Abstract
Agrobacterium tumefaciens can adhere to plant tissues and abiotic surfaces and forms biofilms. Cell surface appendages called pili play an important role in adhesion and biofilm formation in diverse bacterial systems. The A. tumefaciens C58 genome sequence revealed the presence of the ctpABCDEFGHI genes (cluster of type IV pili; Atu0216 to Atu0224), homologous to tad-type pilus systems from several bacteria, including Aggregatibacter actinomycetemcomitans and Caulobacter crescentus. These systems fall into the type IVb pilus group, which can function in bacterial adhesion. Transmission electron microscopy of A. tumefaciens revealed the presence of filaments, significantly thinner than flagella and often bundled, associated with cell surfaces and shed into the external milieu. In-frame deletion mutations of all of the ctp genes, with the exception of ctpF, resulted in nonpiliated derivatives. Mutations in ctpA (a pilin homologue), ctpB, and ctpG decreased early attachment and biofilm formation. The adherence of the ctpA mutant could be restored by ectopic expression of the paralogous pilA gene. The ΔctpA ΔpilA double pilin mutant displayed a diminished biovolume and lower biofilm height than the wild type under flowing conditions. Surprisingly, however, the ctpCD, ctpE, ctpF, ctpH, and ctpI mutants formed normal biofilms and showed enhanced reversible attachment. In-frame deletion of the ctpA pilin gene in the ctpCD, ctpE, ctpF, ctpH, and ctpI mutants caused the same attachment-deficient phenotype as the ctpA single mutant. Collectively, these findings indicate that the ctp locus is involved in pilus assembly and that nonpiliated mutants, which retain the CtpA pilin, are proficient in attachment and adherence.
INTRODUCTION
Agrobacterium tumefaciens is a plant pathogen in the family Rhizobiaceae related to nitrogen-fixing symbionts of leguminous plants. Unlike the rhizobia, A. tumefaciens causes crown gall disease, which is characterized by the formation of neoplastic tumors at sites of bacterial infection, in a wide variety of plants (1). It is well known that A. tumefaciens can genetically transform plants by transferring a segment of its own DNA (T-DNA) into the genomes of plants (1). A. tumefaciens is widely used to engineer transgenic plants in agricultural biotechnology (2).
Cell attachment to tissues and biofilm formation by A. tumefaciens contribute to plant infection and environmental persistence of the pathogen. A. tumefaciens is able to form complex, structured biofilms on both plant roots and abiotic surfaces (3–5). The mechanisms of attachment and biofilm formation in A. tumefaciens are an area of active study. Initially, flagellar motility enables A. tumefaciens to contact target surfaces, including chemotaxis toward attractants produced by wounded plant tissues (6, 7). Cells transiently attach to the surface, and then a more stable association is formed. Early stages of A. tumefaciens biofilms on abiotic surfaces are visible in laboratory cultures in defined growth medium with a glucose or mannitol carbon source after a day of incubation (4). These biofilms are relatively shallow layers of cells, of which a large proportion are attached via a single pole (3, 4). Over several days of incubation, the biofilm structure matures with the formation of multicellular microcolonies (4, 8).
Several adherence factors have been reported to function in attachment and biofilm formation (7, 9–11). Flagellar motility is required for efficient biofilm formation in static culture (7). A. tumefaciens C58 produces multiple exopolysaccharides (EPSs), including the unipolar polysaccharide (UPP), cellulose, and β-1,2-glucans, which can contribute to biofilm formation (9, 10, 12, 13). A calcium-dependent outer membrane protein called rhicadhesin is reported to promote the attachment of bacteria to plant cell walls, but the gene that encodes this activity has never been identified (11).
Pili are filamentous, proteinaceous, extracellular appendages on bacterial surfaces (also known as fimbriae). These structures can function in horizontal gene transfer, motility, and, in many cases, adhesion (14, 15). Several types of pili exist in A. tumefaciens C58. Three forms of conjugal pili are independently involved in conjugal transfer activities of the two endogenous plasmids. The avhB genes located on the pAtC58 plasmid (16) and the trb genes on the Ti plasmid (17) contribute to interbacterial plasmid conjugal transfer. Also encoded by several of the virulence genes in the virB operon located on the Ti plasmid are the T pili, which are involved in the association with plants that leads to T-DNA transfer and genetic transformation. Ti plasmid conjugal transfer and virulence gene expression are tightly regulated on the Ti plasmid (18, 19), and these pili are not produced in standard laboratory culture. In contrast to the Ti plasmid, the pAtC58 plasmid is active for conjugal transfer in culture, and the presumptive pAtC58 conjugal pili could be produced in culture but have never been defined (16).
In addition to these plasmid-encoded conjugal pili in A. tumefaciens, there have also been electron microscopic studies that revealed the presence of what were described as common pili (20). The complete genome sequence of A. tumefaciens C58 (21, 22) revealed a chromosomally located nine-gene cluster on the circular chromosome that was annotated as the ctp (cluster of type IV pili) locus (ctpABCDEFGHI), homologous to genes that encode a subset of type IV pili best characterized as the Cpa pilus cluster of Caulobacter crescentus and the tad (tight adherence) locus in Aggregatibacter actinomycetemcomitans (23, 24). tad-type loci exist in widespread bacteria, including Yersinia pestis, Vibrio cholerae, Mycobacterium tuberculosis, Pseudomonas aeruginosa, and C. crescentus. In the systems characterized, the tad genes are responsible for the biogenesis of Flp (fimbrial low-molecular-weight protein) pili, also known as type IVb subclass pili. Flp-type pili form bundles in A. actinomycetemcomitans (25) but exist as individual filaments in C. crescentus (23). Flp-type pili are essential for biofilm formation and pathogenesis in several different bacteria (23, 26, 27).
In this study, we report that the ctp locus encodes the A. tumefaciens common pili and that all of the ctp locus genes, with the exception of ctpF, are essential for pilus formation. A second unlinked Flp-type pilin gene (pilA) can function interchangeably with ctpA. Our findings indicate that the Ctp pili contribute to the reversible step of surface interactions and also during maturation of the biofilm. A subset of Ctp pilus mutants, however, are activated to bypass the loss of pili and stimulate reversible surface interactions that require the CtpA pilin but not the Ctp pili.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
For the strains and plasmids used in this study, see Table S1 in the supplemental material. The oligonucleotide primers used in this study were obtained from Integrated DNA Technologies (Coralville, IA) (see Table S2 in the supplemental material). All DNA was manipulated by using standard protocols (28). All of the restriction enzymes and molecular biology reagents used were purchased from NEB (Ipswich, MA). DNA sequencing was performed on an ABI 3730 at the Indiana Molecular Biology Institute, Bloomington, IN. DNA was purified with QIAquick Spin kits (Qiagen, Germantown, MD) or E.Z.N.A. Plasmid Miniprep kits (Omega Bio-tek, Norcross, GA) by following the manufacturer's protocols. Plasmid introduction into A. tumefaciens was performed via electroporation (29). Bacteria were usually grown on LB medium for Escherichia coli or AT minimal medium (30) supplemented with 0.5% (wt/vol) glucose and 15 mM ammonium sulfate (ATGN) medium for A. tumefaciens. Sucrose (5%) was used to replace glucose as the sole carbon source (ATSN) during sacB counterselection. The following antibiotic concentrations were used: E. coli, 100 μg ml−1 ampicillin and 25 μg ml−1 kanamycin (Km); A. tumefaciens, 300 μg ml−1 Km. For Plac induction, media were supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG). Unless otherwise noted, reagents, antibiotics, and microbiological media were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO).
Generation of in-frame markerless deletions of ctp locus genes, pilA, and uppC.
DNA fragments of approximately 500 bp flanking the targeted gene were generated by PCR. These fragments were located upstream (amplified by primers 3 and 4 [see Table S2 in the supplemental material]) and downstream (amplified by primers 1 and 2 [see Table S2]) of the gene to be deleted and were designed to precisely remove the coding sequence without affecting adjacent genes and to maintain translational coupling for overlapping genes. Primers 2 and 3 were designed with complementary sequences at the 5′ ends to allow splicing by overlapping extension as described previously (7, 31). Briefly, the flanking amplicons were generated with Phusion high-fidelity DNA polymerase (NEB, Ipswich, MA) and agarose gel purified. The purified fragments were used as primers and templates for five cycles of PCR. A final PCR was performed with 1 μl of the flanking amplicons as the template and primers 1 and 4. The resulting amplicon was cloned into pGEM-T Easy and confirmed by DNA sequencing. The fragment was excised with the appropriate restriction enzymes and ligated into the sacB counterselectable suicide vector pNPTS138 which had been cleaved with the same enzymes. The vector pNPTS138 (M. R. K. Alley, unpublished data) replicates with a colE1 origin that is incapable of replicating in A. tumefaciens and contains a Km resistance (Kmr) marker and the sacB gene, which confers sucrose sensitivity (Sucs). Derivatives of pNPTS138 were introduced into A. tumefaciens by plate mating. The only way for pNPTS138 to confer Kmr on A. tumefaciens is recombination into a stable endogenous replicon. Integrants were selected for by growth on ATGN plates supplemented with Km, and plasmid integration was confirmed by PCR, and sucrose sensitivity was confirmed by patching on ATSN. Excision of the integrated plasmid from the Kmr Sucs clones was facilitated by culturing the recombinants in ATGN without Km overnight twice and then plating them on ATSN. The resulting Sucr colonies were replica plated on ATGN supplemented with Km to confirm the loss of the plasmid marker. Deletion of the appropriate region of DNA was confirmed by PCR and DNA sequencing.
Creation of complementation constructs.
Complementation constructs were prepared by PCR and subsequent cloning of the intact wild-type coding sequences into pSRK-Km or pSRK-Gm (32). The 5′ primers were designed to be cloned into the NdeI site of these vectors (pSRK-Km and pSRK-Gm), directly in frame with the lacZα start codon (see Table S2 in the supplemental material). Coding sequences for ctpA, ctpB, ctpE, and pilA were PCR amplified from A. tumefaciens C58 genomic DNA with Phusion high-fidelity DNA polymerase and the corresponding primers (primers 7 and 8 for ctpA, primers 5 and 6 for pilA, ctpB, ctpE, and ctpG). PCR products were gel purified and ligated into pGEM-T Easy. Inserts were confirmed by DNA sequencing. The coding sequences of ctpA, ctpB, ctpE, ctpG, and pilA were excised by NdeI and HindIII cleavage. All of the excised fragments were ligated with T4 DNA ligase into pSRK-Km(Gm) that had been cleaved with corresponding restriction enzymes. Proper fragment insertion was confirmed by PCR, and constructs were electroporated into A. tumefaciens competent cells.
Construction of lacZ fusions of upstream and intergenic regions of ctp genes and the pilA gene.
The upstream and intergenic regions of the ctpA, ctpB, ctpC, and pilA genes were PCR amplified from A. tumefaciens C58 genomic DNA with Phusion high-fidelity DNA polymerase and the corresponding primers (P1 and P2 for ctpA, pilA, ctpB, ctpC, and ctpE [see Table S2 in the supplemental material]). PCR products were gel purified and ligated into pGEM-T Easy. Inserts were confirmed by DNA sequencing. These DNA fragments were excised and ligated with T4 DNA ligase into pRA301. Proper fragment insertion was confirmed by PCR, and plasmids were electroporated into A. tumefaciens competent cells.
β-Galactosidase assay.
Cultures were prepared for β-galactosidase assay by culturing in ATGN medium. The optical densities at 600 nm (OD600s) of exponential-phase cultures were measured, and they were frozen at −80°C. β-Galactosidase activity was measured as described previously (33). Briefly, cells were permeabilized in Z-buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol brought to pH 7.0) by the addition of 2 drops of 0.05% SDS and 4 drops of chloroform. β-Galactosidase reactions were initiated by the addition of 100 μl of a 4-mg/ml solution of the colorimetric substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) and terminated by the addition of 600 μl of 1 M NaCO2. Intact cells and debris were removed by centrifugation, free ONPG was determined by measuring the A420, and specific activity was reported in Miller units. At least three biological replicates were performed for each strain at each time point.
Transmission electron microscopy (TEM).
Appropriate A. tumefaciens C58 derivatives were grown in ATGN minimal medium at 28°C to an OD600 of ∼1.0 and deposited on carbon-Formvar films on 300-mesh, 3-mm copper grids. A 10-μl volume of a bacterial culture was placed on each grid for 1 min, and then the grids were stabilized with glutaraldehyde for 1 min and stained with 2% uranyl acetate for 1 min. The samples were examined with a JEOL JEM-1010 electron microscope at 80 kV in the Indiana University Electron Microscopy Center.
Static-culture biofilm assay.
Static-culture biofilms were grown on sterile PVC coverslips suspended vertically in the wells of UV-sterilized 12-well polystyrene dishes. Cell cultures were adjusted to an OD600 of 0.05, inoculated into 3 ml of ATGN broth, and then incubated at room temperature for 24 to 72 h. Adherence to PVC coverslips was visualized by staining of rinsed coverslips with 1% crystal violet (CV) for 10 min at room temperature. Quantification of adherent biomass was achieved via solubilization of adsorbed CV in 1 ml of 33% acetic acid. Solubilized CV was measured by determining the A600 to estimate relative amounts of adhered biomass and normalized to the planktonic culture density (OD600).
Short-term binding assay with UPP staining.
Short-term binding assays were conducted by growing cell cultures to an OD600 of ∼0.6. Glass coverslips were placed at the bottom of 2 ml cell culture volumes in 3-ml petri plates for 1 h. Coverslips were removed from the plates, rinsed with water three times for 10 min per rinse. Fluorescent Alexa fluor 594-conjugated wheat germ agglutinin (af-WGA; Invitrogen) at 0.01 mg/ml was then added as a 100-μl pool to one side of the coverslip lying flat, incubated for 30 min, washed thoroughly with water, and mounted on slides for microscopy. Bright-field and epifluorescence microscopy was performed on a Nikon E800 at ×100 magnification. Epifluorescence microscopy used a G-2B filter set (excitation filter, 510 to 560 nm; dichromatic mirror, 575 nm; emission filter, 610 nm). Each strain was tested in triplicate, and fields were analyzed randomly. Cell numbers and lectin labeling were counted manually or automatically with the ImageJ program (NIH).
Plant root binding assay.
An attachment assay with Arabidopsis thaliana ecotype WS used 7- to 10-day-old seedlings grown from seeds surface sterilized with ethanol and planted on half-strength Murashige-Skoog agar medium (34, 35). Roots were cut into 1-cm segments and rinsed in sterile dishes containing 2 ml of 1 mM CaCl2 and 0.4% sucrose and then inoculated with cell suspensions of A. tumefaciens (OD600 of ∼0.01). The appropriate derivative strains of A. tumefaciens C58 used in this assay carried a green fluorescent protein reporter construct under constitutive expression (Ptac-gfpmut3, pJZ383). Four root segments were inoculated per strain. After 48 h of incubation in the dark at room temperature, the root segments were recovered, rinsed and resuspended in fresh calcium chloride-sucrose solution, and sealed under coverslips for hydration. Microscopy was performed with a Nikon A1 confocal microscope at ×60 magnification with a 488-nm laser for excitation and a 500- to 550-nm filter for emission. Images were acquired and analyzed with the Elements software package.
Potato disk tumor assay.
The virulence potential of each strain was evaluated by monitoring the ability to initiate tumor formation on red potato slices (34, 35). One-centimeter-diameter cores were obtained from organic red potatoes that had been washed in distilled water, incubated for 20 min in 10% bleach, and UV irradiated for 20 min. Disks were sliced (0.5 cm) from the center portion of individual cores and placed onto 1.5% agar plates with no additional nutritional supplementation. Overnight cultures of the strains indicated were diluted to an OD600 of 0.6 in ATGN and serially diluted 10- and 100-fold. A 50-μl volume of each dilution were inoculated onto the potato surface and allowed to be absorbed. Plates were sealed with Parafilm and incubated undisturbed for 4 weeks at room temperature. Tumors were enumerated on days 14, 21, and 28. Each strain was tested in three independent experiments containing five technical replicates per experiment per inoculum.
Flow cell biofilm assay.
Flow cell biofilms were cultured as described previously (4). Once-through flow cells were purchased through the Technical University of Denmark and inoculated with diluted overnight cultures (100 μl per flow chamber, final OD600 of 0.05). After inoculation, cells were permitted to attach for 60 min before the flow of ATGN medium with appropriate antibiotics commenced at ∼3 ml/h. Each strain harbored the pJZ383 (Ptac-gfpmut3) plasmid and was inoculated into triplicate chambers. Growing biofilms were examined by confocal microscopy at 24-h intervals for 1 to 5 days postinoculation. Microscopy was performed with a Nikon A1 scanning laser confocal microscope at ×60 magnification with a 488-nm laser for excitation and a 500- to 550-nm filter for emission. Images were acquired with the Elements software package and analyzed with an automated version of the biofilm analysis software COMSTAT (36) called autoCOMSTAT running in MATLAB 2013 (7).
RESULTS
The ctp genes are similar to the tad type IVb pilus assembly locus.
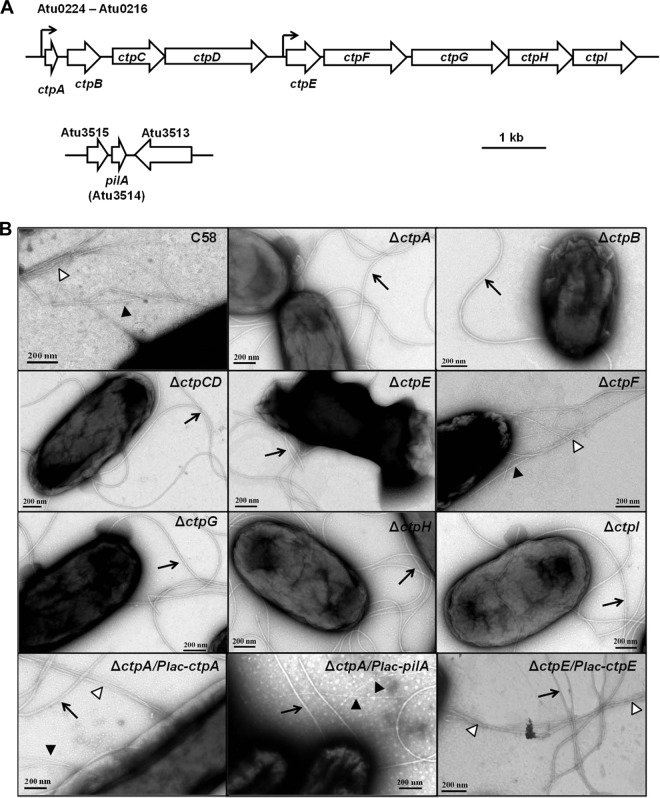
Common pili are readily identified by TEM analysis of A. tumefaciens C58 (Fig. 1), but their genetic basis had not been established. The A. tumefaciens ctpABCDEFGHI (cluster of type IV pili, Atu0216 to Atu0224) locus on the C58 circular chromosome was highly homologous to the tad-type locus from A. actinomycetemcomitans (Table 1) (24, 37). The tad locus has 14 genes, including 2 pilin genes, 2 pseudopilin genes, 1 peptidase gene, and 9 pilus biogenesis and basal body genes. The ctp locus in A. tumefaciens C58 contains one gene that encodes a 63-amino-acid (aa) protein homologous to pilin (ctpA), one presumptive pilin peptidase gene (ctpB), and seven additional genes (ctpCDEFGHI) predicted to make up the pilus assembly and basal body complex (Table 1) (Fig. 2A). The A. tumefaciens ctp gene cluster is also highly homologous to the cpa locus in C. crescentus, which encodes the Cpa pili (Table 1) (24). Orthologous genes in the ctp, tad, and cpa clusters were syntenous, with pairwise amino acid sequence identities of 20 to 60% (Table 1). In addition, there is another gene that encodes a 71-aa protein, is similar to ctpA, is designated pilA (Atu3514, Fig. 2A), and is located on the linear chromosome but is not associated with other tad-type genes. The upstream and intergenic regions of several ctp genes and pilA were used to create plasmid-borne lacZ translational fusions, and putative promoter activities were identified upstream of the ctpA and ctpE genes (Table 1) (Fig. 2A).
FIG 1.
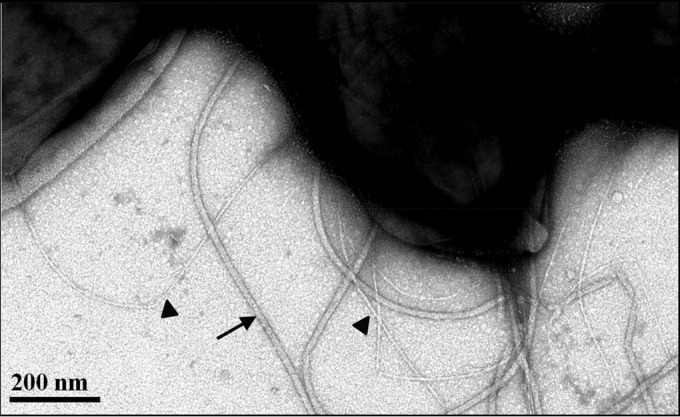
TEM of A. tumefaciens C58. Cells were negatively stained with 2% uranyl acetate and visualized by TEM. Arrow, flagellum; filled triangles, common (Ctp) pili.
TABLE 1.
Percent sequence identities between the products of the ctp gene of A. tumefaciens, the cpa gene of C. crescentus, and the tad gene of A. actinomycetemcomitans and promoter activities of intergenic regions in the ctp cluster
| A. tumefaciens ctp product | C. crescentus cpa product (% identity)a | A. actinomycetemcomitans tad product (% identity)a | Promoter activityb |
|---|---|---|---|
| CtpA | PilA (55.9) | Flp-1 (24.2), Flp-2 (23.4) | 2,099.8 ± 224.5 |
| PilA | PilA (30.4) | Flp-1 (23.9), Flp-2 (28.8) | <1.0 |
| CtpB | CpaA (34.1) | TadV (22.0) | 8.3 ± 4.2 |
| CtpC | CpaB (32.0) | RcpC (17.6) | 10.2 ± 1.1 |
| CtpD | CpaC (30.9) | RcpA (23.5) | NA |
| CtpE | CpaD (27.1) | RcpB (11.2) | 77.4 ± 7.5 |
| CtpF | CpaE (43.9) | TadZ (15.3) | NA |
| CtpG | CpaF (67.3) | TadA (45.7) | NA |
| CtpH | TadB (21.7) | NA | |
| CtpI | TadC (16.7) | NA |
Protein sequence alignment was performed by the ClustalW method with the Lasergene software.
Values are in Miller units. NA, not assayed.
FIG 2.
ctp genes encode the production of common pili. (A) Gene maps of the ctp cluster and the unlinked pilA gene (Atu3514). The ctp cluster contained nine genes (Atu0216 to Atu0224). Arrows indicate promoters in the ctp cluster. (B) TEM of A. tumefaciens C58, ctp mutants, and complemented derivatives. Arrows, flagella; filled triangles, Ctp pili; open triangles, Ctp pilus bundles. Cells were negatively stained with uranyl acetate and visualized by TEM.
The Flp pilin components are a unique subclass within the type IVb pilin family and have several novel features (38). First, the Flp prepilin proteins usually vary from 50 to 80 aa. Second, the signal peptides in Flp prepilin have variable lengths of 10 to 26 aa. Third, a highly conserved “Flp motif” (G/XXXXEY) is situated in the hydrophobic N-terminal domain and is the site at which the prepilin peptidase cleavage generates the mature pilin. Finally, no cysteine residues exist in the Flp prepilin protein. Both CtpA and PilA from A. tumefaciens exhibit these Flp-pilin features, and thus, we hypothesized that they could each function to form Flp pili.
The ctp genes encode the common pili.
Two types of filaments on the cell surface were observed in the wild-type C58 strain under TEM (Fig. 1). The thicker (18- to 20-nm) and more curved filaments were flagella (Fig. 2B, arrows). The other filaments observed were much thinner (∼3 nm thick) and straighter (Fig. 2B, filled triangles), sometimes in large bundles (Fig. 2B, open triangles; see C58 panel for an example). These are similar in size and character to the common pili reported previously (20). In-frame deletions of each ctp gene were constructed in A. tumefaciens C58. The common pili were absent from a TEM analysis of all of the ctp mutants, except the ΔctpF mutant (Fig. 2B). In addition, IPTG-controlled expression of plasmid-borne ctpA and ctpE could complement the nonpiliated phenotypes of the ΔctpA and ΔctpE mutants, respectively (Fig. 2B). These results indicate that the ctp locus is required for elaboration of the common pili, and they are likely to be composed of the CtpA pilin.
A subset of ctp genes is required for biofilm formation.
The in-frame deletion mutants were examined for biofilm formation on coverslips under static conditions (Fig. 3). According to their biofilm phenotypes, ctp mutants were divided into two categories. The ΔctpA, ΔctpB, and ΔctpG mutants were all significantly deficient in biofilm formation. Both the ΔctpA and ΔctpB mutants showed depressed biofilm formation during the entire experiment, whereas the ΔctpG mutant was similar to the wild type in the early stages (12 h) but manifested significantly decreased biofilm formation from 24 to 72 h (see Fig. S1 in the supplemental material). These biofilm-deficient mutants were all designated class I mutants. However, other ctp gene cluster mutants, including the ΔctpCD, ΔctpE, ΔctpF, ΔctpH, and ΔctpI mutants, were equivalent to the wild type in biofilm formation (Fig. 3). This was surprising, as our TEM results indicated that the ΔctpCD, ΔctpE, ΔctpH, and ΔctpI mutants do not produce pili under standard growth conditions (Fig. 2B; the ΔctpF mutation affected neither pilus formation nor biofilm formation). The ctp genes that did not impact biofilm formation were designated class II genes. To evaluate the relationship between class I and II ctp genes, the entire ctp locus (Δctp, Atu0216 to Atu0224) was deleted. The Δctp mutant phenotype was indistinguishable from that of the class I mutants (Fig. 3).
FIG 3.
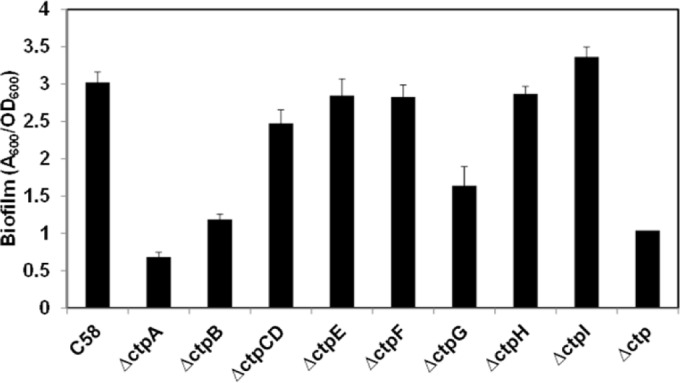
Biofilm formation by A. tumefaciens ctp deletion mutants. Static biofilms of A. tumefaciens C58 and all ctp mutants were incubated for 72 h on coverslips. All bacterial strains grown in shaking liquid culture overnight were diluted to an OD600 of 0.05 and incubated in 12-well plates with PVC coverslips for 72 h at 28°C. After rinsing, coverslips were stained with 1% CV and the adherent biomass was quantified by measuring the absorbance of acetic acid-solubilized CV, normalized for planktonic culture growth (A600/OD600). Error bars show standard deviations of results of assays performed in triplicate.
The biofilm deficiencies of class I mutants were complemented by plasmid-borne expression of the corresponding genes under Plac promoter control induced with IPTG (Fig. 4A), confirming the roles of these individual genes. The ctpA gene encodes the prepilin subunit protein, ctpB encodes a peptidase that presumptively processes the prepilin, and ctpG encodes an ATPase, predicted to be cytoplasmic and to provide the energy for pilus assembly (38, 39). Moreover, ectopic expression of ctpA was unable to suppress the biofilm and short-term attachment deficiency of the ΔctpB mutant (see Fig. S2B in the supplemental material), suggesting that CtpA prepilin was not functional until processed by CtpB.
FIG 4.
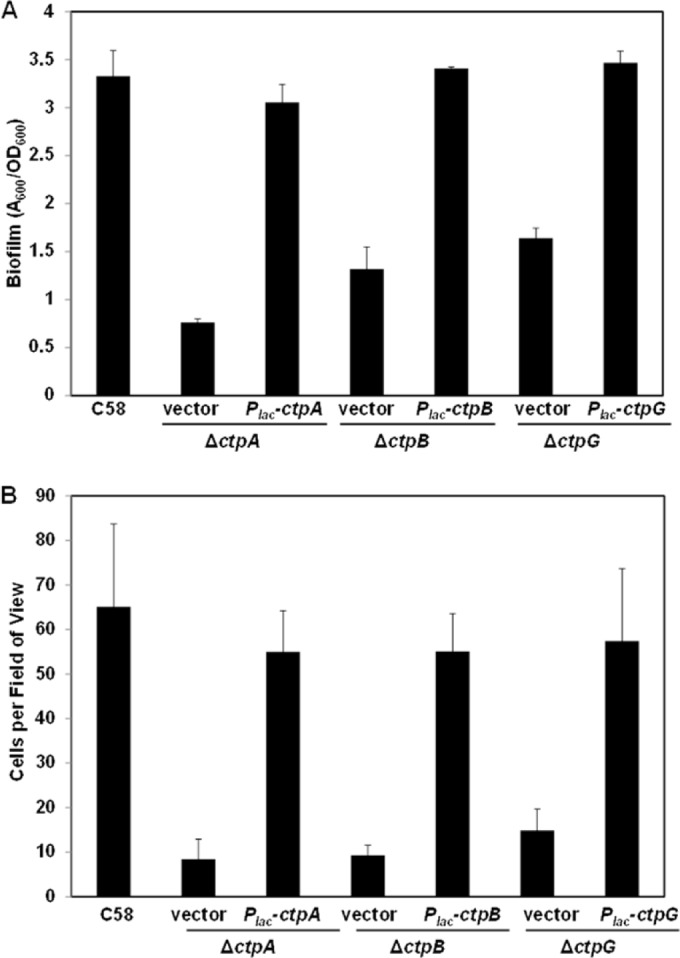
Complementation of class I Ctp mutants. A. tumefaciens mutants were complemented by plasmid-borne, ectopically expressed constructs (Plac-ctpA, Plac-ctpB, and Plac-ctpG). (A) Seventy-two-hour static biofilm assays on coverslips prepared as described in the legend to Fig. 3 and grown with 500 μM IPTG induction. (B) Short-term irreversible binding assays on coverslips with 500 μM IPTG induction. Coverslips were incubated with cell culture for 1 h and then subjected to strong washing. Total numbers of attached cells per field of view (∼3.13 × 104 μm2) were determined by counting with a Nikon E800 in bright-field mode at ×100 magnification. The values are mean results of 10 fields of view, and the error bars show the standard deviations.
A second pilin homologue can function in the place of ctpA.
In addition to the ctp cluster located on the circular chromosome, another Flp-type pilin is located on the linear chromosome and annotated as pilA (Atu3514, Fig. 2A). There are no additional tad-like genes adjacent to pilA. The N-terminal hydrophobic segments of PilA and CtpA are highly similar, and PilA has the well-conserved Flp motif G/ATAIEY (24). These features suggested that CtpA and PilA might have similar functions. In-frame deletion of pilA did not produce any decrease in biofilm formation, and overexpression of pilA in the wild type did not increase biofilm formation (see Fig. S2 in the supplemental material). The ΔctpA ΔpilA double pilin deletion mutant exhibited a biofilm deficiency similar to that of the ΔctpA single mutant. Strikingly, however, overexpression of a plasmid-borne pilA gene from Plac in the ΔctpA mutant could effectively complement the deficiency in biofilm formation and pilus production in the ΔctpA mutant (Fig. 2B; see Fig. S2A in the supplemental material). As with ctpA, ectopic expression of pilA had no effect in a ctpB mutant, suggesting that CtpB is also required to process PilA (see Fig. S2B). Moreover, the promoter activity of the pilA gene upstream region was negligible under standard culture conditions (Table 1), suggesting that the pilA gene is not expressed under minimal-medium standard culture conditions.
Class I ctp genes function in cell-to-surface attachment and also promote biofilm maturation.
We examined the early stages of surface association of the class I ctp mutants. A short-term binding assay was performed with the class I mutants and their complemented derivatives to evaluate the proficiency of their attachment to abiotic surfaces. The three class I mutants displayed pronounced short-term binding deficiencies, and the plasmid-borne copies of the corresponding genes complemented these defects (Fig. 4B).
Furthermore, careful comparison of biofilm formation by the wild type and mutants over time revealed a role for the class I ctp gene products in biofilm maturation. Four linear regression models were fitted to biofilm formation (A600) relative to days of incubation, with P values of <0.05 (see Fig. S3 in the supplemental material). This analysis reflected a linear correlation between incubation time and biofilm formation. With linear regression models, the amount of biofilm formed at 12 h (A600) for the wild-type strain was 0.49, greater than that of the ctpA mutant (0.23), the ctpB mutant (0.18), and the ctpG mutant (0.36), consistent with the surface attachment deficiency of these mutants. In addition, the slopes of linear biofilm accumulation (A600/h), which are indicative of the rate of biomass accumulation in the biofilm, were also significantly greater for the wild-type strain, at 0.77 (A600/h), than for the class I mutants (ΔctpA, 0.11; ΔctpB, 0.28; ΔctpG, 0.35).
Deficiencies in biofilm formation under flowing conditions.
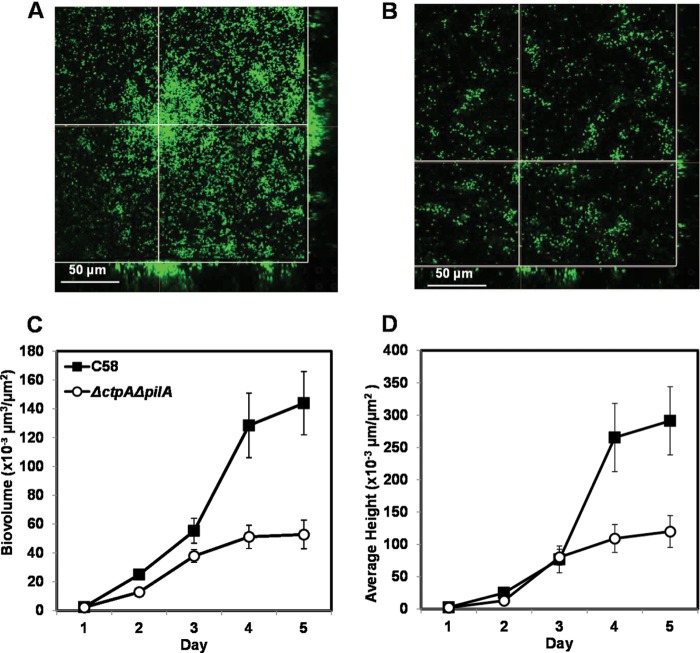
We compared the biofilm formation of wild-type C58 and the ΔctpA ΔpilA double mutant in flow cell chambers under low-shear conditions in which the growing biofilms were continuously washed with fresh medium. As we knew that, when expressed, pilA can function in place of ctpA, we used the double pilin mutant to ensure that the mutant would be nonpiliated regardless of the culture conditions. CSLM analysis of biofilms with derivatives constitutively expressing gfp revealed that, similar to the biofilm deficiency observed in static assays, the ΔctpA ΔpilA double mutant had clearly diminished biofilm formation relative to that of the wild type (Fig. 5). As has been documented previously (4), wild-type C58 developed extensive biofilms and formed large, tall (up to 30 μm) microcolonies (Fig. 5A), whereas the ΔctpA ΔpilA mutant biofilms were sparse with small cell aggregates (Fig. 5B). We examined the confocal image data with an automated version of the COMSTAT biofilm analysis program, autoCOMSTAT (7) to quantitatively analyze the biofilm structure. The ΔctpA ΔpilA mutant had a lower biovolume than the wild type during the entire incubation period (Fig. 5C). Early on, the average heights of the mutant and wild-type biofilms were similar but the wild-type biofilms then increased rapidly and eventually became three times as high as the mutant biofilms (Fig. 5D).
FIG 5.
Flow cell biofilms of wild-type C58 and a ΔctpA ΔpilA double mutant. SCLM images of flow cell biofilms formed by wild-type C58 (A) and the ΔctpA ΔpilA double mutant (B) each harboring a Ptac-gfpmut3 plasmid (pJZ383) after 5 days of incubation with a Nikon A1 SCLM with a 488-nm laser for excitation and a 500- to 550-nm filter for emission. The crossed lines in each image indicate the positions of the sagittal, three-dimensional reconstructions in the side bars. The error bars show the standard errors of the means. The images were rendered with the Nikon Elements software package. The autoCOMSTAT program was used to calculate the biovolume per substratum area (C) and average biofilm height per substratum area (D). The values are averages calculated for 12 to 15 image stacks collected from three channels for each strain per time point.
Biofilm formation on plant roots and tumor formation on potato disks.
We also qualitatively examined binding to A. thaliana root segments suspended in a nonnutritive buffered medium. No consistent difference between the 48-h biofilms formed on plant roots by the ΔctpA mutant and the wild type was observed (see Fig. S4A in the supplemental material). These data suggested that lack of Ctp pili does not cause a significant deficiency in mature biofilm formation on plant roots. However, this method is nonquantitative and provides only a rough measurement of the bacterium-plant interaction, so subtle differences in surface interactions, such as those manifested by the nonpiliated class I mutants, may not be distinguishable.
Moreover, we examined the virulence of ctp mutants. The ΔctpA (class I) and ΔctpCD (class II) mutants were virulent and caused tumor formation in potato disks, with no obvious differences from the wild-type strain in tumor quantity or size (see Fig. S4B in the supplemental material).
Class II ctp mutants are not rescued by other pilin secretion systems but have elevated reversible cell-to-surface attachment.
It was surprising that although class II mutants formed biofilms that were similar to those of the wild type, they did not produce common pili detectable by TEM. These class II ctp genes (ctpCDE and ctpGHI) are predicted to make up the basal body of the Ctp pilus, based on homology to other tad systems (24). It seemed plausible that pilin subunits might be secreted out of the cell through the secretion systems of other pilus types. As mentioned earlier, only the conjugal pili for the pAtC58 plasmid are expected to be produced under standard laboratory conditions (the Ti plasmid T pilus and the plasmid's conjugal pili are both under tight regulation), but it was formally possible that the pAtC58 avhB secretion system or the others might function in the absence of the class II ctp genes (16, 20). Class II mutants were therefore created in the C58p− strain, a derivative cured of the At and Ti plasmids (40). No difference between the C58p− strain and its class II mutants was observed (see Fig. S5 in the supplemental material), excluding the possibility that that pilin subunits were secreted through these plasmid-borne pilus systems.
In addition to mature biofilm formation, we also tested the short-term bacterium-to-surface attachment of class II mutants. A. tumefaciens produces a UPP that is crucial for irreversible bacterial attachment to surfaces (41). The UPP is produced upon surface contact, and it is rarely made by free-swimming cells (42). Bacterium-to-surface attachment and UPP production within 1 h by the wild type and the class II mutants were compared (Fig. 6). Intriguingly, the class II mutants all exhibited greater short-term binding and a greater proportion of cells producing the UPP in 1 h than the wild type.
FIG 6.
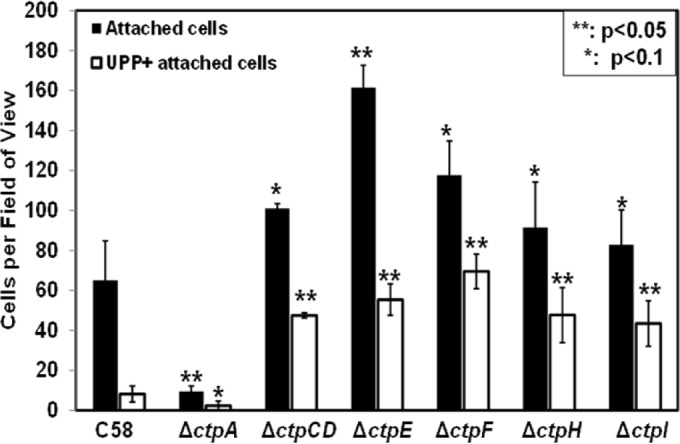
Irreversible attachment of class II Ctp mutants: Coverslips were incubated with cell cultures for 1 h, subjected to robust rinsing, stained with af-WGA for 30 min, and then vigorously rinsed again. The total numbers of attached cells per field of view (∼3.13 × 104 μm2) were determined with a Nikon E800 at ×100 magnification in bright-field mode. Enumeration of UPPs was done by determining the number of af-WGA-stained red fluorescent dots by fluorescence microscopy. The values are mean results of 10 fields of view, and the error bars show the standard deviations.
There are two hypotheses that might explain the enhanced short-term binding of class II ctp mutants. (i) These mutants have greater reversible bacterium-to-surface attachment, resulting in more uniform deployment of the UPP in the colonizing population. (ii) These mutants have more elevated UPP production in planktonic cells, leading to more stable bacterial engagement with the surface. uppC, a UPP biosynthetic gene required for polysaccharide production (Atu1238; which encodes a Wza outer membrane porin for polysaccharide export), was deleted from all of the class II mutants. These mutants cannot produce the UPP but clearly are much more effective at reversible attachment than the ΔuppC single mutant alone is (Fig. 7; see Fig. S6 in the supplemental material). Mutants with the ΔuppC deletion were readily removed from the surface by washing, whereas the wild type was stably attached. These results are consistent with the hypothesis that class II mutants have enhanced reversible bacterium-to-surface interactions.
FIG 7.
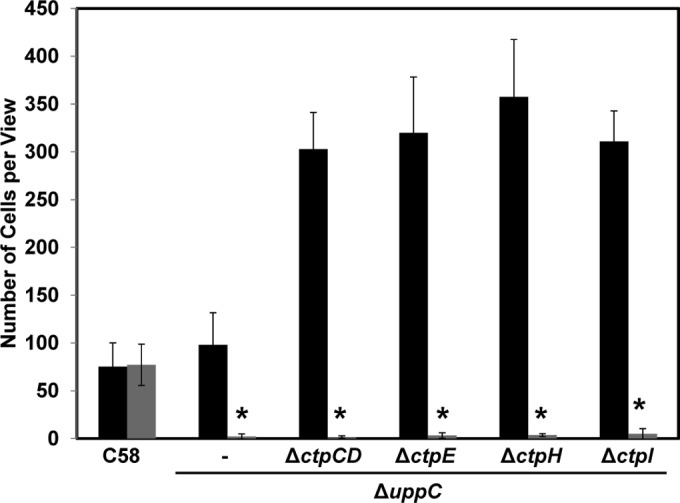
Comparison of reversible and stable surface attachment. Short-term binding assays were performed with A. tumefaciens C58 and ΔuppC single and ΔuppC/class II Ctp double mutants. Coverslips were incubated with cell culture for 1 h and then minimally rinsed (black bars, total numbers of reversible attached cells) or vigorously rinsed (gray bars, total numbers of irreversibly attached cells). Total numbers of attached cells per field of view (∼3.13 × 104 μm2) were determined with a Nikon E800 in bright-field mode at ×100 magnification. The values are mean results of 10 fields of view, and the error bars show the standard deviations. Asterisks show that attached cells were rarely observed.
The ctpA mutation is epistatic over class II ctp mutants.
The class II mutants still express the other components of the Ctp pilus, including the ctpA pilin and its presumptive peptidase ctpB. It seemed likely that the class II mutants, which do not make visible pili and have enhanced reversible surface binding and robust biofilm formation, would be independent of the CtpA pilin. The ΔctpA in-frame deletion was constructed in all of the class II mutants. Surprisingly, the ΔctpA class II double mutants had a biofilm-deficient phenotype similar to that of the ΔctpA single mutant (Fig. 8). Moreover, deletion of ctpA from class II mutants abolished their elevated short-term binding, suggesting that the class II mutants still require the CtpA pilin, even though this protein is not assembled into a visible pilus structure.
FIG 8.
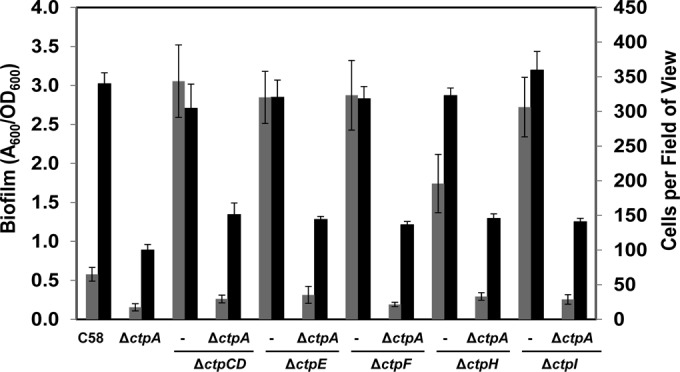
The ctpA mutation is epistatic to class II Ctp mutants. Static biofilm formation and stable short-term surface attachment of C58 and ΔctpA, class II, and ΔctpA/class II Ctp mutants were measured. Gray bars are the total numbers of irreversible attached cells per field of view (∼3.13 × 104 μm2) from short-term binding assays viewed with a Nikon E800 at ×100 magnification in bright-field mode. The values are mean values of 10 fields of view, and the error bars show the standard deviations of the means. Black bars are quantifications of acetic acid-solubilized CV on 72-h coverslip biofilms. Adherent biomass was normalized by growth (A600/OD600), and error bars are standard deviations of results of assays run in triplicate.
DISCUSSION
The common pili of A. tumefaciens have been observed by electron microscopy in previous studies (20). In this study, we have provided evidence that the ctpABCDEFGHI locus (Atu0216 to Atu0224) encodes the production of these common pili and that all of the genes except for ctpF are required to generate the filaments. As with other members of the type IVb pilus subfamily (24), we have found that the Ctp common pili of A. tumefaciens contribute to attachment and biofilm formation. Nonpiliated class I Ctp mutants manifest a consistent and significant deficiency in reversible surface interactions and biofilm formation, although these mutants are not completely deficient. Our observations are consistent with the adhesion phenotype of mutants in the related Cpa pilus from C. crescentus, in which mutants show decreased attachment but are not completely blocked (15). In contrast, tad mutants of A. actinomycetemcomitans have a dramatic loss of attachment, aggregation, and biofilm formation (37, 39). Our findings suggest that in A. tumefaciens, the Ctp pili enhance reversible surface interactions but are not absolutely required for attachment (Fig. 2 and 3). It seems likely that these pili promote initial interactions that eventually result in stable attachment and biofilm formation. As biofilms mature, this activity appears to contribute to cell-cell interactions.
Tad-type pilus systems and the A. tumefaciens Ctp cluster.
The A. actinomycetemcomitans Flp pili were the first discovered type IVb pili and remain the most thoroughly studied (25). In A. actinomycetemcomitans, the tad locus is responsible for the assembly and secretion of the Flp pili. Adherent strains of A. actinomycetemcomitans form copious, long, thick, and bundled Flp pilus fibrils; show a wrinkled colony morphology; form autoaggregates that are difficult to disperse; and tenaciously form biofilms on solid surfaces (27). A. actinomycetemcomitans derivatives that cannot form Flp pili are nonadherent and nonaggregating and produce smooth colonies. In contrast, wild-type A. tumefaciens C58 forms a limited number of Ctp pilus fibrils, has a smooth colony morphology, and aggregates only modestly. The Ctp pili observed by TEM in A. tumefaciens strains were often unbundled and distributed around the cell body. Additional cells and other pili were often observed bound at the tips of the fibers, consistent with their role in attachment and reversible surface interactions. Sporadically, we also observed thick Ctp pilus bundles (indicated by the open triangles in TEM images of the C58 strain in Fig. 2B). Thus, the Ctp pili might also associate to form a matrix of thick, bundled fibrils, binding along their length to bacterial cell surfaces to promote cell accumulation during biofilm maturation. These two hypothesized functions of Ctp pili agree with our results that the Ctp pili function both in reversible cell-to-surface short-term binding at early stages of surface interaction and in late biofilm maturation.
The tad locus in A. actinomycetemcomitans contains 14 genes, flp-1, flp-2, tadV, rcpCAB, and tadZABCDEFG, and independent mutations in these genes caused a nonadherence phenotype and the loss of Flp pilus production in A. actinomycetemcomitans (37, 39). Similar tad loci are found in the genomes of a wide variety of Gram-negative and Gram-positive bacteria (24). In C. crescentus, the Cpa locus contains nine genes that are required for production of the polar Cpa pili that function in adhesion, although other genes may be required as well (23). The Cpa locus is nearly gene-for-gene homologous and syntenous with the Ctp-encoding genes in A. tumefaciens. We determined that all of the A. tumefaciens ctp genes were required for elaboration of Ctp pili, except ctpF and the unlinked pilA gene that is homologous to ctpA.
The ctpF gene was not required for pilus formation, and the ctpF mutant did not influence adherence. The location of ctpF in the middle of the gene cluster (Fig. 2A) suggests that it should be coexpressed with the other ctp genes. In the C. crescentus cpa gene cluster, the cpaE gene is homologous to ctpF (23). CpaE is thought to be required for unipolar localization of pili in C. crescentus. However, the Ctp common pili in A. tumefaciens are distributed over the entire cell body. The CtpF protein must therefore perform some other function in A. tumefaciens that is not fundamentally required for pilus production under laboratory conditions or it is not functional.
The adherence of the pilA mutant was also unaffected, but the ectopic expression of pilA from a plasmid rescued the adherence defect of the ctpA mutant. Although it encodes a protein that is highly similar to the CtpA prepilin, the pilA gene is located on the linear chromosome rather than the circular chromosome on which the ctp locus resides and there are no other tad-type genes on this replicon. These observations suggest that PilA alone can function as a pilin subunit, productively interacting with the Ctp pilus assembly components, but that it may not be fully expressed under laboratory conditions. The plasmid-borne allele expresses pilA from the strong Plac promoter and fuses the start codon of the gene to the efficient lacZ Shine-Dalgarno sequence for translation. It remains unclear why this separate and apparently functional prepilin gene is maintained in the A. tumefaciens genome.
The ctpA pilin and other class I Ctp genes.
Deletion of ctpA, the major pilin subunit gene in A. tumefaciens C58, resulted in a moderate decrease in biofilm formation, an approximately 50% reduction relative to that of the wild type. It seems that the Ctp pili do not play as crucial a role in the adherence-related phenotypes of A. tumefaciens as they do in A. actinomycetemcomitans. It is possible that under laboratory conditions, the ctp pili are not fully expressed. These pili may play a more important role in bacterial attachment under other environmental conditions that lead to more active pilus production. However, as yet, no such conditions that clearly stimulate Ctp pilus elaboration in A. tumefaciens have been identified.
The ctpB gene encodes a homologue of the prepilin peptidase (e.g., TadV) thought to process the Flp-type prepilin to its mature form, by cleaving them at conserved glycine residues at the G/XXXXEY motif in the prepilin N terminus (43). Given that the ctpB deletion mutant has the same level of adhesion defect as the ctpA mutant and overexpression of neither ctpA nor pilA could suppress the ΔctpB mutant biofilm deficiency, it seems highly likely that ctpB recognizes and processes the CtpA prepilin. Likewise, it must be capable of processing the PilA protein. Despite limited overall homology between the CtpA and PilA proteins, they both have the G/XXXXEY motif and the sequence is the same (G/ATAIEY, positions 14 to 20 in CtpA and 29 to 35 in PilA).
Interestingly the ctpG mutant manifests an adhesion deficiency (class I) whereas those with mutations in the other ctp genes flanking it do not (class II). ctpG encodes a predicted ATPase homologous to TadA and CpaF with Walker A and B motifs, proteins required to energize pilus assembly (44). The ctpG mutant is clearly nonpiliated and does manifest an adhesion deficiency consistent with the ctpA and ctpB mutants. Its inability to assemble pili is likely responsible for this defect. Whatever CtpA-dependent mechanism allows the class II mutants to attach efficiently despite the absence of common pili, this either must not be engaged in the ctpG mutant or may require functional CtpG (see below).
Class II Ctp-encoding genes are dispensable for adherence but require the CtpA pilin.
The ctp locus contains only nine genes and lacks homologues of the tadDEFG genes (24). Most of the tad genes are required for adherence in A. actinomycetemcomitans, but in A. tumefaciens, deletion of only the class I gene ctpA, ctpB, or ctpG caused decreases in attachment and biofilm formation. Mutations of the class II genes ctpCD, ctpE, ctpG, ctpH, and ctpI resulted in strains that did not produce pili but were not diminished in adherence and even seemed to accelerate reversible surface attachment. This adherence, however, remained dependent upon ctpA. Thus, it seems that the Ctp pilin subunits can function in an unexpected way, in which they promote adherence even in the absence of the Ctp pili.
Two models seem plausible to explain the class II mutant attachment phenotype. In the first, mature pilin subunits accumulate in the periplasm of class II mutants. In the absence of the remaining pilus biogenesis functions, the accumulated pilin proteins in the periplasm become jammed in one or more export systems. These jammed pilins function as independent adhesins in bacterial attachment, which could compensate for the attachment deficiency due to the lack of pilus formation. The second model still invokes the accumulation of pilins in the periplasm, but in this model, the adherence phenotype is influenced by envelope stress that triggers increased adhesion via other mechanisms and compensates for the lack of pili. This stress response might activate specific downstream regulatory systems that promote cell-to-surface attachment and biofilm formation that is independent of the Ctp pili and does not directly involve CtpA pilin. It could be that this envelope stress requires the presumptive CtpG ATPase since it is required for normal attachment and does not bypass the requirement for pili. CtpG may have a role in the accumulation of CtpA in the periplasm in class II Ctp mutants. These two models are not mutually exclusive, and the observed phenotype of class II Ctp mutants may result from a combination of both processes.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by National Institutes of Health (NIH) grant GM080546 to C.F.
We thank Y. V. Brun, M. E. Winkler, and D. B. Kearns for helpful input and suggestions, as well as Fuqua lab members for extensive discussions. We also acknowledge assistance from the IUB Light Microscopy Imaging Center and the IU Electron Microscopy Center.
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01670-14.
REFERENCES
- 1.Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW. 1977. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11:263–271. 10.1016/0092-8674(77)90043-5 [DOI] [PubMed] [Google Scholar]
- 2.Moore LW, Chilton WS, Canfield ML. 1997. Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl. Environ. Microbiol. 63:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danhorn T, Hentzer M, Givskov M, Parsek MR, Fuqua C. 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186:4492–4501. 10.1128/JB.186.14.4492-4501.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramey BE, Matthysse AG, Fuqua C. 2004. The FNR-type transcriptional regulator SinR controls maturation of Agrobacterium tumefaciens biofilms. Mol. Microbiol. 52:1495–1511. 10.1111/j.1365-2958.2004.04079.x [DOI] [PubMed] [Google Scholar]
- 5.Heindl J, Wang Y, Heckel BC, Mohari B, Feirer N, Fuqua C. 2014. Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front. Plant Sci. 5:176. 10.3389/fpls.2014.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashby AM, Watson MD, Loake GJ, Shaw CH. 1988. Ti plasmid-specified chemotaxis of Agrobacterium tumefaciens C58C1 toward vir-inducing phenolic compounds and soluble factors from monocotyledonous and dicotyledonous plants. J. Bacteriol. 170:4181–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merritt PM, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189:8005–8014. 10.1128/JB.00566-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abarca-Grau AM, Penyalver R, Lopez MM, Marco-Noales E. 2011. Pathogenic and non-pathogenic Agrobacterium tumefaciens, A. rhizogenes and A. vitis strains form biofilms on abiotic as well as on root surfaces. Plant Pathol. 60:416–425. 10.1111/j.1365-3059.2010.02385.x [DOI] [Google Scholar]
- 9.Matthysse AG. 1983. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J. Bacteriol. 154:906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breedveld MW, Miller KJ. 1994. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58:145–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelvin SB. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67:16–37. 10.1128/MMBR.67.1.16-37.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol. Microbiol. 89:929–948. 10.1111/mmi.12321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lassalle F, Campillo T, Vial L, Baude J, Costechareyre D, Chapulliot D, Sham M, Abrouk D, Lavire C, Oger-Desfeux C, Hommais F, Guequen L, Daubin V, Muller D, Nesme X. 2011. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 3:762–781. 10.1093/gbe/evr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542–554. 10.1046/j.1365-2958.2001.02422.x [DOI] [PubMed] [Google Scholar]
- 15.Bodenmiller D, Toh E, Brun Y. 2004. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186:1438–1447. 10.1128/JB.186.5.1438-1447.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Chen Y, Wood DW, Nester EW. 2002. A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J. Bacteriol. 184:4838–4845. 10.1128/JB.184.17.4838-4845.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Bodman SB, McCutchan JE, Farrand SK. 1989. Characterization of conjugal transfer functions of Agrobacterium tumefaciens Ti plasmid pTiC58. J. Bacteriol. 171:5281–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullner KJ, Lara JC, Nester EW. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273:1107–1109. 10.1126/science.273.5278.1107 [DOI] [PubMed] [Google Scholar]
- 19.Cook DM, Li PL, Ruchaud F, Padden S, Farrand SK. 1997. Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J. Bacteriol. 179:1291–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai EM, Chesnokova O, Banta LM, Kado CI. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705–3716. 10.1128/JB.182.13.3705-3716.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D, Sr, Chapman P, Clendenning J, Deatherag G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri P, Raymond C, Rouse R, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olsen MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. 10.1126/science.1066804 [DOI] [PubMed] [Google Scholar]
- 22.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman SB, Cao YW, Askenazi M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slatte S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328. 10.1126/science.1066803 [DOI] [PubMed] [Google Scholar]
- 23.Skerker JM, Shapiro L. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223–3234. 10.1093/emboj/19.13.3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomich M, Planet PJ, Figurski DH. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363–375. 10.1038/nrmicro1636 [DOI] [PubMed] [Google Scholar]
- 25.Inoue T, Tanimoto I, Ohta H, Kato K, Murayama Y, Fukui K. 1998. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42:253–258. 10.1111/j.1348-0421.1998.tb02280.x [DOI] [PubMed] [Google Scholar]
- 26.Rosenfeld JA, Sarkar IN, Planet PJ, Figurski DH, DeSalle R. 2004. ORFcurator: molecular curation of genes and gene clusters in prokaryotic organisms. Bioinformatics 20:3462–3465. 10.1093/bioinformatics/bth427 [DOI] [PubMed] [Google Scholar]
- 27.Inoue T, Ohta H, Kokeguchi S, Fukui K, Kato K. 1990. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 69:13–18. 10.1111/j.1574-6968.1990.tb04167.x [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Mersereau M, Pazour GJ, Das A. 1990. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90:149–151. 10.1016/0378-1119(90)90452-W [DOI] [PubMed] [Google Scholar]
- 30.Tempé J, Petit A, Holsters M, Montagu MV, Schell J. 1977. Thermosensitive step associated with transfer of Ti plasmid during conjugation—possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. U. S. A. 74:2848–2849. 10.1073/pnas.74.7.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warrens AN, Jones MD, Lechler RI. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29–35. 10.1016/S0378-1119(96)00674-9 [DOI] [PubMed] [Google Scholar]
- 32.Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74:5053–5062. 10.1128/AEM.01098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34.Morton ER, Fuqua C. 2012. Phenotypic analyses of Agrobacterium. Curr. Protoc. Microbiol. Chapter 3:Unit 3D.3. 10.1002/9780471729259.mc03d03s25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand VK, Heberlein GT. 1977. Crown gall tumorigenesis in potato tuber tissue. Am. J. Bot. 64:153–158. 10.2307/2442102 [DOI] [Google Scholar]
- 36.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersball BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10):2395–2407 [DOI] [PubMed] [Google Scholar]
- 37.Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, Figurski DH. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J. Bacteriol. 182:6169–6176. 10.1128/JB.182.21.6169-6176.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bentzmann S, Aurouze M, Ball G, Filloux A. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188:4851–4860. 10.1128/JB.00345-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez BA, Kachlany SC, Tomich M, Fine DH, Figurski DH. 2006. Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation. J. Bacteriol. 188:6361–6375. 10.1128/JB.00496-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morton ER, Platt TG, Fuqua C, Bever JD. 2014. Non-additive costs and interactions alter the competitive dynamics of co-occurring ecologically distinct plasmids. Proc. Biol. Sci. 281:20132173. 10.1098/rspb.2013.2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. 2012. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res. Microbiol. 163:674–684. 10.1016/j.resmic.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol. Microbiol. 83:41–51. 10.1111/j.1365-2958.2011.07909.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomich M, Fine DH, Figurski DH. 2006. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J. Bacteriol. 188:6899–6914. 10.1128/JB.00690-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927–5936. 10.1128/JB.183.20.5927-5936.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.