Abstract
CS31A, a K88-related surface antigen specified by the clp operon, is a member of the type P family of adhesive factors and plays a key role in the establishment of disease caused by septicemic and enterotoxigenic Escherichia coli strains. Its expression is under the control of methylation-dependent transcriptional regulation, for which the leucine-responsive regulatory protein (Lrp) is essential. CS31A is preferentially in the OFF state and exhibits distinct regulatory features compared to the regulation of other P family members. In the present study, surface plasmon resonance and DNase I protection assays showed that Lrp binds to the distal moiety of the clp regulatory region with low micromolar affinity compared to its binding to the proximal moiety, which exhibits stronger, nanomolar affinity. The complex formation was also influenced by the addition of PapI or FooI, which increased the affinity of Lrp for the clp distal and proximal regions and was required to induce phase variation. The influence of PapI or FooI, however, was predominantly associated with a more complete shutdown of clp expression, in contrast to what has previously been observed with AfaF (a PapI ortholog). Taken together, these results suggest that the preferential OFF state observed in CS31A cells is mainly due to the weak interaction of the leucine-responsive regulatory protein with the clp distal region and that the PapI homolog favors the OFF phase. Within the large repertoire of fimbrial variants in the P family, our study illustrates that having a fimbrial operon that lacks its own PapI ortholog allows it to be more flexibly regulated by other orthologs in the cell.
INTRODUCTION
Bacterial infection includes a dynamic balance between the requirement of virulence factors that are produced by the microorganism and the production of an antigen recognized by the host immune system (1, 2). Phase variation is an example of the complex regulatory systems that pathogenic strains have developed to change their phenotype in response to environmental signals. This stochastic and reversible switch between an all or none (ON/OFF) phenotype is a key mechanism to generate phenotypic diversity in genetically identical cells, especially to survive in a hostile environment (3, 4). After division, most daughter cells retain the expression phase of the parent, whereas a minority of cells switch to the other phenotype. Despite its energetic cost and the risk of being deleterious to individual cells, phase variation confers a net advantage to the entire population by allowing it to anticipate host environmental changes without having to sense them. Thus, many virulence factors, such as fimbrial adhesins, are subject to phase variation, and pathogenic bacteria coordinately express more than one adhesin by phase variation (5, 6).
The clp-encoded CS31A surface antigen, found in septicemic and enterotoxigenic Escherichia coli strains of bovine and human origins, is required for full bacterial virulence (7, 8). It was first described in E. coli strains recovered from calves with septicemia or diarrhea. It is a capsule-like surface protein, immunologically related to F4 (K88) and F41 fimbriae (8). Experimental infections (9) and epidemiological data (10) suggest a close association of CS31A-producing strains with cases of septicemia (11). Pap-related fimbriae are the most frequent E. coli adhesins associated with human urinary tract infection, but they are also found in strains producing CS31A (12). There is a worldwide distribution of CS31A antigen in bovine E. coli strains (11, 13). CS31A is required for full bacterial virulence in the rat model (J.-P. Girardeau, personal communication), mediates adhesion to Caco-2 and Int-407 cells (14), and protects bacteria from phagocytosis by bovine polymorphonuclear neutrophils (PMNs) (15).
While CS31A is genetically and immunologically related to K88, its expression is controlled by phase variation, like the well-characterized Pap-regulatory family of adhesive factors. It has also been proposed that CS31A depends on the expression of an additional fimbrial gene cluster that is also controlled by phase variation (15–17).
For Pap, expression is inherited in an epigenetic manner (6) because of methylation-controlled modifications within the regulatory region of the pap operon and the presence of the leucine-responsive regulatory protein (Lrp) (6). Lrp controls a large number of operons in E. coli, including those involved in amino acid biosynthesis, amino acid degradation, metabolite transport, and carbon metabolism (18–20). The regulatory region of pap consists of a DNA sequence of approximately 400 bp that is flanked by two divergently transcribed genes, papI and papB (Fig. 1). The intergenic region also includes 6 Lrp binding sites [all containing the consensus sequence GN(2/3)TTT, where N is any nucleotide], as well as 2 GATC sites spaced 102 bp apart (the proximal [GATCprox] and distal [GATCdist] GATC sites). Differences in the methylation status of these sequences determine the binding of Lrp (21, 22). Thus, switching from one phenotype to the other is the result of competition between the binding of Lrp at sites 1 to 3 or sites 4 to 6 and methylation by Dam methyltransferase at the opposite GATC site, which prevents the binding of Lrp (Fig. 1) (for reviews, see references 23, 24, and 25). When GATCdist is fully methylated, Lrp cooperatively binds to sites 1 to 3, maintaining the cells in the OFF state, and when GATCprox is fully methylated, Lrp binds to sites 4 to 6, maintaining the ON state (Fig. 1). Control of pap phase variation also requires the action of PapI, a positive regulator that increases the affinity of Lrp for sites 4 to 6 in vivo (25, 26), as well as PapB, a second specific regulator of the pap operon that coordinates the expression of the pBA and pI promoters (25). Taken together, the combined actions of Lrp, Dam methyltransferase, PapI, and PapB result in a finite probability that each cell will express Pap fimbriae (ON) or not (OFF) just after DNA replication.
FIG 1.
Overview of the clp operon. (A) Schematic representation of the clp operon. The two GATC sites subject to methylation by Dam are indicated by GATCdist and GATCprox. Note the absence of a papI homologue in the clp operon. (B) Comparison of the nucleotide sequences of the regulatory regions of the clp and pap operons. Identical nucleotides in clp and pap are shown as boxed regions. Lrp-binding site positions 1 to 6 in the pap sequence are indicated with underlines, and letters a to g with overlines indicate the positions of Lrp-binding sites in the clp sequence, as identified by DNase I footprint. The regions corresponding to the PapB binding sites and the CAP binding site are represented with dashed underlines, and the putative promoter regions (−10 and −35 sites) with underlines. Red letters indicate GATC sites. Blue and red arrows represent the boundaries of the distal and proximal fragments, respectively, used in Biacore experiments.
Distinct regulatory features can be observed between members of the P-regulatory family. In particular, Lrp activates the expression of some operons, including pap and foo, and represses the expression of others, including clp (15, 16). In addition, for CS31A, striking features include the absence of a PapI homologue encoded by the clp operon and a moderate level of clp transcription. However, the addition of the PapI homologue AfaF in trans promotes clp phase variation (15, 17), confirming that it is required during clp phase variation. Similar to the results for Pap, we have previously shown that CS31A phase variation in the presence of afaF is mediated by the global regulator Lrp and methylation protection of the two clp GATC sites (15, 17). However, CS31A is characterized by a higher level of OFF cells than Pap in the presence of afaF (15).
To date, direct evidence for the interplay between PapI homologues and phase variation has only been described with AfaF, the papI homologue found in afa-3 (15, 17). This mechanism might not mimic what is found in natural isolates, since strains may possess adhesin gene clusters other than afa-3. For instance, Bertin et al. showed that the reference strain 31A encodes not only CS31A but also P fimbriae (12). Whether PapI homologues encoded by other pap-related operons in CS31A strains cross-regulate clp transcription remains to be investigated.
To provide direct evidence as to why clp expression (CS31A) gives rise to a high level of OFF cells compared to pap expression (Pap) and to establish whether PapI homologues influence CS31A phase variation in an identical manner, we analyzed the interactions between Lrp, PapI, and/or FooI and the intergenic regions of the clp operon in our present work; notably, FooI is a PapI homologue whose sequence differs from that of PapI by only a single amino acid modification (D17N).
By combining in vitro binding experiments with genetic phase variation studies, we show that the preferential OFF state observed in CS31A-positive cells is mainly due to a weak interaction of Lrp with the distal region of clp. Moreover, the addition of PapI or FooI (a PapI homolog encoded by the foo operon) promoted CS31A phase variation to similar extents and, due to increased Lrp affinity for both GATCdist and GATCprox, the OFF state surpassed the levels observed previously in the presence of afaF. Furthermore, the overall affinity of the ternary complex (DNA together with Lrp and PapI or FooI) for the clp proximal region is so strong that only a small fraction of cells can switch to the ON state.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strain MC4100.λ6 contains a single chromosomal copy of a clp-lacZYA fusion. ptrf5 and ptrf6, multicopy recombinant plasmids derived from ptrc99A (Pharmacia Biotech) express fooI and papI, respectively, under the control of the trc promoter. Luria-Bertani (LB) broth and M9 minimal broth were prepared as described previously (27, 28). When necessary, these media were supplemented with 100 μg · ml−1 ampicillin and 40 μg · ml−1 kanamycin (unless otherwise noted), 1mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 40 μg · ml−1 5-bromo-4-chloro-3-indolyl-b-d-galactopyranoside (X-Gal). Cultures of strains harboring ptrf5 or ptrf6 were grown in the absence of IPTG to avoid too-high expression of fooI or papI.
Plasmid construction.
For overexpression and purification of the FooI protein, the pET32a-fooI plasmid was constructed by first amplifying the fooI sequence from the chromosome of strain 4787 using the NcoI-fooIrg1F and XhoI-fooIrg1R primers, containing the NcoI and XhoI restrictions sites, respectively. After enzymatic digestion, the fragment was ligated into the NcoI/XhoI restriction sites of the expression plasmid pET32a. The inserts were sequenced for verification after being transformed into E. coli BL21(DE3)pLysS by electroporation. Plasmid pET32a-papI was constructed using the QuikChange site-directed mutagenesis system (Stratagene). Briefly, the expression plasmid pET32a-fooI was extracted from strain E. coli BL21(DE3)pLysS, purified, and used as the template for amplification using the fooIN17D-F and fooIN17D-R primers. PCR-amplified plasmids were purified and digested with the restriction enzyme DpnI to remove any trace of the pET32a-fooI template. The PCR-amplified plasmids were dialyzed and transformed into strain E. coli BL21(DE3)pLysS by electroporation.
Purification of recombinant proteins.
E. coli strain CV1494 was used to purify His6-tagged Lrp as previously described (29). A 1-liter culture of CV1494 was grown in LB broth at 37°C with shaking to an optical density at 600 nm (OD600) of 0.7 to 0.8. IPTG was added to the culture at a final concentration of 0.5 mM for 6 h. Bacteria were harvested, resuspended in 30 ml of Tris-buffered saline (20 mM Tris-HCl [pH 7.4], 0.5 M NaCl), and lysed with a French press and an ultrasonic processor. The soluble fraction was clarified by centrifugation at 15,000 × g for 20 min at 4°C. Proteins were purified using an ÄKTA purifier system with a 1-ml HisTrap HP column (Amersham Biosciences) according to the instructions of the manufacturer. Purified proteins were subsequently dialyzed against TG50ED buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.1 mM dithiothreitol [DTT], 0.1 M NaCl), and the purity of the proteins was confirmed by SDS-PAGE and staining with Coomassie blue.
For purification of FooI and PapI, a single E. coli BL21(DE3)pLysS colony carrying plasmid pET32a-fooI or pET32a-PapI was incubated in LB broth with shaking at 37°C to an OD600 of 0.4 to 0.6. The culture was incubated for 3 h more in the presence of IPTG at a final concentration of 1 mM. Bacteria were harvested, resuspended in 30 ml of Tris-buffered saline (20 mM Tris-HCl [pH 7.4], 0.5 M NaCl), and lysed with a French press and an ultrasonic processor. The soluble fraction was clarified by centrifugation at 15,000 × g for 20 min at 4°C. The His6-Trx-FooI and His6-Trx-PapI fusion proteins were purified using an ÄKTA purifier system using the same procedure as for His6-tagged Lrp. FooI and PapI proteins were removed from their His6-Trx tag after enzymatic digestion using enterokinase (New England BioLabs) in digestion buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 2 mM CaCl2). Proteins were dialyzed against 20 mM 2-(N-morpholino)ethanesulphonic acid (MES), pH 6.0, and purified using an ÄKTA purifier system with a mono S 5/50 GL column (Amersham Biosciences) according to the instructions of the manufacturer. The purity of FooI and PapI proteins was confirmed by SDS-PAGE and staining with Coomassie blue.
Surface plasmon resonance (SPR).
The interactions between 5′ biotinylated, double-stranded oligonucleotides (clp distal region, 120 kDa; clp proximal region, 77 kDa; and clp distal and proximal region, 185 kDa) and three DNA-binding proteins (Lrp, 20 kDa monomer; FooI, 9 kDa; and PapI, 9 kDa) were examined using a Biacore 3000 SPR spectrometer (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Experiments were performed on research-grade streptavidin (SA)-coated sensor chips (XanTec Bioanalytics GmbH, Muenster, Germany) at 25°C using filtered (0.2 μm) and degassed CALVO-P running buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 0.4 mM EDTA, 1 mM MgCl2, 0.1 mM DTT, 0.005% [vol/vol] Tween 20). As recommended by the manufacturer, SA-coated sensor chips were preconditioned with three 1-min pulses of 1 M NaCl in 50 mM NaOH. The protein-grade detergents (Tween 20 and Empigen) were from Anatrace (Maumee, OH, USA), and fatty acid-free bovine serum albumin (BSA) was from Millipore (Billerica, MA, USA); all other chemicals were reagent-grade quality. Purified protein concentrations were determined using the bicinchoninic acid (BCA) assay, and protein purity (>95%) was assessed by SDS-PAGE (12.5% polyacrylamide) under reducing (5% [vol/vol] 2-mercaptoethanol) and nonreducing conditions (data not shown).
To examine binding with Lrp, the appropriate oligonucleotides were annealed and then captured (20 μl/min with 10 nM DNA oligonucleotides in running buffer containing 0.5 M NaCl) to generate low-density surfaces (<100 response units [RU] immobilized). Lrp (0 to 100 nM) or BSA (negative control) was injected over reference (i.e., SA only) and active (i.e., clp DNA fragments) surfaces in tandem using the Kinject mode (25 μl/min for 10 min association and 10 min dissociation). Between sample injections, surfaces were regenerated at 50 μl/min using two 30-s pulses of solution I (1 M NaCl, 0.02% [vol/vol] Empigen in CALVO-P), followed by Extraclean and Rinse procedures.
To examine binding with FooI and PapI, medium-density oligonucleotide surfaces (300 to 400 RU immobilized) were prepared as noted above. FooI or PapI (0 to 15 μM) was injected over reference and active surfaces in tandem using the Kinject mode (5 μl/min × 5 min association plus 10 min dissociation). Between sample injections, surfaces were regenerated as noted above. In additional experiments, Lrp (50 nM) was also injected in the presence of FooI and PapI (0.0 to 5.6 and 0.0 to 8.1 μM, respectively) using the Kinject mode (10 μl/min × 5 min association plus 10 min dissociation).
The mass transport-independent data were doubled referenced (30) and are representative of duplicate injections acquired from three independent trials. For each replicate series, a buffer blank was injected first, the highest titrant concentration second, and serial dilutions thereafter (from the lowest to the highest concentration); comparing responses between the two highest titrant injections verified consistent DNA surface activity throughout each assay. To estimate apparent equilibrium dissociation constants (KD), steady-state binding responses (Req; average RU near the end of the association phase) were plotted as a function of protein concentration (C) and then subjected to nonlinear regression (1-site-specific binding with Hill slope or steady-state affinity model using Prism5 version 5.0c for Mac OS X; GraphPad Software, San Diego, CA, USA). To estimate individual dissociation rate constants (Kd), sensorgram profiles were analyzed using the Fit Kinetics Separate ka/kd tool (BIAevaluation version 4.1). Theoretical binding maxima were predicted using the equation Rmax = (MWA/MWL) × RL × n, where Rmax is the maximal binding response (RU) at saturating protein concentration, MWA is the molecular weight (kDa) of the protein injected in solution, MWL is the molecular weight (kDa) of the DNA fragment immobilized, RL is the amount (RU) of DNA immobilized, and n is the predicted binding stoichiometry (e.g., 1:1).
Footprint analysis.
DNase I footprinting of free DNA and DNA-protein complexes was performed as described previously (31). The DNA fragment corresponding to the clp regulatory region (317 bp) was amplified using primers clp-F (5′-GCGCTACCGTTTTTTGACTCTCCC-3′) and clp-R (5′-GCAGCGAAGATTATCACGATGTTTTATAGCG-3′). A clp DNA fragment end labeled with 32P (125,000 cpm, 0.3 nM) was subsequently incubated in a total volume of 20 μl with 2 μg of salmon sperm DNA and 2 μg of acetylated bovine serum albumin (New England BioLabs) in binding buffer (60 mM Tris-HCl [pH 7.5], 40 mM KCl, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT). After incubation for 10 min at room temperature, 2 μl of DNase I (200 U · ml−1; New England BioLabs) containing 22 mM CaCl2 and 22 mM MgCl2 was added for 4 min (top fragment) and 6 min (bottom fragment). The reaction was stopped by the addition of 100 μl of stop buffer (10% glycerol, 2.5 mM EDTA, 0.6 M ammonium acetate, 2 mg yeast tRNA per ml) to each sample. DNA fragments were precipitated in ethanol, and amounts with equivalent cpm (5 × 104) from each reaction were loaded onto 6% polyacrylamide–7 M urea gels. Maxam-Gilbert A+G reactions (32) were carried out on the appropriate 32P-labeled DNA fragments, and the products loaded alongside the DNase I footprinting reaction mixtures. The gels were dried and analyzed by autoradiography.
Phase variation and measurement of switching frequencies.
The switch frequency was calculated as described previously (15, 33). Strains were inoculated onto LB-kanamycin agar. Colonies were excised from the agar and resuspended in M9 salts containing no carbon source. Appropriate dilutions were subsequently spread onto M9 X-Gal agar containing 0.2% glycerol and, after 36 h of growth at 37°C, plates were photographed using a digital camera. Three colonies showing a uniform Lac+ or Lac− phenotype were excised from the agar, resuspended in M9 salts, and spread onto M9 X-Gal agar as described above. The passage from M9 X-Gal agar to fresh M9 X-Gal agar containing the same source of carbon was repeated twice more to follow the switch frequency through three generations. At least 2,000 colonies were scored for a Lac phenotype as described previously (33), and the switch frequencies were calculated according to the formula (M/N)/g, where M is the number of cells that underwent phase variation, N is the total number of cells evaluated, and g is the total number of generations (estimated to be 20 generations for all the strains tested in our experiments).
β-Galactosidase activity assay.
A single bacterial colony was used to inoculate an overnight culture in M9 glycerol medium. This culture was diluted 50-fold and grown to an OD600 of 0.5 to 0.8 in M9 glycerol medium. The culture was then assayed for β-galactosidase activity as described previously (27). For strains showing phase variation, a blue or white colony was picked from M9 X-Gal glycerol agar plates as starting material for assays of the β-galactosidase activities of ON and OFF cultures, respectively. Each experiment was performed in duplicate at least three times.
RESULTS
Differences in complex formation between Lrp and clp proximal or distal region.
Similar to the intensively studied pap operon, clp phase variation requires Lrp. The Lrp target regulatory region contains two GATC sites separated by 102 or 103 bp, and protection of these sites from methylation is Lrp dependent (15, 17). We hypothesized that the lower affinity of Lrp for the clp distal region than for the clp proximal region is responsible for the CS31A bias toward the OFF phase. To test for these interactions, label-free surface plasmon resonance (SPR) was used to monitor real-time binding of Lrp to unmethylated DNA fragments corresponding to the clp regulatory regions. Biotinylated oligonucleotide duplexes containing the clp distal, clp proximal, or clp distal-proximal region were immobilized on streptavidin-coated sensor chips, and His6-Lrp was injected over the reference and DNA surfaces simultaneously. As shown by the results in Fig. 2, dose-dependent and/or saturable binding of Lrp to the clp regions was detected. In contrast to what has already been described for pap (22, 25), Lrp binding to the clp distal region was noticeably weaker (i.e., slow association and fast dissociation kinetics) than its binding to the clp proximal region (i.e., fast association and slow dissociation kinetics). A hybrid of the distal and proximal kinetics is reflected in the binding of Lrp (i.e., biphasic association and dissociation phases) to the full-length clp fragment.
FIG 2.
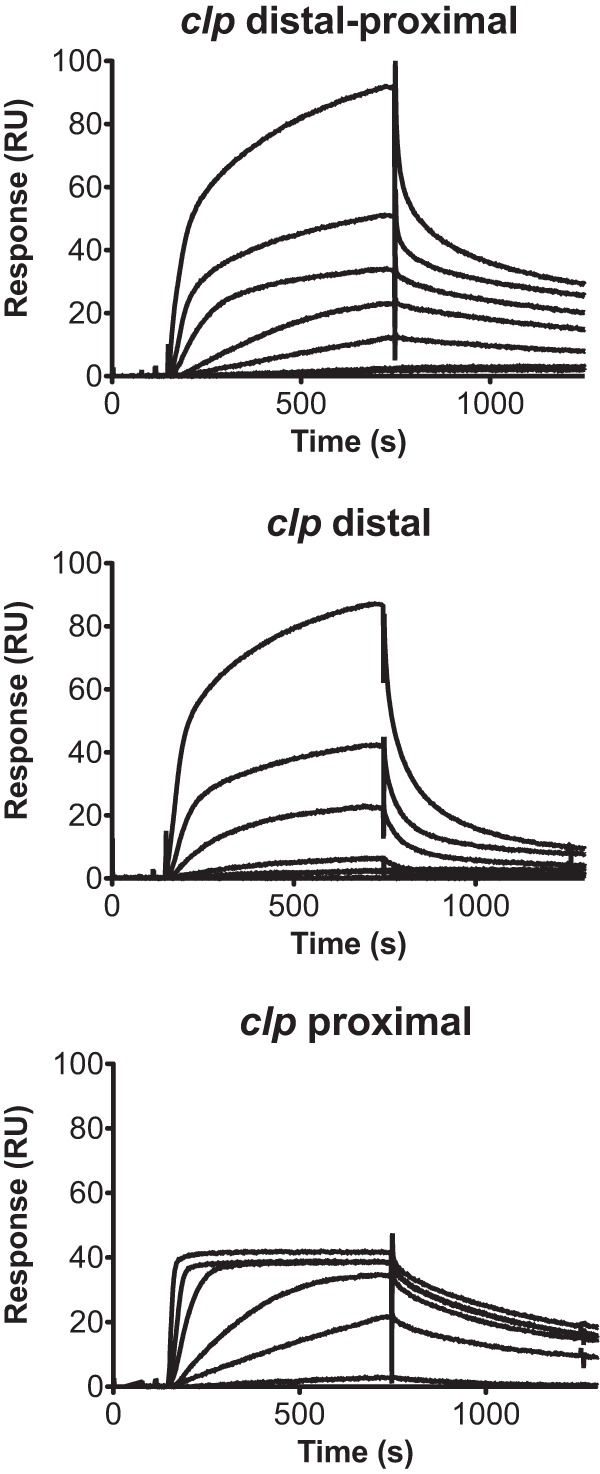
Association and dissociation kinetics of His6-Lrp binding to clp regulatory regions. Representative SPR analyses of Lrp (0 to 100 nM; 2-fold dilution series) injected over clp distal, proximal, and distal-proximal fragments (95, 40, and 100 RU, respectively) at 25 μl/min (10 min association plus 10 min dissociation).
Analysis of the data sets was performed according to previous reports in the literature indicating that Lrp binding is cooperative (i.e., sites 1 to 3 and 4 to 6) (34). Similar to SPR analyses of other cooperative protein-DNA kinetics (35), the clp proximal region titration series fit well to a 1-site model with Hill slope, which indicated positive cooperativity (i.e., Hill coefficients of >1) (see Fig. S1 in the supplemental material). While Lrp binding to the proximal region exhibited low nanomolar affinity (KD, ∼10 nM) (Table 1), binding to the distal region exhibited significantly different micromolar affinity (KD, ∼11 μM). Analogous to other biomolecules where variable binding sites are critical for differential function (36, 37), we propose the following physiological duality with clp: the preferential OFF state in CS31A-positive cells is mainly due to Lrp interacting with the weak-affinity GATCdist site; in the presence of PapI or FooI, however, the increased Lrp affinity involves both the low-affinity GATCdist and high-affinity GATCprox sites. Notably, ternary complex formation between the DNA, Lrp, and the PapI homologue is so strong for the clp proximal region that only a small fraction of cells can switch to the ON state. Since Lrp can exhibit higher affinity for the clp proximal region than for the distal region, this differential property could block the entry of the RNA polymerase at the clpBA promoter, thereby yielding a predominant OFF phase.
TABLE 1.
Apparent equilibrium dissociation rate constants for Lrp, FooI, and PapI binding to clp regulatory regionsa
| clp fragment | Avg KD ± SEM |
||
|---|---|---|---|
| Lrp (nM) | PapI (μM) | FooI (μM) | |
| Distal | 11,000 ± 1000 | 13 ± 1.0 | 14 ± 1.4 |
| Proximal | 10 ± 1 | 17 ± 1.1 | 14 ± 0.7 |
| Distal-proximal | 180 ± 15 | ND | ND |
SPR binding responses (RU, average 710 to 730 s) were plotted as a function of concentration using Prism5 version 5.0c for Mac OS X (GraphPad Software, San Diego, CA, USA). Lrp constants were determined by fitting the experimental data to a 1-site-specific binding with Hill slope model. PapI and FooI constants were determined by fitting the experimental data to a steady-state affinity model. Each titration series was analyzed globally using the specified model, and estimates represent the average results of three independent trials. clp distal-proximal refers to the DNA fragment encompassing the clp distal and the clp proximal regions. ND, not done.
Given that SPR showed that Lrp has a higher affinity for the clp proximal region than for the distal region, we hypothesized that such a difference could be due to differences in the proximal and distal Lrp-binding sites and/or to divergent nucleotide sequences surrounding the GATC sites. In contrast to pap, for which six Lrp-binding sites have been identified, the clp intergenic region contains more GN(2/3)TTT motifs of Lrp-binding sites (Fig. 1). Thus, we used DNase I footprint analysis to identify the Lrp-binding sites within the full-length clp regulatory region. The results in Fig. 3 show that the addition of His6-Lrp was associated with the presence of multiple gel-protected regions, with four corresponding to the proximal region and three to the distal region. GATCprox and GATCdist sites were included within Lrp-binding sites of clp. In addition, DNase protection was observed at a lower concentration of His6-Lrp for the clp proximal region than for the distal region, in agreement with the higher affinity of Lrp for the clp proximal region observed by SPR. In contrast, the pap proximal region was protected at an Lrp concentration 10 times lower than that at which the clp proximal region was protected (38). Thus, this unique pattern of Lrp-binding sites may result in a weak affinity for the clp distal region. In addition, DNase I footprinting shows periodic regions of DNase I protection and hypersensitivity, characteristic of bent DNA (39). This was also previously observed in pap and foo (38).
FIG 3.

Lrp localization on the clp regulatory region. DNase I footprinting assays were performed with increasing amounts of Lrp. First to 4th lanes, DNase I footprinting assays with increasing amounts of Lrp on the clp regulatory region (317 bp) end labeled with 32P on the top (left) and bottom (right) strands (1st lane, DNA alone; 2nd lane, Lrp at 100 nM; 3rd lane, Lrp at 1 μM; 4th lane, Lrp at 5 μM); 5th lane, Maxam-Gilbert A+G reactions. Letters and bars indicate the positions of Lrp-binding sites in the clp sequence.
PapI and FooI direct CS31A phase variation toward the OFF state.
In E. coli K-12, phase variation of the clp operon requires the presence in trans of a PapI homologue (15–17). So far, this has only been demonstrated with AfaF, a PapI homologue that controls the genetically distant operon afa-3 and which is absent in the pathogenic reference strain E. coli 31A (17). However, strain 31A carries a pap fimbrial operon in its genome, so clp phase variation could result from the cross-regulation between clp and pap fimbriae through PapI supplied in trans. To assess whether PapI homologues can influence clp phase variation (as previously demonstrated with AfaF [17]), clp phase variation was measured in vivo by providing FooI or PapI in trans. Strain MC4100.λ6, carrying a chromosomal fusion between the clp regulatory region and the lacZYA reporter system, was transformed with plasmid ptrf5 or ptrf6 (expressing fooI and papI, respectively). The two transformed strains were then plated onto X-Gal M9 agar without IPTG (to avoid a too-high level of PapI or FooI protein in the cell), and the [Lac] phenotype was followed for up to two platings. This medium uses glycerol as the carbon source to avoid catabolite repression, which affects phase variation (28), and has no amino acids (40). While strain MC4100.λ6 led to a uniform phenotype characterized by light-blue colonies (data not shown), transformation with either ptrf5 or ptrf6 led to a majority of white colonies with few dark blue colonies (Fig. 4A and Table 2). Thus, PapI or FooI produced in trans induced phase variation of the clp operon. However, phase variation of clp was biased toward the OFF state by more than an order of magnitude, even when the starting phenotype was the ON state (Table 2). This was also observed when bacteria were grown in M9 glycerol liquid medium, with the β-galactosidase activity being lower in the presence than in the absence of any PapI homologue tested, regardless of the starting phenotype (Fig. 4B). This result is in agreement with the observation that Lrp shows a higher affinity for the clp proximal region than for the clp distal region whether a PapI homologue is present or absent (15, 17). As a result, the low affinity of Lrp for the clp distal region contributes greatly to the low level of ON cells. Interestingly, differences in the ability of PapI and FooI to promote switching toward the ON state were observed. Notably, the rate of switching to ON cells was higher in the presence of FooI (average of 0.8 × 10−3) than in the presence of PapI (average of 0.1 × 10−3) (Table 2). In contrast, no significant difference was seen in ON-to-OFF switching between these two orthologs. Thus, it seems that even if PapI homologues are all able to promote phase variation, they can still differentially influence the level of the ON population.
FIG 4.
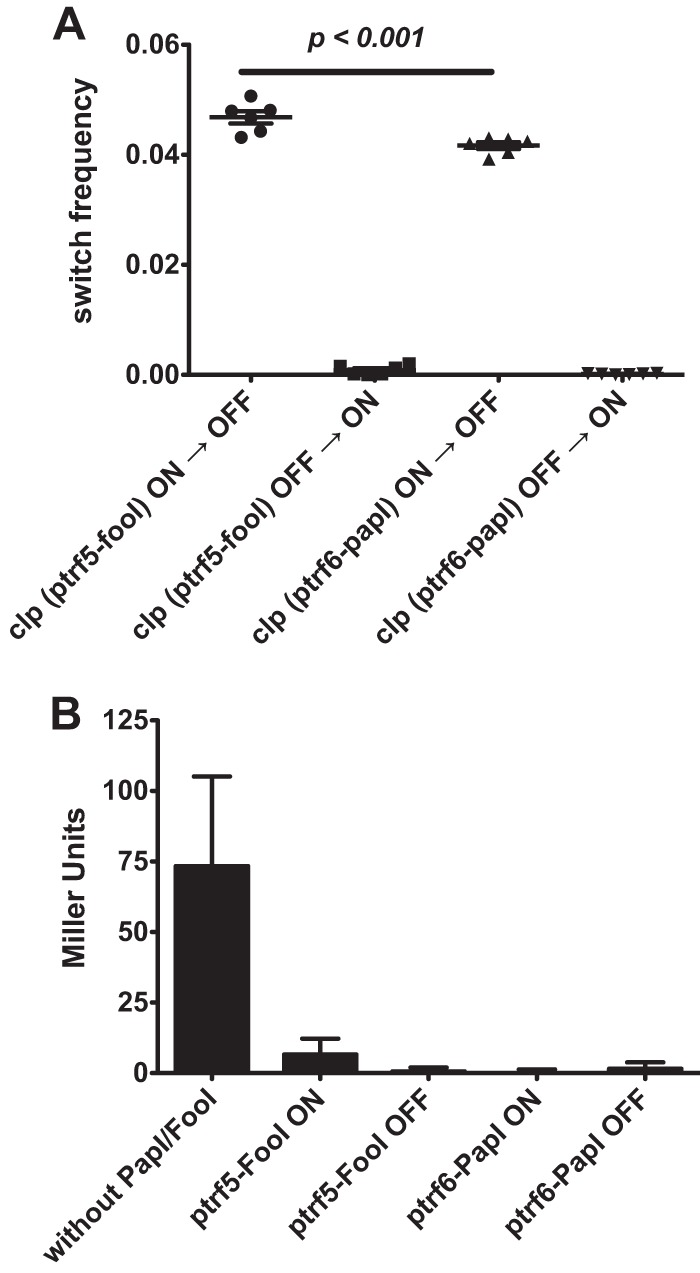
Effects of PapI homologues on clp basal expression. Lac+ and Lac− phenotypes of clp strains expressing papI or fooI. (A) Switch frequencies were calculated using the formula (M/N)/g, where M is the number of cells that underwent phase transition, N the total number of cells evaluated, and g the number of generations, estimated to be 20 generations, that gave rise to the colony. Cells were grown on M9 X-Gal agar containing 0.2% glycerol for 36 h at 37°C. Horizontal bars represent the mean results. (B) Influence of PapI or FooI on the transcription levels of the clp operon as measured by the lacZYA reporter system. Bacteria were grown to an OD600 of 0.5 to 0.8 in M9 glycerol medium. Cultures were then assayed for β-galactosidase activity as described previously (27). For strains showing phase variation, a blue or white colony was picked from M9 X-Gal glycerol agar plates as starting material for assays of β-galactosidase activities of ON and OFF cultures, respectively. β-Galactosidase activities are indicated in Miller units. Bars represent the mean results of at least three independent experiments. Error bars show standard deviations.
TABLE 2.
Effects of PapI homologues on phase variation of E. coli clp-lacZYA fusion lysogen
| Strain and phenotype | Colony | Total no. of colonies counted | No. of [Lac+] colonies/no. of [Lac−] colonies | % of [Lac+] colonies | α or β switch frequencya (× 10−3) |
|---|---|---|---|---|---|
| MC4100.λ6 (ptrf5-fooI) | |||||
| [Lac−] | 1 | 5,889 | 239/5,650 | 4.05 | α Þ 1.0 |
| 2 | 4,838 | 57/4,781 | 1.18 | α Þ 0.6 | |
| [Lac+] | 1 | 4,864 | 155/4,709 | 3.18 | β Þ 49 |
| 2 | 6,664 | 575/6,089 | 8.62 | β Þ 44 | |
| MC4100.λ6 (ptrf6-papI) | |||||
| [Lac−] | 1 | 7,774 | 19/7,755 | 0.20 | α Þ 0.82 |
| 2 | 6,141 | 29/6,112 | 0.40 | α Þ 0.2 | |
| [Lac+] | 1 | 8,701 | 72/8,629 | 0.80 | β Þ 40 |
| 2 | 7,099 | 149/6,950 | 2.10 | β Þ 42 |
The switch frequencies α (OFF to ON) and β (ON to OFF) were calculated using the formula (M/N)/g, where M is the number of cells that underwent phase transition, N is the total number of cells evaluated, and g is the total number of generations (estimated to be 20) that gave rise to the colony. Colony 1 corresponds to the initial plate analyzed; colony 2 corresponds to the second plate analyzed and was obtained directly from colony 1.
PapI and FooI preferentially increase Lrp association with and stability of its binding to the clp proximal region.
To understand why FooI and PapI had such strong influences on the ON-to-OFF switching, we used SPR to investigate the direct binding of FooI or PapI to the clp distal and proximal regions. Rapid-ON, rapid-OFF binding of FooI and PapI to all clp fragments correlated with low micromolar overall binding affinities (Fig. 5 and Table 1), consistent with previous work by Kawamura et al., who showed direct binding of PapI to pap sites 2 and 5 with similarly weak affinities (41). However, no noticeable differences in the binding kinetics were observed between FooI and PapI with all clp fragments (Fig. 5; see also Fig. S2 in the supplemental material). In additional SPR experiments, a fixed concentration of His6-Lrp was injected over the clp distal and proximal regions in the absence or presence of increasing PapI or FooI concentrations. While low micromolar injections of FooI or PapI (i.e., <8 μM) yielded small binding responses to the clp fragments on their own (Fig. 5), Lrp binding was significantly altered in the presence of FooI or PapI (Fig. 6). In a dose-dependent manner, increasing concentrations of FooI or PapI enhanced the overall amounts of Lrp bound in all cases (i.e., increased association). The stability of the resultant clp distal and proximal complexes was also increased (i.e., dissociation rates were approximately 50% lower with 6 μM FooI and 8 μM PapI).
FIG 5.
Association and dissociation kinetics of PapI and FooI when bound to the distal or proximal region of the clp operon. Representative SPR analyses of PapI or FooI (0 to 15 μM; 2-fold dilution series) binding to immobilized clp distal (400 RU) and proximal (300 RU) fragments at 5 μl/min (5 min association and 5 min dissociation).
FIG 6.
PapI and FooI increase both association and stability of Lrp at the clp proximal and distal regions. Representative SPR analyses of 50 nM Lrp binding to immobilized clp (95 RU distal, 40 RU proximal) fragments in the absence (solid lines) and presence (dashed lines) of FooI (0 to 6 μM; 2-fold dilution series) or PapI (0 to 8 μM; 2-fold dilution series).
Since the transcription of papI homologues on a ptrc99A vector is independent of the clp operon, a PapI homologue may contribute to enhancing the binding of Lrp at the clp proximal region, in combination with the action of the Dam methylase. Thus, these results suggest that PapI and FooI have two opposite functions. First, they contribute to switching ON the clp operon by increasing the association kinetics of Lrp for the clp distal region, and second, they strongly maintain cells in the OFF state by forming a particularly stable complex between Lrp and the clp proximal region.
DISCUSSION
The objective of the present study was to determine why clp expression (CS31A) gives rise to a higher level of OFF cells than does pap expression (Pap). Using in vitro techniques, we have demonstrated that the weak micromolar affinity of Lrp for the clp distal region likely contributes to the low level of ON cells observed. Moreover, we have also shown that PapI homologues, namely, FooI and PapI, favor the OFF state during CS31A phase variation, mainly through their capacity to increase the association and stability of Lrp binding to the proximal region of the clp operon.
During phase variation of P fimbriae, the competitive Lrp and Dam assembly on the regulatory region is important for the heritability of the OFF state. In phase OFF cells, Lrp binds the proximal region, which blocks the entry of the RNA polymerase and protects GATCprox from Dam methylation. In phase ON cells, Lrp binds to the distal region and protects GATCdist from methylation. The methylation of the GATCprox site by Dam is required for transition to the phase ON state by specifically blocking PapI-dependent binding of Lrp to promoter proximal sites (25).
The regulation of the clp operon, encoding CS31A, shares similar mechanisms with the regulation of the expression of other P-related fimbrial operons (17). Notably, CS31A production is under phase variation control in strain 31A. The clp gene cluster does not carry any papI homologue, however, even though one is required for phase variation to occur. Only AfaF, a PapI homologue controlling the genetically distant operon afa-3, has been used to observe CS31A phase variation to date. Based on these observations, it was suggested that the presence of an additional pap operon on the chromosome of CS31A-positive strains may supply the PapI required in trans, resulting in clp phase variation (15). We have now used several techniques to demonstrate that PapI homologues (PapI or FooI) can induce phase variation of the clp operon, but overall clp expression is repressed due to their capacity to enhance Lrp binding at the proximal region of the clp operon. Moreover, we show here that the affinity of Lrp for the distal region is significantly lower (103-fold) than its affinity for the proximal region—this distinction contributes greatly to the overall repression of clp. Martin showed that, in the absence of a PapI homologue supplied in trans (i.e., in the absence of phase variation), Lrp protects the GATC sites from methylation and cannot bind methylated GATC sites (17). Here, we provide further information showing that Lrp possesses a lower affinity for GATCdist binding sites than for GATCprox binding sites in the clp regulatory region.
In the Pap model, the low nanomolar Lrp affinity is only 2-fold higher for the proximal sites 1 to 3 than for the distal sites 4 to 6 (25). Moreover, the concentration of Lrp must be relatively low for the system to work properly (42). Binding of Lrp at the proximal sites 1 to 3 reduces by 10-fold the affinity of Lrp for pap at the distal sites 4 to 6. This phenomenon, apparent when Lrp binding on plasmid DNA is analyzed, has been called “mutual exclusion” (25). However, no mutual exclusion sensu stricto could be observed in CS31A, since the Lrp affinity is 103-fold higher for the proximal region than for the distal region. Moreover, our findings are consistent with previous reports (43) that have proposed two distinct interaction states for Lrp, binding to the clp proximal region with low nanomolar affinity (in a range similar to what is shown when Lrp is considered a specific regulator) versus binding to the clp distal region with low micromolar affinity (in the same range as is shown when Lrp is considered a nucleoprotein). As a result, Lrp protects GATCprox from methylation and negatively controls basal clp expression.
By analyzing the phase variation control of F1651, another member of the P family that includes Pap, we showed that a higher level of ON cells for F1651 is due to an altered Lrp binding stability at the DNA repressor sites 1 to 3 (38). Furthermore, we identified nucleotides surrounding the Lrp-binding site 1 that are critical for maintaining a high OFF-to-ON switch rate during F1651 phase variation, as well as for switching Pap fimbriae toward the OFF state by reducing Lrp dissociation from the proximal site. In the case of CS31A, the predominant OFF state is mainly due to the low Lrp affinity for the clp distal region. The presence of an additional Lrp binding site, if not directly involved in Lrp binding, probably influences the interaction of Lrp with the clp regulatory region. This suggests that the interaction of Lrp with its binding sites dictates their accessibility to methylation and, thus, influences the epigenetic process of phase variation. Given that phase variation of P fimbriae involves the action of multiple regulators, both local (e.g., PapB and PapI) and global (e.g., CpxAR and cyclic AMP-catabolite activator protein [cAMP-CAP]) regulators may contribute to phase variation differences between strains (44–47).
In E. coli reference strain 31A, CS31A is encoded by the high-molecular-weight plasmid p31A, which does not harbor a clp that is a specific homologue to papI. It was suggested that the phase variation control observed in wild-type strains could result from cross talk between clp and a chromosomal pap-related operon, as strain 31A encodes P fimbriae (15, 16). In this study, PapI and the homologue FooI restored the bistable state of phase variation when supplied in trans. Similar to pap, the affinity of Lrp for both the distal and proximal regions increases in the presence of PapI or FooI. Moreover, the significantly stronger affinity of the PapI-/FooI-Lrp complex for the proximal region means that only a minority of clp cells is in the ON phase. Furthermore, decreased dissociation of the PapI-/FooI-Lrp complex to the DNA target could also explain why the level of clp operon expression in the OFF phase is even lower than the moderate level observed in the absence of a PapI homologue (i.e., the stable PapI-Lrp complex blocks clp transcription more efficiently than Lrp alone or in the presence of AfaF [15]). Taken together, our results show that, in the case of clp, PapI homologues play a dual role, (i) promoting the ON phase when papI is not expressed (Fig. 7) and (ii) promoting the OFF phase when expressed, thus inducing phase variation in a manner different than what was observed with AfaF. The precise mechanism by which PapI homologues promote the switch from OFF to ON remains unresolved.
FIG 7.
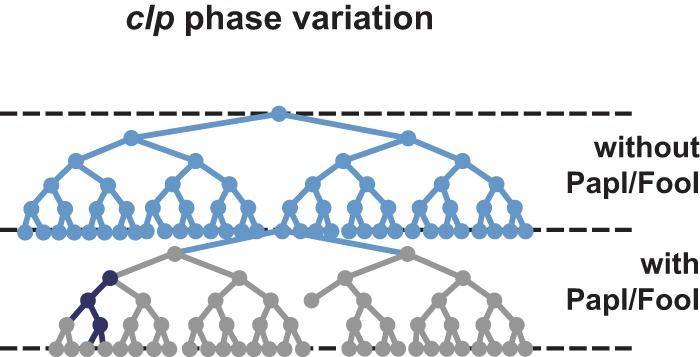
Schematic model representing clp phase variation, depending on the presence of PapI in trans. In this model, the control of clp expression by phase variation depends on the presence in trans of PapI expressed by a pap-related operon in the genome of the CS31A strain. Since the expression of pap fimbriae is also controlled by phase variation, the expression of papI reflects Pap ON bacteria. Thus, the presence/absence of PapI results in cross talk between pap and clp in pap-positive and clp-positive bacteria: for cells in the ON state for pap, PapI is produced, and then clp is mostly repressed, with a few cells that show a high level of expression for clp. Reciprocally, for cells in the OFF state for pap, PapI is not produced, and then clp is expressed at a low level.
It is also noteworthy that the levels of ON colonies can vary greatly depending on which PapI homologue is present in the cell. Indeed, previous studies using AfaF (17) exhibited higher levels of ON cells than we observed in the present study, and our head-to-head comparison between FooI and PapI also demonstrated differences in the levels of ON cells. Considering that at least 16 papI alleles have been identified so far (45) and that CS31A is often associated with the presence of other type P-related fimbriae, such as Pap or F1651 (7, 12, 48, 49), we suggest that PapI homologues can be seen as specific regulators that cocoordinately regulate the expression of more than one fimbrial operon (Fig. 7) (45). Similarly, Totsika et al., studying the cross-activation of pap variants by FooI and PapI, suggested that sequence variation among PapI homologues could affect their ability to activate pap transcription and that they may have evolved in order to prevent cross-activation of related proteins (45).
Strains producing CS31A are often associated with septicemia and have features of extraintestinal E. coli. While virulent extraintestinal pathogenic E. coli strains mainly belong to the phylogenetic groups B2 and D, reference strain CS31A belongs to the less virulent group A and is thus considered to be opportunistic, although it was isolated from a clinical case (9, 50). In certain environments, the expression of pap would reduce that of clp in CS31A. This may enhance its adaptation to fluctuating environments and contribute to its fitness.
Overall, the present study provides novel insights about why clp expression is repressed by Lrp alone and, in the presence of a PapI homolog, has a low switch rate, like pap. This is likely due to the very low affinity of Lrp to the clp distal site. Our data emphasize that the fine molecular interaction of regulatory proteins with their DNA-binding sites influences phase variation and the expression of fimbriae. Within the large repertoire of fimbrial operon variants in the P family, we show that clp, found in septicemic and enterotoxigenic E. coli strains, is an exquisite example of finely tuned regulatory expression that arms the bacterium with strategies for adapting to more than one particular environment. In conclusion, our study exemplifies how bacteria may orchestrate a finely tuned regulatory expression with diverse adhesive factors, thus arming themselves for quick adaptation to changing environments.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to David Low (University of California) for the generous gift of purified PapI and enlightening discussions. We are grateful to Cécile Crost (CRIP), Jean-Pierre Girardeau and George Szatmari for their invaluable advice during this study.
This work was supported by funding from the Centre de Recherche en Infectiologie Porcine (CRIP; Fonds Québécois de la Recherche sur la Nature et les Technologies Regroupements Stratégiques 111946), the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery grants 25120 to J.H. and 262746 to M.M.), and the Canada Research Chair program (to M.M.). SPR infrastructure at Sheldon Biotechnology Centre was provided by Canada Foundation for Innovation and supported by a Research Resource Grant from the Canadian Institutes of Health Research.
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01622-14.
REFERENCES
- 1.Dobrindt U, Hacker J. 2008. Targeting virulence traits: potential strategies to combat extraintestinal pathogenic E. coli infections. Curr. Opin. Microbiol. 11:409–413. 10.1016/j.mib.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 2.Smith JL, Fratamico PM, Gunther NW. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4:134–163. 10.1089/fpd.2007.0087 [DOI] [PubMed] [Google Scholar]
- 3.Acar M, Mettetal JT, van Oudenaarden A. 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 40:471–475. 10.1038/ng.110 [DOI] [PubMed] [Google Scholar]
- 4.Thattai M, van Oudenaarden A. 2004. Stochastic gene expression in fluctuating environments. Genetics 167:523–530. 10.1534/genetics.167.1.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Woude MW. 2006. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 254:190–197. 10.1111/j.1574-6968.2005.00038.x [DOI] [PubMed] [Google Scholar]
- 6.van der Woude MW, Bäumler AJ. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581–611. 10.1128/CMR.17.3.581-611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contrepois M, Fairbrother JM, Kaura YK, Girardeau JP. 1989. Prevalence of CS31A and F165 surface antigens in Escherichia coli isolates from animals in France, Canada and India. FEMS Microbiol. Lett. 50:319–323 [DOI] [PubMed] [Google Scholar]
- 8.Girardeau JP, Der Vartanian M, Ollier JL, Contrepois M. 1988. CS31A, a new K88-related fimbrial antigen on bovine enterotoxigenic and septicemic Escherichia coli strains. Infect. Immun. 56:2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contrepois M, Dubourguier HC, Parodi AL, Girardeau JP, Ollier JL. 1986. Septicaemic Escherichia coli and experimental infection of calves. Vet. Microbiol. 12:109–118. 10.1016/0378-1135(86)90073-8 [DOI] [PubMed] [Google Scholar]
- 10.Mohamed Ou Said A, Contrepois MG, Der Vartanian M, Girardeau JP. 1988. Virulence factors and markers in Escherichia coli from calves with bacteremia. Am. J. Vet. Res. 49:1657–1660 [PubMed] [Google Scholar]
- 11.Mercado EC, Rodríguez SM, D'Antuono AL, Cipolla AL, Elizondo AM, Rossetti CA, Malena R, Méndez MA. 2003. Occurrence and characteristics of CS31A antigen-producing Escherichia coli in calves with diarrhoea and septicaemia in Argentina. J. Vet. Med. B Infect. Dis. Vet. Public Health 50:8–13. 10.1046/j.1439-0450.2003.00610.x [DOI] [PubMed] [Google Scholar]
- 12.Bertin Y, Girardeau JP, Darfeuille-Michaud A, Martin C. 2000. Epidemiological study of pap genes among diarrheagenic or septicemic Escherichia coli strains producing CS31A and F17 adhesins and characterization of Pap31A fimbriae. J. Clin. Microbiol. 38:1502–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fecteau G, Fairbrother JM, Higgins R, Van Metre DC, Paré J, Smith BP, Holmberg CA, Jang S. 2001. Virulence factors in Escherichia coli isolated from the blood of bacteremic neonatal calves. Vet. Microbiol. 78:241–249. 10.1016/S0378-1135(00)00299-6 [DOI] [PubMed] [Google Scholar]
- 14.Jallat C, Darfeuille-Michaud A, Rich C, Joly B. 1994. Survey of clinical isolates of diarrhoeogenic Escherichia coli: diffusely adhering E. coli strains with multiple adhesive factors. Res. Microbiol. 145:621–632. 10.1016/0923-2508(94)90079-5 [DOI] [PubMed] [Google Scholar]
- 15.Crost C, Garrivier A, Harel J, Martin C. 2003. Leucine-responsive regulatory protein-mediated repression of clp (encoding CS31A) expression by L-leucine and L-alanine in Escherichia coli. J. Bacteriol. 185:1886–1894. 10.1128/JB.185.6.1886-1894.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crost C, Harel J, Berthiaume F, Garrivier A, Tessier MC, Rakotoarivonina H, Martin C. 2004. Influence of environmental cues on transcriptional regulation of foo and clp coding for F165(1) and CS31A adhesins in Escherichia coli. Res. Microbiol. 155:475–482. 10.1016/j.resmic.2004.02.005 [DOI] [PubMed] [Google Scholar]
- 17.Martin C. 1996. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and L-alanine at the transcriptional level. Mol. Microbiol. 21:281–292. 10.1046/j.1365-2958.1996.00651.x [DOI] [PubMed] [Google Scholar]
- 18.Hung SP, Baldi P, Hatfield GW. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309–40323. 10.1074/jbc.M204044200 [DOI] [PubMed] [Google Scholar]
- 19.Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:13471–13476. 10.1073/pnas.212510999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho B-K, Barrett CL, Knight EM, Park YS, Palsson BØ. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:19462–19467. 10.1073/pnas.0807227105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braaten BA, Nou X, Kaltenbach LS, Low DA. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577–588. 10.1016/0092-8674(94)90120-1 [DOI] [PubMed] [Google Scholar]
- 22.Nou X, Braaten B, Kaltenbach L, Low DA. 1995. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14:5785–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blomfield IC. 2001. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 45:1–49. 10.1016/S0065-2911(01)45001-6 [DOI] [PubMed] [Google Scholar]
- 24.Casadesús J, Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70:830–856. 10.1128/MMBR.00016-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernday A, Krabbe M, Braaten B, Low D. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. U. S. A. 99(Suppl 4):S16470–S16476. 10.1073/pnas.182427199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Woude M, Braaten B, Low D. 1996. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4:5–9. 10.1016/0966-842X(96)81498-3 [DOI] [PubMed] [Google Scholar]
- 27.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 28.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. Chapter 1:Unit 1.16. 10.1002/0471142727.mb0116s78 [DOI] [PubMed] [Google Scholar]
- 29.Matthews RG, Cui Y, Friedberg D, Calvo JM. 2000. Wild-type and hexahistidine-tagged derivatives of leucine-responsive regulatory protein from Escherichia coli. Methods Enzymol. 324:322–329. 10.1016/S0076-6879(00)24241-9 [DOI] [PubMed] [Google Scholar]
- 30.Myszka DG. 1999. Improving biosensor analysis. J. Mol. Recognit. 12:279–284. [DOI] [PubMed] [Google Scholar]
- 31.Kaltenbach LS, Braaten BA, Low DA. 1995. Specific binding of PapI to Lrp-pap DNA complexes. J. Bacteriol. 177:6449–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxam AM, Gilbert W. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499–560. 10.1016/S0076-6879(80)65059-9 [DOI] [PubMed] [Google Scholar]
- 33.Blyn LB, Braaten BA, White-Ziegler CA, Rolfson DH, Low DA. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Iannolo M, Calvo JM. 2005. Cooperative binding of the leucine-responsive regulatory protein (Lrp) to DNA. J. Mol. Biol. 345:251–264. 10.1016/j.jmb.2004.10.047 [DOI] [PubMed] [Google Scholar]
- 35.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. 2007. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 18:3225–3236. 10.1091/mbc.E07-05-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melman L, Cao ZF, Rennke S, Marzolo MP, Wardell MR, Bu G. 2001. High affinity binding of receptor-associated protein to heparin and low density lipoprotein receptor-related protein requires similar basic amino acid sequence motifs. J. Biol. Chem. 276:29338–29346. 10.1074/jbc.M103717200 [DOI] [PubMed] [Google Scholar]
- 37.Marciniak BC, Pabijaniak M, de Jong A, Duhring R, Seidel G, Hillen W, Kuipers OP. 2012. High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genomics 13:401. 10.1186/1471-2164-13-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graveline R, Mourez M, Hancock MA, Martin C, Boisclair S, Harel J. 2011. Lrp-DNA complex stability determines the level of ON cells in type P fimbriae phase variation. Mol. Microbiol. 81:1286–1299. 10.1111/j.1365-2958.2011.07761.x [DOI] [PubMed] [Google Scholar]
- 39.Hochschild A, Ptashne M. 1986. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell 44:681–687. 10.1016/0092-8674(86)90833-0 [DOI] [PubMed] [Google Scholar]
- 40.Berthiaume F, Crost C, Labrie V, Martin C, Newman EB, Harel J. 2004. Influence of L-leucine and L-alanine on Lrp regulation of foo, coding for F1651, a Pap homologue. J. Bacteriol. 186:8537–8541. 10.1128/JB.186.24.8537-8541.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamura T, Le LUK, Zhou H, Dahlquist FW. 2007. Solution structure of Escherichia coli PapI, a key regulator of the pap pili phase variation. J. Mol. Biol. 365:1130–1142. 10.1016/j.jmb.2006.10.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson SN, Reich NO. 2008. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J. Mol. Biol. 383:92–105. 10.1016/j.jmb.2008.07.086 [DOI] [PubMed] [Google Scholar]
- 43.Peterson SN, Dahlquist FW, Reich NO. 2007. The role of high affinity non-specific DNA binding by Lrp in transcriptional regulation and DNA organization. J. Mol. Biol. 369:1307–1317. 10.1016/j.jmb.2007.04.023 [DOI] [PubMed] [Google Scholar]
- 44.Hernday AD, Braaten BA, Broitman-Maduro G, Engelberts P, Low DA. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16:537–547. 10.1016/S1097-2765(04)00646-X [DOI] [PubMed] [Google Scholar]
- 45.Totsika M, Beatson SA, Holden N, Gally DL. 2008. Regulatory interplay between pap operons in uropathogenic Escherichia coli. Mol. Microbiol. 67:996–1011. 10.1111/j.1365-2958.2007.06098.x [DOI] [PubMed] [Google Scholar]
- 46.van der Woude MW, Braaten BA, Low DA. 1992. Evidence for global regulatory control of pilus expression in Escherichia coli by Lrp and DNA methylation: model building based on analysis of pap. Mol. Microbiol. 6:2429–2435 [DOI] [PubMed] [Google Scholar]
- 47.Weyand NJ, Braaten BA, van der Woude M, Tucker J, Low DA. 2001. The essential role of the promoter-proximal subunit of CAP in pap phase variation: Lrp- and helical phase-dependent activation of papBA transcription by CAP from −215. Mol. Microbiol. 39:1504–1522. 10.1046/j.1365-2958.2001.02338.x [DOI] [PubMed] [Google Scholar]
- 48.Bertin Y, Martin C, Girardeau JP, Pohl P, Contrepois M. 1998. Association of genes encoding P fimbriae, CS31A antigen and EAST 1 toxin among CNF1-producing Escherichia coli strains from cattle with septicemia and diarrhea. FEMS Microbiol. Lett. 162:235–239. 10.1111/j.1574-6968.1998.tb13004.x [DOI] [PubMed] [Google Scholar]
- 49.Cherifi A, Contrepois M, Picard B, Goullet P, de Rycke J, Fairbrother J, Barnouin J. 1990. Factors and markers of virulence in Escherichia coli from human septicemia. FEMS Microbiol. Lett. 58:279–283 [DOI] [PubMed] [Google Scholar]
- 50.Bruant G, Maynard C, Bekal S, Gaucher I, Masson L, Brousseau R, Harel J. 2006. Development and validation of an oligonucleotide microarray for detection of multiple virulence and antimicrobial resistance genes in Escherichia coli. Appl. Environ. Microbiol. 72:3780–3784. 10.1128/AEM.72.5.3780-3784.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





