Abstract
Rac1 regulates lamellipodium formation, myosin II-dependent contractility, and focal adhesions during cell migration. While the spatiotemporal assembly of those processes is well characterized, the signaling mechanisms involved remain obscure. We report here that the cytoskeleton-related Coronin1A and -1B proteins control a myosin II inactivation-dependent step that dictates the intracellular dynamics and cytoskeletal output of active Rac1. This step is signaling-branch specific, since it affects the functional competence of active Rac1 only when forming complexes with downstream ArhGEF7 and Pak proteins in actomyosin-rich structures. The pathway is used by default unless Rac1 is actively rerouted away from the structures by upstream activators and signals from other Rho GTPases. These results indicate that Coronin1 proteins are at the center of a regulatory hub that coordinates Rac1 activation, effector exchange, and the F-actin organization state during cell signaling. Targeting this route could be useful to hamper migration of cancer cells harboring oncogenic RAC1 mutations.
INTRODUCTION
The organization of the F-actin cytoskeleton has to fluctuate along the longitudinal axis of migrating cells to make possible coherent vectorial movements, directional changes in response to sudden alterations in chemical or topological cues, and the preservation of cell integrity (1). Some of the most critical upstream regulators of those processes are members of the Rho GTPase family. Thus, at the leading edge, Cdc42 generates filopodia; RhoA initiates the earliest steps of lamellipodium formation; and Rac proteins, such as Rac1 and RhoG, drive the generation of lamellipodia and membrane ruffling. In areas located away from the leading edge, Rac1 contributes to the regulation of myosin II (MII) contractility, as well as to focal-adhesion maturation and disassembly. In turn, RhoA favors the generation of actomyosin bundles, stress fibers, focal adhesions, and the contractility-driven forces required for trailing-edge detachment (2). The coregulation of these migration phase- and site-specific functions is conditioned by the membrane receptors engaged, the GDP/GTP exchange factors (GEFs) involved in the GTPase activation step, and interactions of GTPases with subcellular-localization-specific tethering factors (1–3). In addition, it relies on both the spectrum and localization of downstream effectors engaged. For example, Rac1 can promote the stimulation of Arp2/3 upon association with the Wave complex at the migration front, leading to both filopodium collapse and lamellipodium formation (4–6). By contrast, it can elicit the growth and stability of F-actin fibers in the same areas when interacting with type I Pak serine/threonine protein kinases (7). This effect can be redirected toward changes in MII contractility and focal-adhesion turnover rates when the interaction of the two proteins occurs in areas behind the migration front (8). Rho GTPase signaling cycles can be further fine-tuned by posttranslational modifications, signaling inputs that regulate GTPase stability at membranes, and feedback loops from other Rho GTPases (2, 9). When the cytoskeletal change has to stop, Rho proteins are inactivated by GTPase-activating proteins and sequestered in heteromolecular complexes with Rho GDP dissociation inhibitors (GDI) (9).
To provide additional flexibility to the system, the cytoskeleton is further regulated by the distal actions of a large number of actin-binding proteins (10). Those include Coronin1A (Coro1A) and Coro1B, two proteins implicated in lamellipodial architecture and dynamics via the regulation of F-actin-bundling processes, Arp2/3 complex inhibition, and activation of the F-actin-severing factor cofilin (11–17). Whereas the first two functions are mediated by direct interactions of Coro1 proteins with F-actin and Arp2/3, the last requires interactions of Coro1B with the Slingshot phosphatase (13). Whether Coro1A also associates with that phosphatase is currently unknown. In addition to these cytoskeletal roles, we have recently shown that Coro1A participates in the induction of serial waves of upstream Rac1 activation during mitogenic responses. This function, which is not shared by Coro1B, is mediated by the association of Coro1A with Pak and RhoGDI/Rac complexes, which, via the Pak-mediated phosphorylation of RhoGDI, promotes the release and subsequent activation of Rac1 (18). This process also requires the interaction of Coro1A with F-actin and ArhGEF7 (also known as β-Pix and Cool1) (18), a catalytically inactive Rac1 GEF that can physically interact with Rac1, Pak, and a variety of focal-adhesion-localized proteins (19). The above observations led us to hypothesize that Coro1A could represent a network hub involved in the coordinated assembly of long-lasting, self-amplifying cycles of Rac1-dependent cytoskeletal change in mitogen-stimulated cells. To investigate this possibility, we decided to monitor the cytoskeletal changes induced by constitutively active versions of Rac1 in cells lacking either the Coro1A or Coro1B protein. These studies led us to discover a Coro1-dependent regulatory route specifically involved in the proper stereospatial organization of the Rac1-dependent cytoskeleton. Unexpectedly, such a function does not seem to be linked to the general F-actin-regulatory properties of those proteins. In contrast, it relies on a Coro1/MII-dependent step that, by modulating the intracellular dynamics of Rac1/ArhGEF7/Pak2 complexes, dictates the overall organization and shape of Rac1-driven peripheral protrusions in cells.
MATERIALS AND METHODS
Cell lines.
Control and CORO1 knockdown cell clones (COS1) and cell pools (HEK 293T and Jurkat) have been described previously (18). However, the designation of the knockdown cell lines has been changed to make the names more intuitive for readers. The new and old (in parentheses) names are KD1A (ACC2-2), KD1B (VOS7-3), 293T.KD1A (VOS6-3), and J.KD1A (VOS2-1) (18). When indicated, cells were incubated with 100 ng/ml epidermal growth factor (EGF) (Millipore; 10 min), 50 μM CK-636 (Sigma; 90 min), 50 μM blebbistatin (Sigma; 2 h), 10 μM Y-27632 (Sigma; 2 h), or 1 μM ML-7 (Sigma; 2 h) under serum-free medium conditions.
Plasmids.
The expression plasmid encoding enhanced green fluorescent protein (EGFP)-Rac1Q61L (pNM42) has been previously described (20). The expression plasmid encoding AU5-Rac1Q61L was a gift from P. Crespo (IBBTEC-Universidad de Cantabria, Santander, Spain). Mammalian expression vectors encoding AU5-Rac1F28L (pRMP46), AU5-Rac1V44S+Q61L (pRMP12), FLAG-RhoGQ61L (pRMP49), and AU5-RhoGQ61L have been described previously (21). The mammalian expression vector for Cherry fluorescent protein (ChFP)-Rac1G12V was provided by M. A. del Pozo (Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain). Mammalian expression vectors encoding EGFP-tagged RhoAQ63L (pNM041), Cdc42Q61L (pNM040), Rac1F37A+Q61L (pMJC6), Rac1Y40C+Q61L (pMJC7), and the Vav1 DH-PH-ZF cassette (pNM103) have been previously described (22–25). The expression vector encoding a myristoylated version of Rac1Q61L (pVOS12) was obtained by ligating a BamHI/EcoRI human RAC1Q61L cDNA fragment from pXRB99 into the BamHI-EcoRI-linearized pCEFL-Myr plasmid (obtained from P. Crespo). The expression plasmid encoding EGFP-Rac1Q61L under the speckle promoter (pVOS35) was generated by amplifying the human RAC1Q61L cDNA contained in pNM42 using primers 5′-GGC CAG ATC TCA GGC CAT CAA GTG TGT GG-3′ (forward) and 5′-GGC CGG ATT CCA ACA GCA GGC-3′ (reverse), digestion of PCR fragments with BglII and BamHI (restriction sites in the oligonucleotides are underlined), and ligation of the PCR product into a BglII/BamHI-linearized pGFP-speckle backbone (provided by M. Vicente-Manzanares, La Princesa University Hospital, Madrid Autonomous University, Madrid, Spain). Coro1AshRNAMUT-EGFP (pACC58), Coro1AshRNAMUT+R29D-EGFP (pACC67), and pCor-RFP (wild-type [WT]) plasmids have been described previously (18). The mammalian expression vector encoding Coro1B-EGFP (pVOS13) was obtained by subcloning an EcoRI/BamHI human CORO1B cDNA fragment obtained from the retroviral vector pCoro1B-GFP (provided by J. Bear, University of North Carolina at Chapel Hill, Chapel Hill, NC) into the EcoRI-BamHI-linearized pEGFP-N2 (Clontech). To generate the vector encoding Coro1BR30D-EGFP (pVOS19), we used the QuikChange kit (Stratagene) to incorporate the R30D mutation into the CORO1B cDNA present in pVOS13. The sequences of the mutagenesis primers were 5′-CCA GTG CTA TGA GGA CAT TGA CGT GTC CCG TGT TAC CTG G-3′ (forward; the mutation is underlined) and 5′-CCA GGT AAC ACG GGA CAC GTC AAT GTC CTC ATA GCA CTG G-3′ (reverse; the mutation is underlined). To generate the plasmid encoding Coro1AshRNAMUT-red fluorescent protein (RFP) (pVOS60), we mutagenized the pCor-RFP (WT) plasmid using oligonucleotides 5′-CCT GGG ACA GTG GCT TCT GTG CCG TAA ACC CTA AGT TTG TGG CCC-3′ (forward) and 5′-GGG CCA CAA ACT TAG GGT TTA CGG CAC AGA AGC CAC TGT CCC AGG-3′ (reverse). This mutagenesis step introduced mutations (underlined) in the 41 (A) and 42 (V) CORO1A codons that make the transcript insensitive to degradation by the CORO1A short hairpin RNA (shRNA) present in KD1A cells (18). To generate the Coro1BshRNAMUT-EGFP (pVOS14)- and Coro1BshRNAMUT+R30D-EGFP (pVOS20)-encoding vectors, we mutated the pVOS13 and pVOS19 vectors, respectively, using oligonucleotides 5′-GGC TGC GAC AAC GTG GTA CTA ATT TGG AAT GTG GGC-3′ (forward) and 5′-GCC CAC ATT CCA AAT TAG TAC CAC GTT GTC GCA GCC-3′ (reverse). Transcripts expressed by those plasmids are insensitive to degradation by the CORO1B shRNA integrated into KD1B COS1 cells due to the introduction of conservative mutations (underlined) at the 158 (L) and 159 (I) CORO1B codons. pEGFP-mDia2 and pRFP-Ruby-N1-Lifeact were provided by M. A. del Pozo. Expression vectors encoding EGFP-cofilinS3A, myosin light chain (MLCT19D+S20D)-EGFP, and ChFP-MIIA were from M. Vicente-Manzanares. The MLC-EGFP (pVOS49) plasmid was generated by site-directed mutagenesis of the pRLCT19A+S20D-EGFP vector (provided by M. Vicente-Manzanares) using oligonucleotides 5′-GCG CCC GCA GCG CGC CAC CTC CAA TGT CTT CGC TAT GTT CG-3′ (forward; the mutations are underlined) and 5′-CGA ACA TAG CGA AGA CAT TGG AGG TGG CGC GCT GCG GGC GC-3′ (reverse; the mutations are underlined). All oligonucleotides were purchased from Invitrogen. All plasmids generated were verified by DNA sequencing of the entire cDNA at the Genomics and Proteomics Facility of our center.
Antibodies and other immunological reagents.
Rhodamine- and Alexa Fluor 635-labeled phalloidin and Alexa Fluor 647-labeled CTxB were obtained from Molecular Probes/Invitrogen. Mouse monoclonal antibodies to AU5 and GFP and rabbit polyclonal antibodies to MIIB were from Covance. Mouse monoclonal antibodies to cortactin and fascin and rabbit polyclonal antibodies to Arpc2 and ArhGEF7 were from Millipore. Rabbit polyclonal antibodies to the FLAG epitope, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phospho-MLC, phosphocofilin, and RFP were obtained from Sigma, Santa Cruz Biotechnology, Rockland, Cell Signaling, and Thermo Scientific, respectively. Mouse monoclonal antibodies to Vasp, Rac1, and IQGAP1 and rabbit polyclonal antibody to caveolin were from BD Transduction Laboratories. Mouse monoclonal antibodies to Pak2 and Coro1A were from Cell Signaling and Sigma, respectively. Mouse monoclonal antibodies to tubulin α were ordered from Calbiochem. Rabbit polyclonal antibodies to phospho-Erk were from Cell Signaling. When required, appropriate Cy2-, Cy3-, and Cy5-labeled secondary antibodies were used (Jackson ImmunoResearch). In the case of immunoblotting, horseradish peroxidase-coupled secondary antibodies were used (GE Healthcare Life Biosciences).
Immunofluorescence experiments.
To monitor the distribution of the proteins under analysis, transfected cells were fixed in 3.7% formaldehyde in a phosphate-buffered saline (PBS) solution for 15 min. The cells were then permeabilized in 0.5% Triton X-100 in TBS-T (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Tween 20) for 10 min, washed three times with TBS-T, blocked in 2% bovine serum albumin in TBS-T for 30 min, and stained using the appropriate stepwise addition of primary and secondary antibodies diluted in blocking solution for 1 h each. To visualize the F-actin cytoskeleton, the above-mentioned preparations were subsequently incubated with either rhodamine- or Alexa Fluor 635-labeled phalloidin diluted 1:100 in blocking solution for 30 min. To detect lipid rafts, Alexa Fluor 647-conjugated CTxB was added to cultured cells (0.5 μg/ml) for 2 min prior to fixation. The stained preparations were mounted on microscope slides using Mowiol (Calbiochem). Unless otherwise indicated, confocal images shown in the figures were taken in cell sections showing enriched F-actin structures and therefore may not correspond to the same positions in the cell lines compared. Confocal images were collected using a Leica SP5 confocal microscope with a 63× objective (Leica). When required, triple colocalization of proteins was analyzed by using the ColocalizeRGB plug-in in ImageJ software (version 1.43u). This plug-in finds the colocalized pixels of three separate channels from three-color immunofluorescence microscopy images. When numerical values are provided, they refer to the percentage of a given phenotype in the total population of fluorescent-protein-expressing cells. In histograms, we defined as “spread” cells displaying the typical morphology of COS1 cells. “Ruffles” refers to cells exhibiting the characteristic large membrane ruffles typically present in cells expressing constitutively active Rac1 versions. A score was considered positive when those structures occupied more than half of the cell perimeter. We did not include in this category the small peripheral ruffles that are occasionally seen in untransfected COS1 cells. “Lamella-like” refers to cells displaying unpolarized cytoplasmic extensions that were supported by radial phalloidin- and Rac1-decorated F-actin filaments. Although there were differences in terms of overall cell size, this parameter was not quantified. Quantification was done using 150 cells in three independent experiments.
Immunoblotting.
Protein samples were denatured by boiling in SDS-PAGE sample buffer, separated electrophoretically, and transferred onto nitrocellulose filters using the iBlot dry blotting system (Invitrogen) according to the manufacturer's instructions. Membranes were blocked in 5% nonfat dry milk in TBS-T for 1 h and then incubated overnight at 4°C with the indicated primary antibodies diluted in blocking buffer. After extensive washes with TBS-T, the membrane was incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution) for 1 h at room temperature. Immunoreacting bands were developed using a standard chemiluminescence method (enhanced chemiluminescence [ECL]; GE Healthcare Life Biosciences).
Transient transfections.
In the case of standard immunofluorescence studies with the COS1 and HEK 293T cell lines, cells were grown on 6-well plates and transfected using Lipofectamine 2000 (Invitrogen). To that end, 1 μg of the appropriate plasmid DNA and 3 μl of Lipofectamine 2000 were diluted separately in 100 μl of serum-free Dulbecco's modified Eagle's medium (DMEM), and the two solutions were mixed, incubated for 20 min at room temperature, and added to the cells. Except where otherwise indicated, transfected cells were trypsinized 24 h later and seeded onto 0.001% polylysine (Sigma)-, 10 μg/ml fibronectin (Sigma)-, or 10 μg/ml laminin (Invitrogen)-coated coverslips and fixed 24 h later. In the case of small interfering RNA (siRNA)-based studies, 30 to 40% confluent COS1 cells were transfected with the appropriate siRNA (60 mM) using Lipofectamine 2000 (Invitrogen) as described above. For coimmunoprecipitation experiments, 30 to 40% confluent COS1 cells were transfected using Lipofectamine 2000 in 10-cm dish plates. To this end, 2 μg of the indicated plasmids and 6 μl of Lipofectamine 2000 were diluted separately in 200 μl of serum-free DMEM and added to the cells as described above. Cells were harvested 36 to 48 h after the transfection step, as indicated (18).
In the case of Jurkat T cells, transfections were performed using the Neon transfection system (Life Technologies) following the manufacturer's protocols. To this end, 20 μg of plasmid DNA was added to 2 × 106 cells diluted in 2 ml of R buffer (included in the Neon transfection system kit, catalogue number MPK10096; Life Technologies), and the mixture was subjected to two 20-ms electroporation cycles at 1.7 mV. The cells were then transferred to standard culture media for 36 h and then plated onto 0.001% polylysine-coated coverslips, allowed to settle for 20 min, fixed, and stained.
Video recording.
Subconfluent cells plated on 35-mm plastic plates were transfected with pNM42 (encoding EGFP-Rac1) and pRFP-Ruby N1-Lifeact plasmids and analyzed 24 h later using a 20× objective (Olympus) in a Delta Vision microscope (Image Solutions). Images were taken using Resolve3D softWoRx-Aquire software (version 5. 5. 0). The capture settings were 0.3- and 1-s exposure times for EGFP-Rac1Q61L and Lifeact fluorescence, respectively, using 1 s of interval time. The videos were played at 10 frames/s.
siRNAs.
For knockdown purposes, we used pools of the three Stealth siRNA target sequences (Invitrogen) available for human PAK2 (catalog numbers HSS107578, HSS107579, and HSS10780), ARHGEF7 (catalog numbers HSS113107, HSS113108, and HSS113109), and CTTN (catalog numbers HSS103231, HSS103232, and HSS103233) transcripts. As a control, we used a scrambled Stealth siRNA (catalog number 12935-300).
Coimmunoprecipitation experiments.
Cells cultured in 100-mm dishes were washed with chilled PBS and disrupted in 1 ml of lysis buffer composed of 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 1 mM Na3VO4, 10 mM β-glycerophosphate, and a protease inhibitor cocktail (cOmplete; Roche). The lysates were incubated on ice for 10 min and precleared by centrifugation at 14,000 rpm for 10 min at 4°C. The supernatants obtained were immunoprecipitated with the GFP-Trap reagent (ChromoTek) for 1 h, and the immunocomplexes were collected by centrifugation, washed, separated electrophoretically, and analyzed by immunoblotting as indicated in the figures. Analyses of total cellular lysates were performed in parallel to detect the expression of the indicated proteins.
Image processing.
Images were assembled and processed for final figure presentation using Canvas 9.0.4 (Deneba Systems).
Statistical analyses.
Data from at least three independent experiments were analyzed using the Student t test. P values of ≤0.05 were considered statistically significant.
RESULTS
Coro1-deficient cells show abnormal shapes and contractile properties.
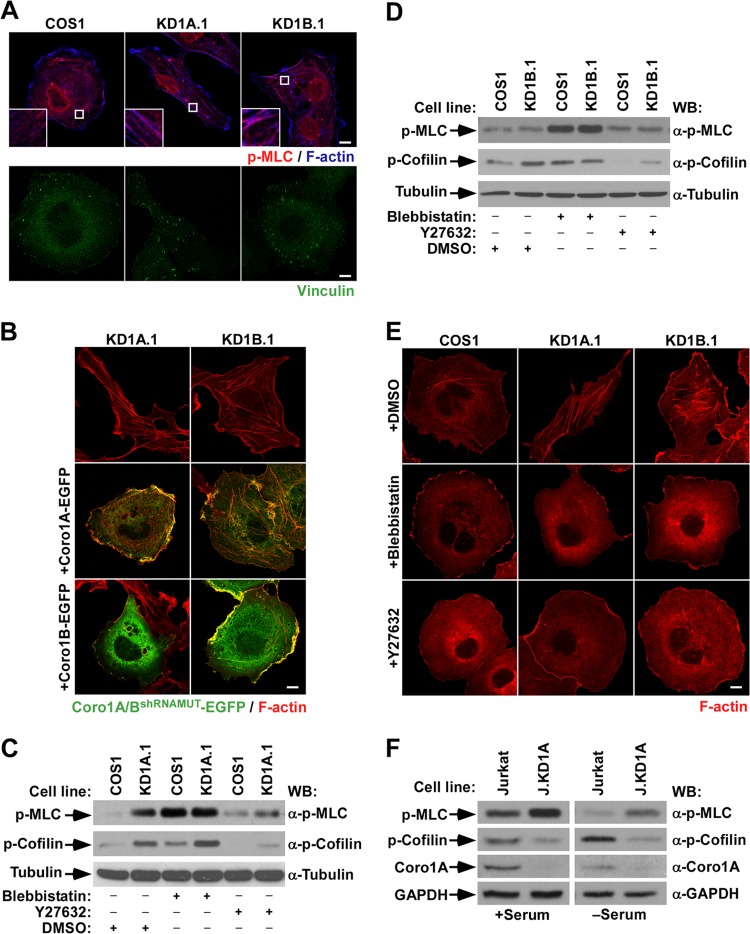
During a previous study, we generated COS1 cells lacking expression of either Coro1A (KD1A.1 and KD1A.2 cells) or Coro1B (KD1B.1 and KD1B.2 cells) using a lentiviral shRNA approach (18). A similar method was utilized to generate CORO1A-deficient human HEK 293T embryonic kidney (293T.KD1A.1 and 293T.KD1A.2) and T-lymphoblastoid Jurkat (J.KD1A) cells (18). Effective protein depletion was confirmed in each case using immunoblotting techniques (18). When analyzed by confocal microscopy, we observed that the stable depletion of Coro1A and, to a much lesser extent, of Coro1B promotes the acquisition of contractile features (Fig. 1A), as evidenced by the detection in knockdown cells of phospho-MLC-decorated stress fibers (Fig. 1A), enlarged focal adhesions (Fig. 1A), and diminished cell areas (14.9 ± 2.9 and 28.5 ± 8.3 μm2 in CORO1A and CORO1B knockdown cells versus 36.1 ± 7.2 μm2 in control cells; P ≤ 0.001, n = 50 cells for each type) (Fig. 1A). It should be noted, however, that the morphological features induced by the single depletion of each Coro1 protein are different. For example, CORO1A knockdown cells consistently display elongated shapes and contain thin stress fibers, usually oriented along the longitudinal axis. In contrast, Coro1B-deficient cells are more rounded and contain stress fibers thicker and shorter than those found in Coro1A-depleted cells (Fig. 1A). We found similar morphologies in independent knockdown clones for each of the proteins (examples are shown in Fig. 2A), indicating that these phenotypes are the consequence of the intrinsic lack of each Coro1 protein. Consistent with this, we could rescue normal morphologies upon expression of the appropriate shRNA-resistant CORO1 transcript in knockdown cells (Fig. 1B). Interestingly, we observed that the ectopic expression of Coro1B can rescue the morphological phenotype of both CORO1A and CORO1B knockdown cells (Fig. 1B). However, Coro1A can rescue only the defects found in the former cell line (Fig. 1B). This result rules out the possibility that such morphological changes can be derived from the Rac1 activation defects previously found in CORO1A knockdown COS1 cells (18). In addition to these morphological alterations, we observed that CORO1A knockdown cells have elevated amounts of both phospho-MLC and phosphocofilin compared to controls (Fig. 1C). In contrast, CORO1B knockdown COS1 cells show only a marked upregulation in the amount of phosphocofilin (Fig. 1D). Despite these signaling differences, these defects are clearly MII and Rock dependent, because parental cell-like shapes can be restored in those cells using short-term treatments with either an MII (blebbistatin) or a Rock (Y-27632) inhibitor (Fig. 1E). Thus, it is possible that the lack of Coro1B promotes either small or subcellular-localization-specific alterations in MII activity that cannot be reliably detected using standard immunoblot analyses. Consistent with this, we observed that Y-27632 reduces the phosphorylation levels of both phospho-MLC and phosphocofilin in CORO1A knockdown cells (Fig. 1C) and of phosphocofilin in CORO1B-deficient cells (Fig. 1D). We could not determine the effect of blebbistatin because its use promotes MLC hyperphosphorylation in all cell lines tested (Fig. 1C and D). Although it is prima facie counterintuitive, such an effect has been reported by others (6, 26). The implication of Coro1A in the steady-state MLC activation status is cell type independent, because Jurkat cells lacking the protein also display larger amounts of phospho-MLC that in this case are not associated with parallel elevations in phosphocofilin (Fig. 1F).
FIG 1.
Unregulated myosin II-dependent contractility contributes to the cytoskeletal phenotype of CORO1 knockdown cells. (A) Representative images of the indicated cells stained with antibodies to phospho-MLC (top row, red signals) and Alexa Fluor 635-labeled phalloidin (top row, blue signals) and with vinculin (bottom row, green signals). In the top row, magnifications of the boxed cell areas are shown in the insets. Color codes for fluorescence signals are indicated. Colocalization of phospho-MLC and F-actin is shown in purple (top row). Scale bars, 10 μm. (B and E) Representative images of the cytoskeletal phenotype exhibited by CORO1 knockdown cells (top) upon ectopic expression of the indicated Coro1-EGFP proteins (B, left) and drug treatments (E, left). In panel B, colocalization of Coro1 and F-actin is shown in yellow. Scale bars, 10 μm. (C, D, and F) Total cellular extracts from the indicated cell lines and experimental conditions were analyzed by Western blotting (WB) to detect the amount of phospho (p)-MLC (top gels), phosphocofilin (second gels from top), and Coro1A (F, third gel from top). The loading controls used were tubulin α (C and D, bottom) and GAPDH (F, bottom). The antibodies used in the immunoblot analyses are indicated on the right.
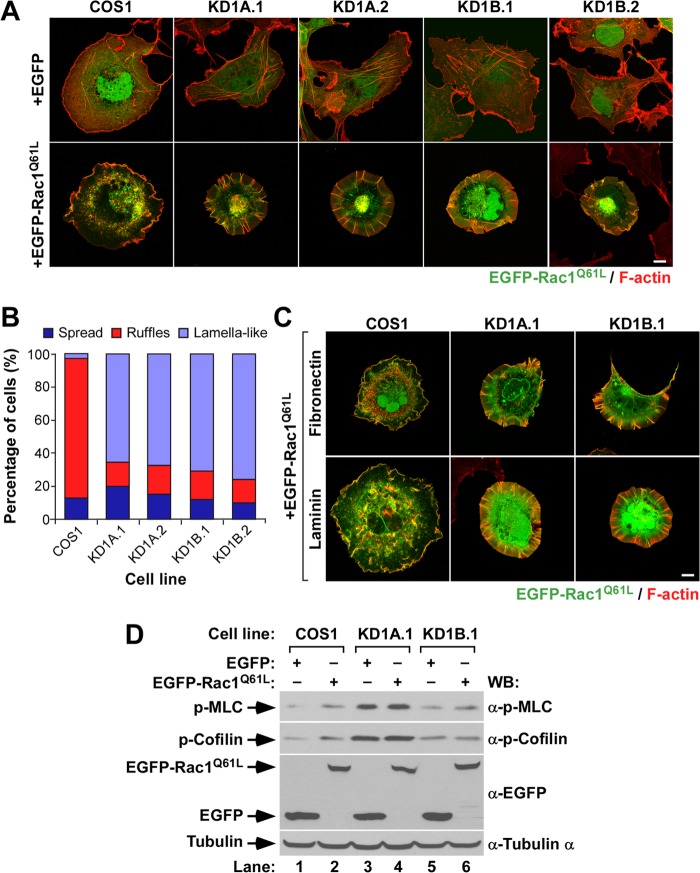
FIG 2.
Abnormal Rac1 cytoskeletal signaling in CORO1 knockdown cells. (A and C) Representative images of the cytoskeletal phenotype exhibited by EGFP-expressing (A) and EGFP-Rac1Q61L-expressing (A and C) cell lines attached to polylysine-coated (A), fibronectin-coated (C), or laminin-coated (C) coverslips. The cell lines are indicated at the top. Ectopically expressed proteins are indicated at the left. Colocalization areas of active Rac1 with F-actin are shown in yellow. Scale bars, 10 μm. (B) Quantification of the types of cytoskeletal change induced by ectopically expressed EGFP-Rac1Q61L in the indicated cells attached to polylysine-coated coverslips. (D) Total cellular extracts from the indicated cell lines and experimental conditions were analyzed by Western blotting to detect the amounts of phospho-MLC, phosphocofilin, and EGFP proteins expressed. Tubulin α was used as a loading control.
Coro1 proteins play critical roles in Rac1-dependent cytoskeletal signaling.
To investigate the role of Coro1 proteins downstream of active Rac1, we next evaluated the cytoskeletal changes induced by the transient expression of constitutively active Rac1 (with a Q61L mutation) in CORO1A and CORO1B knockdown COS1 cells. To facilitate the visualization of transfected cells, Rac1 was tagged at its N terminus with EGFP. Upon transfection, the cells were trypsinized; plated on polylysine-, fibronectin-, or laminin-coated coverslips; stained with fluorochrome-labeled phalloidin; and analyzed by confocal microscopy. Irrespective of the substrate used during the plating step, we observed that constitutively active Rac1 proteins cannot generate peripheral ruffles when expressed in any of the CORO1A and CORO1B knockdown COS1 cells used in these experiments (Fig. 2A to C). In contrast, they induce the formation of extended lamellas characterized by the presence of radial extensions of poorly branched F-actin filaments that in many cases are decorated with active Rac1 (Fig. 2A and C). These filaments are present within the cell perimeter but in some cases form small, filopodium-like protrusions that extend outward from those limits. The expression of EGFP-Rac1Q61L does not have any effect on the large amounts of phospho-MLC (Fig. 2D, top gel, lanes 3 and 4) and phosphocofilin (Fig. 2D, second gel from top, lanes 3 to 6) previously seen in Coro1-depleted cells. It does elicit, however, an increase in the phosphorylation of cofilin (Fig. 2D, second gel from top, lanes 1 and 2) and, to a much lesser extent, of MLC (Fig. 2D, top gel, lanes 1 and 2) when expressed in control cells.
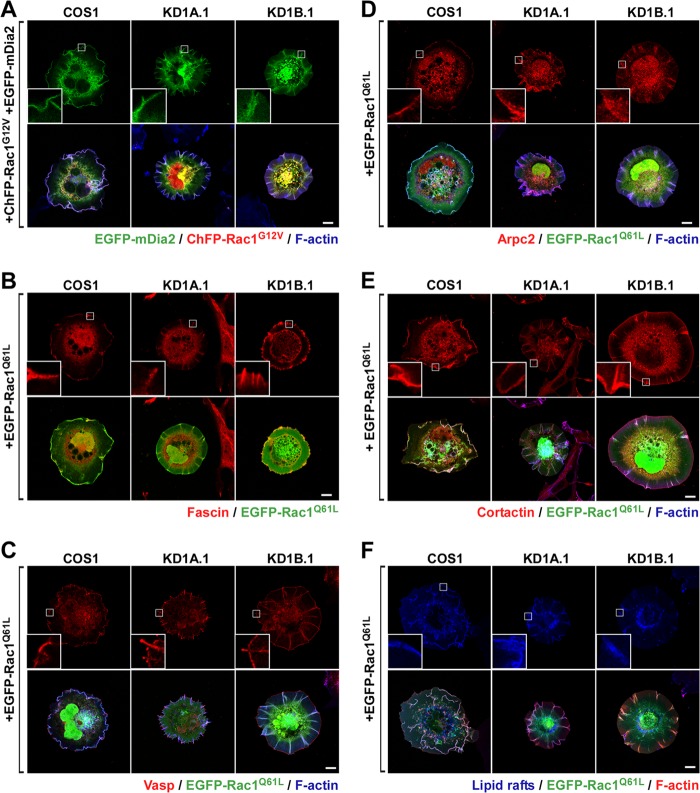
Time-lapse microscopy analyses revealed that these F-actin filaments are rather dynamic, since they sprout from many independent cell areas, grow centripetally, and eventually collapse. The last step takes place when F-actin filaments reach the cell edge or, alternatively, when they detach from the substratum and undergo a rearward movement using the most proximal end as a hinge (see Video S1 in the supplemental material). At the time of collapse, cells form short-lived, small, ruffle-like structures distinct from the large and long-lasting retrograde ruffles typically seen in control cells (see Video S2 in the supplemental material). Confirming the confocal microscopy results, we observed that EGFP-Rac1Q61L colocalizes with these F-actin structures during their entire assembly-disassembly cycle (see Video S1 in the supplemental material). Further immunostaining experiments indicated that the F-actin ribs present in Rac1-expressing CORO1 knockdown cells contain typical filopodial markers, such as mDia2, fascin, and Vasp (Fig. 3A to C). These structures are also decorated with Arp2/3 complexes and cortactin in the absence of any evident F-actin branching (Fig. 3D and E), a Rac1-dependent feature previously described by others in filopodia (27). In vivo staining of cells with Clostridium botulinum toxin B revealed that the majority of lipid rafts cosegregate with these Rac1/F-actin-rich structures present in knockdown cells (Fig. 3F).
FIG 3.
Abnormal Rac1 cytoskeletal signaling in CORO1 knockdown cells. (A to F) Representative images of the subcellular localization of the indicated cytoskeletal proteins (A to E) and CTxB-stained lipid rafts (F) in parental and CORO1-depleted cells (top rows) expressing the indicated ectopic proteins (left). With the exception of Rac1 and mDia2, all signals correspond to endogenous proteins. The insets are enlarged images of the boxed cell areas. Scale bars, 10 μm.
Coro1 proteins do not work redundantly downstream of GTP-bound Rac1.
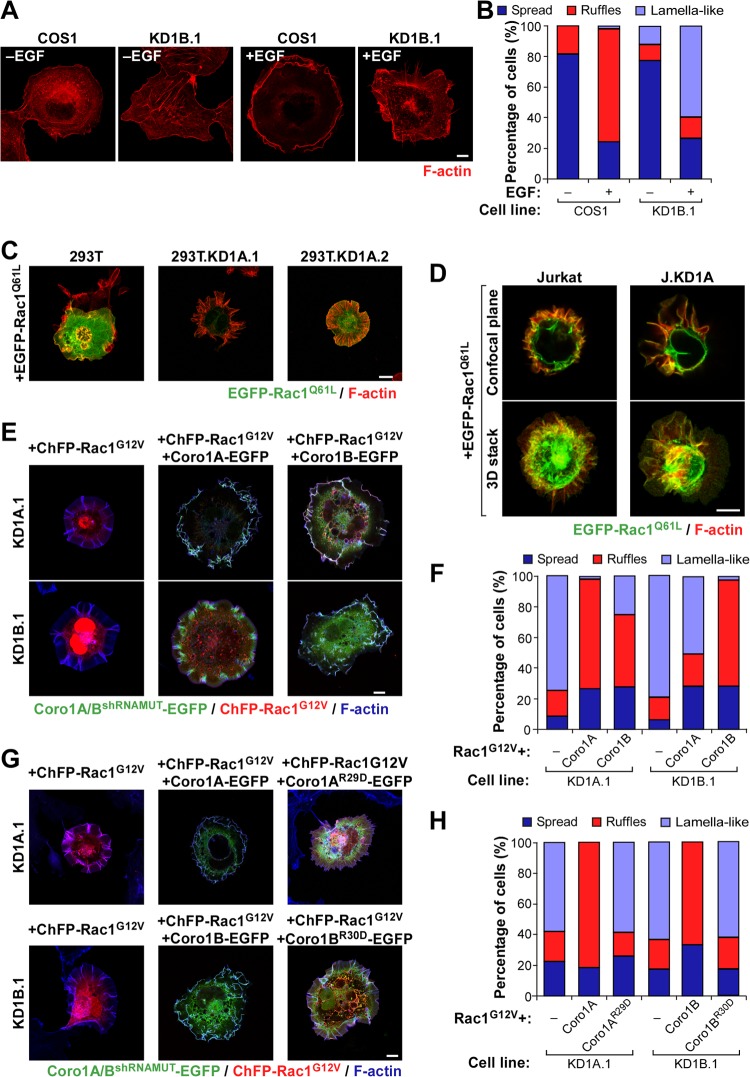
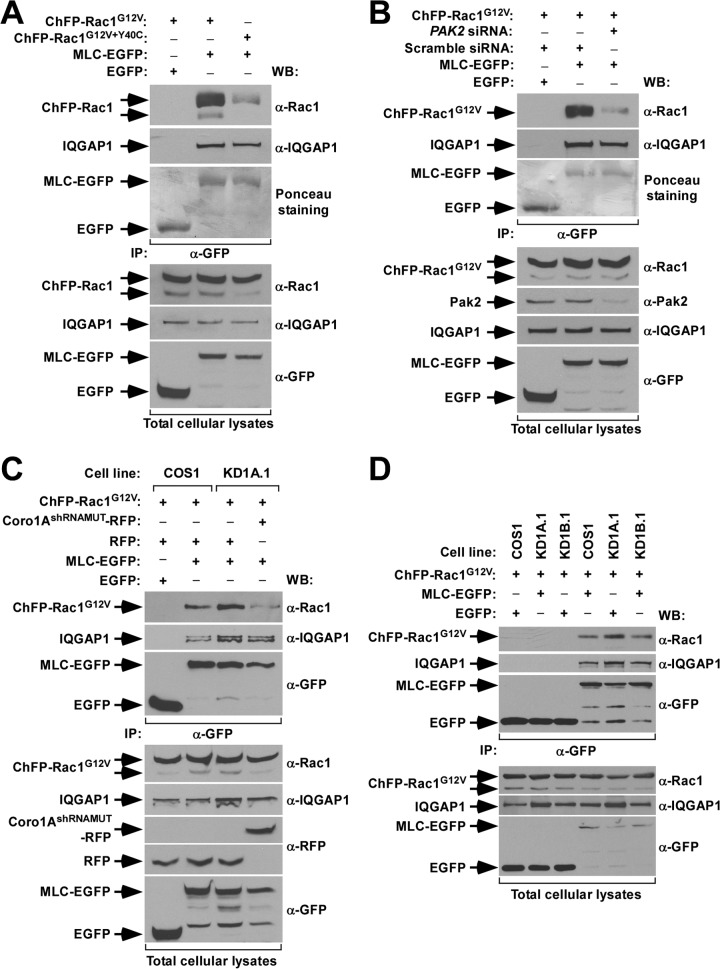
A number of independent experiments confirmed the physiological relevance of the above observations. First, we could observe the formation of abnormal cytoskeletal responses in nontransfected, serum-starved CORO1B knockdown cells upon stimulation with EGF. Although milder than those exhibited under conditions of ectopic Rac1 expression, these alterations included the generation of extended lamellar areas, the presence of radial projections of very thin F-actin fibers, the occasional generation of filopodia, and a lack of robust membrane ruffling (Fig. 4A and B). These results indicate that Coro1B is also involved in the cytoskeletal program of endogenous Rac1. CORO1A knockdown COS1 cells were not used in these experiments because, unlike the case of the Coro1B-depleted counterparts, they cannot activate Rac1 due to the disruption of the previously described upstream Coro1A/RhoGDI/GDP-Rac1/ArhGEF7/Pak2 signaling complex (18). Second, we demonstrated, using Coro1-deficient pools of HEK 293T (Fig. 4C) and Jurkat (Fig. 4D) cells, that the effect of the Coro1 depletion on Rac1Q61L cytoskeleletal signaling is cell type independent. Finally, using reconstitution experiments, we confirmed that these defects are intrinsic to the lack of these proteins. In particular, we found that Coro1A-EGFP can restore normal ruffling activity when coexpressed in CORO1A knockdown cells with a constitutively active, ChFP-tagged version of Rac1 (with a G12V mutation) (Fig. 4E and F). Ectopic Coro1A, however, cannot rescue normal ruffling in CORO1B-deficient cells (Fig. 4E and F). In contrast, the coexpression of Coro1B-EGFP does rescue such activity in both Coro1A- and Coro1B-depleted cells (Fig. 4E and F). These results indicate that the abnormal cytoskeletal response of those cells is not due to shRNA off-target effects and, more importantly, that the two Coro1 proteins work nonredundantly in this process. Interestingly, the rescue properties of Coro1 proteins observed in this assay are similar to those found in the restoration of the morphological shape of cells that had not been transfected with active Rac1 (Fig. 1), suggesting that these two processes can be mechanistically intertwined (see below). These experiments also revealed that actin binding-deficient versions of Coro1A (with an R29D mutation) (18) and Coro1B (with an R30D mutation) (12) show no rescue activity (Fig. 4G and H), indicating that the participation of Coro1 proteins in this biological process requires their normal interaction with the cytoskeleton.
FIG 4.
Cell-type-independent and nonredundant implication of Coro1 proteins in Rac1 cytoskeletal signaling. (A and B) Representative images (A) and quantification (n = 3; 150 cells/condition) (B) of F-actin organization induced by EGF in the indicated cell lines. Scale bar, 10 μm. (C and D) Examples of the cytoskeletal changes induced by ectopic EGFP-Rac1Q61L in the indicated HEK 293T (C) and Jurkat (D) cells. In the case of Jurkat cells, a confocal plane and a Z-stack 3-dimensional (3D) reconstruction are shown. 293T.KD1A.1 and 293T.KD1A.2 are two independent pools of CORO1A knockdown HEK 293T cells. J.KD1A is a pool of CORO1A knockdown Jurkat cells. Colocalization areas of Rac1 with F-actin are shown in yellow. Scale bars, 10 (C) and 5 (D) μm. (E to H) Representative images (E and G) and quantification (n = 3; 150 cells/condition) (F and H) of the cytoskeletal phenotype exhibited by Coro1-depleted COS1 cells (E and G, left) upon ectopic expression of the indicated combinations of ChFP-Rac1G12V and Coro1-EGFP proteins. Coro1A/BshRNAMUT-EGFP refers to the ectopic protein that cannot be targeted by the shRNA harbored in knockdown cells. In panels E and G, colocalization areas of Rac1 with Coro1 proteins and F-actin are shown in white. Scale bars, 10 μm.
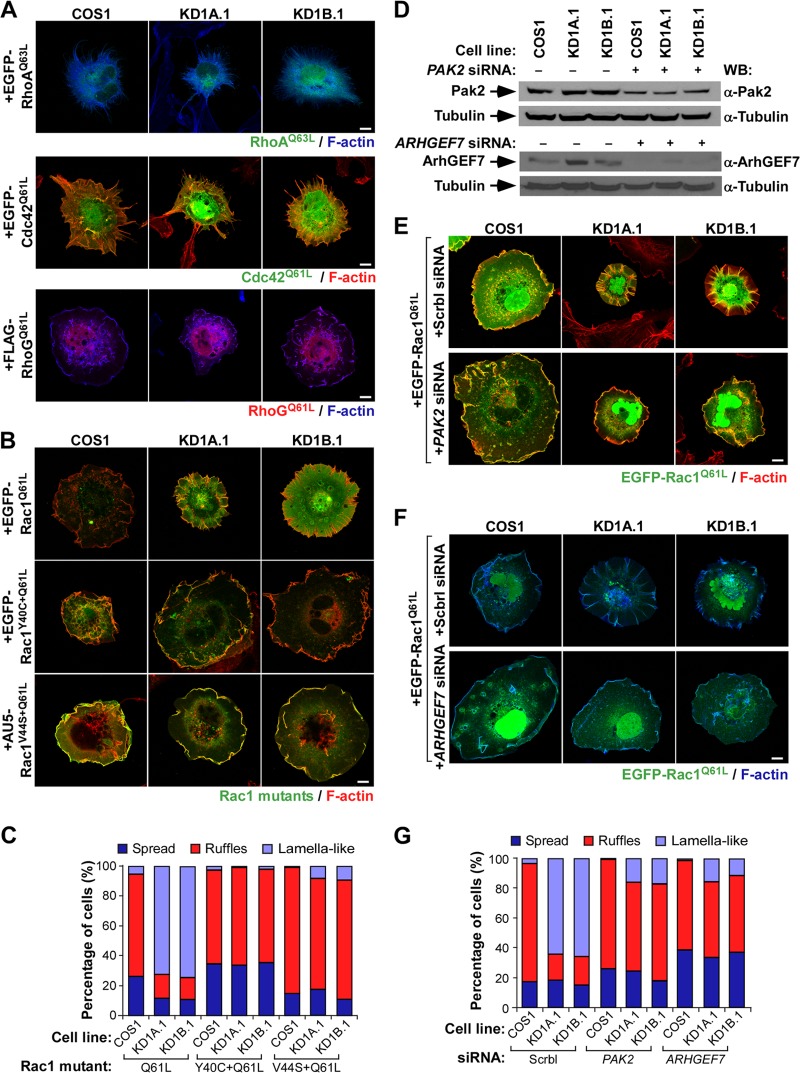
Coro1 proteins control cytoskeletal change in a GTPase- and Pak-specific manner.
The cytoskeletal defects observed are Rac1 specific, because Coro1-depleted COS1 cells undergo control cell-like cytoskeletal changes when constitutively expressing active versions of EGFP-RhoA (with a Q63L mutation), EGFP-Cdc42 (with a Q61L mutation), or FLAG-tagged RhoG (with a Q61L mutation) (Fig. 5A). The results obtained with the last protein represented a seminal clue to us, because it is known that this GTPase can promote Rac1Q61L-like cytoskeletal changes (21, 28) (Fig. 5A) despite the fact that it cannot physically interact with the Rac1 downstream elements Pak (21, 28) and ArhGEF7 (V. Ojeda and X. R. Bustelo, unpublished data). This led us to surmise that Coro1 proteins could be specifically involved in the regulation of the Pak/ArhGEF7 downstream signaling branch of Rac1. Consistent with this idea, we found that EGFP-Rac1Q61L proteins containing single point mutations (Y40C and V44S) that eliminate the interaction with Pak family proteins (21, 29) induce normal cytoskeletal change when expressed in Coro1-depleted cells (Fig. 5B and C). To further corroborate this hypothesis, we decided to investigate whether the siRNA-mediated elimination of either PAK2 or ARHGEF7 mRNA could restore ruffling responses in Rac1Q61L-expressing knockdown cells. We chose the PAK2 transcript for these experiments because it is the main type I Pak family member expressed in COS1 cells (18). We found that the depletion of either of these two transcripts (Fig. 5D) restores normal EGFP-Rac1Q61L-driven cytoskeletal effects in both CORO1A and CORO1B knockdown cells (Fig. 5E to G), indicating that the cytoskeletal defects present in these cells are due to alterations in the signaling competence of active Rac1/ArhGEF7/Pak2 complexes rather than to the impairment of canonical F-actin- and Arp2/3-related roles of Coro1 proteins.
FIG 5.
The implication of Coro1 proteins is specific for the Rac1/Pak2 signaling branch. (A) Cytoskeletal changes induced in the indicated cell lines by representative members of the Rho family. Scale bars, 10 μm. (B and C) Representative images (B; scale bar, 10 μm) and quantification (n = 3; 150 cells/condition) (C) of cytoskeletal changes elicited by the indicated Rac1 proteins in control and Coro1-depleted cells. Colocalization regions of Rac1 and F-actin are shown in yellow. (D) Western blot analysis showing the siRNA-mediated depletion of endogenous Pak2 and ArhGEF7 in the indicated COS1 cell lines. Expression of tubulin α in each sample was used as a loading control. (E to G) Representative images (E and F; scale bars, 10 μm) and quantification (n = 3; 150 cells/condition) (G) of cytoskeletal changes induced by EGFP-Rac1Q61L upon depletion of Pak2 (E and G) and ArhGEF7 (F and G) in the indicated cell lines. In panels E and F, cell lines and transfected molecules are shown at the top and left of the images, respectively. Colocalization regions of Rac1 and F-actin are shown in either yellow (E) or light blue (F). Scrbl, scrambled.
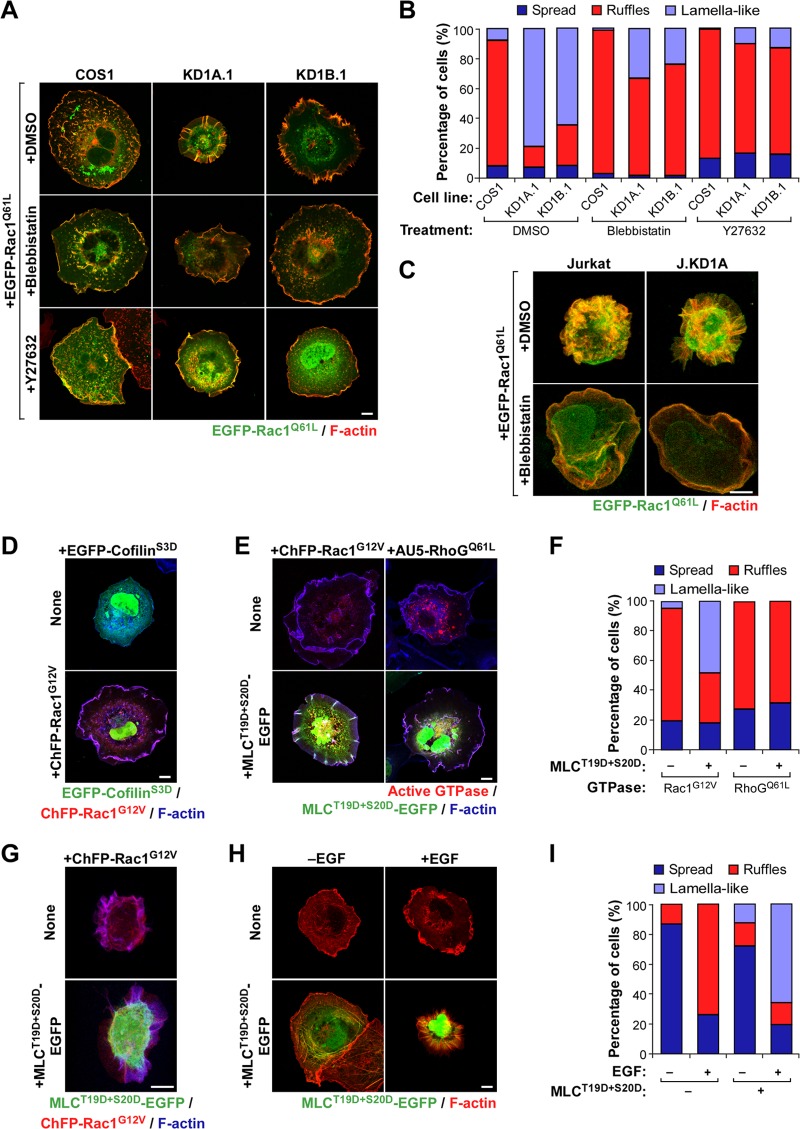
Coro1 proteins control Rac1 cytoskeletal output via MII inhibition.
The similar rescue properties shown by shRNA-resistant Coro1 proteins in both nontransfected (Fig. 1) and Rac1-transfected (Fig. 4E and F) knockdown cells suggested to us that the basal and Rac1-dependent cytoskeletal defects exhibited by these cells could stem from a common signaling cause. Consistent with this idea, we observed that CORO1 knockdown cells maintain clear signs of enhanced actomyosin contractility even when expressing constitutively active Rac1 (Fig. 1 to 5). Furthermore, and similar to results obtained with nontransfected cells (Fig. 1), we could also restore normal cytoskeletal changes in EGFP-Rac1Q61L-expressing CORO1 knockdown COS1 (Fig. 6A and B) and Jurkat (Fig. 6C) cells using short-term treatments with either blebbistatin or Y-27632. To further assess the implication of cofilin and MII in the cytoskeletal phenotype of Coro1-depleted cells, we decided to test whether phosphomimetic mutant versions of either cofilin (S3D; inactive protein) or MLC (T19D and S20D; active protein) could reproduce the effects of the Coro1 depletion in parental cells. EGFP-cofilinS3D does not induce any obvious cytoskeletal dysfunction in ChFP-Rac1G12V-expressing parental cells (Fig. 6D), indicating that inefficient cofilin inactivation cannot generate per se the cytoskeletal phenotype of CORO1 knockdown cells. In contrast, MLCT19D+S20D-EGFP does elicit such defects when coexpressed with active Rac1 in both parental COS1 (Fig. 6E and F) and Jurkat (Fig. 6G) cells. This mutant protein also promotes a change in the normal cytoskeletal program of endogenous Rac1 in EGF-stimulated COS1 cells (Fig. 6H and I), leading to the generation of cytoskeletal structures quite similar to those found in EGF-stimulated CORO1B knockdown cells (Fig. 4A). A similar effect has been described previously in other cell types (30). These effects are Rac1 specific, because MLCT19D+S20D-EGFP does not alter the cytoskeletal change typically induced by RhoGQ61L in parental COS1 cells (Fig. 6E and F). Collectively, these results suggest that Coro1 proteins contribute to Rac1 downstream signaling in an actomyosin-dependent manner.
FIG 6.
MII is involved in the Coro1-dependent Rac1 cytoskeletal route. (A and B) Representative images (A) and quantification (n = 3; 150 cells/condition) (B) of the effects of the indicated drugs in EGFP-Rac1Q61L-expressing COS1 cell lines. In panel A, areas of colocalization of Rac1 and F-actin are shown in yellow. Scale bar, 10 μm. DMSO, dimethyl sulfoxide. (C) Confocal Z-stack 3D reconstruction of the effects of blebbistatin in the indicated EGFP-Rac1Q61L-expressing wild-type (Jurkat) and CORO1A knockdown (J.KD1A) Jurkat cells. The color codes are as in panel A. Scale bar, 5 μm. (D) Cytoskeletal changes elicited by EGFP-cofilinS3D alone (top) or in combination with ChFP-Rac1G12V (bottom) in parental COS1 cells. Colocalization between Rac1 and F-actin is shown in purple. Scale bar, 10 μm. (E and F) Representative images (E) and quantification (n = 3; 150 cells/transfection) (F) of the effects of the indicated combinations of proteins on the cytoskeleton of parental COS1 cells. In panel E, colocalization areas of transfected proteins with F-actin are shown in either pink (top) or white (bottom). Scale bar, 10 μm. (G) Confocal Z-stack 3D reconstruction of the effect of MLCT19D+S20D-EGFP on the cytoskeletal organization of ChFP-Rac1G12V-expressing Jurkat cells. The confocal color codes are as in panel D. Scale bar, 5 μm. (H and I) Representative images (H; scale bar, 10 μm) and quantification (n = 3; 150 cells/condition) (I) of the effects of ectopic MLCT19D+S20D-EGFP in the cytoskeleton of nonstimulated and EGF-stimulated parental COS1 cells. The color codes are as in panel A.
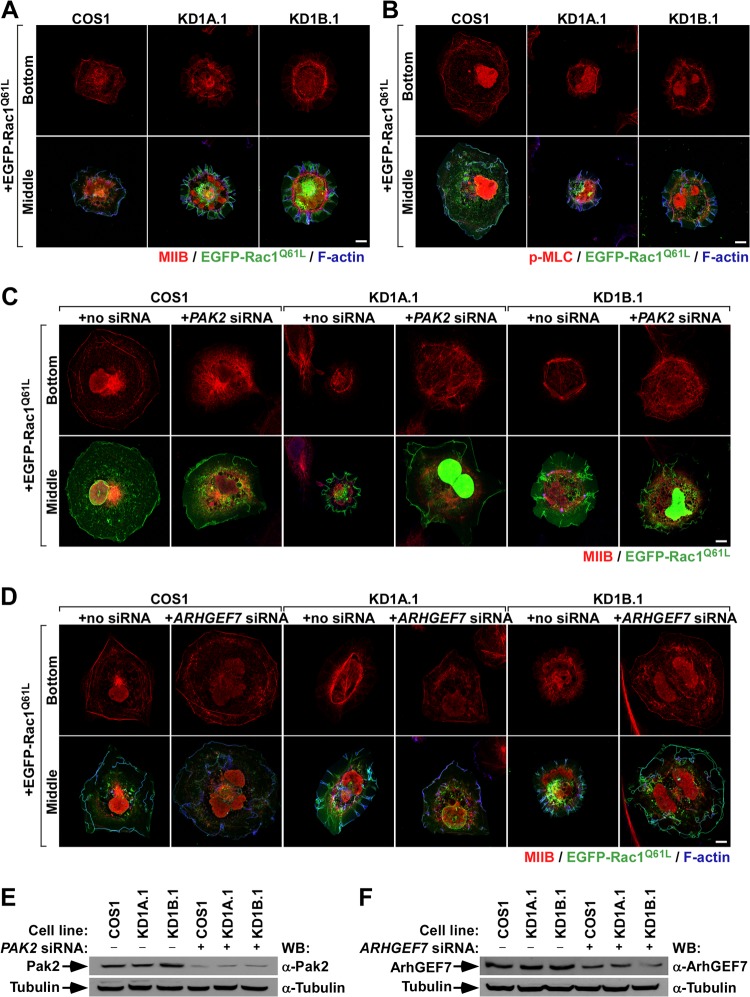
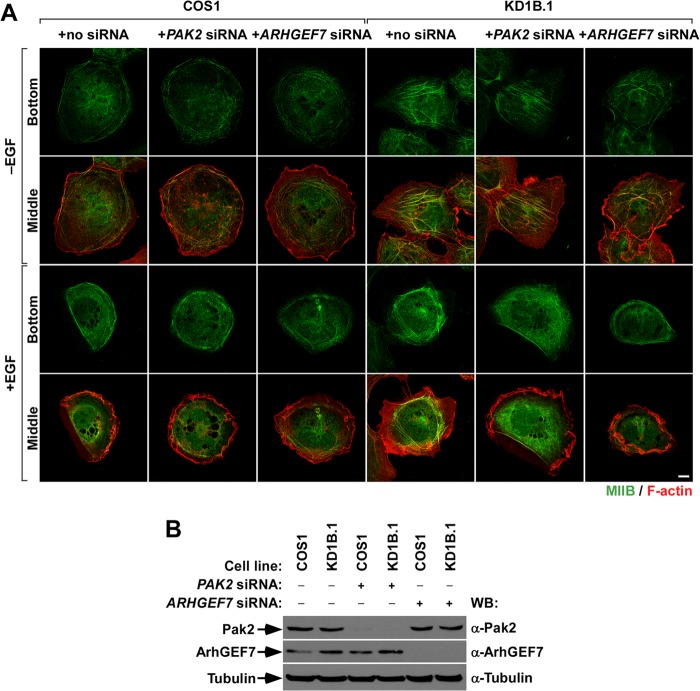
Coro1 proteins regulate actomyosin dynamics during Rac1-dependent responses.
The above observations led us to investigate in more detail the status of the MLC route in CORO1 knockdown cells. Using confocal microscopy, we observed that EGFP-Rac1Q61L promotes the localization of both endogenous MIIB (Fig. 7A, top row) and phospho-MLC (Fig. 7B, top row) in cell-substrate interface areas located underneath regions of active membrane ruffling in the case of control cells. In contrast, the two proteins are clearly localized away from membrane ruffles (Fig. 7A and B, bottom row). However, in Coro1-depleted cells expressing active Rac1, we found that MIIB and phospho-MLC localize preferentially in the actomyosin ring and, to a lesser extent, in the proximal region of the F-actin ribs generated in those cells (Fig. 7A and B). This is not due to the known lack of MIIA expression in COS cells (31), because the coexpression of the protein with EGFP-Rac1Q61L does not rescue normal cytoskeletal responses in CORO1 knockdown cells (Ojeda and Bustelo, unpublished). Furthermore, ectopically expressed MIIA shows subcellular distributions quite similar to those found in the case of MIIB and phospho-MLC in both control and knockdown cells (Ojeda and Bustelo, unpublished). Linking this differential distribution with the cytoskeletal dysfunctions found in CORO1 knockdown cells, we observed that the siRNA-mediated depletion of endogenous transcripts for either Pak2 or ArhGEF7 restores control cell-like MIIB (Fig. 7C to F) and phospho-MLC (Ojeda and Bustelo, unpublished) subcellular localization patterns in EGFP-Rac1Q61L-expressing CORO1 knockdown cells. These data can be extrapolated to the behavior of endogenous Rac1, because we also detected abnormal localization of MIIB in the case of nontransfected, EGF-stimulated CORO1B knockdown cells (Fig. 8A, third row from top, compare the first and fourth images from left). Likewise, we could induce parental-cell-like intracellular localizations of MIIB (Fig. 8A, third row from top, compare images 4 to 6 from left), as well as proper EGF-induced ruffling activity (Fig. 8A, bottom row, compare images 4 to 6 from left) when either Pak2 or ArhGEF7 protein was siRNA depleted in CORO1B knockdown cells. The effective reduction of Pak2 (Fig. 8B, top gel) and ArhGEF7 (Fig. 8B, second gel from top) in the appropriate experimental samples was confirmed by immunoblotting. These results suggest that MII complexes cannot be efficiently inactivated in CORO1 knockdown cells, leading to the formation of stable actomyosin structures and a total change in the cytoskeletal program of active Rac1.
FIG 7.
Coro1 depletion blocks actomyosin ring disassembly in Rac1-expressing COS1 cells. (A and B) Intracellular distribution of endogenous MIIB (A) and phospho-MLC (B) in the indicated EGFP-Rac1Q61L-expressing cell lines. Confocal images were taken at sections corresponding to the bottom and intermediate (Middle) cell areas along the perpendicular axis. Scale bar, 10 μm. (C and D) Effects of endogenous Pak2 (C) and ArhGEF7 (D) protein depletion on the subcellular distribution of MIIB in the indicated COS1 cell lines. Confocal images were taken as for panels A and B (left). The siRNA and Rac1 protein transfected are indicated at the top and left, respectively. Scale bars, 10 μm. (E and F) Western blot analysis showing the depletion of Pak2 (E) and ArhGEF7 (F) proteins with the indicated experimental conditions and cell lines. The amount of tubulin α in each sample was used as a loading control.
FIG 8.
Actomyosin ring disassembly is also blocked in EGF-stimulated Coro1B-depleted cells. (A) Effects of endogenous Pak2 and ArhGEF7 protein depletion on the intracellular distribution of MIIB in the indicated COS1 cell lines under the indicated stimulation conditions. (B) Western blot analysis showing the depletion of Pak2 and ArhGEF7 proteins with the indicated experimental conditions and cell lines. The amount of tubulin α was used as a loading control.
Coro1 proteins regulate the release of Rac1/ArhGEF7/Pak2 complexes from actomyosin.
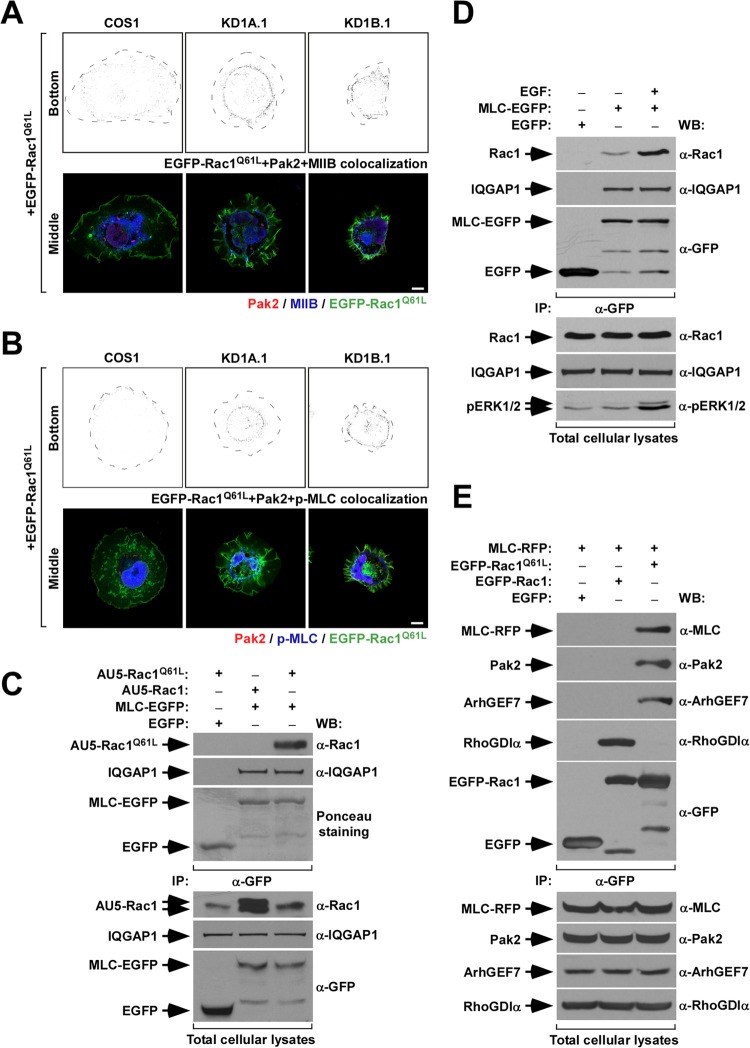
Taking into consideration the results described above, we decided to investigate whether the abnormal cytoskeletal organization shown by CORO1 knockdown cells could be the result of trapping the GTP-Rac1/Pak2/ArhGEF7 complex in actomyosin structures. To test this hypothesis, we first compared the subcellular localizations of EGFP-Rac1Q61L, Pak2, MIIB, and phospho-MLC in both control and CORO1 knockdown cells. To avoid mislocalization of proteins due to overexpression, we used a “speckle” plasmid that, due to a partial truncation of the cytomegalovirus promoter, expresses very small amounts of Rac1 (32). Upon transfection, cells were fixed and incubated with antibodies to endogenous Pak2, MIIB, and phospho-MLC, and subcellular-localization patterns were monitored in different confocal cell layers. We found that triple-positive pixels for EGFP-Rac1Q61L, Pak2, and MIIB were mostly restricted to the cell-substrate interfaces located just behind the cell edge in the case of control cells (Fig. 9A, top left). This localization resembles the typical distribution found for nascent adhesions in protruding cell areas. In contrast, the three proteins colocalized in the characteristic actomyosin ring present in CORO1 knockdown cells (Fig. 9A, top, middle and right). We obtained similar results when the colocalization of EGFP-Rac1Q61L, Pak2, and phospho-MLC was studied in these cells (Fig. 9B).
FIG 9.
Defective subcellular localization of Rac1/Pak2/MII complexes in CORO1 knockdown COS1 cells. (A and B) (Top rows) Colocalization (gray dots) of EGFP-Rac1Q61L and endogenous Pak2 with either endogenous MIIB (A) or phospho-MLC (B) in the cell-substrate interfaces of the indicated cell lines. Cell edges are depicted as broken black lines. (Bottom rows) Distribution of the proteins in the upper confocal sections of the cells under study. Scale bars, 10 μm. (C and D) Anti-EGFP immunoprecipitates (IP) from COS1 cells transiently expressing the indicated combinations of proteins (C and D) and under the indicated stimulation conditions (D) were analyzed by Western blotting to detect the amount of coimmunoprecipitated ectopic (C) or endogenous (D) Rac1 and endogenous IQGAP1 under each experimental condition. As a control, filters were either stained with Ponceau solution prior to the immunoblotting step (C) or immunoblotted with antibodies to EGFP to visualize the immunoprecipitated EGFPs (D). In panel D, the bottom gel shows an immunoblot of lysates with an antibody to phospho-Erk to demonstrate the effective activation of COS1 by the EGF treatment in these experiments. (E) Anti-EGFP immunoprecipitates from COS1 cells expressing the indicated proteins were analyzed by Western blotting to detect the presence of ectopic MLC, endogenous Pak2, ArhGEF7, and RhoGDIα. As a control, filters were immunoblotted with antibodies to EGFP to visualize the immunoprecipitated EGFPs. (C to E, bottom) The amounts of proteins present in lysates prior to the immunoprecipitation step were determined by Western blotting of aliquots of the total cell lysates used in the experiments.
Corroborating these immunolocalization-based data, we found that MLC-EGFP can interact in vivo with both ectopically and endogenously expressed Rac1 (Fig. 9C and D, top). Such interaction is highly dependent on the GTP-bound status of Rac1, since it is detected preferentially when lysates are from cells that either ectopically express Rac1Q61L (Fig. 9C, top) or that had been subjected to EGF stimulation (5.58 ± 3.36 versus 1 found in EGF-stimulated and nonstimulated cells; P ≤ 0.05; n = 3) (Fig. 9D, top). As a control, we observed that MLC-EGFP coimmunoprecipitated similar amounts of IQGAP, an MLC binding protein identified in proteomic experiments (Ojeda and Bustelo, unpublished), regardless of the ectopic Rac1 protein (Fig. 9C, second gel from top) or stimulation conditions (Fig. 9D, second gel from top) used in the experiments. The preferential interaction between MLC and Rac1Q61L was also observed using reverse immunoprecipitation experiments with EGFP-Rac1 proteins (Fig. 9E, top). Using this approach, we also detected the presence of endogenous Pak2 (Fig. 9E, second gel from top) and ArhGEF7 (Fig. 9E, third gel from top) in those complexes. However, RhoGDIα is excluded from them (Fig. 9E, fourth gel from top). Conversely, EGFP–wild-type Rac1 interacts preferentially with the last protein while it shows no detectable binding to MLC-RFP, Pak2, or ArhGEF7 (Fig. 9E). All these interactions are specific, since they cannot be detected in immunoprecipitates obtained using the nonchimeric EGFP (Fig. 9C to E, top gels).
The formation of active Rac1/MLC complexes requires Pak2, as evidenced by the lack of efficient coimmunoprecipitation of the Pak binding-deficient Rac1G12V+Y40C protein with MLC-EGFP (0.11 ± 0.02 versus 1 found in Rac1G12V+Y40C- and Rac1G12V-expressing cells; P ≤ 0.001; n = 3) (Fig. 10A, top). A similar reduction in the amount of GTP-bound Rac1/MLC-EGFP complexes was observed when Pak2 was siRNA depleted in wild-type COS1 cells (0.3 ± 0.1 versus 1 found in PAK2 siRNA- and scrambled-siRNA-transfected cells; P ≤ 0.001; n = 3) (Fig. 10B, top). We also found that the interaction of active Rac1 with MLC-EGFP takes place at higher levels in CORO1A knockdown than in control cells (2.6 ± 0.6 versus 1; P ≤ 0.01; n = 3) (Fig. 10C, top). This enhanced association is lost upon reexpressing Coro1A in knockdown cells (0.70 ± 0.11 versus 1 in control cells; P ≤ 0.01; n = 3) (Fig. 10C), confirming that the above-mentioned results are intrinsic to the absence of Coro1A in those cells. In contrast, we observed similar amounts of MLC-Rac1Q61L interaction when using CORO1B-deficient COS1 cells (Fig. 10D, top). The variations in the amounts of MLC-EGFP/Rac1 complexes formed are specific, because the interaction of MLC-EGFP with IQGAP1 does not significantly change under each of the above-described experimental conditions (Fig. 10A to D, second gels from top). The detected coimmunoprecipitations are also specific, since they do not show up when MLC-EGFP is replaced by the nonchimeric EGFP in these experiments (Fig. 10A to D, top). These observations indicate that Coro1 proteins, by favoring an MII inactivation step, are important for the disassembly of GTP-Rac1/Pak2/ArhGEF7 complexes from actomyosin structures.
FIG 10.
Enhanced formation of Rac1/Pak2/MII complexes in CORO1A knockdown COS1 cells. (A to D) Anti-EGFP immunoprecipitates from parental (A to D) and CORO1 knockdown (C and D) COS1 cells transiently expressing the indicated combinations of proteins (A to D) and siRNAs (B) were analyzed by Western blotting to detect the amount of coimmunoprecipitated ectopic Rac1 and endogenous IQGAP1 under each experimental condition. (A and B) As a control, filters were stained with Ponceau solution prior to the immunoblotting step in some experiments to visualize the immunoprecipitated EGFPs. (C and D) Alternatively, the lysates were immunoblotted with antibodies to EGFP. Aliquots of the same cell lysates were analyzed by Western blotting to evaluate the expression of the indicated proteins prior to the immunoprecipitation step (bottom). Note that in panels C and D the increased amounts of immunoprecipitated IQGAP1 in lanes 3 and 5, respectively, are probably due to the slight variations in the amount of input lysate before the immunoprecipitation step (fifth gel from top).
Subcellular relocalization of Rac1 rescues membrane ruffling in Coro1-depleted cells.
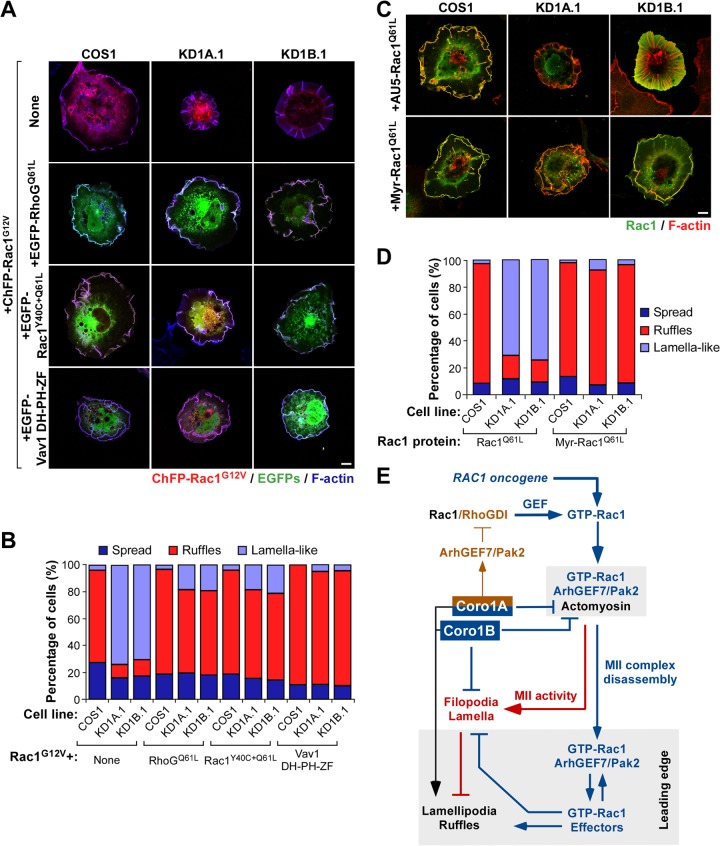
Given the results indicating that the active Rac1/Pak2/ArhGEF7 complexes are probably trapped in actomyosin structures present in Coro1-deficient cells, we speculated that normal cytoskeletal responses could potentially be restored in those cells if such trapping was bypassed by the engagement of other intracellular signals or, alternatively, by artificially rerouting active Rac1 to the plasma membrane. Taking into account that RhoGQ61L could circumvent the Coro1-like effects induced by constitutively active MLCT19D+S20D-EGFP in parental COS1 cells (Fig. 5 and 6), we first decided to investigate whether that GTPase could also restore Rac1-dependent cytoskeletal responses in CORO1A and CORO1B knockdown cells. Coexpression experiments with EGFP-RhoGQ61L and ChFP-Rac1G12V in the cells confirmed that this is the case (Fig. 11A and B). We also found similar rescues when ChFP-Rac1G12V was coexpressed with either the Pak binding-deficient EGFP-Rac1Y40C+Q61L protein or an oncogenic version of the Vav1 GEF (Fig. 11A and B), indicating that the actomyosin trap can be inactivated when pools of either ectopic or endogenous Rac1 molecules are activated at the plasma membrane. These signals must be Pak2 independent, given the rescue effect observed with both RhoGQ61L and Rac1Y40C+Q61L mutants (Fig. 11A and B). This interpretation is also in agreement with the observation that the Pak2 depletion restores per se effective membrane ruffling in Rac1Q61L-expressing Coro1-deficient cells (Fig. 5D to G). These results also indicate that active Rac1 does not have a dominant role over other extrinsic, Coro1-independent routes that trigger Rac1-like cytoskeletal changes in cells. Finally, we tested whether the forced redirection of active Rac1 toward the plasma membrane could bypass the actomyosin trap and, therefore, the Coro1 deficiency. To this end, we compared the cytoskeletal changes elicited by a myristoylated version of Rac1Q61L in both control and Coro1-depleted COS1 cells. Unlike the case of EGFP-Rac1Q61L, we found that the myristoylated version of Rac1Q61L showed plasma membrane localization and elicited ruffling activity irrespective of the expression status of Coro1 proteins in COS1 cells (Fig. 11C and D). These results confirm the idea that Coro1 proteins play specific roles in the intracellular dynamics of the GTP-Rac1/Pak2/ArhGEF7 complex.
FIG 11.
Subcellular relocalization of Rac1 rescues membrane ruffling in Coro1-depleted cells. (A to D) Representative images (A and C) and quantification (n = 3; 150 cells/transfection) (B and D) of the cytoskeletal changes induced by the indicated combinations of GTPases (A to D) and the catalytic DH-PH-ZF region of the Vav1 Rho GEF (A and B) in parental and CORO1A and CORO1B knockdown cells. In panel A, colocalization of the indicated Rho and Vav1 proteins with both ChFP-Rac1G12V and F-actin is shown in white. In panel C, colocalization of the Rac1 proteins and F-actin is shown in yellow. Scale bars, 10 μm. (E) Integrated view of the results reported. The Coro1-dependent route described is indicated by blue (functional steps) and red (dysfunctional steps induced upon Coro1 depletion) lines. The previously described Coro1A-dependent loop involved in optimal Rac1 activation is shown with brown lines. The intrinsic role of Coro1 in F-actin regulation is shown in black. Activation and inactivation steps are indicated by arrows and blunted lines, respectively. MII-rich and leading-edge areas are shaded.
DISCUSSION
Our results have revealed that Rac1 requires a Coro1- and MII-dependent step in order to translocate to the plasma membrane and elicit its typical effects on F-actin polymerization and membrane ruffling (Fig. 11E). Interestingly, we have found that this new regulatory step exclusively affects the signaling output of the active Rac1/Pak2/ArhGEF7 complexes and not other Rac1-dependent signaling branches. Coro1 proteins work nonredundantly in this process, as evidenced by the development of a phenotype upon depletion of each of them. Furthermore, we observed that the ectopic expression of Coro1A did not restore normal cytoskeletal responses in CORO1B knockdown cells. Coro1B did rescue normal cytoskeletal responses when overexpressed in CORO1A knockdown cells. However, since these cells still express amounts of endogenous Coro1B comparable to those present in control cells (18), we believe that such rescue activity could probably derive from the stimulation of an MII-independent step downstream of Coro1A, such as a direct effect on F-actin filament stability. Indeed, the expression of actin-severing proteins, such as active forms of both gelsolin and cofilin, can rescue normal morphology in Coro1-deficient COS1 cells (Ojeda and Bustelo, unpublished). In favor of this possibility, we have observed that CORO1A and CORO1B knockdown COS1 cells are rather different in terms of shape, basal amounts of phospho-MLC, or the ability to favor the overall formation of active Rac1/MLC complexes in vivo. The lack of functional redundancy between the two proteins is not surprising, since we have previously shown that Coro1A, but not Coro1B, is in charge of engaging the F-actin/ArhGEF7/Pak2/F-actin loop that favors the Rac1 activation step during cell signaling (18) (Fig. 11E). These observations also suggest that some of these regulatory actions, particularly the Coro1B-dependent ones, could be exerted by other Coro1 family or cytoskeleton-regulatory proteins in some specific cell lineages. We surmise that this is possibly the case in T lymphocytes, because they exhibit Coro1b transcript levels approximately 50-fold lower than those for the Coro1a mRNA (Ojeda and Bustelo, unpublished).
Disruption of this Coro1-dependent step leads to the sequestration of active Rac1/Pak2/ArhGEF7 complexes in the actomyosin ring and the formation of large lamella-like extensions sustained by radially projected F-actin bundles. Interestingly, most of the lipid rafts present in CORO1 knockdown cells are also in close association with F-actin rib structures, a feature that probably contributes to further limiting the diffusion of Rac1 away from those structures and, at the same time, to promoting the rapid internalization of the GTPase from other plasma membrane areas (18, 33). All these processes are mutually dependent, since we can restore normal Rac1 cytoskeletal responses in CORO1 knockdown cells by using MII inhibitors and strategies that reroute active Rac1 away from the actomyosin trap. This new regulatory step has been found in a number of independent cell types and during cytoskeletal responses elicited by endogenous Rac1, suggesting its physiological relevance. Collectively, these data indicate that Coro1 proteins, and in particular Coro1A, play a hub-like role critical for coupling Rac1 activation and downstream, Rac1-dependent cytoskeletal outputs in mitogen-stimulated cells (Fig. 11E).
It has recently been reported that coronins may regulate MII disassembly in Dictyostelium discoideum by inhibiting the basal stimulation status of Rac1 and Pak (34). Such a role seems to be mediated by the independent interaction of a Cdc42- and Rac1-interactive binding (CRIB)-like domain present in the β-propeller domain of most coronin family members with inactive versions of both Rac1 and Pak1 (34). According to this model, the elimination of coronins would lead to the release of such inhibitory action, increased Rac1 and Pak1 activities, the phosphorylation-mediated inactivation of the myosin heavy chain kinase (MHCK) by activated Pak1, and, finally, the elimination of the MHCK-mediated disassembly of MII complexes in cells (34). Although bearing some similarity to the results reported here (i.e., the two mechanisms must result in increased MII activity), we believe that our data are mechanistically incompatible with this report. Thus, in contrast to the CRIB-based model, we have demonstrated before that the elimination of Coro1A from COS1 cells leads to the inhibition, not the stimulation, of Rac1 activity (18). In addition, the elimination of Coro1B affects the cytoskeletal output of constitutively active Rac1 despite inducing no obvious change in the stimulation levels of the endogenous GTPase (18). The morphology exhibited by untransfected CORO1A and CORO1B knockdown cells is also quite different from the phenotype expected in cells displaying high levels of Rac1 activity, and certainly, constitutively active Rac1 does not elicit a CORO1 knockdown-like cytoskeletal phenotype when ectopically expressed in parental cells. Moreover, we have demonstrated that the Coro1A-mediated inactivation step reported here is dependent on the presence of active Rac1, not on interactions of Coro1A proteins with inactive Rac1, as proposed in the CRIB model (34). In this context, it is also unclear how the coronin CRIB-like domain would be required to inhibit Rac1, considering that the GDP-bound versions of the GTPase are fully trapped in RhoGDI complexes in nonstimulated cells (35). Given that Coro1A and Coro1B share a CRIB domain (34), the CRIB-based model cannot explain the lack of rescue of the cytoskeletal programs of Coro1-deficient cells expressing Coro1A mutant proteins containing fully functional CRIB domains (i.e., Coro1AR29D and Coro1BR30D). Although outside the scope of this study, it would be interesting to investigate in the near future the actual implication of Coro1 proteins in the regulation of MII activity and/or the stability of actomyosin complexes. The MII contractile cycle is known to be primarily controlled by the combined actions of an MLC kinase and an MLC phosphatase that in turn are regulated via phosphorylation-dependent mechanisms by a number of upstream elements, including the RhoA-stimulated Rock family of serine/threonine kinases (36). Direct regulation of MLC kinase by Coro1 proteins can be excluded, because the pharmacological inactivation of the kinase does not rescue normal Rac1-dependent cytoskeletal responses in CORO1 knockdown cells (Ojeda and Bustelo, unpublished). Due to this, Coro1 proteins may be involved in the negative control of Rock (37) or MLC phosphatase or MII enzyme activities. Alternatively, they could regulate more downstream regulatory steps, such as the destabilization of MII-based structures. The latter model, for example, would justify the results showing that active RhoG can promote membrane ruffling even when cells express the nonregulatable MLCT19D+S20D mutant protein. If this were the case, this process has to be Pak2 independent, given the rescue observed in the subcellular localization of active Rac1 by GTPase versions that cannot stimulate Pak2, such as RhoG and EGFP-Rac1Y40C+Q61L.
In addition to the intrinsic interest of this new Coro1- and MII-dependent regulatory mechanism controlling the signaling-branch-specific output of Rac1, we believe that our results provide useful information about other collateral issues related to the spatiotemporal organization of Rac1-dependent signaling in cells. For example, they reveal that active Rac1 molecules probably utilize this new route by default to regulate actomyosin complexes in a Pak-dependent manner and that only upon the Coro1-mediated MII inactivation step will the active GTPase/Pak2/ArhGEF7 complexes be redirected to the plasma membrane to undergo effector exchange and, subsequently, trigger lamellipodia and membrane ruffling formation. The present results also indicate that wild-type Rac1 can be rerouted away from this default pathway, depending on the upstream GEFs engaged during cell stimulation or, alternatively, through signaling inputs derived from concurrently activated Rho proteins (i.e., RhoG). In contrast, they showed that constitutively active Rac1 mutants seem to preferentially follow this default route and therefore that the Coro1 route is particularly critical for proper Rac1-dependent signaling under such conditions. This indicates that the pharmacological targeting of this route could be an interesting avenue to block the migration of cancer cells bearing oncogenic RAC1 gene mutations, a particularly intriguing observation given the recent discovery of this type of mutations in human tumors (38–40).
The implication of MII in this new Coro1-dependent route also provides information about its potential implication in other cytoskeleton-related cell responses. Thus, ample evidence indicates that MII is at the center of the inhibitory interplay established between focal and nascent adhesions during cell signaling. Current models indicate that focal-adhesion-localized MII complexes coordinate such interplay using an “at a distance” mechanism that limits the formation of Rac1-dependent nascent adhesions in other subcellular areas. Although not yet fully characterized, recent findings suggest that such action is mediated by sequestration of Rac1 GEFs, a mechanism that obviously restricts the activation of Rac1 in cell regions located outside focal adhesions. There are conflicting reports, however, on whether such a process relies on the nonspecific binding of MII to many Rac1 GEFs or, alternatively, on the specific trapping of ArhGEF7 (30, 41). In addition to the problem associated with the quite different mechanistic scenarios offered by these two models, the latter omits the known fact that ArhGEF7 is a catalytically inactive GEF (19). In this context, our results indicate that the role of MII in this process may be more complex than previously anticipated, as it may also contribute to sequestering active Rac1/Pak/ArhGEF7 complexes in actomyosin structures. Furthermore, they suggest that ArhGEF7 may act in this process in the context of active Rac1/Pak/ArhGEF7 complexes rather than as a Rac1 GEF per se, a model that fits well the known biochemical properties of the protein.
Recent reports have revealed that mutations in the CORO1A gene cause severe combined T-cell immunodeficiencies (42, 43). These diseases have been attributed to conventional actions of the protein in F-actin and Arp2/3 regulation that contribute, either directly or indirectly, to the engagement of polarization, migration, homing, signaling, and survival responses in mature T cells (44–46). However, others have challenged this view, suggesting that many of the prosurvival functions of the protein are F-actin and Arp2/3 independent (47). Based on the present data, it would be interesting to consider the hypothesis that some of the dysfunctions could be mediated by unregulated MII activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Blázquez and personnel of the CIC Microscopy and Genomics and Proteomics Units for technical assistance and M. Vicente-Manzanares and M. Dosil for comments on the manuscript.
X.R.B.'s work has been funded by the Spanish Ministry of Economy and Competitiveness (SAF2009-07172, SAF2012-3171, RD06/0020/0001, and RD12/0036/0002), the Castilla-León Autonomous Government (CSI101U13), and the Asociación Española Contra el Cáncer. V.O.'s position has been supported by a JAE-Predoc contract (CSIC) and grant no. RD12/0036/0002. Spanish funding is cosponsored by the European Regional Development Fund.
V.O. carried out experimental work, analyzed data, edited figures, and wrote initial manuscript drafts. A.C.-C. generated reagents used in the work. X.R.B. directed the work, analyzed data, and wrote the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00347-14.
REFERENCES
- 1.Parsons JT, Horwitz AR, Schwartz MA. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell. Biol. 11:633–643. 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridley AJ. 2011. Life at the leading edge. Cell 145:1012–1022. 10.1016/j.cell.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 3.Pertz O. 2010. Spatio-temporal Rho GTPase signaling—where are we now? J. Cell Sci. 123:1841–1850. 10.1242/jcs.064345 [DOI] [PubMed] [Google Scholar]
- 4.Beli P, Mascheroni D, Xu D, Innocenti M. 2008. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat. Cell Biol. 10:849–857. 10.1038/ncb1745 [DOI] [PubMed] [Google Scholar]
- 5.Miki H, Yamaguchi H, Suetsugu S, Takenawa T. 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408:732–735. 10.1038/35047107 [DOI] [PubMed] [Google Scholar]
- 6.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. 2012. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148:973–987. 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, Waterman-Storer C, Danuser G, Bokoch GM. 2007. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell 13:646–662. 10.1016/j.devcel.2007.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delorme-Walker VD, Peterson JR, Chernoff J, Waterman CM, Danuser G, DerMardirossian C, Bokoch GM. 2011. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J. Cell Biol. 193:1289–1303. 10.1083/jcb.201010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustelo XR, Sauzeau V, Berenjeno IM. 2007. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays 29:356–370. 10.1002/bies.20558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. 2003. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83:433–473 [DOI] [PubMed] [Google Scholar]
- 11.Cai L, Holoweckyj N, Schaller MD, Bear JE. 2005. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J. Biol. Chem. 280:31913–31923. 10.1074/jbc.M504146200 [DOI] [PubMed] [Google Scholar]
- 12.Cai L, Makhov AM, Bear JE. 2007. F-actin binding is essential for coronin 1B function in vivo. J. Cell Sci. 120:1779–1790. 10.1242/jcs.007641 [DOI] [PubMed] [Google Scholar]
- 13.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. 2007. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128:915–929. 10.1016/j.cell.2007.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai L, Makhov AM, Schafer DA, Bear JE. 2008. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134:828–842. 10.1016/j.cell.2008.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oku T, Kaneko Y, Murofushi K, Seyama Y, Toyoshima S, Tsuji T. 2008. Phorbol ester-dependent phosphorylation regulates the association of p57/coronin-1 with the actin cytoskeleton. J. Biol. Chem. 283:28918–28925. 10.1074/jbc.M709990200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galkin VE, Orlova A, Brieher W, Kueh HY, Mitchison TJ, Egelman EH. 2008. Coronin-1A stabilizes F-actin by bridging adjacent actin protomers and stapling opposite strands of the actin filament. J. Mol. Biol. 376:607–613. 10.1016/j.jmb.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uetrecht AC, Bear JE. 2006. Coronins: the return of the crown. Trends Cell Biol. 16:421–426. 10.1016/j.tcb.2006.06.002 [DOI] [PubMed] [Google Scholar]
- 18.Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, Couceiro JR, Pimentel-Muinos FX, Del Pozo MA, Bustelo XR. 2011. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 30:3913–3927. 10.1038/emboj.2011.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberger G, Kutsche K. 2006. AlphaPIX and betaPIX and their role in focal adhesion formation. Eur. J. Cell Biol. 85:265–274. 10.1016/j.ejcb.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 20.Ruiz S, Santos E, Bustelo XR. 2007. RasGRF2, a guanosine nucleotide exchange factor for Ras GTPases, participates in T-cell signaling responses. Mol. Cell. Biol. 27:8127–8142. 10.1128/MCB.00912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prieto-Sanchez RM, Bustelo XR. 2003. Structural basis for the signaling specificity of RhoG and Rac1 GTPases. J. Biol. Chem. 278:37916–37925. 10.1074/jbc.M301437200 [DOI] [PubMed] [Google Scholar]
- 22.Caloca MJ, Zugaza JL, Matallanas D, Crespo P, Bustelo XR. 2003. Vav mediates Ras stimulation by direct activation of the GDP/GTP exchange factor Ras GRP1. EMBO J. 22:3326–3336. 10.1093/emboj/cdg316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauzeau V, Sevilla MA, Montero MJ, Bustelo XR. 2010. The Rho/Rac exchange factor Vav2 controls nitric oxide-dependent responses in mouse vascular smooth muscle cells. J. Clin. Invest. 120:315–330. 10.1172/JCI38356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuebel KE, Movilla N, Rosa JL, Bustelo XR. 1998. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 17:6608–6621. 10.1093/emboj/17.22.6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zugaza JL, Lopez-Lago MA, Caloca MJ, Dosil M, Movilla N, Bustelo XR. 2002. Structural determinants for the biological activity of Vav proteins. J. Biol. Chem. 277:45377–45392. 10.1074/jbc.M208039200 [DOI] [PubMed] [Google Scholar]
- 26.Goeckeler ZM, Bridgman PC, Wysolmerski RB. 2008. Nonmuscle myosin II is responsible for maintaining endothelial cell basal tone and stress fiber integrity. Am. J. Physiol. Cell Physiol. 295:C994–C1006. 10.1152/ajpcell.00318.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston SA, Bramble JP, Yeung CL, Mendes PM, Machesky LM. 2008. Arp2/3 complex activity in filopodia of spreading cells. BMC Cell Biol. 9:65. 10.1186/1471-2121-9-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wennerberg K, Ellerbroek SM, Liu RY, Karnoub AE, Burridge K, Der CJ. 2002. RhoG signals in parallel with Rac1 and Cdc42. J. Biol. Chem. 277:47810–47817. 10.1074/jbc.M203816200 [DOI] [PubMed] [Google Scholar]
- 29.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. 1996. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science 274:1374–1376. 10.1126/science.274.5291.1374 [DOI] [PubMed] [Google Scholar]
- 30.Lee CS, Choi CK, Shin EY, Schwartz MA, Kim EG. 2010. Myosin II directly binds and inhibits Dbl family guanine nucleotide exchange factors: a possible link to Rho family GTPases. J. Cell Biol. 190:663–674. 10.1083/jcb.201003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicente-Manzanares M, Koach MA, Whitmore L, Lamers ML, Horwitz AF. 2008. Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 183:543–554. 10.1083/jcb.200806030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crivat G, Taraska JW. 2012. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 30:8–16. 10.1016/j.tibtech.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA. 2004. Integrins regulate Rac targeting by internalization of membrane domains. Science 303:839–842. 10.1126/science.1092571 [DOI] [PubMed] [Google Scholar]
- 34.Swaminathan K, Muller-Taubenberger A, Faix J, Rivero F, Noegel AA. 2014. A Cdc42- and Rac-interactive binding (CRIB) domain mediates functions of coronin. Proc. Natl. Acad. Sci. U. S. A. 111:E25–E33. 10.1073/pnas.1315368111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DerMardirossian C, Bokoch GM. 2005. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 15:356–363. 10.1016/j.tcb.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 36.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell. Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rana MK, Worthylake RA. 2012. Novel mechanism for negatively regulating Rho-kinase (ROCK) signaling through Coronin1B protein in neuregulin 1 (NRG-1)-induced tumor cell motility. J. Biol. Chem. 287:21836–21845. 10.1074/jbc.M112.346114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R. 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44:1006–1014. 10.1038/ng.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. 2013. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc. Natl. Acad. Sci. U. S. A. 110:912–917. 10.1073/pnas.1220895110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. 2012. A landscape of driver mutations in melanoma. Cell 150:251–263. 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo JC, Han X, Hsiao CT, Yates JR, III, Waterman CM. 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13:383–393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiow LR, Roadcap DW, Paris K, Watson SR, Grigorova IL, Lebet T, An J, Xu Y, Jenne CN, Foger N, Sorensen RU, Goodnow CC, Bear JE, Puck JM, Cyster JG. 2008. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat. Immunol. 9:1307–1315. 10.1038/ni.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moshous D, Martin E, Carpentier W, Lim A, Callebaut I, Canioni D, Hauck F, Majewski J, Schwartzentruber J, Nitschke P, Sirvent N, Frange P, Picard C, Blanche S, Revy P, Fischer A, Latour S, Jabado N, de Villartay JP. 2013. Whole-exome sequencing identifies Coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated B-cell lymphoproliferation. J. Allergy Clin. Immunol. 131:1594–1603. 10.1016/j.jaci.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mugnier B, Nal B, Verthuy C, Boyer C, Lam D, Chasson L, Nieoullon V, Chazal G, Guo XJ, He HT, Rueff-Juy D, Alcover A, Ferrier P. 2008. Coronin-1A links cytoskeleton dynamics to TCR alpha beta-induced cell signaling. PLoS One 3:e3467. 10.1371/journal.pone.0003467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller P, Massner J, Jayachandran R, Combaluzier B, Albrecht I, Gatfield J, Blum C, Ceredig R, Rodewald HR, Rolink AG, Pieters J. 2008. Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat. Immunol. 9:424–431. 10.1038/ni1570 [DOI] [PubMed] [Google Scholar]
- 46.Foger N, Rangell L, Danilenko DM, Chan AC. 2006. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science 313:839–842. 10.1126/science.1130563 [DOI] [PubMed] [Google Scholar]
- 47.Mueller P, Liu X, Pieters J. 2011. Migration and homeostasis of naive T cells depends on coronin 1-mediated prosurvival signals and not on coronin 1-dependent filamentous actin modulation. J. Immunol. 186:4039–4050. 10.4049/jimmunol.1003352 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.