting with indicated antisera. PA, photoadduct. (C) Immunoisolation using anti-Tom22 or preimmune serum with digitonin-solubilized mitochondria. Load and unbound (Unb.), 5%; eluate (E), 100%. An asterisk indicates a cross-reactive band. (D) Isolated wild-type mitochondria were subjected to photo-cross-linking as in panel B. Proteins were isolated through the use of Tom22 antiserum as in panel C and analyzed via Western blotting using streptavidin-horseradish peroxidase (SA-HRP). PA, photoadduct. (E) Assay performed as in panel B, utilizing the indicated strains. PA, photoadduct. A dot indicates a cross-reactive band. WT, wild type.
Further investigating Tom40's presequence interaction, we performed in organello photo-cross-linking in selected mutants corresponding to the established presequence-binding sites of Tom5 and Tom22IMS. This allowed us to explore the possibility of indirect Tom40 photoadduct formation due to presequence binding in the immediate vicinity, as the para-benzoylphenylalanine (BPA) group is known to exhibit substantial flexibility in its active radius (3 to 15 Å) (15, 54, 55). The deletion of neither Tom5 nor Tom22IMS affected the formation of the Tom40 photoadduct (Fig. 1E). Taken together, these analyses depicted Tom40 as a predominant presequence interactor of the TOM complex and demonstrated its functionality independent of neighboring presequence-binding domains.
Tom40 contains distinct presequence probe-interacting sites.
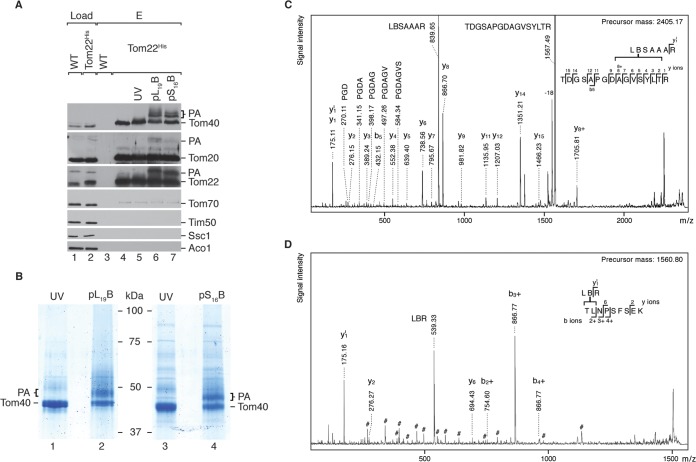
A considerable advantage of BPA-based cross-linking is that this photophore forms stable C-C bonds (54), allowing for the identification of the corresponding cross-linked residue via mass spectrometry. In order to apply this technique, the isolation of the target protein at a preparative scale is required; therefore, large-scale isolations of the TOM complex were performed from mitochondria isolated from a strain containing a chromosomally integrated His10 tag at the C terminus of Tom22 (43). Under in vitro conditions, when presequence probes were incubated with purified TOM complex, we achieved efficient photoadduct formation (Fig. 2A). Hence, this approach was exploited to obtain preparative amounts of endogenous Tom40 photoadduct bands. As seen in Fig. 2B (lanes 2 and 4), both pL19B and pS16B produced significant photoadducts upon cross-linking to isolated TOM complex. When photoadduct bands were subjected to in-gel tryptic digestion and analyzed via liquid chromatography (LC)-MALDI-MS/MS, cross-links to three Tom40 tryptic fragments were identified (Fig. 2C and D and 3A to C). One site could be resolved to the single-residue level (M94) (Fig. 3A), mainly based on the characteristic fragmentation pattern of the Met side chain when cross-linked to BPA (15, 56). The other two cross-link-containing tryptic fragments were observed to reside at a considerable distance from M94 and were refined to the heptapeptide 228AGVSYLT234 and the dipeptide 182TL183 (Fig. 2C and D and 3B and C). In these cases, the unexpected instability of the newly formed bond between BPA and Tom40 under the conditions of mass spectrometric sequencing prevented the assignment of the cross-linking site at the single-residue level.
FIG 2.
Identification of Tom40's presequence-interacting regions. (A) Ni-NTA–agarose was incubated with digitonin-treated mitochondria isolated from wild-type or Tom22His strains. Eluates were subjected to photo-cross-linking with presequence probes where indicated and analyzed via Western blotting using specified antisera. Load, 5%; eluate (E), 100%. PA, photoadduct. (B) Purified TOM complex from Tom22His mitochondria was subjected to presequence probe photo-cross-linking as in panel A and analyzed via SDS-PAGE and Coomassie blue staining, followed by in-gel digestion prior to LC-MALDI-MS/MS. PA, photoadduct. (C and D) Fragment ion mass spectra of presequence probe cross-linked Tom40 peptides. The b- and y-ions are indicated by b+ and y+ when carrying a cross-link to tryptic fragments of pS16B or pL19B. Note their high abundance when fragmentation occurred N terminally of Pro or C terminally of Asp, which is in agreement with the preferential cleavage of these peptide bonds during mass spectrometric sequencing. Fragment ions resulting from cleavage of the cross-link bond are labeled with the respective tryptic peptide sequence. (C) Tom40220–235 cross-linked to pL19B18–24; [M+H]+obs, 2,405.17; [M+H]+calc, 2,405.17; internal fragments with N-terminal Pro are indicated; the y8+-ion identifies 228AGVSYLT234 as the minimal cross-linking site. (D) Tom40182–190 cross-linked to pS16B15–17; [M+H]+obs, 1,560.80; [M+H]+calc, 1,560.81; the b+-ion series identifies 182TL183 as the minimal cross-linking site; the pound signs indicate signals corresponding to fragmentation of another nearly isobaric Tom40 peptide coisolated in the precursor selection window.
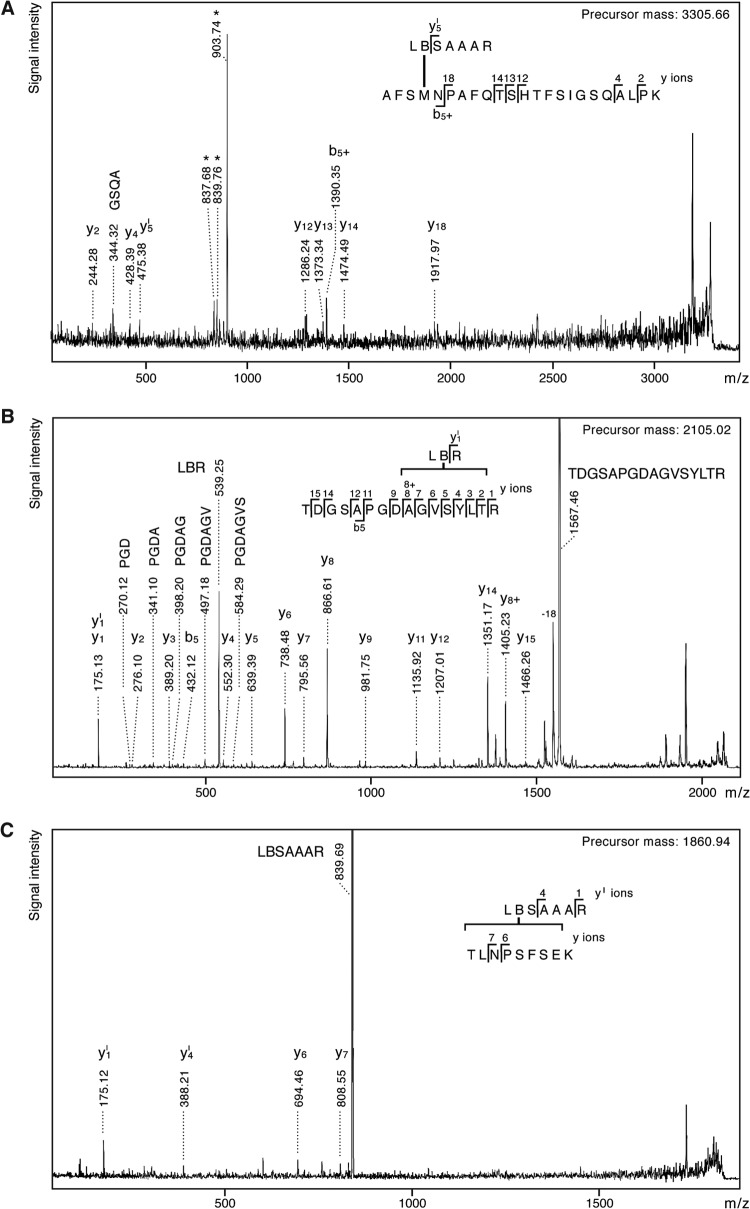
FIG 3.
Fragment ion mass spectra of presequence probe cross-linked Tom40 peptides. The b- and y-ions are indicated by b+ and y+ when carrying a cross-link to tryptic fragments of pS16B or pL19B. Note their higher abundance when fragmentation occurred N terminally of Pro or C terminally of Asp, which is in agreement with the preferential cleavage of these peptide bonds during mass spectrometric sequencing. Fragment ions resulting from cleavage of the cross-link bond are labeled with the respective tryptic peptide sequence. Internal fragments with N-terminal Pro are indicated. (A) Tom4091–113 cross-linked to pL19B18–24; [M+H]+obs, 3,305.66; [M+H]+calc, 3,305.64. The b5+-ion together with the indicative triplet signal (marked by asterisks) identified M94 as the cross-linking site. (B) Tom40220–235 cross-linked to pS16B15–17; [M+H]+obs, 2,105.02; [M+H]+calc, 2,105.03; the y8+-ion identifies 228AGVSYLT234 as the minimal cross-linking site. (C) Tom40182–190 cross-linked to pL19B18–24; [M+H]+obs, 1,860.94; [M+H]+calc, 1,860.95.
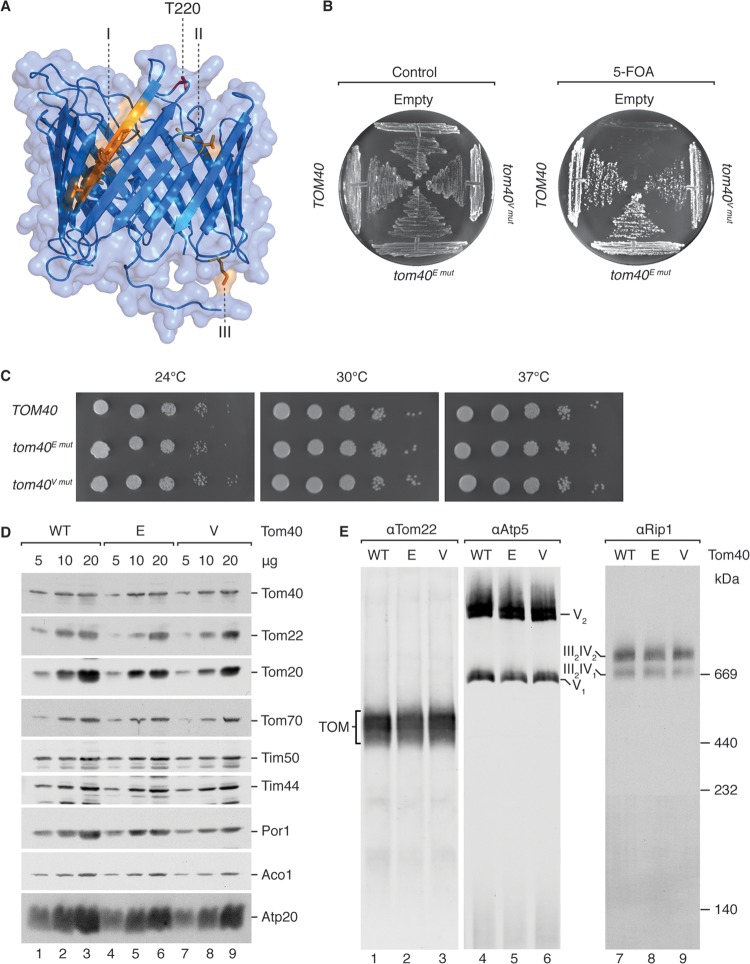
Fortuitously, recent cross-linking data as well as a study utilizing mass spectrometry joined with limited proteolysis have provided support for a Tom40 homology model based upon the mouse VDAC structure (37, 40). Together, these studies empowered us to examine the photo-cross-linking sites within a structural context (Fig. 4A). The placement of the identified presequence-interacting sites within the Tom40 homology model is strongly suggestive of two presequence recognition regions (Fig. 4A, I and II [region one] and III [region two]), corresponding to opposite ends of the barrel. Importantly, one of the two regions was easily envisaged as two photoadduct-containing peptides were found within the vicinity of each other (Fig. 4A, I and II). Further investigation of the amino acid residues residing between these two cross-linking sites led us to T220 (Fig. 4A, highlighted in red), as it was previously shown to be phosphorylated in mitochondria (38).
FIG 4.
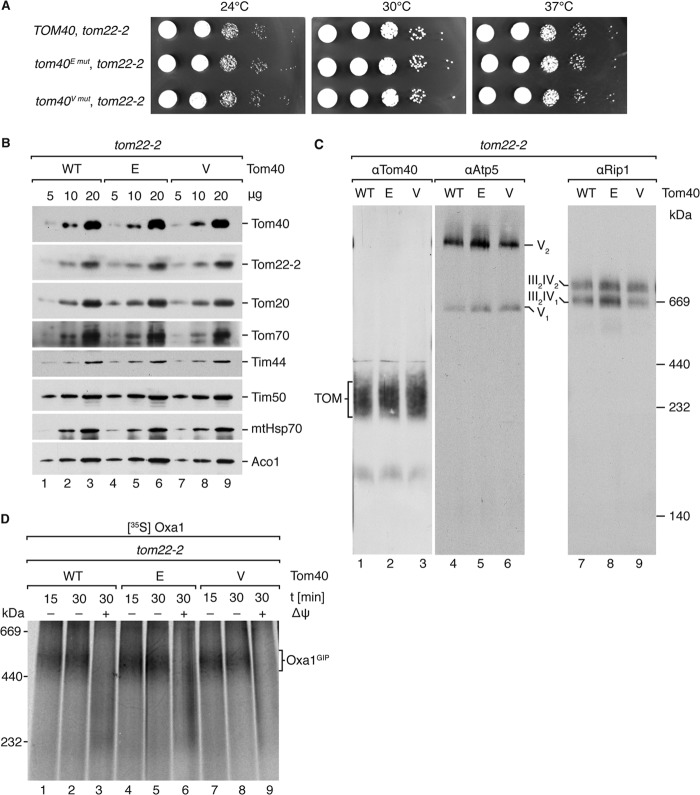
Tom40 T220 phosphomutants tom40E mut and tom40V mut exhibit no growth phenotype and wild-type-like TOM stability. (A) Lateral view of Tom40 homology model based upon the mouse VDAC structure (PDB ID 3EMN). Photo-cross-linked residues or fragments are in orange, and T220 is in red. I, 228AGVSYLT234; II, 182TL183; III, M94. (B) Yeast cells containing a chromosomal deletion of TOM40, complemented by wild-type TOM40 on a URA3 selection plasmid, were transformed with plasmids containing TOM40 alleles as indicated and subjected to plasmid loss on 5-FOA-containing medium. (C) Serial dilutions (10-fold) of the indicated strains were incubated at the indicated temperature for 3 days on nonfermentable medium. (D) Western blot analysis of steady-state protein levels in isolated mitochondria from the indicated strains after SDS-PAGE using specified antisera. (E) BN-PAGE Western blot analysis of steady-state complex levels with Tom22, Atp5, and Rip1 antisera using 40 μg, 15 μg, and 15 μg isolated mitochondrial protein, respectively.
TOM complex stability remains unaltered in Tom40 phosphomimetic (T220E) and phosphoblock (T220V) mutants.
With efforts to validate the newly identified presequence-interacting region, while remaining as close to physiological conditions as possible, we created two Tom40 T220 point mutants, mimicking a stable phosphorylation (T220E) or preventing phosphorylation (T220V). Although the implementation of targeted phosphomimetic mutations certainly falls short of replicating true phosphorylation events, their use is supported by a wide scope of literature. To this end, a tom40Δ yeast strain carrying wild-type TOM40 on a plasmid with URA3 as a selectable marker (40, 41) was transformed with either an empty plasmid or a plasmid carrying wild-type TOM40, tom40E mut, or tom40V mut. 5-FOA selection for loss of the URA3 plasmid demonstrated the viability of the tom40E mut and tom40V mut mutants as wild-type-like colony formation was observed (Fig. 4B). Growth tests on nonfermentable media revealed no growth phenotype at all tested temperatures (Fig. 4C). Moreover, we isolated mitochondria from these mutant strains and examined various steady-state protein levels via Western blotting (Fig. 4D). All proteins analyzed were shown to exhibit wild-type-like levels, especially Tom40 and other TOM subunits.
We analyzed the stability of the mutant TOM complex by migration on BN-PAGE, as previous analyses showed that mutations in Tom40 often led to destabilization of the TOM complex (35, 36, 57, 58). In our analyses, TOM stability was not affected, as complexes containing the mutant versions Tom40E and Tom40V exhibited wild-type migration patterns and amounts (Fig. 4E). Additionally, the migration of control proteins from complex III (Rip1) and complex V (Atp5) was not affected.
Analysis of TOM translocation intermediates in Tom40 T220 mutants.
To assess if the newly identified presequence interactions of Tom40 were affected in the mutants, in organello photo-cross-linking was performed in wild-type, Tom40E, and Tom40V mitochondria, utilizing pL19B (Fig. 5A). Photoadduct formation was severely compromised in Tom40E mitochondria, while Tom40V was left unaffected compared to the wild type. Importantly, the photoadducts of the TIM23 presequence receptors Tim50 and Tim23 were not attenuated in these mitochondria, indicating that in all samples the presequence peptides crossed the outer membrane and reached the intermembrane space (15, 17, 19, 20, 59, 60). Taken together, these findings confirmed the mass spectrometrically delineated Tom40 photo-cross-linking localization data and firmly establish Tom40 as a presequence interactor in mitochondria.
FIG 5.
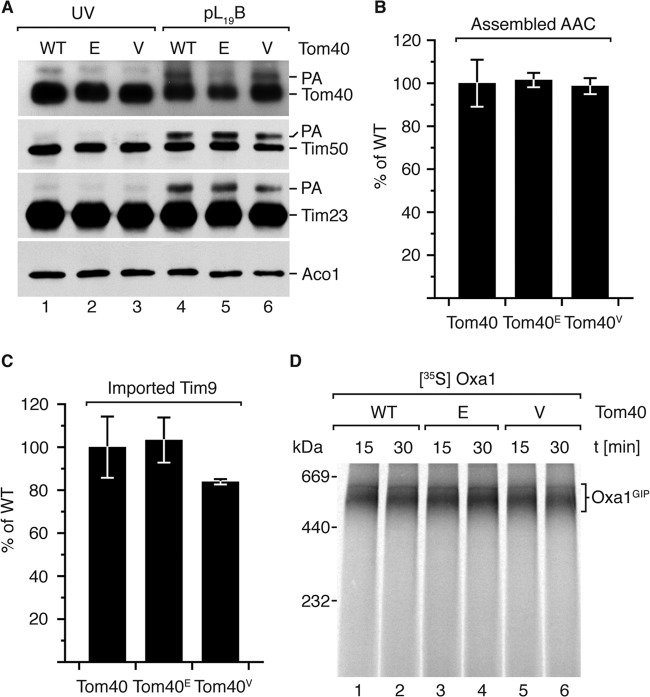
Photo-cross-linking and import analysis in isolated Tom40E and Tom40V mitochondria. (A) In organello photo-cross-linking analysis in isolated Tom40E and Tom40V mitochondria, visualized via Western blotting using the indicated antisera. PA, photoadduct. (B) Radiolabeled AAC was imported for 15 min into isolated mitochondria from the indicated strains, followed by BN-PAGE. Fully assembled AAC dimer was quantified through digital autoradiography and is presented as percentage of wild type (n = 3; standard error of the mean). (C) Radiolabeled urea-denatured Tim9 was imported for 3 min into isolated mitochondria and resolved via reducing SDS-PAGE. Imported Tim9 was quantified and is shown as percentage of wild type (n = 3; standard error of the mean). (D) Radiolabeled OxaI was imported in the absence of Δψ for the indicated times and subsequently analyzed by BN-PAGE and autoradiography.
Examining the presequence pathway specificity of the Tom40E and Tom40V mutant mitochondria, we monitored the import along other import pathways. Transport along the carrier pathway was assessed by importing radiolabeled ADP/ATP carrier (AAC) into isolated mitochondria. Import and assembly of AAC into the membrane-embedded dimer were resolved via BN-PAGE. Quantification of the assembled AAC dimer showed that mutant mitochondria were not compromised in their ability to import substrates of the carrier pathway compared to the wild type (Fig. 5B). Furthermore, Tim9, a substrate of the MIA pathway destined for the inner membrane space, was imported into mutant mitochondria, resolved via reducing SDS-PAGE, and quantified within the linear import phase (Fig. 5C). Upon comparison of the Tom40 mutants to the wild type, no significant difference was found. The Tom40V mutant mitochondria exhibited a slight reduction in Tim9 import, which was within the error of the wild-type control. Collectively, transport along the carrier and MIA pathways through the TOM complex in both the Tom40E and Tom40V mutants demonstrates essentially unaltered TOM functionality.
In order to monitor presequence-mediated translocation selectively over the TOM complex, we followed the formation of the productive OxaIGIP intermediate by importing radiolabeled OxaI precursor into mutant mitochondria with a dissipated Δψ, followed by BN-PAGE (14, 61). Comparing the relative levels of OxaIGIP-intermediate formation in Tom40E and Tom40V mutant mitochondria with those in the wild type, no significant differences were observed (Fig. 5D). The unaltered precursor translocation across the TOM complex, in conjunction with the abovementioned presequence probe photo-cross-linking data (Fig. 5A), is suggestive of either compensation from other TOM presequence interaction sites and/or late-stage (post-OxaIGIP-intermediate formation) involvement of the Tom40 binding site under evaluation.
Investigating Tom40's presequence-interacting region in the context of a Tom22IMS mutant.
In order to investigate a potential functional relationship between the established presequence-binding role of Tom22IMS and the newly identified Tom40 presequence-interacting region, we created a series of double mutants. tom40Δ yeast strains containing plasmids encoding Tom40, Tom40E, or Tom40V were transformed with an integration cassette giving rise to the previously characterized chromosomal deletion mutant tom22-2, coding for Tom22 lacking the IMS domain (12). The double mutants were respiration competent and did not exhibit a growth phenotype at all tested temperatures (Fig. 6A). Moreover, we isolated mitochondria from the double mutant cells. These mitochondria showed no alteration of tested mitochondrial proteins at steady state as assessed via Western blotting (Fig. 6B). As with the Tom40 T220 point mutants, TOM stability within the double mutants, as assessed via BN-PAGE migration, was unaltered compared to the corresponding tom22-2 single mutant strain (Fig. 6C).
FIG 6.
tom40E mut and tom40V mut alleles exhibit unaltered growth and protein levels in the tom22-2 background. (A) Serial dilutions (10-fold) of the tested strains were incubated at the indicated temperature for 3 days on nonfermentable medium. (B) Western blot analysis of steady-state protein levels in isolated mitochondria from the indicated strains via SDS-PAGE using specified antisera. (C) BN-PAGE Western blot analysis of steady-state complex levels using antisera for Tom40, Atp5, and Rip1 using 40 μg, 15 μg, and 15 μg isolated mitochondrial protein, respectively. (D) Radiolabeled OxaI was imported in the presence or absence of Δψ into mitochondria isolated from the indicated strains and analyzed by BN-PAGE and autoradiography.
Tom22IMS has previously been implicated in the import of presequence-containing substrates (12, 13, 29, 62, 63). The deletion of Tom22IMS slightly reduces import kinetics along the presequence pathway, and accordingly it has been shown to negatively affect the production of the OxaIGIP intermediate (14, 62). Therefore, we evaluated possible combinatorial functional effects of Tom22IMS deletion in the Tom40E and Tom40V mutants employing OxaIGIP intermediate formation. In the tom22-2 background, both Tom40E and Tom40V mutants behaved like the single tom22-2 mutant with regard to OxaIGIP generation (Fig. 6D). Taken together, the unaltered TOM intermediate formation across all examined Tom40 mutant mitochondria is suggestive of a later-stage protein transport role of the Tom40 presequence interaction site.
In summary, our analyses showed reduced presequence interaction with Tom40 in Tom40E mitochondria but wild-type-like precursor accumulation in the TOM complex. In contrast, Tom40V displayed wild-type-like endpoint presequence photoadducts as well as wild-type-like precursor accumulation in the TOM complex. Importantly, the removal of the IMS domain of Tom22 within the Tom40 phosphomutant backgrounds failed to impact the formation of the OxaI TOM translocation intermediate (Fig. 5D and 6D). Thus, we speculated that the Tom40 presequence-interacting region under evaluation functionally impacts protein transport downstream of the stable OxaIGIP intermediate.
Tom40E and Tom40V exhibit opposing import kinetics for matrix transport.
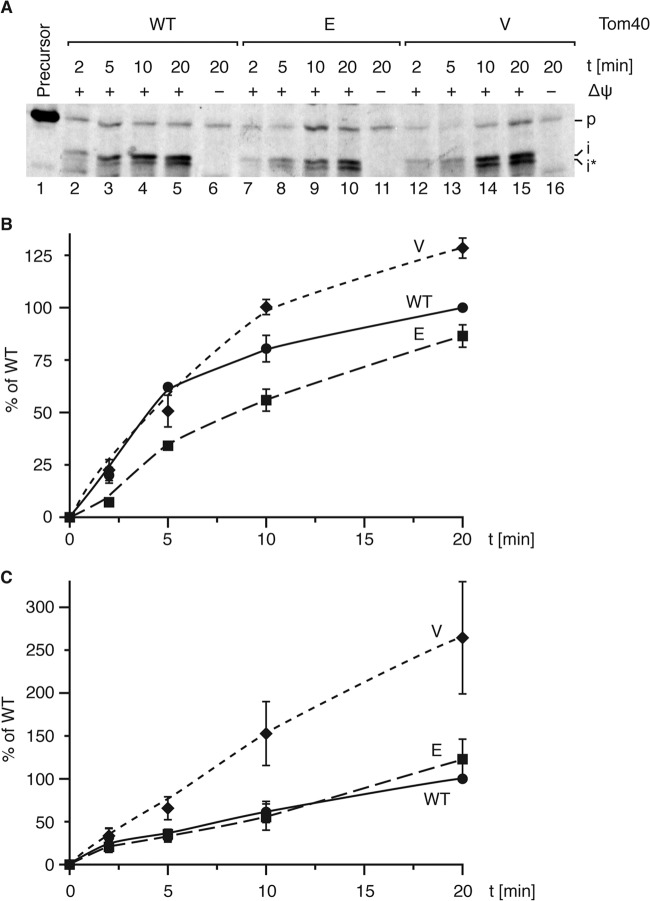
To address the role of Tom40's presequence interaction in the context of matrix transport along the presequence pathway, we challenged the Tom40T220 mutant mitochondria and performed in vitro imports with the purified matrix-destined substrate b2 (167)Δ-dihydrofolate reductase (DHFR), consisting of an N-terminal portion of cytochrome b2 fused to mouse dihydrofolate reductase. Comprehensive kinetic import analysis revealed opposing phenotypes for the mutant versions of Tom40 (Fig. 7A and B). Tom40E mitochondria exhibited retarded import during the early, linear phase of the transport reaction. Subsequently, the kinetics reached near-wild-type levels while the overall import rate remained below that of the wild-type control over the course of the experiments (Fig. 7B). In contrast, Tom40V mitochondria displayed wild-type-like import kinetics at first but appeared to remain in the linear phase for a longer period of time, resulting in a ∼20% increase in overall presequence import capacity during the time course.
FIG 7.
Tom40E and Tom40V mitochondria exhibit antagonistic Tom22IMS-related matrix import kinetics. (A) Purified b2 (167)Δ-DHFR was imported into isolated mitochondria from the indicated strains, proteinase K treated, and analyzed via Western blotting employing anti-DHFR antiserum. Precursor lane represents 15% of the input. p, precursor; i and i*, processed intermediates. (B) Quantification of import reactions as described for panel A. The processed intermediates are shown as percentage of wild type at 20 min (n = 3; standard error of the mean). (C) Purified b2 (167)Δ-DHFR was imported into mitochondria isolated from Tom40 mutant strains in the tom22-2 background as in panel A, and quantification was performed as for panel B (n = 6; standard error of the mean).
The deletion of Tom22IMS in the Tom40E and Tom40V mutant backgrounds had no significant effect on the formation of the OxaIGIP intermediate compared to the tom22-2 single mutant (Fig. 6D). However, due to the observed opposing import kinetics within the Tom40 phosphomutants, we then assessed matrix import via the presequence pathway in the double mutants using purified b2 (167)Δ-DHFR (Fig. 7C), as performed in the TOM40 single mutants. In the tom22-2 background, the Tom40E mutant exhibited tom22-2-like import kinetics, while the Tom40V mutant displayed an increase in matrix import capacity (Fig. 7C). Interestingly, the subtle reduction in matrix import kinetics observed in the single mutant Tom40E mitochondria (Fig. 7B) was alleviated through the deletion of Tom22IMS. Conversely, the increase in matrix import capacity seen in Tom40V single mutant mitochondria was exacerbated with the deletion of the IMS domain of Tom22, resulting in a significant increase in matrix import (Fig. 7C). Accordingly, the reduction in import efficiency observed in Tom40E mitochondria is functionally related to the IMS domain of Tom22, as its absence leads to tom22-2-like import kinetics. Moreover, the newly identified Tom40 presequence interaction site can also function independently of Tom22IMS, as evidenced by the persistent increase in matrix import kinetics irrespective of the IMS domain of Tom22.
Together, these data integrate presequence recognition by Tom40 functionally into the presequence pathway. Modifications of Tom40's presequence recognition motif alter import rates and modulate matrix import. Furthermore, these findings indicate a link between Tom40's presequence-binding capacity and the Tom22IMS-domain for precursor transfer from the TOM complex to the inner membrane presequence translocase.
DISCUSSION
In the present study, we identified Tom40 as a predominant presequence-interacting subunit of the TOM complex. We were able to map two presequence-interacting regions, corresponding to either side of the β-barrel, in agreement with Tom40's observed cis and trans presequence-binding sites (23, 25, 27, 29, 64). Previous works indicated the existence of specific regions involved in presequence transport through Tom40. However, mutational analyses mostly led to secondary effects on TOM stability/assembly. Case in point, a previous genetic screen identified a TOM40 allele that displayed mainly presequence-specific sorting defects but at the same time was compromised in TOM stability (35). Several mutants with attenuated presequence-specific import defects were also identified in Neurospora crassa TOM40 (36). Moreover, the recent description of a conserved polar groove uniquely present in Tom40, as opposed to other β-barrel outer membrane proteins, was speculated to be involved in presequence interaction (37). Here, we used a biochemical strategy to identify and characterize presequence-interacting regions in isolated Tom40. One of the two presequence-interacting regions found by our approach was defined to a higher degree due to the identification of multiple cross-linked peptides within the immediate vicinity (228AGVSYLT234 and 182TL183). Intriguingly, this presequence-interacting region coincides with the previously speculated area (β-sheets 8 to 11) (37).
An analysis of the TOM phosphoproteome had uncovered a multitude of phosphorylated residues within Tom40 (38). T220, the target of a currently elusive kinase, lies in the middle of one of the presequence-interacting regions defined here (Fig. 4A, T220 highlighted in red). Using phosphomimetic mutant versions of Tom40, we confirmed a presequence interaction site and found that Tom40E mutant mitochondria displayed selectively compromised Tom40 presequence-cross-linking. It is important to note that the photo-cross-linking assay would suggest that more than straightforward ionic forces are at play as the net positively charged presequence would be electrically attracted to the Tom40E mutant, albeit a distinct reduction in presequence binding is seen in this mutant. This observation is supported by past investigations into the mode of presequence binding at Tom40, where moderate affinity was still detected at 1 M NaCl (31).
Utilizing import assays in Tom40E and Tom40V mutant mitochondria, functional characterization of the newly mapped Tom40 presequence interaction site revealed unaltered import flux along both the carrier and MIA pathways, as well as in the formation of a productive TOM intermediate (OxaIGIP). However, when full transport of a matrix-targeted precursor was assessed, the import kinetics of Tom40E and Tom40V mutant mitochondria displayed opposing phenotypes, seen as diminished and escalated import kinetics, respectively. In this regard, the antagonistic import phenotypes strongly support a physiological significance of the implemented Tom40 T220 point mutants. Moreover, interpreting Tom40E and Tom40V mutant mitochondrial presequence probe photo-cross-linking data in the light of the import findings, it is apparent that this assay is blind to combinatorial effects involving a multitude of interaction sites, almost entirely lacks the capacity for kinetic resolution, and yet is irrefutably invaluable in obtaining spatial information.
Our findings place the import-mediating actions of the newly characterized Tom40 presequence interaction site functionally downstream of OxaIGIP intermediate formation. Moreover, our data suggest that the presequence-binding domains of Tom22IMS and the presented Tom40 site are to some degree functionally related. While the T-to-V exchange at position 220 allows for more efficient precursor transport independent of Tom22IMS, a T-to-E exchange renders protein transport dependent on Tom22IMS. We conclude that the precursor can be passed via both recognition sites in the wild-type context during transit toward the matrix. The T-to-E exchange in Tom40 leads to a loss of presequence recognition and concomitantly directs precursor transport via Tom22IMS. This conclusion is in agreement with the observation that Tom40E mutant mitochondria display decreased import compared to wild-type mitochondria containing uncompromised variants of both presequence interaction sites. In contrast, the T-to-V exchange allows for efficient Tom22IMS-independent transport.
The implications of a selective presequence import effect due to the adjustment of a single residue's phosphostatus strongly advocate the existence of a presequence import-modulating system, involving the presented Tom40 presequence interaction site. Such mitochondrial regulatory networks have been previously presented and intimately involve the TOM complex (38, 65).
Finally, our presequence mapping data have laid the groundwork for further mutational analyses with the aim of creating specific Tom40 mutants imparting a true block of this presequence-binding site. These mutants would significantly further the quest toward an unambiguous step-by-step delineation of outer membrane presequence passage.
ACKNOWLEDGMENTS
We are indebted to O. Lytovchenko for helpful discussion. We thank Klaus Neifer and Lars van Werven for expert technical support.
This work was supported by the Deutsche Forschungsgemeinschaft, SFB860, the Göttingen Graduate School for Neurosciences and Molecular Biosciences, and the Max Planck Society (P.R.). J.M. is a doctoral student of the Ph.D. program Molecular Biology—International Max Planck Research School and the Göttingen Graduate School for Neurosciences and Molecular Biosciences (GGNB) (DFG grant GSC 226/1) at the Georg August University Göttingen. The research of L.W. and A.C. was supported by the Foundation for Polish Science-Welcome Program cofunded by the EU within the European Regional Development Fund.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Dudek J, Rehling P, van der Laan M. 2013. Mitochondrial protein import: common principles and physiological networks. Biochim. Biophys. Acta 1833:274–285. 10.1016/j.bbamcr.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 2.Endo T, Yamano K, Kawano S. 2011. Structural insight into the mitochondrial protein import system. Biochim. Biophys. Acta 1808:955–970. 10.1016/j.bbamem.2010.07.018 [DOI] [PubMed] [Google Scholar]
- 3.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644. 10.1016/j.cell.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neupert W, Herrmann JM. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749. 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- 5.Vögtle F-N, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickamnn A, Pfanner N, Meisinger C. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139:428–439. 10.1016/j.cell.2009.07.045 [DOI] [PubMed] [Google Scholar]
- 6.Brix J, Dietmeier K, Pfanner N. 1997. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272:20730–20735. 10.1074/jbc.272.33.20730 [DOI] [PubMed] [Google Scholar]
- 7.Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100:551–560. 10.1016/S0092-8674(00)80691-1 [DOI] [PubMed] [Google Scholar]
- 8.Saitoh T, Igura M, Obita T, Ose T, Kojima R, Maenaka K, Endo T, Kohda D. 2007. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 26:4777–4787. 10.1038/sj.emboj.7601888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamano K, Yatsukawa Y-I, Esaki M, Hobbs AEA, Jensen RE, Endo T. 2008. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283:3799–3807. 10.1074/jbc.M708339200 [DOI] [PubMed] [Google Scholar]
- 10.Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T. 2011. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U. S. A. 108:15179–15183. 10.1073/pnas.1105921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietmeier K, Hönlinger A, Bömer U, Dekker PJ, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. 1997. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 388:195–200. 10.1038/40663 [DOI] [PubMed] [Google Scholar]
- 12.Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N. 1997. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 17:6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. 1998. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the “acid chain” hypothesis. EMBO J. 17:3886–3898. 10.1093/emboj/17.14.3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazier AE, Chacinska A, Truscott KN, Guiard B, Pfanner N, Rehling P. 2003. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell. Biol. 23:7818–7828. 10.1128/MCB.23.21.7818-7828.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P. 2011. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 195:643–656. 10.1083/jcb.201105098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lytovchenko O, Melin J, Schulz C, Kilisch M, Hutu DP, Rehling P. 2013. Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 32:886–898. 10.1038/emboj.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marom M, Dayan D, Demishtein-Zohary K, Mokranjac D, Neupert W, Azem A. 2011. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286:43809–43815. 10.1074/jbc.M111.261040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman B, Kawano S, Yunoki-Esaki K, Anzai T, Endo T. 2014. NMR analyses on the interactions of the yeast Tim50 C-terminal region with the presequence and Tim50 core domain. FEBS Lett. 588:678–684. 10.1016/j.febslet.2013.12.037 [DOI] [PubMed] [Google Scholar]
- 19.Bauer MF, Sirrenberg C, Neupert W, Brunner M. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33–41. 10.1016/S0092-8674(00)81320-3 [DOI] [PubMed] [Google Scholar]
- 20.Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082. 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- 21.Vestweber D, Brunner J, Baker A, Schatz G. 1989. A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature 341:205–209. 10.1038/341205a0 [DOI] [PubMed] [Google Scholar]
- 22.Baker KP, Schaniel A, Vestweber D, Schatz G. 1990. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348:605–609. 10.1038/348605a0 [DOI] [PubMed] [Google Scholar]
- 23.Rapaport D, Neupert W, Lill R. 1997. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem. 272:18725–18731 [DOI] [PubMed] [Google Scholar]
- 24.Gaikwad AS, Cumsky MG. 1994. The use of chemical cross-linking to identify proteins that interact with a mitochondrial presequence. J. Biol. Chem. 269:6437–6443 [PubMed] [Google Scholar]
- 25.Mayer A, Neupert W, Lill R. 1995. Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell 80:127–137. 10.1016/0092-8674(95)90457-3 [DOI] [PubMed] [Google Scholar]
- 26.Juin P, Thieffry M, Henry JP, Vallette FM. 1997. Relationship between the peptide-sensitive channel and the mitochondrial outer membrane protein translocation machinery. J. Biol. Chem. 272:6044–6050. 10.1074/jbc.272.9.6044 [DOI] [PubMed] [Google Scholar]
- 27.Rapaport D, Mayer A, Neupert W, Lill R. 1998. cis and trans sites of the TOM complex of mitochondria in unfolding and initial translocation of preproteins. J. Biol. Chem. 273:8806–8813. 10.1074/jbc.273.15.8806 [DOI] [PubMed] [Google Scholar]
- 28.Rapaport D, Künkele KP, Dembowski M, Ahting U, Nargang FE, Neupert W, Lill R. 1998. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Mol. Cell. Biol. 18:5256–5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanamori T, Nishikawa S, Nakai M, Shin I, Schultz PG, Endo T. 1999. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 96:3634–3639. 10.1073/pnas.96.7.3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stan T, Ahting U, Dembowski M, Künkele KP, Nussberger S, Neupert W, Rapaport D. 2000. Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J. 19:4895–4902. 10.1093/emboj/19.18.4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon DM, Wang J, Amutha B, Pain D. 2001. Self-association and precursor protein binding of Saccharomyces cerevisiae Tom40p, the core component of the protein translocation channel of the mitochondrial outer membrane. Biochem. J. 356:207–215. 10.1042/0264-6021:3560207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395:516–521. 10.1038/26780 [DOI] [PubMed] [Google Scholar]
- 33.Künkele KP, Juin P, Pompa C, Nargang FE, Henry JP, Neupert W, Lill R, Thieffry M. 1998. The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore. J. Biol. Chem. 273:31032–31039 [DOI] [PubMed] [Google Scholar]
- 34.Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S. 2001. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 153:1151–1160. 10.1083/jcb.153.6.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabriel K, Egan B, Lithgow T. 2003. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22:2380–2386. 10.1093/emboj/cdg229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman EL, Taylor RD, Go NE, Nargang FE. 2006. Effect of mutations in Tom40 on stability of the translocase of the outer mitochondrial membrane (TOM) complex, assembly of Tom40, and import of mitochondrial preproteins. J. Biol. Chem. 281:22554–22565. 10.1074/jbc.M601630200 [DOI] [PubMed] [Google Scholar]
- 37.Gessmann D, Flinner N, Pfannstiel J, Schlösinger A, Schleiff E, Nussberger S, Mirus O. 2011. Structural elements of the mitochondrial preprotein-conducting channel Tom40 dissolved by bioinformatics and mass spectrometry. Biochim. Biophys. Acta 1807:1647–1657. 10.1016/j.bbabio.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 38.Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C. 2011. Regulation of mitochondrial protein import by cytosolic kinases. Cell 144:227–239. 10.1016/j.cell.2010.12.015 [DOI] [PubMed] [Google Scholar]
- 39.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu J, Wenz L-S, Zerbes RM, Oeljeklaus S, Bohnert M, Stroud DA, Wirth C, Ellenrieder L, Thornton N, Kutik S, Wiese S, Schulze-Specking A, Zufall N, Chacinska A, Guiard B, Hunte C, Warscheid B, van der Laan M, Pfanner N, Wiedemann N, Becker T. 2013. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154:596–608. 10.1016/j.cell.2013.06.033 [DOI] [PubMed] [Google Scholar]
- 41.Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, Wiedemann N. 2008. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132:1011–1024. 10.1016/j.cell.2008.01.028 [DOI] [PubMed] [Google Scholar]
- 42.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972. [DOI] [PubMed] [Google Scholar]
- 43.Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N. 2001. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol. Cell. Biol. 21:2337–2348. 10.1128/MCB.21.7.2337-2348.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. [DOI] [PubMed] [Google Scholar]
- 45.Meisinger C, Pfanner N, Truscott KN. 2006. Isolation of yeast mitochondria. Methods Mol. Biol. 313:33–39. 10.1385/1-59259-958-3:033 [DOI] [PubMed] [Google Scholar]
- 46.Jahn O, Eckart K, Brauns O, Tezval H, Spiess J. 2002. The binding protein of corticotropin-releasing factor: ligand-binding site and subunit structure. Proc. Natl. Acad. Sci. U. S. A. 99:12055–12060. 10.1073/pnas.192449299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahting U, Thun C, Hegerl R, Typke D, Nargang FE, Neupert W, Nussberger S. 1999. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147:959–968. 10.1083/jcb.147.5.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker PJ, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408–5419. 10.1093/emboj/16.17.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wrobel L, Trojanowska A, Sztolsztener ME, Chacinska A. 2013. Mitochondrial protein import: Mia40 facilitates Tim22 translocation into the inner membrane of mitochondria. Mol. Biol. Cell 24:543–554. 10.1091/mbc.E12-09-0649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuhoff V, Arold N, Taube D, Ehrhardt W. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262. 10.1002/elps.1150090603 [DOI] [PubMed] [Google Scholar]
- 51.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ujwal R, Cascio D, Colletier J-P, Faham S, Zhang J, Toro L, Ping P, Abramson J. 2008. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U. S. A. 105:17742–17747. 10.1073/pnas.0809634105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 54.Dormán G, Prestwich GD. 1994. Benzophenone photophores in biochemistry. Biochemistry 33:5661–5673. 10.1021/bi00185a001 [DOI] [PubMed] [Google Scholar]
- 55.Wittelsberger A, Thomas BE, Mierke DF, Rosenblatt M. 2006. Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBS Lett. 580:1872–1876. 10.1016/j.febslet.2006.02.050 [DOI] [PubMed] [Google Scholar]
- 56.Rodríguez-Castañeda F, Maestre-Martínez M, Coudevylle N, Dimova K, Junge H, Lipstein N, Lee D, Becker S, Brose N, Jahn O, Carlomagno T, Griesinger C. 2010. Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity. EMBO J. 29:680–691. 10.1038/emboj.2009.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rapaport D, Taylor RD, Käser M, Langer T, Neupert W, Nargang FE. 2001. Structural requirements of Tom40 for assembly into preexisting TOM complexes of mitochondria. Mol. Biol. Cell 12:1189–1198. 10.1091/mbc.12.5.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor RD, McHale BJ, Nargang FE. 2003. Characterization of Neurospora crassa Tom40-deficient mutants and effect of specific mutations on Tom40 assembly. J. Biol. Chem. 278:765–775. 10.1074/jbc.M208083200 [DOI] [PubMed] [Google Scholar]
- 59.Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. 2009. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184:129–141. 10.1083/jcb.200808068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de la Cruz L, Bajaj R, Becker S, Zweckstetter M. 2010. The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 19:2045–2054. 10.1002/pro.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120:817–829. 10.1016/j.cell.2005.01.011 [DOI] [PubMed] [Google Scholar]
- 62.Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C. 2003. Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22:5370–5381. 10.1093/emboj/cdg532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolliger L, Junne T, Schatz G, Lithgow T. 1995. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 14:6318–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki H, Kadowaki T, Maeda M, Sasaki H, Nabekura J, Sakaguchi M, Mihara K. 2004. Membrane-embedded C-terminal segment of rat mitochondrial TOM40 constitutes protein-conducting pore with enriched beta-structure. J. Biol. Chem. 279:50619–50629. 10.1074/jbc.M408604200 [DOI] [PubMed] [Google Scholar]
- 65.Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalińska M, Guiard B, Pfanner N, Meisinger C. 2013. Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metab. 18:578–587. 10.1016/j.cmet.2013.09.006 [DOI] [PubMed] [Google Scholar]