Abstract
The phosphoinositide 3-kinase (PI3K)/PTEN (phosphatase and tensin homolog) pathway is one of the central routes that enhances cell survival, division, and migration, and it is frequently deregulated in cancer. PI3K catalyzes formation of phosphatidylinositol 3,4,5-triphosphate [PI(3,4,5)P3] after cell activation; PTEN subsequently reduces these lipids to basal levels. Activation of the ubiquitous p110α isoform precedes that of p110β at several points during the cell cycle. We studied the potential connections between p110α and p110β activation, and we show that cell stimulation promotes p110α and p110β association, demonstrating oligomerization of PI3K catalytic subunits within cells. Cell stimulation also promoted PTEN incorporation into this complex, which was necessary for PTEN activation. Our results show that PI3Ks dimerize in vivo and that PI3K and PTEN activities modulate each other in a complex that controls cell PI(3,4,5)P3 levels.
INTRODUCTION
The phosphoinositide 3-kinase (PI3K)/PTEN axis regulates cell survival, division, and migration. PI3Ks are lipid kinases conserved throughout evolution that catalyze phosphorylation of the third position of the inositol ring of phosphatidylinositol (PI). The PI3K family is subdivided into three classes, of which only class I generates phosphatidylinositol 3,4,5-triphosphate [PI(3,4,5)P3] and, after SH2 domain-containing inositol 5′-phosphatase action, PI(3,4)P2 (1–3). These plasma membrane components initiate signaling pathways that lead to cell activation and are downregulated by the PTEN phosphatase (1–5). Class IA PI3Ks are heterodimers composed of a 110-kDa catalytic subunit and a regulatory subunit: p85α (and its alternative spliced forms, p55α or p50α), p85β, or p55γ (1–5). p85α and p85β are ubiquitous, whereas p55γ is expressed only in certain tissues (1–5). Although four genes encode p110 subunits, only p110α and p110β are expressed ubiquitously in mammals and are more important in cancer (1–5). p110α and p110β are activated by receptor tyrosine kinases, although p110β can also be activated through G protein-coupled receptors (GPCRs) (6, 7). After activation, PI3K regulates processes such as cell cycling and survival (8–10).
PI3K activity increases after stimulation of growth factor (GF) receptors in early G1 phase and again in advanced G1. The first PI3K peak corresponds mainly to p110α activation, followed by a minor p110β activity peak; p110α is also activated before p110β in late G1-S-phase entry (8, 9). p110α is found in the cytoplasm, whereas p110β shuttles between the cytosol and nucleus and accumulates in the nucleus in S phase (8–10). The distinct p110α and p110β localizations and activation kinetics might reflect nonredundant roles for each isoform. p110α controls early G1 events, whereas p110β regulates S-phase progression (8–10); p110α is involved in cell responses to insulin and angiogenesis (11, 12), whereas p110β affects nuclear processes such as DNA replication and repair (9–13). p110α is activated again at the beginning of mitosis, followed by that of p110β (14). Activation of p110α thus precedes that of p110β at several points in cell cycle progression (8, 9, 13, 14), although it is not known whether p110α regulates p110β activation. The PI3K activation-induced increase in PI(3,4,5)P3 levels is subsequently reduced to basal levels by PTEN (15–17). Various mechanisms regulate PTEN activity (16, 17), although it is unclear how its activation is linked to that of PI3K.
Given that p110α activation precedes that of p110β, we postulated that p110α activity might be necessary for p110β activation. We found that after cell stimulation, p110α associated with p110β, and formation of this complex controlled optimal p110β activation. PTEN was also incorporated into this complex, and its association with p110α/p110β regulated its phosphatase activity. The results show that PI3K catalytic subunits oligomerize within cells. We propose that this complex controls optimal activation of p110β and PTEN, suggesting PI3K/PTEN cooperation in the temporal control of cell PI(3,4,5)P3 and PI(3,4)P2 levels.
MATERIALS AND METHODS
Cell lines, cell culture, plasmids, siRNA, and reagents.
p110αflox/flox mouse embryonic fibroblasts (MEF) and p110β-deficient MEF were kindly donated by J. J. Zhao and T. M. Roberts (Dana-Farber Cancer Institute, Boston, MA) (18, 19). Wild-type (WT), p85β−/−, and p85α+/− MEF were obtained from p85β−/− and p85α+/− mice (20, 21), which were donated by D. Fruman (University of California, Irvine, CA). MEF were prepared as described previously (8, 18, 19). U2OS, PC3, 293T, NIH 3T3, and MEF cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal bovine serum, 2 mM glutamine, 10 mM HEPES, 100 IU/ml penicillin, and 100 μg/ml streptomycin. pSG5-p110α, -myc-p110α, -myc-p110β, -p85α, and -p85β are described elsewhere (8, 22). His-tagged wild-type hp110β was donated by B. Vanhaesebroeck (Barts Cancer Institute, Cancer Research UK, London, United Kingdom); the cDNA encoding V12-Ras was donated by J. Downward (London Research Institute, London, United Kingdom). Recombinant hemagglutinin (rHA)-p85α, rHA-p85β, and rHA-SH3-BcR (SH3, Src-homology domain 3; BcR, breakpoint cluster region homology domain), and p50α were described previously (23). LacZ and E160-p85α lentiviral vectors were donated by G. Mills (M. D. Anderson Cancer Center, Houston, TX) (24). Murine p110β shRNA was from Origene, and p110α shRNA has been described elsewhere (8). Small interfering RNA (siRNA) for p85α or -β, human p110 isoforms, and PTEN and scrambled siRNA were custom-made (Invitrogen). PIK75 and TGX221 are described elsewhere (8). Okadaic acid and the PTEN inhibitor bpV(pic) were from Calbiochem.
Antibodies, Western blotting, immunoprecipitation, and in vitro transcription/translation.
Blots were probed with the following antibodies (Ab): anti-Myc tag (9B11; Cell Signaling), anti-pan-p85 and anti-histone (Upstate Biotechnology, Millipore), anti-β-actin (Sigma), anti-Akt, anti-pAkt(Thr308), and anti-pAkt(Ser473) (Cell Signaling). Anti-HA was from Covance, and anti-p85β was described previously (22). Anti-His monoclonal antibody (MAb) was from Clontech, and anti-PCNA was from BD Transduction Labs; antibodies to Sin1 and p400 were from Bethyl Laboratories and Abcam, respectively. Horseradish peroxidase-conjugated secondary Ab were from Dako Cytomations. We used anti-p110α C73F8 MAb (Cell Signaling) for all purposes, anti-p110β C33D4 MAb (Cell Signaling) for human cells, and S-19 Ab (Santa Cruz Biotechnology) for murine p110β. We used anti-PTEN 6H2.1 MAb (Millipore) for Western blotting (WB) of whole-cell lysates and goat polyclonal Ab N-19 (Santa Cruz Biotechnology) for immunoprecipitations. For optimal detection of immunoprecipitated PTEN, WB was performed using anti-PTEN MAb 138G6 (Cell Signaling). An enhanced chemiluminescence kit was from GE Healthcare. Cells were lysed in a detergent buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 10 mM KCl, 0.5% Nonidet P-40) with protease inhibitors (11836170001; Roche Applied Science) and incubated at 4°C for 1 h, followed by centrifugation (9,300 × g, 10 min). Western blotting and immunoprecipitation were performed as described previously (8, 23). Specific cDNAs were transcribed and translated in vitro in the presence of [35S]Met using the TNT T7-coupled reticulocyte lysate system (Promega).
Transfection, subcellular fractionation, immunofluorescence, and PI3K assay.
Cells were transfected with Lipofectamine Plus (Qbiogene) and cultured 48 h before analysis; for siRNA transfection, we used Lipofectamine Max (Qbiogene). For subcellular fractionation, cells that were transfected and lysed as described above were cultured for the indicated times/conditions, and cytoplasmic, nuclear, and chromatin fractions were isolated as described previously (25). Immunofluorescence analysis with green fluorescent protein (GFP)–Btk-PH was performed as described previously (10). For PI3K assays, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (9). PI3K was immunoprecipitated with anti-p110α and anti-p110β antibodies. p110α and p110β immunoprecipitates were extensively washed and resuspended in 45 μl of 50 mM HEPES (pH 7.5) containing purified PI (0.5 mg/ml; Avanti Polar Lipids). For the kinase reaction mixture (50 μl, final volume), we used 5 μl 10× kinase buffer (10 μCi [32P]ATP, 100 mM MgCl2, and 200 μM unlabeled ATP). Reaction mixtures were incubated (15 min, 37°C), and reactions were terminated by addition of 1 mM HCl (100 μl). Phosphoinositides were extracted in methanol-chloroform (1:1 [vol/vol]; 200 μl, final volume). Radiolabeled products and nonradioactive PI(3)P, PI(3,4)P2, and PI(3,4,5)P3 standards (Echelon Biosciences) were resolved in silica gel plates; before autoradiography, the positions of phosphoinositide standards was determined with iodine vapor. The signal intensities of PI(3)P spots produced in this assay were quantitated by using ImageJ software, the background signal for control immunoprecipitates was subtracted, and the values were divided by the signal intensity of the p85 loading control. For PI3K assays performed using gel filtration fractions (see Fig. 4C, below), PI(3)P signal was normalized to that of p110α or p110β loading. To compare p110α or p110β activity in different conditions and assays, the distinct values obtained were referred to p110α or p110β maximal activity (considered 100%).
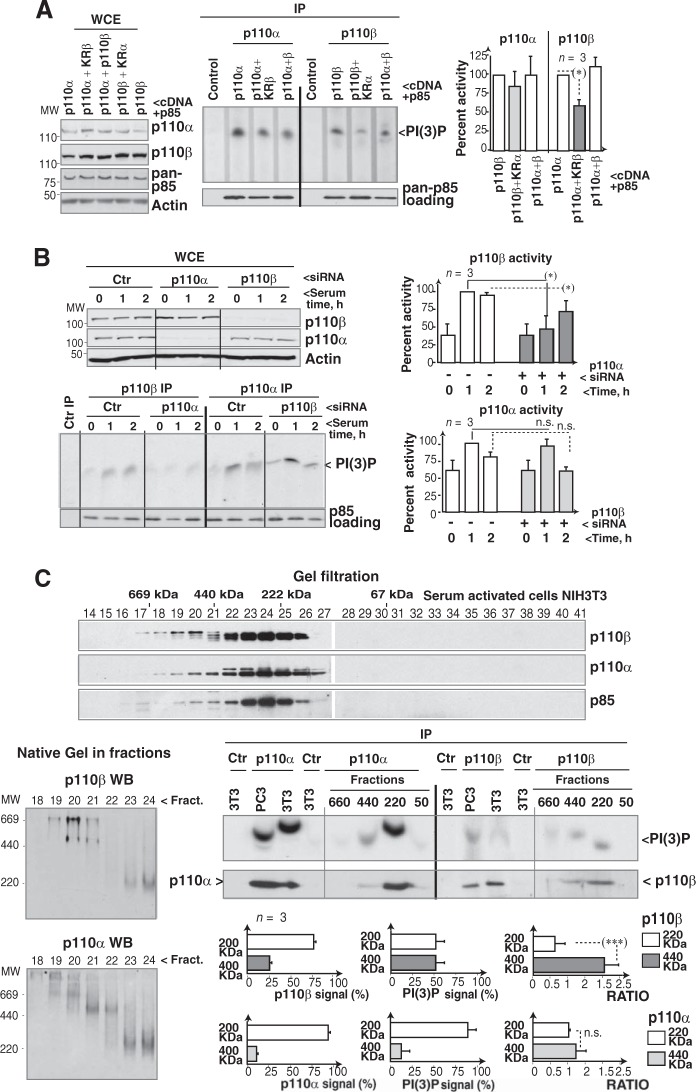
FIG 4.
Interference with p110α expression or activity reduces p110β activity. (A) NIH 3T3 cells were transfected with control, K802R-p110α, and/or K805R-p110β cDNA combined 1:1 with HA-p85α cDNA (36 h). Whole-cell extracts (WCE) were analyzed by WB. Extracts (500 μg) were also immunoprecipitated with p110α or p110β Ab, and the amount of precipitated PI3K was tested by WB with a pan-p85 Ab. PI3K activity was tested in vitro. We used PI as the substrate for immunopurified PI3K, since its PI(3)P product is resistant to PTEN (potentially present in the complex). For controls, cell extracts were incubated with protein A. Graphs show the signal intensity of each PI(3)P spot relative to the maximum for p110α or p110β (100%; n = 3). (B) U2OS cells were transfected with control p110α or p110β siRNA (48 h). Cells were incubated without serum (19 h) and then stimulated with medium plus 10% serum (1 or 2 h). Extracts were analyzed as described for panel A. (C) Serum-starved NIH 3T3 cells were stimulated with serum (10%; 1 h), and extracts were separated by gel filtration. Fractions were resolved by SDS-PAGE and blotted with the indicated antibodies. A fixed volume from these fractions (20 μl) was resolved with 6% native gel electrophoresis and analyzed by WB. We also immunoprecipitated p110α or p110β from NIH 3T3 or PC3 cell extracts (controls) or from the indicated pooled fractions and tested PI3K activity as described for panel A. Graphs show the percentage of p110 in each fraction compared to the total (p110 in all fractions as 100%; left), the amount of PI(3)P signal in each fraction [relative to total PI(3)P signal as 100%; center], and the ratios of these values (right). *, P < 0.05; ***, P < 0.001; n.s., not significant (Student's t test).
Cell cycle, gel filtration, and native gel electrophoresis.
Cell cycle analysis was performed as described previously (8). 293T cells, PC3, and NIH 3T3 cells were lysed as described above. Extracts were applied to a Superdex 200 column (Amersham Pharmacia) and washed with lysis buffer (0.5 ml/min, 4°C); after the void volume, 0.5-ml fractions were collected. Blue native gel electrophoresis was performed using whole-cell extracts or gel filtration fractions as described previously (26).
PTEN activity assay.
For the PTEN activity assay, cells were lysed as described above and immunoprecipitated with anti-PTEN Ab plus protein G (4°C, 19 h). Immunoprecipitates were washed and used for WB and phosphatase assays (27). For the phosphatase assay, immunoprecipitates were washed and resuspended in enzyme reaction buffer (50 mM Tris [pH 8], 50 mM NaCl, 10 mM dithiothreitol, 10 mM MgCl2) with 3 μM PI(3,4,5)P3 (Echelon Biosciences; 30 min, 37°C); reactions were terminated with malachite green reagent (100 μl; Echelon Biosciences). Free phosphate levels were measured in an enzyme-linked immunosorbent assay (ELISA) reader as the absorbance at 620 nm (A620). Phosphate released during the reaction was extrapolated from a titration curve obtained using dilutions of a phosphate standard provided by the kit manufacturer. To compare different assays, PTEN activity for each time and transfection condition was calculated as a percentage of the maximal PTEN activity (A620 minus background) normalized to the maximal signal in each experiment (considered 100%).
Statistical analyses and quantitation.
Cell cycle plots were analyzed with the Cytomics CxP software. Fluorescence and WB signal intensity were measured with ImageJ software (NIH). To quantitate p110 WB signals of SDS-PAGE-resolved p110 immunoprecipitates, we measured the signal intensity in each band and divided this value by the corresponding signal intensity of the p85 loading control. This number was expressed as a percentage relative to the maximum value in each experiment (100%). For cells with lower p110 levels than control cells (after p85 subunit deletion or depletion [see Fig. 3B and C]), the p110β WB signal of SDS-PAGE-resolved p110α immunoprecipitates was first normalized to that of the loading control (immunoprecipitated p110α), and the resulting values were normalized for p110β cellular levels (as determined by WB for whole-cell extracts). Statistical significance was evaluated using Prism5 V.5.0 software.
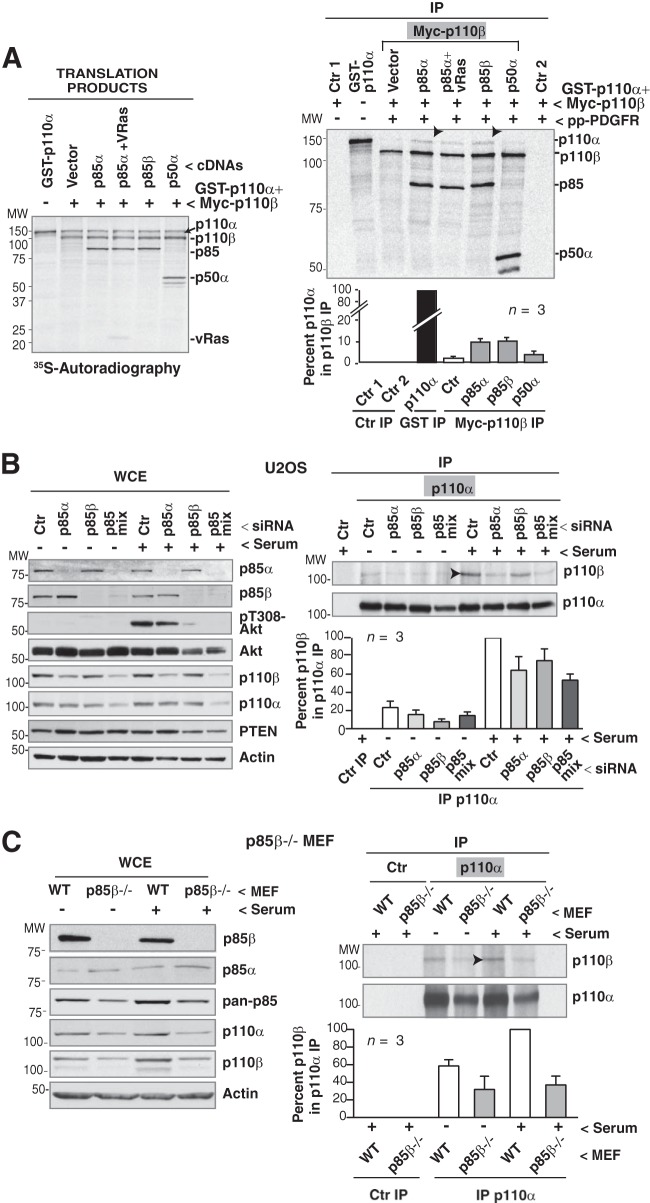
FIG 3.
p85 mediates p110α/p110β association. (A) cDNAs encoding GST-p110α (135 kDa) and Myc-p110β (110 kDa) in combination with different cDNAs as indicated were transcribed/translated in vitro in the presence of [35S]methionine. Products were immunopurified using anti-Myc Ab and incubated with or without a PDGF receptor phosphopeptide (pp-PDGFR) or v-HRas (indicated) for 30 min. Translation products (left) and complexes (right) were resolved by SDS-PAGE and analyzed by autoradiography. The graph shows the percentage of p110α signal in complex with Myc-p110β relative to maximum (GST-p110α purified from a similar amount of translation product). (B) U2OS cells were transfected with different siRNA (48 h), incubated without serum (19 h), and stimulated with 10% serum (1 h). Whole-cell extracts (WCE; 50 μg) or immunoprecipitated proteins (500 μg) were analyzed by WB. The graph shows the amount of p110β in complex with p110α, normalized to the amount of immunoprecipitated p110α and to p110β cellular levels (determined by WB of total extracts). Normalized values are shown relative to the maximum p110β signal (p110β amount in complex with p110α in serum-stimulated control siRNA transfected cells as 100%; means ± standard errors of the means [SEM]; n = 3). The immunoprecipitation control was cell extract plus protein A. (C) WT or p85β-deficient MEF were processed and analyzed as described for panel B. In panels A, B, and C, arrowheads indicate the maximum p110 oligomer detected in the experiment.
RESULTS
Endogenous p110α associates with p110β.
p110α is activated before p110β at the G0/G1 transition, as well as in the G1/S and G2/M transitions (8, 9, 13, 14). We analyzed whether p110α associated with p110β and regulated its activity. To study associations of endogenous proteins, we immunoprecipitated p110β from extracts of exponentially growing 293T cells (1 mg) and tested for associated p110α by WB; we used p110α immunoprecipitated from 200 μg as a positive control. The assay showed that a fraction of p110α was present in complex with p110β (Fig. 1A). The proportion of p110α in p110β immunoprecipitates could not be calculated accurately, since the antibody used to immunoprecipitate p110β differed from that of the positive control (anti-p110α). To compare experiments, we normalized the p110α signal in complex with p110β (and that of the p110α positive control) to the p85 signal. As most cellular p110 is in complex with p85 (28), p85 measurement estimates the amount of p85/p110 in different immunoprecipitates when the same Ab is used. p110α signal intensity was ∼40% in p110β precipitates compared to that of the p110α positive control, indicating that anti-p110β coimmunoprecipitated a relatively large amount of p110α (Fig. 1A). In the reciprocal experiment, anti-p110α antibody precipitated less p110α/p110β complex (discussed below), although this experiment confirmed that a fraction of p110α associated with p110β (Fig. 1A).
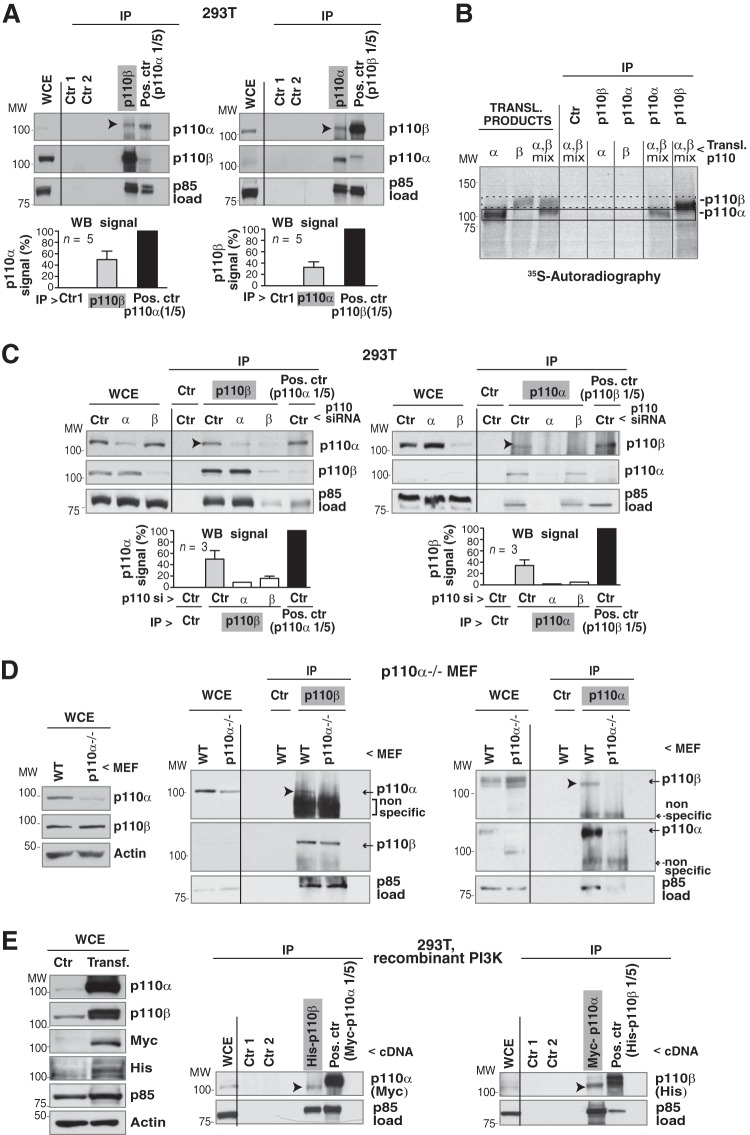
FIG 1.
Endogenous p110α and p110β form a complex. (A) Endogenous p110α or p110β was immunoprecipitated from 293T cell extracts (1 mg) by using specific Ab. WB was used to test for associated p110. A smaller amount (1/5) of whole-cell extract (WCE; 200 μg) was used to immunoprecipitate p110β or p110α positive controls. Negative controls included protein A plus Ab (Ctr 1) and extract plus protein A (Ctr 2); WCE (50 μg) and immunoprecipitates (IP) were analyzed by WB. Graphs show the percentage of p110α signal in the p110β immunoprecipitates and in the p110α positive control, normalized for p85 signal, which estimates the amount of PI3K precipitated under both conditions. Normalized signals are shown as a percentage of maximal signal (p110α positive control, 100%; mean ± standard error of the mean, n = 5). The percentage of p110β in p110α immunoprecipitates was calculated similarly. (B) cDNAs encoding p110α, p110β, or both were transcribed/translated in vitro in the presence of [35S]methionine. Translation products or immunoprecipitated products were visualized by autoradiography. As controls, translation products were incubated with protein A. Frame with solid line, p110α signal; frame with dashed line, p110β signal. (C) 293T cells were transfected with control, p110α, or p110β siRNA (72 h). p110β, p110α, and positive controls were immunoprecipitated from WCE as described for panel A. The negative control contained cell extract plus protein A. The graphs are similar to those for panel A. n = 3. (D) p110αflox/flox MEF were infected with adenoviral Cre (72 h); pik3ca gene deletion was 80% efficient. Cre-infected or WT p110α MEF were lysed and tested as described for panel A. The figure shows a representative experiment (n = 3). (E) 293T cells were transfected with cDNAs encoding Myc-p110α, His-p110β, HA-p85α, and HA-p85β (48 h). WCE (50 μg) was tested by WB or immunoprecipitated (1 mg) using anti-His Ab (for p110β); associated p110α was analyzed with WB. For the positive control, we immunoprecipitated Myc-p110α-transfected cell extract (200 μg) with anti-Myc Ab. Negative controls were as described for panel A; the reciprocal assay was performed similarly. For panels A, C, D, and E, empty lanes rule out spillover from adjacent lanes. Arrowheads indicate the p110 isoform (p110α or p110β) in complex with the other. MW, molecular weight.
We tested for antibody specificity by using p110α and p110β separately transcribed and translated in vitro. Anti-p110α Ab did not immunoprecipitate the p110β-translated product (and vice versa), indicating antibody specificity (Fig. 1B). When cDNAs encoding p110α and p110β were translated together, neither anti-p110α nor -p110β Ab precipitated the p110α/p110β complex, suggesting that the purified isoforms do not associate in vitro under these conditions (Fig. 1B). To confirm Ab specificity, we reduced p110β or p110α expression by transfecting specific or control (scrambled sequences) siRNA in 293T cells (72 h) and tested for complex formation with p110α. The p110α coimmunoprecipitated in complex with p110β was reduced after p110α silencing, showing specificity of p110α detection; anti-p110α Ab precipitated trace amounts of p85 and did not immunoprecipitate associated p110β (Fig. 1C). Immunoprecipitation of p110β in p110β-depleted cells did not precipitate p110α or p110β or p110β-associated p85 (Fig. 1C); results were similar in U2OS cells (see Fig. S1A in the supplemental material). Quantification of WB signals confirmed reduction of p110α in complex with p110β (and vice versa) after p110α (or p110β) knockdown (Fig. 1C).
We also analyzed p110α/p110β oligomers in MEF, which do not express p110α (19). We infected p110αflox/flox MEF with adenoviral Cre twice (72 h) and used WB to estimate pik3ca gene deletion efficiency (∼80%). Although pik3ca gene deletion was partial, it eliminated p110α signal in complex with p110β and vice versa (Fig. 1D). In p110β-deficient MEF, alone or reconstituted with WT p110β (18), immunoprecipitation of p110β followed by WB of p110α confirmed complex formation in controls but not in p110β-deficient MEF (see Fig. S1B in the supplemental material). The p110α/p110β complex was thus only detectable in MEF that express p110α and p110β, confirming Ab specificity.
We verified the p110α/p110β association by using recombinant proteins. Cotransfection of Myc-p110α, His-p110β, and p85 allowed complex isolation using anti-tag Ab. Anti-His Ab was less efficient than anti-Myc Ab, as determined by the low p85 levels precipitated, but immunoprecipitation with either Ab confirmed the p110α/p110β association (Fig. 1E; see also Fig. S1C in the supplemental material).
Cell activation controls p110α/p110β dimerization.
p110α and p110β are activated sequentially after GF receptor stimulation at several points during cell cycle progression (8). To test whether cell activation triggered p110α/p110β association, we used NIH 3T3 cells, which can be arrested in G0 by serum deprivation (19 h) and induced to enter the cell cycle by serum addition (1 h) (8). We immunoprecipitated p110α or p110β in extracts of quiescent or serum-stimulated cells and examined associated proteins by WB. The p110α/p110β association was minimal in quiescence but was induced after cell activation (Fig. 2A). Serum stimulation also induced p110α/p110β complex formation in freshly isolated primary MEF (Fig. 2B) and in human U2OS cells (see Fig. S1D in the supplemental material).
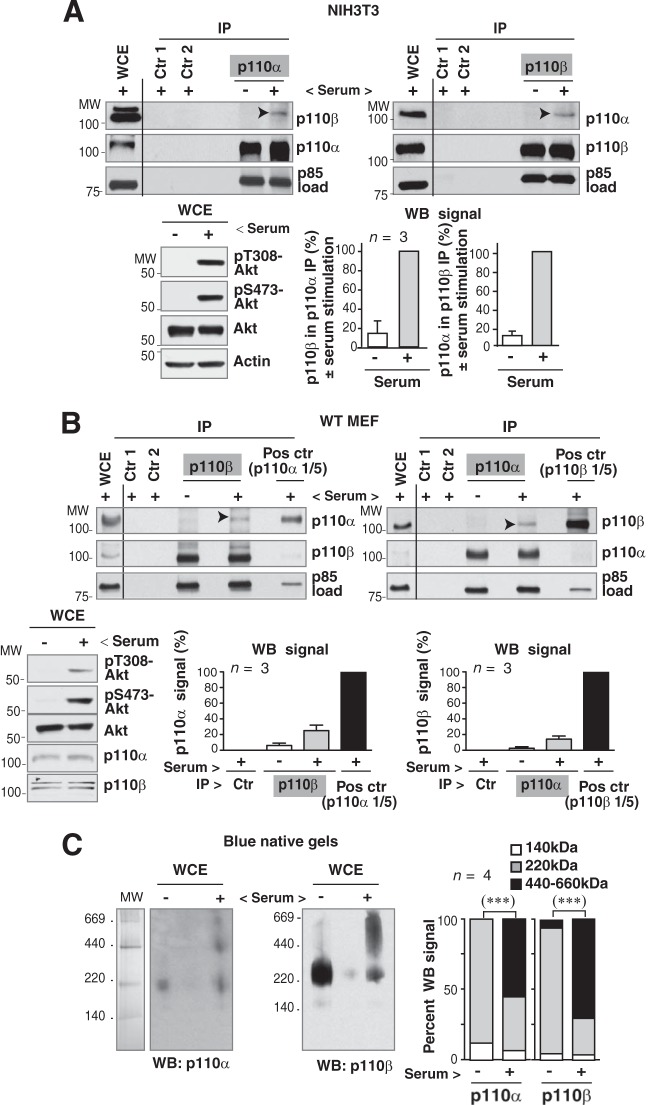
FIG 2.
p110α expression regulates p110β activation. (A) NIH 3T3 cells were synchronized in G0 by serum deprivation (19 h) and activated by serum addition (1 h). Cell activation was confirmed by WB analysis of pAkt levels in whole-cell extracts (WCE; 50 μg). WCE (70 μg) and extracts (1 mg) immunoprecipitated with p110α or p110β Ab were also analyzed by WB. Negative controls were similar to those described for Fig. 1A. Graphs show the percentage of p110β signal in immunoprecipitates of p110α in quiescent cells compared to that for serum-stimulated cells (100%); the graph is similar for p110α (means ± standard errors of the means [SEM], n = 3). (B) WT MEF were serum deprived and activated as described for panel A; cell activation was confirmed by WB analysis of pAkt. Endogenous p110α or p110β was immunoprecipitated from extracts (1 mg) by using specific Ab, and associated p110 was analyzed by WB. A smaller amount (1/5) of WCE (200 μg) was used to immunoprecipitate positive controls. Negative controls were similar to those described for Fig. 1A; WCE (50 μg) and immunoprecipitates were analyzed by WB. Graphs show the percentage of p110α signal in immunoprecipitates of p110β normalized to p85 levels and relative to the maximum (p110α immunoprecipitated from 1/5 WCE as 100%) and a similar representation for p110β signal in immunoprecipitates of p110α (mean ± SEM; n = 3). In panels A and B, empty lanes rule out spillover from adjacent lanes. Arrowheads indicate the p110α or p110β complex. (C) Cell extracts from quiescent and serum-stimulated (1 h) 293T cells were resolved with 4 to 16% blue native gel electrophoresis. The lane containing molecular weight (MW) markers was Coomassie blue stained; the remaining gel was analyzed by WB using anti-p110α or anti-p110β Ab. The graph shows the proportion of p110α or p110β in the fractions compared to signal in the entire lane (means ± SEM; n = 4). ***, P < 0.001, Pearson's chi-square test.
To identify endogenous p110α/p110β complexes by an alternative method, we used blue native gel electrophoresis, which allows detection of protein complexes in total cell extracts (without prior immunoprecipitation). Extracts of quiescent and serum-stimulated 293T cells were resolved by 4 to 16% blue native electrophoresis. Under native conditions, protein complex mobility differed from that in SDS-PAGE. In quiescent cells, the majority of p110α and p110β signal was observed near 220 kDa, corresponding to a p85:p110 heterodimer; in contrast, after 1 h of cell activation, significant fractions of p110α and p110β were found at ∼440 and ∼660 kDa (Fig. 2C). The proportions of p110α and p110β with a high molecular mass identified under native conditions (∼50% of total p110α or p110β) were greater than those from the immunoprecipitation, suggesting that the latter underestimates p110α/p110β complexes in activated cells.
p85 mediates PI3K oligomerization.
Purified transcribed and translated p110α and p110β did not associate in vitro (Fig. 1B), although they exhibit PI3K activity (29), suggesting that incorrect folding of p110α or p110β is not the cause of defective association. We therefore postulated that p110α/p110β association can be mediated by additional proteins. As most cellular p110 is associated with the p85 regulatory subunit (28), we tested whether addition of p85 and platelet-derived growth factor receptor diphosphopeptide (pp-PDGFR), which mimics receptor activation (30), induced p110α/p110β complex formation in vitro. p110α and p110β have similar molecular masses; to improve resolution in WB, we used a fusion protein with glutathione S-transferase for p110α (GST-p110α), which has a mass of 135 kDa. The combination of p85 (p85α or p85β) and the pp-PDGFR peptide triggered p110α/p110β complex formation (Fig. 3A). Class I p110 has a Ras-binding domain, and v-Ras mediates optimal p110α activation (31); nonetheless, addition of v-Ras to the p85 plus pp-PDGFR peptide mixture did not further increase p110α/p110β association (Fig. 3A). These results showed that in the presence of pp-PDGFR and p85, p110α and p110β associate in vitro, although the amount of complex was lower than that in living cells (Fig. 1A).
Most cellular p110 is in complex with p85 (28); since p85 molecules dimerize through their N-terminal SH3-BcR domains (32), p85 dimers might mediate the p110α/p110β association. To test this, we used p50α, a splice form of p85α that has the two SH2 domains and the inter-SH2 region, responsible for p110 association, but lacks the SH3-BcR region needed for p85 dimerization (32). Transcribed/translated p50α plus pp-PDGFR peptide failed to mediate p110α/p110β association (Fig. 3A), suggesting that p85 dimerization through N-terminal SH3-BcR domains is necessary for p110α/p110β complex formation.
To confirm p85 involvement in complex formation, we depleted cells of p85α or p85β. p85 depletion reduced p110 stability (Fig. 3B), as reported previously (33). To distinguish between reduced complex formation and destabilization, we measured the p110β signal in complex with p110α and normalized this signal to the amount of immunoprecipitated p110α and to p110β cellular levels. Depletion of p85α or p85β decreased p110α/p110β complex formation (Fig. 3B). We also studied complex formation in p85-deficient MEF. The low p110 levels in p85α−/− MEF (20) hampered interpretation of results; we used p85β−/− MEF (21). p85β−/− MEF showed less p110β in complex with p110α than control MEF (Fig. 3C). As in knockdown cells, p85β deletion reduced p110 levels; we normalized the p110β signal in complex with p110α to the amount of immunoprecipitated p110α and to cell p110β levels (Fig. 3C).
We found that a fraction of p110α associated with p110β following serum stimulation in human and murine cells (including primary cells), as detected by immunoprecipitation/WB and in native electrophoresis of whole-cell extracts. The findings that p85 but not p50α induces p110α/p110β association in vitro and that p85 deletion or knockdown reduces complex formation indicated that p110α and p110β association is mediated, at least in part, by p85.
p110α regulates p110β activation.
To determine whether p110α regulates p110β activity, we interfered with cellular p110α activity by overexpressing an inactive p110α form (K802R-p110α). Cells in exponential growth were cotransfected with various combinations of WT p110α, His-WT-p110β, Myc-K802R-p110α, and Myc-K805R-p110β and mixed 1:1 with HA-p85α to stabilize p110 expression (48 h) (33). Recombinant protein expression was analyzed via WB; p110 was expressed at levels similar to that of endogenous protein (Fig. 4A). We immunopurified p110α (with anti-p110α Ab) or p110β (with anti- p110β Ab) and confirmed equal enzyme levels by using a pan-p85 Ab in the WB assay, which allowed evaluation of p110α and p110β immunoprecipitates with the same Ab. Although PI(4,5)P2 is the physiological PI3K substrate in vivo, we used PI as the substrate of p110α and p110β, as these immunoprecipitates were not anticipated to have perceptible amounts of 4- or 5-PI-kinases. PI is an efficient substrate in vitro, with the advantage that its product, PI(3)P, is resistant to PTEN action (27). Whereas inactive p110β did not markedly affect p110α activity, inactive p110α reduced p110β activity (Fig. 4A). KR-p110α only partially reduced p110β activity (∼40%), since KR-p110α expression was within the range of the endogenous protein (9). KR-p110β had no effect on p110α activity (Fig. 4A), suggesting that p110α activity regulates that of p110β but not vice versa.
To test the effect of reducing p110α expression on p110β activation, we transfected U2OS cells with p110α or p110β siRNA (72 h) and incubated them in serum-free medium followed by serum stimulation (1 or 2 h). We immunoprecipitated p110α or p110β and compared p110β activity in control and p110α-depleted cells, and vice versa. We checked silencing efficiency via WB; whereas p110β depletion did not notably affect p110α activation, p110α silencing decreased serum-induced p110β activity (Fig. 4B), confirming a p110α effect on p110β activation.
Gel filtration can resolve ∼220-kDa p85/p110 heterodimers from higher-mass p85/p110 oligomers; we used this method to study p110β activity in oligomers in serum-stimulated NIH 3T3 cells. Although the entire gel could not be visualized in the linear range, since different fractions had distinct amounts of p85 and p110, longer exposure times permitted detection of p110α and p110β at ∼220 kDa and at higher masses (∼440 kDa and ∼660 kDa) (Fig. 4C). To test whether p110α and p110β form part of complexes with distinct molecular masses, we analyzed these fractions by native electrophoresis. The analysis identified p110α and p110β in protein complexes, with masses near 220, 440, and 660 kDa (Fig. 4C), which confirmed p85/p110 oligomer formation in serum-stimulated cells. We also immunoprecipitated p110α and p110β from high- and low-mass fractions and performed PI3K assays. The amount of p110 in the ∼660-kDa fraction was very low, making quantitation difficult. The analysis of the ∼220- and ∼440-kDa fractions showed that, whereas the specific activity of p110α was similar at both molecular masses, p110β was more active in the ∼440-kDa fraction, suggesting that p110β was more active in the oligomeric fraction (Fig. 4C).
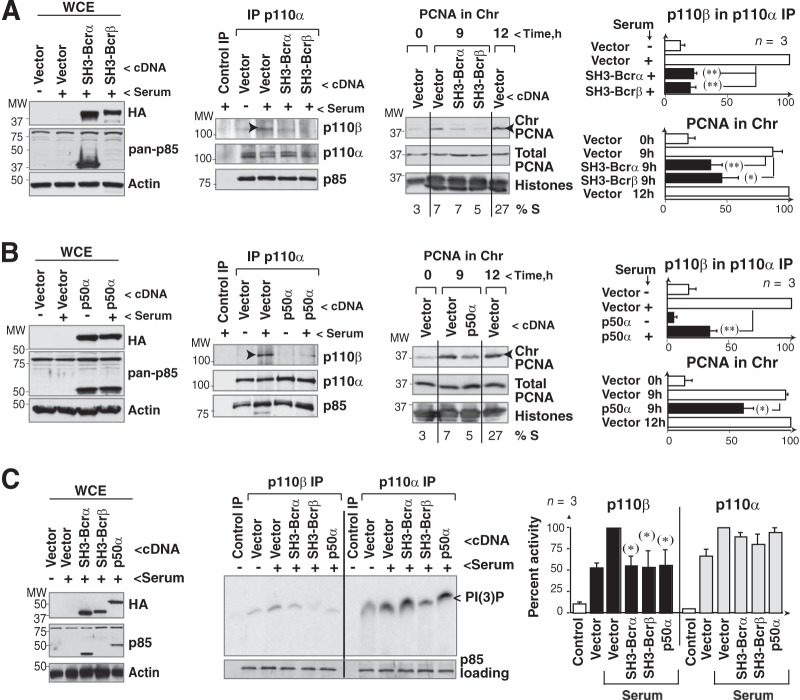
For an alternative approach to show that p110α/p110β association affects p110β activity, we analyzed the effect of disrupting p110α/p110β complexes on PCNA loading onto chromatin, a p110β-regulated process (9). SH3-BcR domains associate between themselves in vitro but do not bind to p110 (1, 32) and thus should not form p110 oligomers. Similarly, p50α (which lacks the SH3-BcR region) associates with p110 but is unable to bind another p85 molecule (32). Expression of p50α or SH3-BcR reduced p110α/p110β complex formation as well as PCNA loading onto chromatin (Fig. 5A and B). The percentage of cells in S phase was similar in control and p50α- or SH3-BcR-expressing cells (Fig. 5A and B; see also Fig. S2 in the supplemental material), ruling out a delay in the G1-S transition as the cause of defective PCNA recruitment onto chromatin.
FIG 5.
Interference with p110α/p110β association reduces p110β activity. (A and B) NIH 3T3 cells were transfected with SH3-BcRα, SH3-BcRβ (A) or p50α cDNA (B) (24 h), and protein expression was analyzed by WB. The anti-pan-p85 Ab did not recognize SH3-BcRβ. Cells were also incubated without serum (19 h) and stimulated by serum addition (1 h), and extracts were immunoprecipitated with anti-p110α Ab. We used protein A plus cell extract as a control. p110β in complex with p110α was analyzed by WB. NIH 3T3 cells were also collected at 9 or 12 h post-serum addition (S-phase entry; percent indicated) and fractionated as cytosolic, nuclear, and chromatin extracts, and the amount of PCNA in chromatin (Chr) was determined by WB. Graphs show the percentage of p110β in complex with p110α relative to the maximum (in controls, indicated with arrowheads, 100%) and that of PCNA bound to chromatin relative to the maximum (with control cells at 12 h as 100%, indicated with arrowheads; means ± standard errors of the means [SEM], n = 3). (C) p110α or p110β was immunoprecipitated from extracts of cells transfected as described for panels A and B; the amount of precipitated PI3K was tested by WB with an anti-pan-p85 Ab. PI3K activity was tested in vitro. WCE were also analyzed by WB. The graph shows PI(3)P signal relative to the maximum for p110α or p110β (100%; mean ± SEM, n = 3). *, P < 0.05; ** P < 0.01 (Student's t test).
We also transfected NIH 3T3 cells with p50α or SH3-BcR to determine their effect on p110β activity. After serum stimulation, p110α activity was unaffected by expression of p50α, SH3-BcRα, or SH3-BcRβ. In contrast, p110β immunoprecipitates from p50α- or SH3-BcR-expressing cells showed lower PI3K activities (Fig. 5C), which indicated that the p110β association with p110α regulates optimal p110β activation. These results showed that serum stimulation induces p110β association with and activation by p110α.
p110β is also activated by GPCRs, such as sphingosine-1-P (S1P) or lysophosphatidic acid (LPA) (6, 7). In contrast to PDGF or serum, S1P alone does not trigger cell cycle entry efficiently (34). To determine whether p110β activation is also linked to that of p110α after GPCR stimulation, we compared Akt activation by PDGF and S1P in p110α or p110β knocked-down cells. PDGF induced greater Akt activation than S1P, which required longer exposure times for pAkt detection. Moreover, whereas early PDGF-induced Akt activation was sensitive to p110α and p110β silencing, S1P-induced pAkt was more sensitive to p110β knockdown (see Fig. S1E in the supplemental material). At later times, p110α or p110β silencing triggered a more sustained PDGF-induced Akt activation than did controls (see below). The modest defect in pAkt levels after p110α silencing in S1P-stimulated cells supports the idea that p110β activation by GPCR is less dependent on p110α than activation via tyrosine kinase receptors.
PTEN expression is not essential for p110α/p110β association.
Based on gel filtration results, p110α and p110β had molecular masses corresponding to p85/p110 heterodimers (∼220 kDa) and higher (∼440 and ∼660 kDa) (Fig. 4). PTEN and p110 isoforms have reported to coelute at ∼660 kDa in gel filtration fractions (35), although it is unknown whether cell activation induces formation of PI3K and PTEN complexes or whether PTEN activity is altered in the ∼660-kDa fraction.
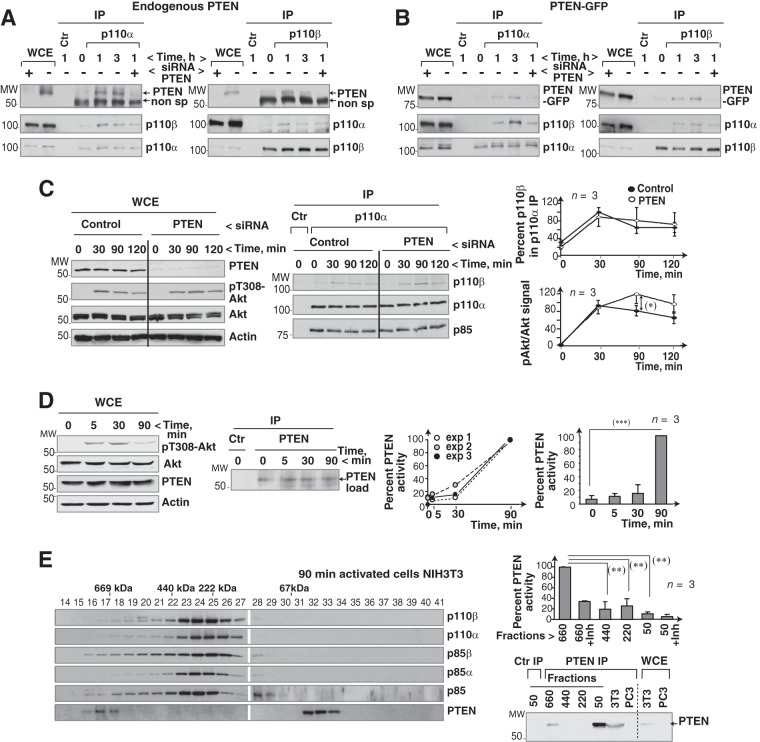
We tested whether cell activation triggered the PTEN association with p110α and p110β. Incubation of NIH 3T3 cells with serum induced PTEN binding to p110α and p110β (Fig. 6A). Although the mobility of the PTEN band was similar to that of IgG, the PTEN signal appeared specific, as it was reduced by PTEN knockdown (Fig. 6A). We also transfected 293T cells with a previously described PTEN-GFP construct (36) with lower mobility (∼80 kDa) than PTEN. PTEN-GFP is functionally active, as it reduces PI(3,4,5)P3 and pAkt levels in cells (36). 293T cells expressing PTEN-GFP were serum stimulated; we used WB to test for PTEN in p110α or p110β immunoprecipitates. Cell activation induced p110α or p110β association with PTEN-GFP (Fig. 6B). PTEN siRNA partially reduced PTEN-GFP levels in whole-cell extracts and decreased PTEN-GFP signal in complex with p110α or p110β (Fig. 6B). To study the influence of PTEN expression on p110α/p110β association, we used gel filtration and resolved extracts of 293T cells and of PTEN-deficient PC3 cells. As controls for the gel filtration experiment, we confirmed that the p400 ATPase eluted in the ∼660-kDa fraction and that the Sin1 protein eluted as a monomer and in high-mass fractions (see Fig. S3 in the supplemental material), as reported elsewhere (37, 38). p110α and p110β appeared in the ∼660-kDa fractions in 293T cells but not in PC3 cells, although p110α immunoprecipitation and WB analysis of p110β showed that both cell types had p110α/p110β complexes in the ∼440-kDa fraction (see Fig. S3).
FIG 6.
PTEN binds to the p110α/p110β complex. (A) 293T cells were incubated in serum-free medium (2 h) and stimulated with serum. Prior to stimulation, some of the cells were transfected with PTEN-specific siRNA (48 h). Extracts were immunoprecipitated with anti-p110α or anti-p110β Ab and tested for associated PTEN by WB. (B) 293T cells were transfected with PTEN-GFP (48 h) and control or PTEN-specific siRNA (48 h); cells were then incubated without serum (2 h) and serum stimulated for the indicated times. We tested PTEN-GFP in p110α or p110β immunoprecipitates by WB. For controls, extracts were incubated with an irrelevant Ab. (C) U2OS cells were transfected with control or PTEN siRNA (48 h) and activated as described for panel A for the indicated times. Transfection efficiency and pAkt levels were analyzed by WB. p110β in complex with p110α was analyzed by WB. Graphs show the percentage of p110β in complex with p110α relative to the maximum (100%) and that of pAkt signal normalized to Akt levels and relative to the maximum (100%) (mean ± SEM, n = 3). (D) To determine optimal PTEN activation time, we incubated NIH 3T3 cells without serum (19 h) and stimulated them with serum for various times. Extracts were analyzed by WB or immunoprecipitated with anti-PTEN Ab and tested in an in vitro phosphatase assay using PI(3,4,5)P3 as the substrate. Phosphate release was analyzed, using an ELISA reader, based on the A620. Values after background subtraction were normalized to PTEN loading. The left graph shows the percentage of PTEN activity relative to the maximum (A620 at 90 min; 100%) in three independent experiments; the right graph shows mean ± SEM PTEN activity (n = 3). Maximum phosphate released at 90 min was ∼103 pmol. (E) Extracts of serum-stimulated NIH 3T3 cells (10%; 90 min) were resolved by gel filtration and analyzed by WB. PTEN was immunoprecipitated from different fractions (as indicated) and analyzed by WB and in a PTEN activity assay. For the ∼50- and ∼660-kDa fractions, duplicate precipitates were tested in the presence of a PTEN inhibitor (bpV, 100 nM). The graph shows PTEN activity for each condition relative to the maximum (100%, 90 min in the ∼660-kDa fraction; mean ± SEM, n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test).
To confirm that PTEN is not needed for p110α/p110β association in an alternative approach, we reduced PTEN with siRNA and tested for p110α/p110β complex formation by immunoprecipitation and WB. PTEN silencing was efficient and induced prolonged Akt activation but did not reduce p110α/p110β association (Fig. 6C). The data suggest that cell activation induces PTEN incorporation into the p110α/p110β complex, but PTEN is not needed for p110α/p110β complex formation.
p110α/p110β regulates PTEN activity.
We evaluated whether PTEN incorporation into p110α/p110β complexes alters PTEN activity. To determine PTEN activation kinetics, we compared the activity of immunoprecipitated PTEN (from quiescent or serum-stimulated NIH 3T3 cells) in a PTEN phosphatase assay using PI(3,4,5)P3 as the substrate. The PTEN activity assay consistently showed a marked increase in PTEN activity at 90 min post-serum activation, which coincided with a pAkt decrease (Fig. 6D).
To compare the activity of monomeric PTEN with that of PTEN in the ∼660-kDa fraction, we used gel filtration to resolve 90-min serum-stimulated NIH 3T3 cell extracts and compared the immunopurified PTEN activities in low- and high-mass fractions. In the ∼660-kDa fraction, PTEN phosphatase activity was significantly greater than that in fractions of monomeric PTEN (∼50 to 60 kDa) (Fig. 6E), suggesting greater PTEN activity in the high-mass fraction containing p110α/p110β complexes. We checked that PTEN activity in the ∼660-kDa fraction was sensitive to PTEN inhibitors (Fig. 6E).
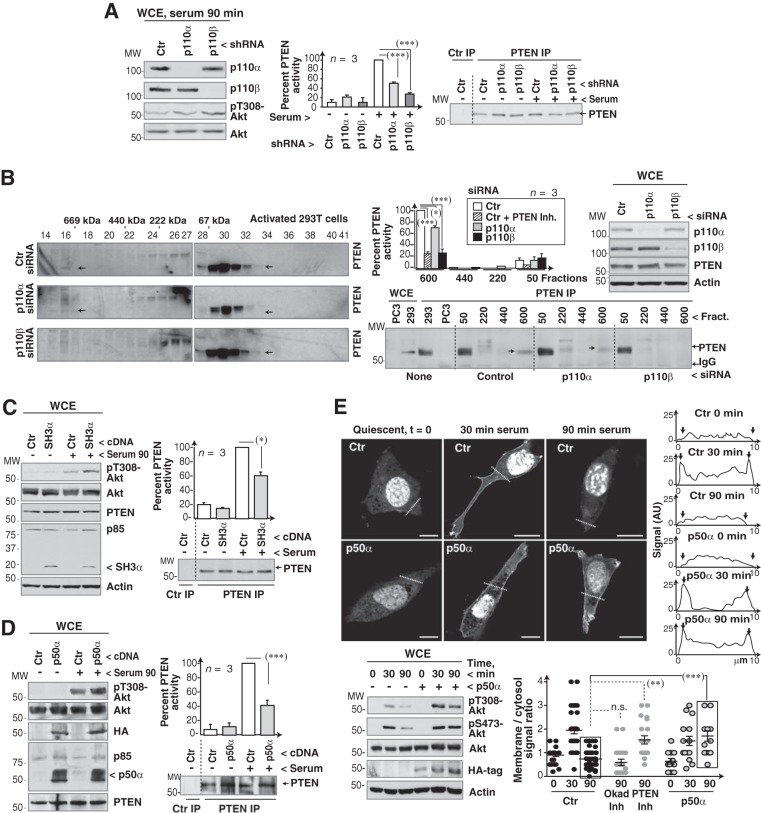
To test whether the p110α/p110β complex affects PTEN activity, we depleted cells of p110α or p110β and determined the activity of immunopurified PTEN. In serum-stimulated cells (90 min), p110α or p110β silencing led to decreased PTEN activity (Fig. 7A). Moreover, although p110α and p110β silencing reduced early Akt activation, pAkt levels were prolonged at late times after serum addition and more markedly in the case of p110β knockdown (Fig. 7A; see also Fig. S4A in the supplemental material); results were similar after cell activation with PDGF (see Fig. S1E in the supplemental material). p110α or p110β inhibition using PIK75 and TGX221 at selective doses (8, 9) reduced the pAkt peak at 30 min and also PTEN activity at 90 min post-serum stimulation (see Fig. S4B in the supplemental material). PIK75 and TGX221 treatments also reduced PTEN activity in primary MEF (see Fig. S4C). Serum induced at late times points higher pAkt levels in p110α−/− and p110β−/− MEF than in controls (see Fig. S4D). Thus, p110α and p110β regulate PTEN activity in normal cells.
FIG 7.
The p110α/p110β complex regulates PTEN activation. (A) NIH 3T3 cells (murine) were transfected with the indicated shRNA (48 h) and serum stimulated (90 min). Extracts were analyzed by WB or immunoprecipitated with anti-PTEN Ab. PTEN activity was assayed in vitro using PI(3,4,5)P3 as the substrate, and PTEN levels in the precipitates were tested by WB. The graph shows PTEN activity normalized to PTEN levels and compared to the maximum (90 min for serum-stimulated control cells; means ± standard errors of the means [SEM], n = 3). (B) 293T cells were transfected with control, p110α, or p110β siRNA (48 h), incubated in serum-free medium (20 h), and serum stimulated (90 min). Extracts were resolved by gel filtration, and PTEN distribution and activity in the fractions from control and p110α- or p110β-depleted cells were analyzed as described for panel A; the graph is also similar to that in panel A. Knockdown efficiency was confirmed by WB. (C and D) 293T cells (C) or NIH 3T3 cells (D) were transfected with the indicated cDNAs (48 h) and serum stimulated (90 min). Extracts were analyzed by WB or immunoprecipitated with anti-PTEN Ab and examined as described for panel A. (E) NIH 3T3 cells transfected with GFP-Btk-PH and control or p50α cDNA (48 h) were incubated in serum-free medium (2 h) and serum stimulated for 30 or 90 min. In some control cells, we added PTEN inhibitor (bpV, 100 nM) for the last 60 min prior to collection (at 90 min). Cells were analyzed by confocal microscopy (central z-sections) to visualize the PI(3,4,5)P3 location. Graphs (right) show the signal intensity (in arbitrary units [AU]) along the dashed lines of representative cells; arrowheads in the graphs mark the cell membrane. The graph (bottom) shows membrane signal in an area surrounding the plasma membrane (after subtracting background), divided by the signal for a similar area in cytosol (background subtracted). Each dot represents a cell (n = 24) of a representative experiment of six. Cell extracts were analyzed by WB. **, P < 0.01; ***, P < 0.001; n.s., not significant (Student's t test).
We also analyzed the effect of p110α or p110β silencing on PTEN activity in low- and high-mass fractions from human cells. p110β knockdown, more markedly than that of p110α, reduced PTEN incorporation and activity in the ∼660-kDa fraction (Fig. 7B). Thus, p110α and p110β modulate PTEN incorporation into the ∼660-kDa oligomer and regulate its activity.
Depletion of p85α or p85β, which reduces p110α/p110β complex formation (Fig. 3), also decreased PTEN phosphatase activity (Fig. S4E in the supplemental material). Since p85α is proposed to activate PTEN by direct interaction (39), the reduction in PTEN activity could be due to defective p110α/p110β complex formation or to less monomeric p85-PTEN binding. To dissect these two possibilities and test whether the p110α/p110β oligomer influences PTEN activity, we analyzed the effect of expressing p85 forms that interfere with p110α/p110β complex formation but do not bind directly to PTEN. As PTEN interacts with p85 via its BcR domain (39), we expressed p50α or an SH3 fragment of p85. We used the natural p85α truncation mutant (E160-p85α), found in endometrial cancer, which encompasses the p85 SH3 domain, does not bind PTEN, and impairs p85 dimerization (24). E160-p85α expression at levels similar to that of endogenous p85 reduced PTEN activity and enhanced pAkt levels in 90 min serum-stimulated cells (Fig. 7C).
Expression of p50α, which does not bind directly to PTEN but impairs the p110α/p110β association (Fig. 5), also inhibited PTEN activity and blocked Akt downregulation at late times (90 min) (Fig. 7D). To further evaluate the effect of expressing p50α on PTEN activity, we analyzed pAkt levels and PI(3,4,5)P3 localization at the plasma membrane at several times post-serum stimulation. We transfected NIH 3T3 cells with the GFP-Btk-PH domain (PH domain of Btk), which specifically binds PI(3,4,5)P3 (40). The membrane signal of GFP-Btk-PH was low in quiescence, increased after serum addition (at 30 min), and decreased by 90 min, coinciding with a pAkt decrease. The reduction in membrane PI(3,4,5)P3 at 90 min post-serum stimulation was sensitive to PTEN inhibitors but not to a PP2A inhibitor (100 nM okadaic acid) (Fig. 7E; see also Fig. S4F in the supplemental material). Whereas p50α expression did not affect plasma membrane PI(3,4,5)P3 levels at 30 min poststimulation, it blocked the reduction in membrane PI(3,4,5)P3 levels at 90 min (Fig. 7E). Accordingly, p50α expression did not reduce, but slightly enhanced, early Akt activation and blocked pAkt level reductions at late times after serum stimulation (90 min) (Fig. 7E), consistent with a PTEN activation defect. These results suggest that serum stimulation induces formation a p110α/p110β complex that incorporates PTEN; interference with this complex reduces PTEN activation.
DISCUSSION
The observation that p110α activation precedes that of p110β at several points in cell cycle progression prompted us to study the potential contribution of p110α in p110β activation. Serum activation of cells promoted formation of a complex between p110α and p110β, as demonstrated by immunoprecipitation, gel filtration, and native gel electrophoresis. Interference with endogenous p110α expression or activity impaired optimal p110β activation in serum-treated cells, suggesting a link between p110α and p110β activation. Disruption of the p110α/p110β complex reduced p110β activity, which confirmed that optimal p110β activation requires p110α association. Cell activation also induced PTEN binding to p110α and p110β. Incorporation into this complex enhanced PTEN phosphatase activity, since interference with complex formation reduced PTEN activity and led to prolonged Akt activation and PI(3,4,5)P3 persistence at the membrane at late times poststimulation. These findings indicated that PI3K oligomerization modulates p110α, p110β, and PTEN activation.
Dimerization of receptors or of signaling molecules is a powerful cell strategy to modulate receptor activation, signal amplitude, enzyme activation, and substrate specificity (41–43). Here, we report in vivo dimerization of p110α and p110β. Only fractions of cellular p110α and p110β form part of this complex. Based on coimmunoprecipitation from human cell extracts, we estimated that ∼40% of cellular p110β is in complex with p110α. The anti-p110α Ab precipitated the complex less efficiently; this antibody is less effective in immunoprecipitation and might recognize a region involved in p110α/p110β association. Analysis of this complex by using native gel electrophoresis showed higher percentages of oligomeric p110α and p110β (∼50%), suggesting that immunoprecipitation underestimates the amount of complex. We detected the complex in human and murine cell lines and in primary MEF. Specificity of the antibodies used to detect complexes was confirmed by using purified transcribed/translated p110α and p110β forms as well as by p110α and p110β depletion (in cell lines) or deletion (in MEF). In many cases, only a fraction of a signaling molecule is activated after cell stimulation, for example, Ras and Cdc42 (22, 44). Moreover, immunofluorescence and fractionation studies show that only a fraction of PI3K translocates to the cell membrane and responds to GF receptor activation (9, 45–47).
We found that p110α associates with and regulates p110β activity after cell stimulation, suggesting that the p110α/p110β complex is not constitutive but is linked to the cell activation process. PI3K heterocomplexes have been reported in vitro (30) and, in the case of p85, by using recombinant bacterial proteins and in insect cells (32). Nonetheless, p110 intermolecular associations were not previously studied in cells. The existence of these complexes is supported by the NSH2-iSH2-p85α/p110α crystal structure, which shows a region of p110 intermolecular contacts (48, 49). We found that interference with p110α expression or activity reduced serum activation of p110β. p110β activity was less dependent on p110α in cells activated via GPCR with S1P, a less potent stimulus than serum, which alone does not trigger cell cycle entry (34); this suggests that p110β can also be activated (at least by GPCR) independently of p110α. The need for p110α/p110β association for optimal p110β activation explains the late p110β activation kinetics compared to p110α in the G1 and S phases and in mitosis (8, 9, 13, 14).
One mechanism by which p110α binding to p110β could regulate p110β activity is direct p110α phosphorylation of p110β, since class IA enzymes have protein kinase activity (50). Our in vitro kinase assays using wild-type p110α kinase and an inactive p110β form (as the substrate) excluded this possibility (data not shown). Large conformational changes during p110 activation might also explain p110β activation when in complex with p110α. Conformational changes in p110 were first postulated to explain that p85/p110 binding to activated receptors enhances p110 activation (51). Binding of an SH2 domain to the PDGF receptor alters SH2 conformation (52). Moreover, comparison of the p110α crystal structure alone, bound to an inhibitor, or with an active form (His1047-p110α), showed the pronounced conformational difference between active and inactive p110α molecules (48, 53–55). Given these differences and the evolutionary conservation of class IA p110 subunits, p110β association with serum-activated p110α might trigger an activating conformational change in p110β; this hypothesis requires structural confirmation.
We showed that p85 subunits facilitate p110α/p110β complex formation. Since SH3-BcR domains associate among themselves (32), it is tempting to speculate that p85 dimerization drives PI3K oligomerization. PI3K complex formation might be triggered by molecule proximity on the membrane; nonetheless, at early time points, insulin receptor stimulation triggers greater activation of p50α/p110 than of p85α/p110 complexes (56), suggesting that p50α/p110 translocates correctly to the membrane. Nonetheless, p50α expression impaired p110α/p110β complex formation, which rules out the possibility that simple membrane proximity triggers p110 dimerization. SH2-SH2 domains bind more efficiently to Tyr-phosphorylated peptides than full-length p85 (23); p50α might thus mediate greater p110 activation, as it lacks the N-terminal p85 domains that mask SH2-SH2 interaction with Tyr-phosphorylated residues. This suggests a mechanism by which p85 can induce p110α/p110β complex formation after cell activation. p85 binding to receptors could involve a conformational change (as for isolated SH2 domains [52]) that alters p85 and unmasks the SH2-SH2 region (for phosphoresidue binding) as well as the SH3-BcR region (for p85-p85 intermolecular associations). As most cellular p85 is in complex with p110, SH3-BcR region-mediated p85 dimerization would bring p110α and p110β into this complex (28). p110α association with p110β could involve additional intermolecular interactions since complexes formed more efficiently in cells than in vitro. Nevertheless, for the in vitro assay we used translated p85, p110α, and p110β in the presence of pp-PDGFR peptide; these conditions do not represent the full cascade of cellular events triggered by serum stimulation in vivo.
Cell activation also induced PTEN association with p110α and p110β. Analysis of gel filtration data suggests that p110α/p110β/PTEN complexes elute in the ∼660-kDa fraction, as the ∼660-kDa p110α and p110β signals were lost in PTEN-deficient cells and that of PTEN (at ∼660 kDa) was reduced by p110α or p110β silencing. Three approaches indicated that PTEN is activated in this complex. First, the PTEN fraction in the oligomer at ∼660 kDa had markedly higher activity than monomeric PTEN. Determination of PTEN activity in the ∼660-kDa complex presented some limitations, as it required extract fractionation and was carried out using a fraction with relatively low PTEN levels (Fig. 6D). Nevertheless, in support of our data, nonphosphorylated PTEN is detected in the ∼660-kDa gel filtration fraction (35) and dephosphorylation is a PTEN activation mechanism (16). Second, depletion of any PI3K component of the complex (p85α, p85β, p110α, or p110β) reduced PTEN activity. Compared to controls, PI3K-depleted cells (p110α or p110β) showed lower PTEN activity and higher pAkt levels at late times (90 min post-PDGF or serum activation), suggesting that p110α and, more markedly, p110β control PTEN activation after cell stimulation.
The third approach that showed regulation of PTEN activation by p110α/p110β complex was interference with complex formation by expression of p50α or of an SH3 truncation mutant (E160-p85α) that reduced PTEN activity and prolonged Akt activation. The p85 BcR region mediates its binding to PTEN (39). p50α and the SH3 truncation mutant thus cannot affect PTEN by direct binding. In contrast, since p50α reduces complex formation (Fig. 5) and E160-p85α impairs p85 dimerization (24), p50α and the SH3 truncation mutant might act by reducing PI3K/PTEN oligomer formation. In the case of p50α, we showed that PTEN inhibition coincides with a defect in downregulation of membrane PI(3,4,5)P3 levels at late time points after serum stimulation, as determined using the Btk-PH domain. Reduction in membrane PI(3,4,5)P3 at 90 min is probably due to PTEN activation, as this is the time of maximal PTEN activity, although we cannot rule out the possibility that an increase in phospholipase C activity at 90 min (which would enhance IP4 levels) could also induce Btk-PH detachment from the membrane (57). The observation that p50α expression inactivated PTEN would help to explain the functional advantage of tumors with increased levels of p55γ, whose domains are similar to those of p50α (58).
The proposal that the p110α/p110β complex regulates PTEN activity contrasts with a previously suggested role for monomeric p85α in PTEN binding and activation (39). The monomeric p85α/PTEN model does not explain our finding that expression of p50α or the E160-p85α mutant impaired PTEN activity without binding directly to PTEN. Direct PTEN regulation by p85 is supported by data showing that liver-specific p85α-deficient mice generate an aggressive hepatocellular carcinoma, and their hepatocytes show reduced PTEN activity and prolonged Akt activation (59, 60). Nonetheless, these findings also support the role proposed for p85 in mediating PI3K/PTEN oligomerization. The mixture of purified PTEN with purified p85 enhances PTEN activity (39); it is nonetheless difficult to envision the cellular scenario represented in this assay, as p85 is not found as a monomer in cells (28). The in vitro activation of PTEN by p85 (39) might mimic the action of endogenous p85 in the oligomeric PTEN at ∼660 kDa. Indeed, those authors showed that only the BcR domain binds to PTEN in pulldown assays, whereas in cells only the ΔSH3-BcR deletion but not the BcR deletion impaired PTEN activation (39), suggesting that the ΔSH3-BcR deletion (similar to p50α) might reduce PTEN activation due to its ability to interfere with PI3K oligomerization.
We propose that, rather than PTEN regulation by monomeric p85α, a PI3K p85/p110α/p110β oligomer binds to and regulates PTEN activity. This model is supported by the finding that the E160-p85 and p50α forms do not bind PTEN but impair p85 dimerization and reduce PTEN activity. PTEN regulation by the p110α/p110β complex is also consistent with the finding that GST-PTEN pulls down p85 as well as p110 from cell extracts (24, 32, 39).
During the course of our study, PTEN was reported to form dimers. In these dimers, an inactive PTEN form can inhibit an associated WT PTEN molecule, which explains the increased tumor formation by inactive PTEN compared to heterozygous loss of PTEN (61). These findings do not contradict our results; those authors isolated dimers of ∼100 kDa but also detected PTEN in the ∼660-kDa fraction. Compared with cells that express a single WT PTEN allele, cells with one WT and one inactive PTEN allele showed a marked increase in pAKT levels. The difference in activity of PTEN isolated from ∼50- and ∼100-kDa fractions was nonetheless minor (they did not test the ∼660-kDa fraction) (61). In light of our results, we consider that PTEN might be incorporated as a dimer in the ∼660-kDa oligomer, where it is fully activated.
Our results show that serum stimulation triggers p110α/p110β complex formation. Only fractions of cellular p110α and p110β are found in this complex, suggesting that only part of the p110α and p110β molecules modulate receptor-induced regulation of PI(3,4,5)P3 levels at the cell membrane; this is the case for other signaling molecules that only mobilize a small proportion of the protein to the plasma membrane during activation. The p110α and p110β association appears to be functionally relevant, as the complex modulates p110β activity and its disruption markedly affects PTEN, as indicated by extended Akt activation, PI(3,4,5)P3 persistence at the cell membrane, and reduced phosphatase activity. Sequential activation of PI(3,4,5)P3-modifying enzymes in this complex coincides with the physiological order of p110α, p110β, and PTEN triggering after cell activation, suggesting that the function of this oligomer is the temporal control of cell PI(3,4,5)P3 levels.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Balla for the GFP-PH-Btk plasmid, S. Alvira, C. Hernández, and L. Sanz for technical support with gel filtration experiments, M. C. Moreno-Ortíz for technical support with flow cytometry, and C. Mark for editorial assistance.
V.P.-G. held a predoctoral FPI fellowship from the Spanish Ministry of Science and Innovation. This work was financed in part by grants from the Spanish Ministry of Science and Innovation (SAF2010-21019), the Network of Cooperative Research in Cancer (RD07/0020/2020 and RD12/0036/0059), and the Madrid regional government (BMD-2502).
Footnotes
Published ahead of print 23 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00167-14.
REFERENCES
- 1.Vanhaesebroeck B, Waterfield MD. 1999. Signaling by distinct classes of phospho-inositide 3-kinases. Exp. Cell Res. 253:239–354. 10.1006/excr.1999.4701 [DOI] [PubMed] [Google Scholar]
- 2.Vogt PK, Kang S, Elsliger MA, Gymnopoulos M. 2007. Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem. Sci. 32:342–349. 10.1016/j.tibs.2007.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Parsons R. 2004. Human cancer, PTEN and the PI-3 kinase pathway. Semin. Cell Dev. Biol. 15:171–176. 10.1016/j.semcdb.2003.12.021 [DOI] [PubMed] [Google Scholar]
- 4.Courtney KD, Corcoran RB, Engelman JA. 2010. The PI3K pathway as drug target inhuman cancer. J. Clin. Oncol. 28:1075–1083. 10.1200/JCO.2009.25.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iglesias-Bartolome R, Martin D, Gutkind JS. 2013. Exploiting the head and neck cancer oncogenome: widespread PI3K-mTOR pathway alterations and novel molecular targets. Cancer Discov. 3:722–735. 10.1158/2159-8290.CD-13-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K, Vanhaesebroeck B. 2008. The p110β isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110γ. Proc. Natl. Acad. Sci. U. S. A. 105:8292–8297. 10.1073/pnas.0707761105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murga C, Fukuhara S, Gutkind JS. 2000. A novel role for phosphatidylinositol 3-kinase beta in signaling from G protein-coupled receptors to Akt. J. Biol. Chem. 275:12069–12073. 10.1074/jbc.275.16.12069 [DOI] [PubMed] [Google Scholar]
- 8.Marqués M, Kumar A, Cortés I, González-García A, Hernández C, Moreno-Ortiz MC, Carrera AC. 2008. PI 3-kinase p110α and p110β regulate cell cycle entry, exhibiting distinct activation kinetics in G1 phase. Mol. Cell. Biol. 28:2803–28014. 10.1128/MCB.01786-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marqués M, Kumar A, Poveda AM, Zuluaga S, Hernández C, Jackson S, Pasero P, Carrera AC. 2009. Specific function of phosphoinositide 3-kinase beta in the control of DNA replication. Proc. Natl. Acad. Sci. U. S. A. 106:7525–7530. 10.1073/pnas.0812000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Fernandez-Capetillo O, Carrera AC. 2010. Nuclear PI 3-kinase beta controls double-strand break DNA repair. Proc. Natl. Acad. Sci. U. S. A. 107:7491–7496. 10.1073/pnas.0914242107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, Salpekar A, Pearce W, Meek S, Millan J, Cutillas PR, Smith AJ, Ridley AJ, Ruhrberg C, Gerhardt H, Vanhaesebroeck B. 2008. Angiogenesis selectively requires the p110α isoform of PI3K to control endothelial cell migration. Nature 453:662–666. 10.1038/nature06892 [DOI] [PubMed] [Google Scholar]
- 12.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. 2006. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature 441:366–370. 10.1038/nature04694 [DOI] [PubMed] [Google Scholar]
- 13.Kumar A, Redondo-Muñoz J, Perez-García V, Cortes I, Chagoyen M, Carrera AC. 2011. Nuclear but not cytosolic phosphoinositide 3-kinase beta has an essential function in cell survival. Mol. Cell. Biol. 31:2122–2133. 10.1128/MCB.01313-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silió V, Redondo-Muñoz J, Carrera AC. 2012. Phosphoinositide 3-kinase β regulates chromosome segregation in mitosis. Mol. Biol. Cell 23:4526–4542. 10.1091/mbc.E12-05-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. 1998. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc. Natl. Acad. Sci. U. S. A. 95:13513–13518. 10.1073/pnas.95.23.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barber DF, Alvarado-Kristensson M, González-García A, Pulido R, Carrera AC. 2006. PTEN regulation, a novel function for the p85 subunit of PI3K. Sci. STKE 362:pe49. 10.1126/stke.3622006pe49 [DOI] [PubMed] [Google Scholar]
- 17.Song MS, Salmena L, Pandolfi PP. 2012. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13:283–296. 10.1038/nrm3330 [DOI] [PubMed] [Google Scholar]
- 18.Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, Zhao JJ. 2008. Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature 454:776–779. 10.1038/nature07091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM. 2006. The p110α isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 103:16296–16300. 10.1073/pnas.0607899103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fruman DA, Snapper SB, Yballe CM, Davidson L, Yu JY, Alt FW, Cantley LC. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283:393–397. 10.1126/science.283.5400.393 [DOI] [PubMed] [Google Scholar]
- 21.Deane JA, Trifilo MJ, Yballe CM, Choi S, Lane TE, Fruman DA. 2004. Enhanced T cell proliferation in mice lacking the p85beta subunit of phosphoinositide 3-kinase. J. Immunol. 172:6615–6625. 10.4049/jimmunol.172.11.6615 [DOI] [PubMed] [Google Scholar]
- 22.Cortés I, Sánchez-Ruíz J, Zuluaga S, Calvanese V, Marqués M, Hernández C, Rivera T, Kremer L, González-García A, Carrera AC. 2012. p85β phosphoinositide 3-kinase subunit regulates tumor progression. Proc. Natl. Acad. Sci. U. S. A. 109:11318–11323. 10.1073/pnas.1118138109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alcázar I, Cortés I, Zaballos A, Hernández C, Fruman DA, Barber DF, Carrera AC. 2009. p85β PI3K regulates CD28 coreceptor function. Blood 113:3198–3208. 10.1182/blood-2008-04-152942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, Ju Z, Cantley LC, Scherer SE, Liang H, Lu KH, Broaddus RR, Mills GB. 2011. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 1:170–185. 10.1158/2159-8290.CD-11-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Méndez J, Stillman B. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. 10.1128/MCB.20.22.8602-8612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swamy M, Siegers GM, Minguet S, Wollscheid B, Schamel WW. 2006. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci. STKE 345:pl4. 10.1126/stke.3452006pl4 [DOI] [PubMed] [Google Scholar]
- 27.McConnachie G, Pass I, Walker SM, Downes CP. 2003. Interfacial kinetic analysis of the tumour suppressor phosphatase PTEN: evidence for activation by anionic phospholipids. Biochem. J. 371:947–955. 10.1042/BJ20021848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. 2007. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc. Natl. Acad. Sci. U. S. A. 104:7809–7814. 10.1073/pnas.0700373104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiménez C, Hernández C, Pimentel B, Carrera AC. 2002. The p85 regulatory subunit controls sequential activation of PI3K by Tyr kinases and Ras. J. Biol. Chem. 277:41556–41562. 10.1074/jbc.M205893200 [DOI] [PubMed] [Google Scholar]
- 30.Layton MJ, Harpur AG, Panayotou G, Bastiaens PI, Waterfield MD. 1998. Binding of a diphosphotyrosine-containing peptide that mimics activated platelet-derived growth factor receptor beta induces oligomerization of phosphatidylinositol 3-kinase. J. Biol. Chem. 273:33379–33385. 10.1074/jbc.273.50.33379 [DOI] [PubMed] [Google Scholar]
- 31.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. 2007. Binding of ras to phosphoinositide 3-kinase p110α is required for ras-driven tumorigenesis in mice. Cell 129:957–968. 10.1016/j.cell.2007.03.051 [DOI] [PubMed] [Google Scholar]
- 32.Harpur AG, Layton MJ, Das P, Bottomley MJ, Panayotou G, Driscoll PC, Waterfield MD. 1999. Intermolecular interactions of the p85α regulatory subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 274:12323–12332. 10.1074/jbc.274.18.12323 [DOI] [PubMed] [Google Scholar]
- 33.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. 1998. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110α catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 18:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata Y, Partridge TA, Matsuda R, Zammit PS. 2006. Entry of muscle satellite cells into the cell cycle requires sphingolipid signaling. J. Cell Biol. 174:245–253. 10.1083/jcb.200605028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabinovsky R, Pochanard P, McNear C, Brachmann SM, Duke-Cohan JS, Garraway LA, Sellers WR. 2009. p85 associates with unphosphorylated PTEN and the PTEN-associated complex. Mol. Cell. Biol. 29:5377–5388. 10.1128/MCB.01649-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacalle RA, Gómez-Moutón C, Barber DF, Jiménez-Baranda S, Mira E, Martínez C, Mañes S. 2004. PTEN regulates motility but not directionality during leukocyte chemotaxis. J. Cell Sci. 117:6207–6215. 10.1242/jcs.01545 [DOI] [PubMed] [Google Scholar]
- 37.Illingworth RS, Botting CH, Grimes GR, Bickmore WA, Eskeland R. 2012. PRC1 and PRC2 are not required for targeting of H2A.Z to developmental genes in embryonic stem cells. PLoS One 7:e34848. 10.1371/journal.pone.0034848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Q, Inoki K, Ikenoue T, Guan KL. 2006. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20:2820–2832. 10.1101/gad.1461206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chagpar RB, Links PH, Pastor MC, Furber LA, Hawrysh AD, Chamberlain MD, Anderson DH. 2010. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U. S. A. 107:5471–5476. 10.1073/pnas.0908899107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito K, Scharenberg AM, Kinet JP. 2001. Interaction between the Btk PH domain and PtdIns-3,4,5-trisphosphate directly regulates Btk. J. Biol. Chem. 276:16201–16206. 10.1074/jbc.M100873200 [DOI] [PubMed] [Google Scholar]
- 41.Mellado M, Rodríguez-Frade JM, Mañes S, Martínez-A C. 2001. Chemokine signaling and functional responses: the role of receptor dimerization and TK pathway activation. Annu. Rev. Immunol. 19:397–421. 10.1146/annurev.immunol.19.1.397 [DOI] [PubMed] [Google Scholar]
- 42.Casar B, Pinto A, Crespo P. 2008. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol. Cell 31:708–721. 10.1016/j.molcel.2008.07.024 [DOI] [PubMed] [Google Scholar]
- 43.Freeman AK, Ritt DA, Morrison DK. 2013. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol. Cell 49:751–758. 10.1016/j.molcel.2012.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villalonga P, López-Alcalá C, Bosch M, Chiloeches A, Rocamora N, Gil J, Marais R, Marshall CJ, Bachs O, Agell N. 2001. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 21:7345–7354. 10.1128/MCB.21.21.7345-7354.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balbis A, Baquiran G, Bergeron JJ, Posner BI. 2000. Compartmentalization and insulin-induced translocations of insulin receptor substrates, phosphatidylinositol 3-kinase, and protein kinase B in rat liver. Endocrinology 141:4041–4049. 10.1210/endo.141.11.7774 [DOI] [PubMed] [Google Scholar]
- 46.Sasaki AT, Chun C, Takeda K, Firtel RA. 2004. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167:505–518. 10.1083/jcb.200406177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto K, Lapetina EG, Moxham CP. 1992. Insulin like growth factor-I induces limited association of PI3K to its receptor. Endocrinology 130:1490–1498. 10.1210/endo.130.3.1311242 [DOI] [PubMed] [Google Scholar]
- 48.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. 2007. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science 318:1744–1748. 10.1126/science.1150799 [DOI] [PubMed] [Google Scholar]
- 49.Scott JD, Pawson T. 2009. Cell signaling in space and time: where proteins come together and when they're apart. Science 326:1220–1224. 10.1126/science.1175668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foukas LC, Beeton CA, Jensen J, Phillips WA, Shepherd PR. 2004. Regulation of PI 3-kinase by its intrinsic serine kinase activity in vivo. Mol. Cell. Biol. 24:966–975. 10.1128/MCB.24.3.966-975.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beeton CA, Das P, Waterfield MD, Shepherd PR. 1999. The SH3 and BH domains of the p85α adapter subunit play a critical role in regulating class Ia phosphoinositide 3-kinase function. Mol. Cell. Biol. Res. Commun. 1:153–157. 10.1006/mcbr.1999.0124 [DOI] [PubMed] [Google Scholar]
- 52.Shoelson SE, Sivaraja M, Williams KP, Hu P, Schlessinger J, Weiss MA. 1993. Specific phosphopeptide binding regulates a conformational change in the PI 3-kinase SH2 domain associated with enzyme activation. EMBO J. 12:795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. 2009. A frequent kinase domain mutation that changes the interaction between PI3Kα and the membrane. Proc. Natl. Acad. Sci. U. S. A. 106:16996–17001. 10.1073/pnas.0908444106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hon WC, Berndt A, Williams RL. 2012. Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene 31:3655–3666. 10.1038/onc.2011.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. 2012. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110a (PIK3CA). Proc. Natl. Acad. Sci. U. S. A. 109:15259–15264. 10.1073/pnas.1205508109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inukai K, Funaki M, Ogihara T, Katagiri H, Kanda A, Anai M, Fukushima Y, Hosaka T, Suzuki M, Shin BC, Takata K, Yazaki Y, Kikuchi M, Oka Y, Asano T. 1997. p85α gene generates three isoforms of regulatory subunit for phosphatidylinositol 3-kinase (PI 3-Kinase), p50α, p55α, and p85α, with different PI 3-kinase activity elevating responses to insulin. J. Biol. Chem. 272:7873–7882. 10.1074/jbc.272.12.7873 [DOI] [PubMed] [Google Scholar]
- 57.Cozier GE, Lockyer PJ, Reynolds JS, Kupzig S, Bottomley JR, Millard TH, Banting G, Cullen PJ. 2000. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J. Biol. Chem. 275:28261–28268. 10.1074/jbc.M000469200 [DOI] [PubMed] [Google Scholar]
- 58.Hu J, Xia X, Cheng A, Wang G, Luo X, Reed MF, Fojo T, Oetting A, Gong J, Yen PM. 2008. A peptide inhibitor derived from p55PIK phosphatidylinositol 3-kinase regulatory subunit: a novel cancer therapy. Mol. Cancer Ther. 7:3719–3728. 10.1158/1535-7163.MCT-08-0499 [DOI] [PubMed] [Google Scholar]
- 59.Taniguchi CM, Tran TT, Kondo T, Luo J, Ueki K, Cantley LC, Kahn CR. 2006. Phospho-inositide 3-kinase regulatory subunit p85α suppresses insulin action via positive regulation of PTEN. Proc. Natl. Acad. Sci. U. S. A. 103:12093–12097. 10.1073/pnas.0604628103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Alemán JO, Luo J, Stephanopoulos G, Weissleder R, Cantley LC, Kahn CR. 2010. The PI 3-kinase regulatory subunit p85α can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 70:5305–5315. 10.1158/0008-5472.CAN-09-3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, Knoblauch N, Guarnerio J, Ito K, Turka LA, Beck AH, Pinton P, Bronson RT, Wei W, Pandolfi PP. 2014. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell 157:595–610. 10.1016/j.cell.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.