Abstract
The brucellae are the etiological agents of brucellosis, a worldwide-distributed zoonosis. These bacteria are facultative intracellular parasites and thus are able to adjust their metabolism to the extra- and intracellular environments encountered during an infectious cycle. However, this aspect of Brucella biology is imperfectly understood, and the nutrients available in the intracellular niche are unknown. Here, we investigated the central pathways of C metabolism used by Brucella abortus by deleting the putative fructose-1,6-bisphosphatase (fbp and glpX), phosphoenolpyruvate carboxykinase (pckA), pyruvate phosphate dikinase (ppdK), and malic enzyme (mae) genes. In gluconeogenic but not in rich media, growth of ΔppdK and Δmae mutants was severely impaired and growth of the double Δfbp-ΔglpX mutant was reduced. In macrophages, only the ΔppdK and Δmae mutants showed reduced multiplication, and studies with the ΔppdK mutant confirmed that it reached the replicative niche. Similarly, only the ΔppdK and Δmae mutants were attenuated in mice, the former being cleared by week 10 and the latter persisting longer than 12 weeks. We also investigated the glyoxylate cycle. Although aceA (isocitrate lyase) promoter activity was enhanced in rich medium, aceA disruption had no effect in vitro or on multiplication in macrophages or mouse spleens. The results suggest that B. abortus grows intracellularly using a limited supply of 6-C (and 5-C) sugars that is compensated by glutamate and possibly other amino acids entering the Krebs cycle without a critical role of the glyoxylate shunt.
INTRODUCTION
Brucella is a genus of Gram-negative bacteria that includes the causative agents of brucellosis, a worldwide-extended zoonosis severely affecting animal production and human welfare. Three species, Brucella abortus, B. melitensis, and B. suis, are the most common causes of brucellosis in domestic livestock and humans. These brucellae can grow both in vitro and within host cells, and their pathogenicity results largely from their capacity to escape prompt detection by innate immunity and the use of a type IV secretion system to reach the replicative niche, an endoplasmic reticulum-derived vacuole (1–6). In this compartment, these bacteria multiply extensively, which shows their ability to efficiently use substrates provided by the host. However, there is only sparse information on either the nature of these substrates or the metabolic pathways used in the replicative niche (7).
In vitro, most strains of B. abortus, B. melitensis, and B. suis grow in several simple chemically defined media (8). Among these, Gerhardt's medium contains glycerol, lactate, glutamate, and mineral salts plus nicotinic acid, thiamine, pantothenic acid, and biotin as growth factors. This medium supports growth better than other simple defined media, including those that provide glucose as the C source (8), and this has been attributed to the ability of these bacteria to use glutamate very efficiently through the tricarboxylic acid (TCA) cycle (9). Indeed, growth in this medium is clear proof of the ability of these bacteria to carry out gluconeogenesis in vitro. Concerning the catabolism of glucose, it has been accepted that it proceeds through the pentose phosphate pathway (in conjunction with TCA), rather than through the glycolysis or Entner-Doudoroff pathway (Fig. 1). This view, although supported by early radiorespirometric and enzymatic studies (10, 11) performed with the attenuated B. abortus S19 vaccine, is not consistent with the presence of the genes putatively encoding all the enzymes of the Entner-Doudoroff pathway (12). It is possible that the multiple defects in S19 (13) and/or the experimental conditions in those early experiments precluded detection of key enzymes of the Entner-Doudoroff pathway (12).
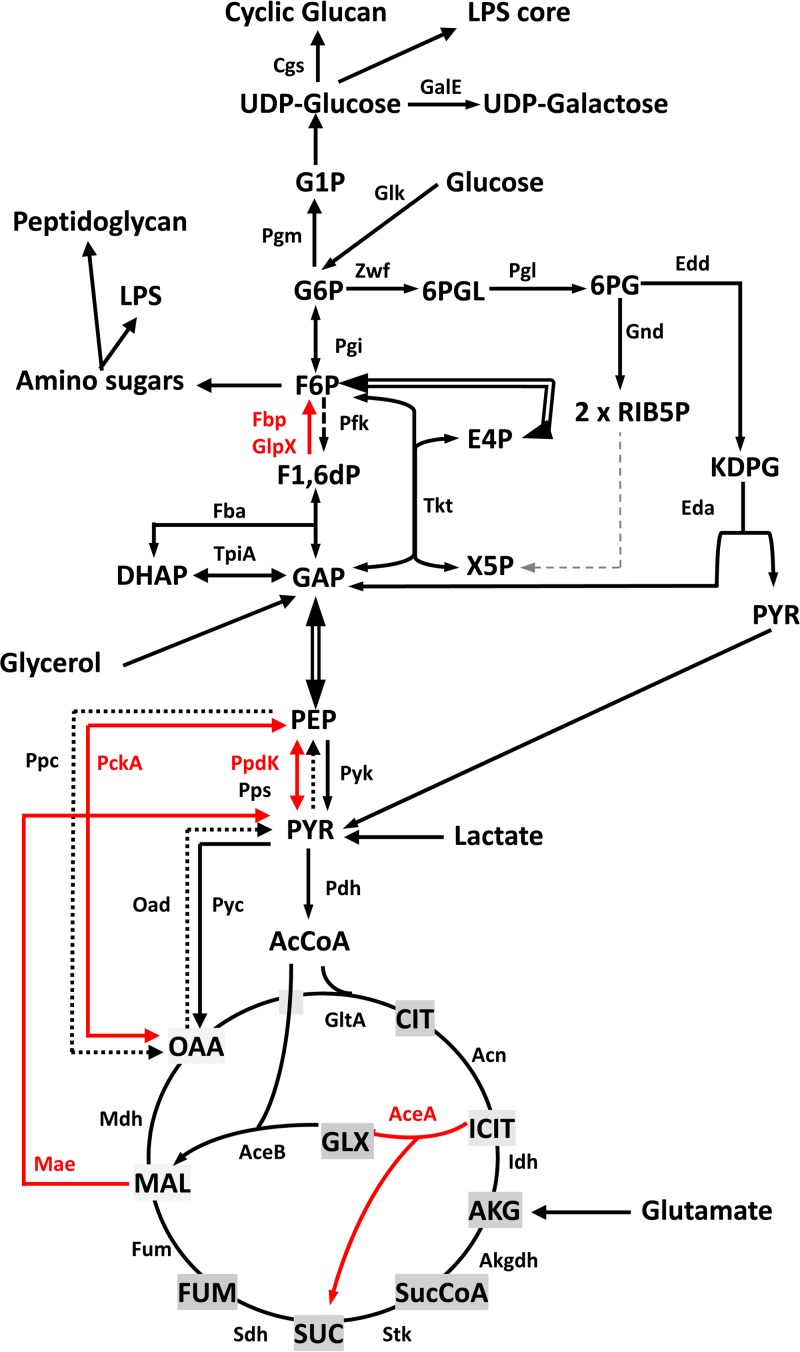
FIG 1.
Conventional central metabolic pathways (glycolysis, gluconeogenesis, Entner-Doudoroff, pentose-phosphate, TCA cycle, and glyoxylate) of bacteria. Dashed arrows indicate steps for which no putative genes can be identified in B. abortus. Red arrows indicate the steps studied in this work. AcCoA, acetyl-CoA; AceA, isocitrate lyase; AceB, malate synthase; Acn, aconitate hydratase; AKG, α-ketoglutarate; Akgdh, α-ketoglutarate dehydrogenase; Cgs, cyclic-glucan synthesis; CIT, citrate; DHAP, dihydroxyacetone phosphate; Eda, keto-deoxy-phosphogluconate aldolase; Edd, 6-phospho-d-gluconate dehydratase; E4P, erythrose-4-phosphate; Fba, fructose bisphosphate aldolase; Fbp, fructose-1,6-bisphosphatase; Fum, fumarase; FUM, fumarate; F1,6dP, fructose-1,6-bisphosphate; F6P, fructose-6-phosphate; GalE, UPD-glucose-4 epimerase; GAP, glyceraldehyde-3-phosphate; Gnd, 6-phosphogluconate dehydrogenase; Glk, glucokinase; GlpX, fructose-1,6-bisphosphatase; GltA, citrate synthase; GLX, glyoxylate; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; ICIT, isocitrate; Idh, isocitrate dehydrogenase; KDPG, 2-keto-3-deoxyphosphogluconate; LPS, lipopolysaccharide; Mae, malic enzyme; MAL, malate; Mdh, malate dehydrogenase; OAA, oxaloacetate; Oad, oxaloacetate decarboxylase; Pfk, phosphofructokinase; PckA, phosphoenol pyruvate carboxykinase; Pdh, pyruvate dehydrogenase; PEP, phosphoenol pyruvate; Pgi, phosphoglucose isomerase; Pgl, lactonase; Pgm, phosphoglucomutase; Ppc, phosphoenol pyruvate carboxylase; PpdK, pyruvate phosphate dikinase; Pps, phosphoenol pyruvate synthase; Pyc, pyruvate carboxylase; Pyk, pyruvate kinase; PYR, pyruvate; RIB5P, ribulose-5-phosphate; Sdh, succinate dehydrogenase; Stk, succinyl-CoA synthetase; SUC, succinate; SucCoA, succinyl-CoA; Tkt, transketolase; TpiA, triose phosphate isomerase; X5P, xylulose-5-phosphate; Zwf, glucose-6-phosphate dehydrogenase; 6PG, 6-phosphogluconate; 6PGL, 6-phosphogluconolactone.
Concerning intracellular metabolism, some data come from randomly obtained mutants that show attenuation in human or mouse macrophages or in HeLa cells. Genes (and their putative functions) identified in this manner include, in B. suis 1330, a gguA homologue (gluP, presumably involved in sugar uptake), some erythritol catabolism genes, gnd (6-phosphogluconate dehydrogenase, required for using glucose through the pentose phosphate pathway), rbsK (ribose kinase), pyc (anaplerotic pyruvate carboxylase), pgi (phosphoglucose isomerase), and genes related to the biosynthesis of amino acids (14–16); in B. melitensis 16M, dbsA and ugpA (ribose and glycerol-3-phosphate transporters, respectively), glpD (glycerol-3-phosphate dehydrogenase), and some erythritol catabolism genes (17, 18); and in B. abortus 2308, gluP (previously shown to encode an active glucose and galactose transporter in this species [19]), gnd (6-phosphogluconate dehydrogenase), gltD (glutamate synthase), and gcvB (glycine dehydrogenase) (20). Also in B. abortus 2308, dxs (an isoprenoid biosynthesis transketolase) and mocC (rhizopine or inositol catabolism) were identified as expressed in macrophages using a fluorescent reporter (21).
Proteomic analyses have also provided clues to the metabolism of brucellae in the host. Al Dahouk et al. (22) found that forty-eight hours after infection of mouse macrophages with B. suis 1330, there was an important reduction of proteins putatively involved in energy, protein, and nucleic acid metabolism. Some exceptions were ribitol kinase, glyceraldehyde-3-phosphate-dehydrogenase (glyceraldehyde-3-P-dehydrogenase), and the isocitrate lyase (AceA) of the glyoxylate cycle. However, other studies in B. suis 1330 do not support the use of the glyoxylate cycle within host cells (15). Lamontagne et al. (23) analyzed B. abortus 2308 protein expression 3, 20, and 44 h after infection of RAW 264.7 macrophages. They found that multiple proteins associated with sugar uptake, the TCA cycle, the pentose phosphate shunt, and the subsequent generation of pyruvate were downregulated 3 h after infection. At 24 h, several proteins involved in sugar metabolism and transport were also reduced. Enzymes associated with protein and amino acid catabolism were mainly increased early (3 h) but also 24 h after infection, when bacteria were already in vacuoles derived from the endoplasmic reticulum. This was also the case for enzymes involved in glutamate synthesis, suggesting conversion of amino acids to glutamate and α-ketoglutarate. Accordingly, amino acid-based alternatives may be the preferred solution for B. abortus to derive precursors for the TCA cycle and ancillary routes during the midpoint time course of infection. At later times, the same authors observed an increase in proteins involved in transport, suggesting that the endoplasmic reticulum is able to supply at least some of the substrates required for bacterial growth. Likewise, the pentose phosphate shunt seemed to partially resume its functions.
Although the information provided by these studies is valuable, the central metabolic pathways used by Brucella during infection remain unclear. The results are contradictory in some cases, as for the glyoxylate cycle or the metabolic activity in cells. In addition, some studies suggest the availability of sugars in the replicative niche, whereas others indicate that amino acids could be the preferred C source in vivo, which may require a gluconeogenic metabolism. Indeed, apparently conflicting data may result from the use of different host cell lines, different times of analysis, polarity of mutations, and other experimental conditions. Moreover, there might be some variation among B. suis 1330, B. melitensis 16M, and B. abortus 2308, as suggested by the known differences in oxidative rates of sugars and amino acids (24). In this work, we attempt to address some aspects of the central metabolic pathways used by B. abortus in the host. For this purpose, we focused our research on genes putatively involved in classical gluconeogenesis, the anabolic pathways bridging the TCA cycle and the triose-phosphate pathway, and the glyoxylate cycle. We constructed in-frame mutations in genes coding for key enzymes and tested the mutants in complex and chemically defined media and for multiplication within cultured cells and for persistence in the mouse model (25). Together with some of the previous analyses, our observations suggest a model of B. abortus metabolism in which, although TCA supplies molecules necessary for biosynthesis and subsequent growth, the classical fructose-1,6-bisphosphatases Fbp and GlpX are not necessary and 6- and/or 5-C molecules for polymer biosynthesis are obtained mostly from the intracellular milieu.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material, and their origin and characteristics are described in previous works (26–28). The strains resulting from the genetic manipulations described below were characterized according to standard Brucella typing procedures, i.e., colonial morphology after 3 days of incubation at 37°C, crystal violet-oxalate exclusion, urease, acriflavine agglutination, sensitivity to Tb, Wb, Iz, and R/C phages, agglutination with anti-A and anti-M monospecific sera, CO2 and serum dependence, and susceptibility to thionin blue, fuchsin, and safranin (24). Bacteria were routinely grown in standard peptone-yeast extract-glucose broth (bioMérieux) or in this medium supplemented with agar (TSA). The following antibiotics were used at the indicated concentrations: kanamycin (Km; 50 μg/ml), nalidixic acid (Nal; 25 μg/ml), chloramphenicol (Cm; 20 μg/ml) and/or gentamicin (Gm; 100 μg/ml or 25 μg/ml) (all from Sigma). When needed, medium was supplemented with 5% sucrose (Sigma). All strains were stored at −80°C in skim milk (Scharlau). To study the phenotype of the metabolic mutants, peptone-yeast extract-glucose or the medium of Gerhardt and Wilson (referred to here as glutamate-lactate-glycerol) was used (29). The components (for 1 liter) of the latter were glycerol (30 g), lactic acid (5 g), glutamic acid (5 g), thiamine (0.2 mg), nicotinic acid (0.2 mg), pantothenic acid (0.04 mg), biotin (0.0001 mg), K2HPO4 (10 g), Na2S2O3 · 5H2O (0.1 g), MgSO4 (10 mg), MnSO4 (0.1 mg), FeSO4 (0.1 mg), and NaCl (7.5 g). The pH was adjusted to 6.8 to 7.
Growth measurements.
Inocula preconditioned to the conditions in the test medium (peptone-yeast extract-glucose, glutamate-lactate-glycerol, glycerol-glutamate, glycerol-lactate, or glutamate-lactate) were prepared as follows. First, the strains to be tested were inoculated into 10 ml of peptone-yeast extract-glucose in a 50-ml flask and incubated at 37°C with orbital shaking for 18 h. These exponentially growing bacteria were harvested by centrifugation, resuspended in 5 ml of the test medium at an optical density at 600 nm (OD600) of 0.1, and incubated at 37°C with orbital shaking for 18 h. Then, these preconditioned bacteria were harvested by centrifugation, resuspended at an OD600 of 0.1 (0.05 starting in the Bioscreen apparatus) in the same test medium in Bioscreen multiwell plates (200 μl/well), and cultivated in a Bioscreen C (Lab Systems) apparatus with continuous shaking at 37°C. Absorbance values at 420 to 580 nm were automatically recorded at 0.5-h intervals over a 120- to 300-h period. All experiments were performed in triplicate. Controls with culture medium and no bacteria were included in all experiments.
DNA manipulations.
Genomic sequences were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). Searches for DNA and protein homologies were carried out using the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) and the European Molecular Biology Laboratory (EMBL)–European Bioinformatics Institute server (http://www.ebi.ac.uk/). Primers were synthesized by Sigma-Genosys (Haverhill, United Kingdom). DNA sequencing was performed by the Servicio de Secuenciación del Centro de Investigación Médica Aplicada (Pamplona, Spain). Restriction-modification enzymes were used under the conditions recommended by the manufacturer. Plasmid and chromosomal DNA were extracted with QIAprep spin miniprep (Qiagen) and Ultraclean microbial DNA isolation kits (Mo Bio Laboratories), respectively. When needed, DNA was purified from agarose gels using the QIAquick gel extraction kit (Qiagen).
In-frame fbp and glpX deletion mutants were constructed by PCR overlap using genomic DNA of B. abortus 2308 as the DNA template. Primers were designed using the B. abortus 2308 sequences available in KEGG (http://www.genome.jp/kegg/). For the construction of the fbp mutant, two PCR fragments were generated: oligonucleotides fbp-F1(5′-GTAGCCAAAAAGCCCAGGT-3′) and fbp-R2 (5′-GCCAACCAGAACCAGAGGA-3′) were used to amplify a 203-bp fragment including codons 1 to 14 of the fbp open reading frame (ORF) as well as a 161-bp fragment upstream of the fbp start codon, and oligonucleotides fbp-F3 (5′-TCCTCTGGTTCTGGTTGGCGTGGCCGAAGAGGTGGATA-3′) and fbp-R4 (5′-CATTTGCCGCTTCCATGA-3′) were used to amplify a 193-bp fragment including codons 327 to 341 of the fbp ORF and a 148-bp fragment downstream of the fbp stop codon. Both fragments were ligated by PCR using oligonucleotides fbp-F1 and fbp-R4 for amplification and the complementary regions between fbp-R2 and fbp-F3 for overlapping. The resulting fragment, containing the fbp deletion allele, was cloned into pCR2.1 (Invitrogen) to generate plasmid pAZI-1, sequenced to ensure that the reading frame was maintained, and subcloned into the BamHI and the XbaI sites of the suicide plasmid pJQKm (30). The resulting mutator plasmid (pAZI-2) was introduced into B. abortus 2308 by conjugation (26). Integration of the suicide vector was selected by Nal and Km resistance, and the excisions (generating both the fbp mutant [BABΔfbp] and a sibling revertant strain carrying an intact gene [BABfbp-sibling revertant]) were then selected by Nal and sucrose resistance and Km sensitivity. The resulting colonies were screened by PCR with primers fbp-F1 and fbp-R4, which amplified a fragment of 396 bp in the mutant and a fragment of 1,332 bp in the sibling revertant strain. The mutation resulted in the loss of about 98% of the fbp ORF, and the mutant strain was called BABΔfbp.
The glpX mutant was constructed in a similar way. Primers glpX-F1 (5′-ACGGTGATTCTGGTGACACA-3′) and glpX-R2 (5′-CGAGCTCCAGTGTGAGAATG-3′) were used to amplify a 576-bp fragment including 61 bp of the glpX ORF as well as 515 bp upstream of the glpX start codon, and primers glpX-F3 (5′-CATTCTCACACTGGAGCTCGATACGACAGATCCGGACGAG-3′) and glpX-R4 (5′-CATCATACAGTTGCCGATGG-3′) were used to amplify a 574-bp fragment including 371 bp of the glpX ORF and 203 bp downstream of the glpX stop codon. Both fragments were ligated by overlapping PCR using primers glpX-F1 and glpX-R4, and the fragment containing the deletion allele was cloned into pCR2.1 to generate plasmid pAZI-3, sequenced to confirm that the glpX ORF had been maintained, and subcloned in pJQKm to produce the mutator plasmid pAZI-4. This plasmid was then introduced into B. abortus 2308, and the deletion mutant generated by allelic exchange was selected by Nal and sucrose resistance and Km sensitivity and by PCR using oligonucleotides glpX-F1 and glpX-R4, which amplified a fragment of 1,150 bp in the deletion strain and a fragment of 1,705 bp in the BABglpX-sibling revertant strain. The mutation resulted in the loss of approximately 56% of the glpX ORF, and the mutant was called BABΔglpX.
To construct the double mutant BABΔfbpΔglpX, the mutator plasmid pAZI-4 was introduced into strain BABΔfbp. After allelic exchange, the double mutant was selected as described above using primers glpX-F1 and glpX-R4.
BABΔaceA was constructed using the same strategy. Oligonucleotides aceA-F1 (5′-TGACAAGATATCGCCAAAACAC-3′) and aceA-R2 (5′-CGAAGGGATGAGGCTGTAAA-3′) amplified a 238-bp fragment, including codons 1 to 10 of the aceA ORF and 208 bp upstream of the aceA start codon. Oligonucleotides aceA-F3 (5′-TTTACAGCCTCATCCCTTCGGAAACCGCACAGTTCAAGC-3′) and aceA-R4 (5′-GGATCAAGAGATCACCCCAGT-3′) amplified a 278-bp fragment including codons 420 to 430 of the ORF aceA and 245 bp downstream of the aceA stop codon. Both fragments were ligated by overlapping PCR using oligonucleotides aceA-F1 and aceA-R4. The PCR product was cloned into pCR2.1 to generate pAZI-7, sequenced, and subcloned into pJQKm to produce the suicide plasmid pAZI-8. B. abortus 2308 mutants were selected by PCR using oligonucleotides aceA-F1 and aceA-R4. PCR products were 1,743 bp in the BABaceA-sibling revertant strain and 738 bp in BABΔaceA. This mutation eliminated 78% of the aceA ORF.
For the construction of the pckA mutant, oligonucleotides pckA-F1 (5′-TGTTTGCAGTTTTCCACACC-3′), pckA-R2 (5′-AATCGAAGCGGCCTTATTGT-3′), pckA-F3 (5′-ACAATAAGGCCGCTTCGATTGACGGCTCGCTGAACAAT-3′), and pckA-R4 (5′-TCTTGCGATAACAGCCAAAA-3′) were used. Primers pckA-F1 and pckA-R2 amplified a 219-bp fragment which included codons 1 to 13 of the pckA ORF and 180 bp upstream of the pckA start codon. Primers pckA-F3 and pckA-R4 amplified a 319-bp fragment including the last 37 codons of the pckA ORF and 208 bp downstream of the pckA stop codon. Both PCR products were ligated by overlapping PCR using pckA-F1 and pckA-R4, cloned into pCR2.1 to generate plasmid pAZI-5, and subsequently subcloned into the BamHI and the XbaI sites of the suicide plasmid pJQKm. The resulting mutator plasmid, pAZI-6, was introduced into B. abortus 2308, where it was integrated into the chromosome. A second recombination generated the excision of the plasmid. The resulting colonies were screened by PCR (with pckA-F1 and pckA-R4) amplifying a fragment of 538 bp in the mutant and a fragment of 1,864 bp in the sibling revertant strain. The mutant strain was called BABΔpckA and lacked the 71.14% of the pckA ORF.
BABΔppdK was constructed using primers ppdK-F1(5′-CTCCCGATTCATTTTTCACG-3′) and ppdK-R2 (5′-TGCTCATTTCAGCCAGGTT-3′) to amplify a 288-bp fragment including the first 103 bp of the ppdK ORF as well as 185 bp upstream of the ppdK start codon and primers ppdK-F3 (5′-AACCTGGCTGAAATGAGCACGGGTCTCGACTATGTGTCC-3′) and ppdK-R4 (5′-TCAACGCATCAAAGCAGAAG-3′) to amplify a 220-bp fragment including the last 86 bp of the ppdK ORF and 134 bp downstream of the ppdK stop codon. Both fragments were ligated by overlapping PCR using primers ppdK-F1 and ppdK-R4, and the fragment obtained, containing the deletion allele, was cloned into pCR2.1 to generate pMZI-1, sequenced to confirm that the reading frame had been maintained, and subcloned into pJQKm to produce the mutator plasmid pMZI-2. This plasmid was introduced into B. abortus 2308, and both the deletion mutant and the sibling revertant strain generated by allelic exchange were selected by Nal and sucrose resistance and Km sensitivity and by PCR using ppdK-F1 and ppdK-R4, which amplified a fragment of 508 bp in BABΔppdK and a fragment of 2,983 bp in BABppdK-sibling revertant strain. The mutation generated resulted in the loss of the 93% of ppdK.
Primers ppdKII-F1 (5′-CTCCCGATTCATTTTTCACG-3′), ppdKII-R2 (5′-CTGCTCATTTCAGCCAGGTT-3′), ppdKII-F3 (5′-AACCTGGCTGAAATGAGCAGCGGGTCTCGACTATGTGTCC-3′), and ppdKII-R4 (5′-TCAACGCATCAAAGCAGAAG-3′) were used to obtain the mutator plasmid pAZI-10. This plasmid was introduced into B. abortus 2308 to obtain a ppdK mutant that maintained only the 34 first amino acids of PpdK (BABΔppdK-II). This mutant had the same phenotype as the one previously described. Thus, the mutator plasmid pAZI-10 was also introduced into strain BABΔpckA carrying the pckA mutation to obtain the double mutant BABΔpckAΔppdK.
BABΔmae was constructed using primers mae-F1 (5′-TATGACGGCGCACTTGTCTA-3′) and mae-R2 (5′-TCGGATAGCGATGGAAGAAC-3′) to amplify a 341-bp fragment including 76 bp of the mae ORF as well as 265 bp upstream of the mae start codon, and primers mae-F3 (5′-GTTCTTCCATCGCTATCCGAGCGAAGCCAATCTTCTGGTA-3′) and mae-R4 (5′-CGCCATAAAACGAACCTCAA-3′) to amplify a 376 bp including 227 bp of the mae ORF and 149 bp downstream of the mae stop codon. Both fragments were ligated by overlapping PCR using primers mae-F1 and mae-R4, and the fragment obtained, containing the deletion allele, was cloned into pCR2.1 to generate pMZI-3, sequenced to confirm that the mae ORF had been maintained, and subcloned into pJQKm to produce the mutator plasmid pMZI-4. This plasmid was then introduced into B. abortus 2308, and the mutant and sibling revertant strains generated by allelic exchange were selected by Nal and sucrose resistance and Km sensitivity and by PCR using oligonucleotides mae-F1 and mae-R4, which amplified a fragment of 717 bp in the deleted strain and a fragment of 2,739 bp in the sibling revertant strain. The mutation generated resulted in the loss of the 87% of the mae ORF.
To check the different mutations, we used internal primers (gene-R5) hybridizing in the deleted regions.
For complementation, a plasmid carrying ppdK was constructed using the Gateway cloning Technology (Invitrogen). Since the sequences of ppdK from B. abortus and B. melitensis are 99% identical, the clone carrying ppdK was extracted from the B. melitensis ORFeome, and the ORF was subcloned into plasmid pRH001 (31) to produce plasmid pAZI-19. This plasmid was introduced into BABΔppdK by mating with Escherichia coli S17 λpir, and the conjugants harboring this plasmid (designated BABΔppdK pAZI-19) were selected by plating the mating mixture onto tryptic soy agar (TSA) plates containing Nal and Km (TSA-NalKm). For the construction of BABΔfbpΔglpX pAZI-21, fbp was amplified from BAB-parental using primers fbp-Fp (5′-GGGATCCATGCTTCTGAAAGGGTGGTACCG-3′) and fbp-R4 (5′-CATTTGCCGCTTCCATGA-3′) and cloned into pCR2.1. The resulting plasmid was sequenced, and the fbp gene was subcloned into the BamHI and the XhoI sites of the vector pBBR1MCS1. The resulting plasmid (pAZI-21) was introduced into BABΔfbpΔglpX by conjugation (see above). BABΔfbpΔglpX(pAZI-23) was constructed by following the same strategy and using primers glpX-F1 (5′-ACGGTGATTCTGGTGACACA-3′) and glpX-R4 (5′-CATCATACAGTTGCCGATGG-3′) to amplify glpX.
Gene expression studies.
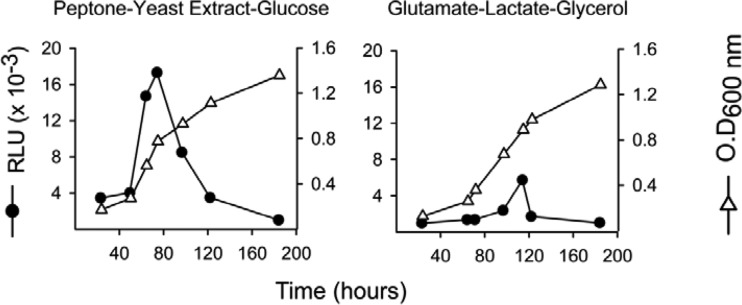
To determine whether aceA was expressed in vitro, its promoter was fused with the luciferase reporter gene. To this end, aceAp-F (5′-GGGATCCTAGTTGCGCTCGATCAGATT-3′) and aceAp-R (5′-TTCTAGACATTTCGGTGTCTCCTCGT-3′) (containing BamHI and XbaI sites [underlined], respectively) were used to amplify a 382-bp region containing the ATG and the aceA promoter from B. abortus 2308 genomic DNA. This PCR product was verified by electrophoresis and ligated into the vector pGEM-T Easy (Promega), thereby originating plasmid pAZI-17. Then, the insert was digested with BamHI and XbaI and ligated to pSKOriTKmluxAB to generate plasmid pAZI-18. This plasmid was introduced into E. coli S17λ pir Nals and then transferred by conjugation to B. abortus 2308 Nalr Kms. Cells were plated on TSA-NalKm. Positive clones gave a 510-bp band when verified by PCR using aceAp-F and luxAB-R. The resulting strain was called B. abortus pBABaceA-luxAB. To measure luciferase activity, fresh B. abortus pBABaceA-luxAB cells were adjusted to an OD600 of 0.4 in saline, and finally, 50 or 200 μl was added to flasks with 10 ml of peptone-yeast extract-glucose or glutamate-lactate-glycerol, respectively. Growth was followed by measuring absorbance at OD600, and 1-ml aliquots were taken at selected intervals to measure the luminescence in relative luminescence units (RLU) after addition of 100 μl ethanol-decanal (1:1).
Cell infections and intracellular trafficking.
In vitro infection assays were performed in RAW 264.7 macrophages (ATCC TIB-71) and HeLa cells (ATCC CCL-2) cultured in Dulbecco's modified Eagle medium (DMEM; Gibco) with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco), 1% (vol/vol) l-glutamine (200 nM; Sigma-Aldrich), and 1% (vol/vol) nonessential amino acids (Gibco). Then, 24-well plates were seeded with 1 × 105 cells/well, and macrophages and HeLa cells were infected at multiplicities of infection of 50:1 and 200:1 (bacteria to cells), respectively, by centrifuging the plates at 400 × g for 10 min at 4°C. After incubation for 15 min at 37°C under a 5% CO2 atmosphere, extracellular bacteria were removed with four DMEM washes followed by Gm treatment (100 μg/ml) for 90 min. Then, fresh medium supplemented with 25 μg/ml of Gm was added and cultures were incubated. At 2, 24, and 48 h, cells were washed three times with 100 mM phosphate-buffered saline (pH 7.2), lysed with 0.1% (vol/vol) Triton X-100 in phosphate-buffered saline, and plated on TSA to determine the number of intracellular bacteria. All experiments were performed in triplicate (32). Results were expressed as means and standard errors (n = 3) of individual log10 CFU/well. Statistical comparison of means was performed by a one-way analysis of variance (ANOVA) followed by Fisher's protected least significant differences (PLSD) tests (33).
For immunofluorescence microscopy, RAW 264.7 macrophages and HeLa cells were grown on coverslips and inoculated with bacteria as described above. Cells were fixed in 3% paraformaldehyde in 100 mM phosphate-buffered saline (pH 7.2) at 37°C for 10 min. Cells were washed twice with phosphate-buffered saline and permeabilized with 0.1% (vol/vol) Triton X-100 and 3% bovine serum albumin (Sigma), for 30 min. Coverslips were incubated with primary antibodies for 45 min at room temperature, washed three times in the same phosphate-buffered saline supplemented with 3% bovine serum albumin, and then incubated with the appropriate secondary antibodies. Coverslips were washed three times with phosphate-buffered saline and once with H2O and mounted on glass slides using Mowiol 4-88. Samples were examined and images acquired using a Leica TCS SP5 laser scanning confocal microscope at UNAmur (Namur, Belgium). The primary antibodies used for immunofluorescence microscopy were rabbit anti-calnexin (SPA-860; Stressgen) and a mouse anti-S-LPS monoclonal antibody (A76/12G12/F12). The secondary antibodies were donkey anti-rabbit IgG conjugated to Alexa Fluor 488 (Invitrogen) and goat anti-mouse IgG conjugated to Alexa Fluor 546 (Invitrogen). For lysosomal labeling, the primary antibody used was rat anti-mouse (Developmental Studies Hybridoma Bank [DSHB]), and the secondary antibody was goat anti-rat IgG conjugated to Alexa Fluor 633 (Invitrogen).
Assays in mice.
Female BALB/c mice (Charles River, France) were kept in cages with water and food ad libitum and accommodated under P3 biosafety containment conditions for 2 weeks before and during the experiments in the facilities of the Instituto de Agrobiotecnología (registration code ES/31-2016-000002-CR-SU-US). The animal handling and other procedures were in accordance with the current European (directive 86/609/EEC) and Spanish (RD 53/2013) legislations, supervised by the Animal Welfare Committee of the Universidad Pública de Navarra, and authorized by the competent authority of Gobierno de Navarra. To prepare inocula, TSA-grown bacteria were harvested, adjusted spectrophotometrically (OD600 = 0.170) in 10 mM phosphate-buffered saline (pH 6.85), and diluted in the same diluent up to approximately 5 × 105 CFU/ml (exact doses were assessed retrospectively). For each bacterial strain, five mice were intraperitoneally inoculated with 0.1 ml/mouse, and the number of CFU in spleen was determined at different weeks postinoculation as described previously (33). The identity of the spleen isolates was confirmed by PCR. The individual number of CFU/spleen was normalized by logarithmic transformation, and the mean log CFU/spleen values and the standard deviations were calculated for each group of mice (n = 5). Statistical comparisons were performed by a one-way ANOVA followed by Fisher's protected least significant differences (PLSD) tests (33).
RESULTS
Dysfunction of B. abortus fbp, glpX, ppdK, and mae but not of pckA or aceA homologues affects growth on gluconeogenic substrates in vitro.
The conversion of fructose-1,6-bisphosphate into fructose-6-P mediated by the cognate bisphosphatases (FBPases) is the only irreversible step among those taking part in gluconeogenesis in Brucella (Fig. 1). Therefore, FBPase activity is strictly necessary for growth under gluconeogenic conditions.
As in E. coli and Salmonella, the B. abortus 2308 genome contains two ORFs for putative FBPases: BAB2_0364 and BAB1_1292. The former is predicted to encode a protein of 340 amino acids that belongs to the class I FBPases (Fbp), and the latter is predicted to encode a 328-amino-acid protein orthologous to E. coli GlpX, an FBPase of class II (Fig. 1). In E. coli, Fbp is connected to the production of fructose-6-phosphate for nucleotide, polysaccharide, and aromatic amino acid biosynthesis. GlpX, on the other hand, belongs to an operon (glpFKX) related to phospholipid biosynthesis (34). The putative B. abortus glpX, however, seems isolated and not part of any obvious operon.
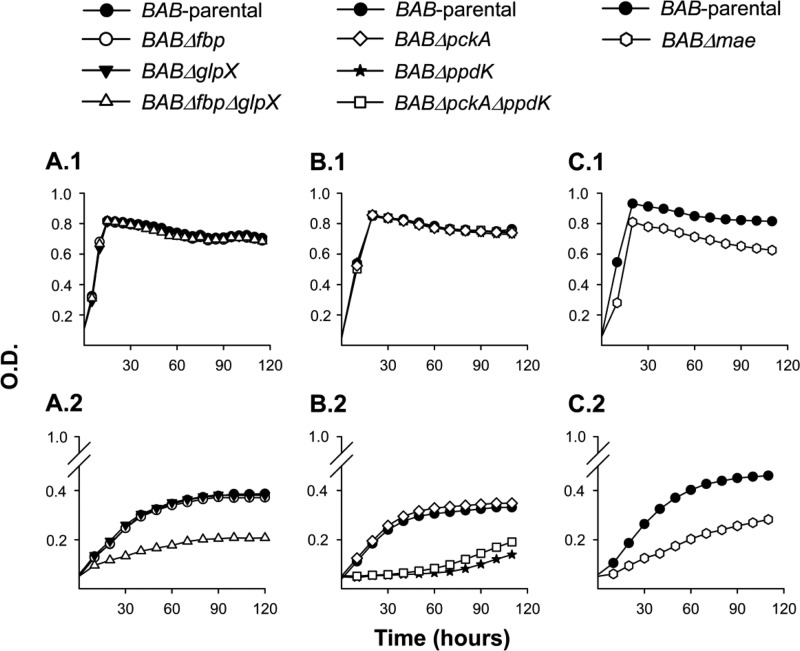
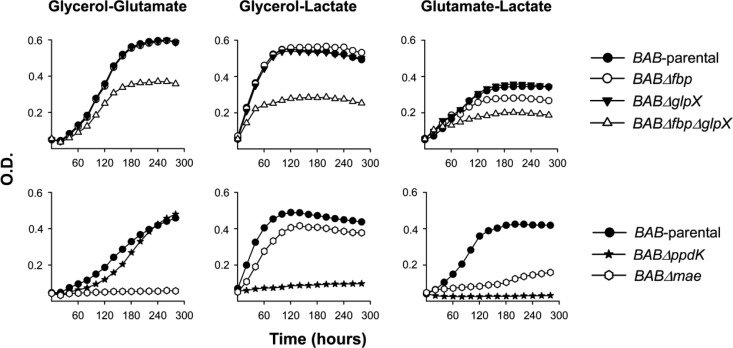
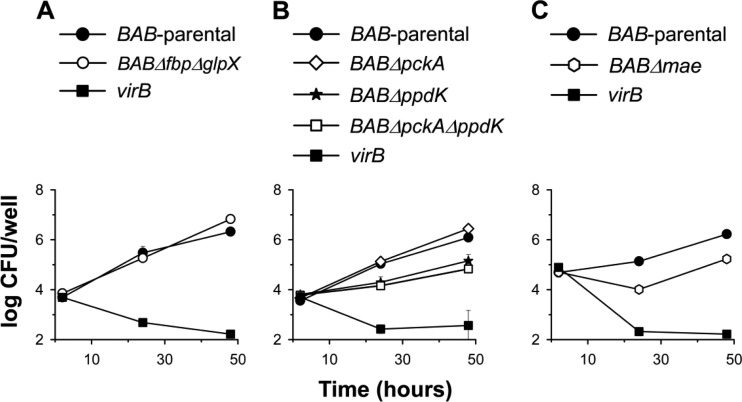
We constructed mutants carrying in-frame deletions in the putative fbp (BABΔfbp mutant) and glpX (BABΔglpX) as well as in both genes (BABΔfbpΔglpX) and tested their growth in a complex (peptone-yeast extract-glucose) medium and in the chemically defined medium of Gerhardt and Wilson (glutamate-lactate-glycerol). In the complex medium, the three mutants grew with generation times and final yields similar to those of B. abortus 2308 (BAB-parental) (Fig. 2A.1). BAB-parental showed reduced growth rates and final yields in glutamate-lactate-glycerol, as expected, and we obtained identical results with BABΔfbp and BABΔglpX (Fig. 2A.2). On the other hand, BABΔfbpΔglpX exhibited a markedly lower increase in turbidity in the minimal medium (Fig. 2A.2). Complementation with plasmid pAZI-21 carrying fbp or with pAZI-23 carrying glpX restored the ability to grow in glutamate-lactate-glycerol to levels close to that of the parental strain (see Fig. S3 in the supplemental material). These experiments strongly suggest that B. abortus Fbp and GlpX are functional, an interpretation reinforced by their reciprocal complementation (i.e., Fbp complemented BABΔglpX, and conversely, GlpX complemented BABΔfbp). We then tested combinations of two C sources (glycerol-glutamate, glycerol-lactate, and glutamate-lactate). Whereas BABΔglpX was not affected, BABΔfbp showed retarded growth only in the absence of glycerol (i.e., in glutamate-lactate) (Fig. 3, top). Since GlpX is the FBPase remaining in BABΔfbp and this mutant grew normally when glycerol was present, this result suggests that, as in E. coli (34), GlpX is related to glycerol metabolism. Finally, although retarded and diminished, the double BABΔfbpΔglpX mutant still showed significant growth (Fig. 3, top).
FIG 2.
Growth curves in peptone-yeast extract-glucose (A.1, B.1, and C.1) and glutamate-lactate-glycerol (A.2, B.2, and C.2) of BAB-parental and the mutants BABΔfbp, BABΔglpX, BABΔfbpΔglpX, BABΔpckA, BABΔppdK, BABΔpckAΔppdK and BABΔmae. Each point represents the mean ± standard error (error bars are within the size of the symbols) of optical density (O.D.) values for triplicate samples. The experiment was repeated three times with similar results.
FIG 3.
Growth curves in glycerol-glutamate, glycerol-lactate, and glutamate-lactate of BAB-parental and the mutants BABΔfbp, BABΔglpX, BABΔfbpΔglpX, BABΔppdK, and BABΔmae. Each point represents the mean for triplicate samples (error bars are within the size of the symbols). The experiment was repeated three times with similar results.
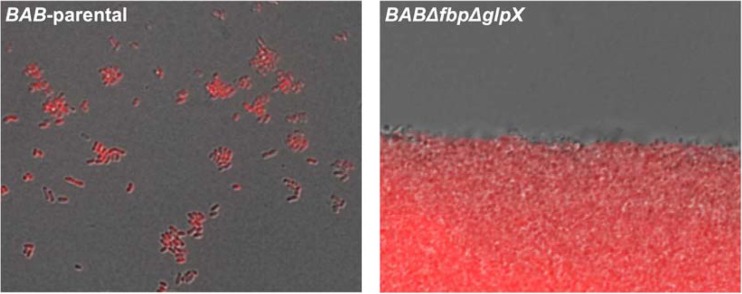
During these experiments we noticed that BABΔfbpΔglpX-inoculated broths did not produce homogeneous growth. This was clearly observed when the double mutant was grown in glutamate-lactate-glycerol in side-arm flasks instead of the automated Bioscreen system. Under these conditions, the bacteria formed macroscopic aggregates (Fig. 4), settling on the bottom of the flasks, which indicated a profound surface modification consistent with an altered biosynthesis of envelope molecules. In summary, although Fbp and GlpX deficiency did not abrogate bacterial multiplication, these enzymes were required not only for full growth but also for production of normal cells.
FIG 4.
Immunofluorescence analysis of BAB-parental and BABΔfbpΔglpX grown in glycerol-lactate-glutamate. Cells were labeled with anti-smooth-lipopolysaccharide mouse monoclonal antibody A76/12G12/F12 and Alexa Fluor 546-conjugated goat anti-mouse immunoglobulin.
Next, we investigated the pathways that supply pyruvate or PEP for gluconeogenesis. For this purpose, we carried out a genomic search for homologues of the genes encoding the enzymes connecting the TCA cycle and the triose-phosphate pathway in bacteria (Fig. 1). We identified homologues of pdh (pyruvate dehydrogenase), pyk (pyruvate kinase), pckA (phosphoenolpyruvate carboxykinase), ppdK (pyruvate-phosphate dikinase), pyc (pyruvate carboxylase), and mae (malic enzyme) but not of oad (oxaloacetete decarboxylase), pps (phosphoenolpyruvate synthase), or ppc (phosphoenolpyruvate carboxylase) (Fig. 1). Since Pyc, Pdh, and Pyk catalyze irreversible catabolic steps, we studied the pckA, ppdK, and mae homologues.
B. abortus 2308 BAB1_2091 is annotated as a pseudogene in some databases (http://www.genome.jp/kegg/) but not in others (http://biocyc.org.). This ORF encodes a protein of 491 amino acids that has 77% similarity to the PckA of the phylogenetically related Agrobacterium tumefaciens, a protein of 536 amino acids known to be functional (35). However, B. abortus BAB1_2091 is separated by a stop codon from an intergenic region that together with BAB1_2090 encodes the last 45 amino acids present in A. tumefaciens PckA. The frameshift that generates this stop codon is present also in all B. abortus strains sequenced so far (http://www.genome.jp/kegg/). Nevertheless, B. abortus PckA conserves the IGGTSYAGE-KKS domain (amino acids 190 to 202, where “-” indicates any amino acid) specifically required for the carboxykinase activity (36) as well as the phosphate binding site (G--G-GKT; amino acids 236 to 243) and ATP and metal binding sites, so that its functionality is not obviously compromised. In contrast, the putative B. abortus 2308 PpdK (encoded by BAB1_0525) represents a complete protein of 887 amino acids with the PEP-binding (amino acids 19 to 376) and the TIM barrel (amino acids 530 to 883) domains characteristic of PEP-utilizing enzymes.
To test whether these ORF-encoded enzymes are required for growth under gluconeogenic conditions, we constructed the nonpolar mutants BABΔpckA and BABΔppdK. Moreover, since both PckA and Ppdk catalyze reactions eventually leading to PEP (Fig. 1), we excluded their reciprocal complementation by constructing the double mutant BABΔpckAΔppdK. We then compared the growth of BAB-parental and the mutants in peptone-yeast extract-glucose and in glutamate-lactate-glycerol. Whereas we did not observe differences in the growth of BABΔpckA and BAB-parental in these two media, both BABΔppdK and BABΔpckAΔppdK had markedly reduced growth in glutamate-lactate-glycerol (Fig. 2B). Complementation with plasmid pAZI-19 carrying ppdK restored the phenotype (see Fig. S1 in the supplemental material), and the sibling revertant control (see Materials and Methods) conserved the wild-type phenotype (data not shown). These results strongly suggest that PpdK is functional and that, consistent with the frameshift in pckA, PckA does not synthesize PEP from oxaloacetate in B. abortus. The possibility that the role of B. abortus PckA is not detectable under these in vitro conditions seems less likely.
We followed a similar strategy to study the putative mae. BAB1_1036 encodes a protein of 774 amino acids annotated as a NADP (or NAD)-dependent enzyme involved in malate metabolism (http://www.genome.jp/kegg/; http://biocyc.org/). The N-terminal domain (amino acids 28 to 160) and the NADP (or NAD)-binding domain (amino acids 172 to 409) characteristic of the malic enzyme are conserved in the BAB1_1036 predicted protein. Mutant BABΔmae (with a deletion in the region encoding amino acids 26 to 673) displayed a small reduction in growth in peptone-yeast extract-glucose and a more marked one in glutamate-lactate-glycerol (Fig. 2C). The impairment was not as accentuated as that of BABΔppdK (Fig. 2, compare panels B and C). In these experiments, mae mutants with the above-described phenotype were consistently obtained, and the control sibling revertants with the wild-type phenotype were recovered after the last recombination event (see Materials and Methods; also, see Fig. S2 in the supplemental material). Taken together, these results support the hypothesis that B. abortus Mae supplies pyruvate for PpdK to produce PEP for gluconeogenesis, lactate being a complementary source of pyruvate in glutamate-lactate-glycerol (Fig. 1). We tested this hypothesis further by using media containing only two of the three C substrates of this chemically defined medium. BABΔmae did not grow in glycerol-glutamate and showed optimal growth in glycerol-lactate and reduced growth in glutamate-lactate (Fig. 3, bottom). These results are fully consistent with the predicted role of Mae (Fig. 1) and suggest that although B. abortus 2308 can use glycerol, glutamate, and lactate, provision of the latter cannot completely replace the Mae pathway. This interpretation is also supported by the fact that whereas PpdK dysfunction in BABΔppdK severely impaired growth in glycerol-lactate and glutamate-lactate, this mutant grew in glycerol-glutamate (Fig. 3, bottom), two substrates able to act as sources of PEP and pyruvate through the triose-P and Mae pathways, respectively (Fig. 1). Indeed, all these results are in agreement with the early studies that led to the formulation of the simple medium of Gerhardt and Wilson as well as with the nutritional studies that showed the preferential use of glutamate and the complementary role of glycerol and lactate in B. abortus (9). Also, the demonstration by Marr et al. (37) of the ability of B. abortus to generate pyruvate (and alanine) from glutamate supports an activity of Mae in this bacterium (Fig. 1).
Since the putative Mae was active in B. abortus 2308, we investigated whether malate replenishment could occur through the classical glyoxylate pathway (Fig. 1) or the PEP-glyoxylate cycle. The latter cycle combines the operation of PckA with the glyoxylate cycle enzymes and operates in E. coli under conditions of glucose limitation (38). In these pathways, isocitrate lyase (AceA) cleaves isocitrate to yield glyoxylate and succinate, and a malate synthase (AceB) condenses glyoxylate and acetyl coenzyme A (acetyl-CoA) to produce malate (39–41). The genome of B. abortus 2308 carries only one putative aceA (ORF BAB1_1631) and one putative aceB (ORF BAB1_1663). The predicted AceA is a protein of 429 amino acids with 61% identity and 76% similarity to E. coli AceA, and it conserves the amino acids required for the enzymatic activity and the assembly of the tetrameric enzyme (42–47). The predicted AceB has 728 amino acids with 59% identity and 74% similarity to E. coli malate synthase G, and it conserves the catalytic site and the amino acids interacting with acetyl-CoA (48, 49). Accordingly, we constructed a nonpolar BABΔaceA mutant with truncations of the 409 central amino acids. This mutant did not show growth differences relative to BAB-parental in peptone-yeast extract-glucose and glutamate-lactate-glycerol (data not shown), even though these media contain acetogenic substrates (glucose, glycerol, lactate, serine, threonine, and alanine) (50). Since the genomic analysis strongly suggests the presence of the glyoxylate cycle, we examined this point further by constructing a luciferase reporter under the control of the AceA promoter. Although growth curves were similar, luciferase activity was considerably higher in peptone-yeast extract-glucose than in glutamate-lactate-glycerol (Fig. 5), as would be expected if the glyoxylate cycle becomes active depending on the abundance of acetogenic substrates. In such a case, the lack of a change in phenotype in complex media could be explained if the glyoxylate cycle plays only a subsidiary role in this rich medium, and the experiments do not rule out the possibility that it becomes important under other nutritional conditions.
FIG 5.
Luciferase expression under the control of B. abortus aceA promoter in peptone-yeast extract-glucose and glutamate-lactate-glycerol. The results are representative of three experiments (RLU, relative luminescence units).
B. abortus strains with mutations in ppdK and mae but not in fbp, glpX, pckA, or aceA show lower multiplication rates in macrophages.
B. abortus is characteristically able to multiply intracellularly in professional phagocytes (51). We thus investigated the ability of the above-described mutants to multiply in macrophages using the parental strain and the attenuated virB mutant (unable to reach the replicating vacuole) as controls. Figure 6A shows that BABΔfbpΔglpX replicated like BAB-parental, even though these bacteria differed in growth under gluconeogenic conditions in vitro (see above). BABΔppdK (Fig. 6B) and BABΔmae (Fig. 6C) multiplied in macrophages, although at lower overall rates than BAB-parental both 24 h (P < 0,0001) and 48 h (P < 0,0001) after infection. On the other hand, mutation of pckA had no effect either by itself or combined with the ppdK deletion (Fig. 6B).
FIG 6.
Intracellular multiplication in RAW 264.7 macrophages of BAB-parental and the mutants BABΔfbpΔglpX, BABΔpckA, BABΔppdK, BABΔpckAΔppdK, and BABΔmae (the virB mutant is an attenuated mutant used as a control). Values are means ± standard errors for triplicate infections, and the results shown are representative of three independent experiments.
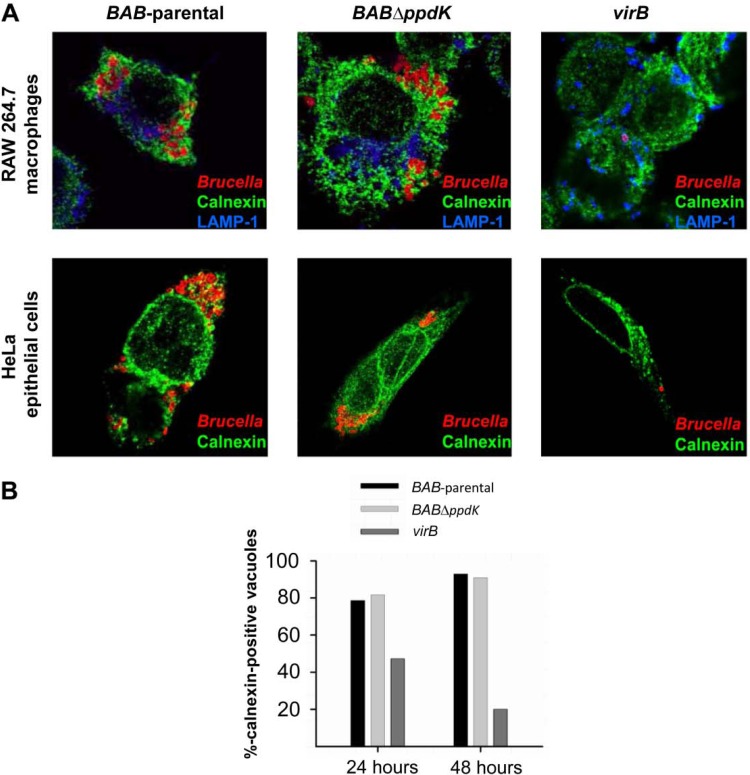
These results suggest that, albeit impaired in growth, BABΔppdK and BABΔmae are still able to reach the replicative intracellular niche. Since Mae and PpdK belong to the same pathway, we selected BABΔppdK (the mutant blocked in the upper step of the pathway; Fig. 1) to confirm that the metabolic dysfunction did not prevent these bacteria from reaching the endoplasmic reticulum-derived replicating niche. Figure 7A shows that, in contrast to the virB mutant, BABΔppdK and the parental bacteria were similar in intracellular distribution.
FIG 7.
Trafficking of BAB-parental, BABΔppdK, and the virB mutant in macrophages and HeLa cells. (A) Confocal images of infected RAW 264.7 macrophages and HeLa cells labeled with monoclonal antibodies to either calnexin (green) or LAMP-1 (blue) 24 h after infection (bacteria are immunostained in red). (B) Percentage of calnexin-positive vacuoles of HeLa cells that contain bacteria at 24 and 48 h postinfection.
B. abortus can also penetrate and multiply in epithelial cells (51). We found that the behavior of BABΔppdK in macrophages was reproduced in HeLa cells (Fig. 7A and B). In addition, we found that the numbers of CFU of the mutant and BAB-parental in HeLa cells did not differ 2 h after infection (data not shown), indicating that they were similar with regard to penetration.
B. abortus strains with mutations in ppdK and mae but not in pckA, fbp, glpX, or aceA are attenuated in mice.
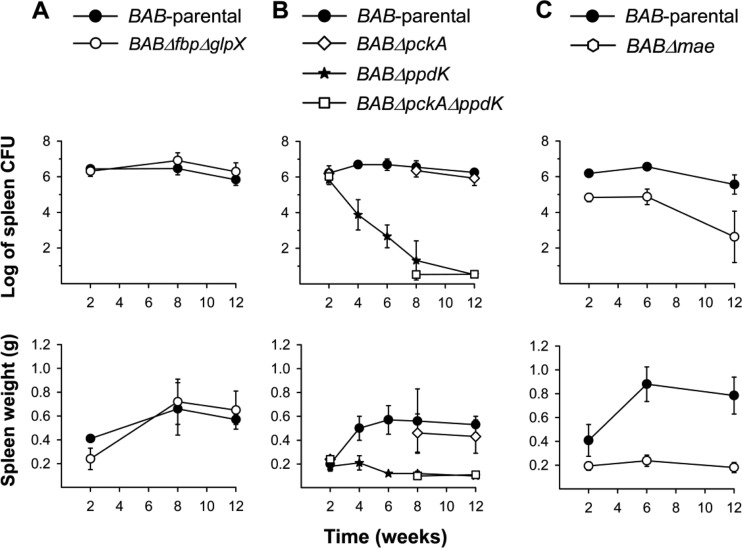
Virulent B. abortus is able to establish spleen infections in mice that characteristically develop in four phases: (i) onset phase (spleen colonization; first 48 h); (ii) acute phase (from the third day to weeks 2 to 4), when bacteria reach maximal numbers; (iii) chronic steady phase (weeks 2 to 4 to 12), where the bacterial numbers plateau; and (iv) chronic declining phase, during which brucellae are eliminated. The ability to induce a marked splenomegaly is also a characteristic of virulent brucellae (25). Using this model, we first studied the BABΔfbpΔglpX mutant and found that it did not differ from BAB-parental in either the CFU/spleen profile or the splenomegaly induced (Fig. 8A). Similarly, we did not observe attenuation for BABΔpckA (Fig. 8B), which is remarkable because pckA expression increases in B. abortus with mutations in BvrR/BvrS, a master regulator of B. abortus virulence (52). Nevertheless, this result is consistent with the genomic features of pckA and with the above-described experiments in vitro and in macrophages.
FIG 8.
Bacterial multiplication (top) and spleen weights (bottom) generated in BALB/c mice by BAB-parental and the mutants BABΔfbpΔglpX (A); BABΔpckA, BABΔppdK, and BABΔpckAΔppdK (B); and BABΔmae (C). Each point is the mean ± standard deviation (n = 5) of the log CFU or grams per spleen. Statistical differences relative to BAB-parental were significant (P < 0.001) from week 4 onwards for BABΔppdK and from week 2 onwards for BABΔmae. BABΔpckAΔppdK was also attenuated at weeks 8 and 12 (P < 0.001) (weeks 4 and 6 were not tested).
BABΔppdK failed to reach the chronic steady phase typical of virulent brucellae, yielding significantly lower CFU counts after week 2 (Fig. 8B). Clearly indicative of the strong attenuation of BABΔppdK, we did not recover any bacteria from the spleens of 3 out of the 5 mice examined at postinfection week 12, and this mutant induced less splenomegaly than the virulent bacteria (Fig. 8B). Consistent with the observations that showed no additive effect of the mutations of ppdK and pckA in macrophages or in vitro, the results for BABΔpckAΔppdK in mice paralleled those for BABΔppdK (Fig. 8B). BABΔmae produced a CFU/spleen profile that differed from those of both BAB-parental and BABΔppdK. Although not affected in the first 48 h (onset phase; not shown) this mutant showed a lower multiplication rate during the acute phase (Fig. 8C) that was reminiscent of the lower multiplication rates observed in macrophages. Strikingly, even though CFU/spleen numbers were lower than those of the wild-type strains, BABΔmae produced a chronic steady phase with reduced splenomegaly.
Finally, we tested BABΔaceA in mice. In a first experiment, the mutant did not show attenuation at weeks 2, 8, and 12 (data not shown). It has been reported that isocitrate lyase is essential for Salmonella persistence in mice during chronic infection but dispensable for acute lethal infection (53). Similarly, isocitrate lyase is dispensable in the acute phase of Mycobacterium tuberculosis infection in lung macrophages of mice but facilitates persistence during the chronic phase (54). Accordingly, we repeated the mouse infections and determined the number of BABΔaceA CFU in spleens 16 and 24 weeks later. However, we did not find any difference between this mutant and the parental strain (data not shown). Therefore, even though we did not rule out the possibility that the glyoxylate cycle plays a role under conditions different from those tested in vitro, we concluded that this shunt is not essential for B. abortus multiplication and persistence in the laboratory models used. Since the PEP-glyoxylate pathway relies on both AceA and PckA, this conclusion can be extended to this cycle.
DISCUSSION
B. abortus lacks the genes necessary for the metabolism of glycogen and poly-beta-hydroxyalkanoates, the two C reserve materials used by prokaryotes (55). Thus, these bacteria depend on nutrients provided by the host to multiply intracellularly, and accordingly, they need a supply of (at least) 6 C skeletons to carry out gluconeogenesis. To investigate these possibilities, we deleted ORFs that could encode enzymes of critical steps of gluconeogenesis, or of steps providing the necessary precursors. The genomic characteristics of the ORFs analyzed and the phenotypes observed in vitro support the hypothesis that they encode FBPases, a pyruvate phosphate dikinase, and a malic enzyme of B. abortus. Moreover, the analyses in cells show that the proteins coded for by ppdK and mae become necessary once the replicative niche is reached, as expected for metabolic mutants.
Since FBPases are essential for gluconeogenesis, the observation that growth in gluconeogenic media was not abolished in the Fbp-GlpX double mutant is intriguing, and several hypotheses can be considered to explain these results. The existence of a third FBPase is the first and most obvious possibility. So far, five different types of FBPases (I to V) in prokaryotes have been described (56). Whereas FBPases of classes IV and V are restricted to Archaea and their close hyperthermophilic Aquifex bacterial group, many bacteria have dual combinations of class I (Fbp homologues), class II (GlpX homologues), and class III FBPases (34, 56). However, a genomic search for Bacillus subtilis YydE homologues (the prototype of class III FBPases [57]) in the Brucellaceae revealed only an imperfect match (a hypothetical protein of 218 amino acids with a 32% identity with the 671 amino acids in YydE) in Ochrobactrum anthropi and none in Brucella. This is in agreement with the fact that no bacterial genome has been described as carrying a combination of classes I and III (34). Also, it has been reported that some E. coli strains carry two class II FBPases (GlpX and YggF [56]), but genomic analysis of all phosphatases in B. abortus failed to identify clear candidates for any phosphatase close to GlpX and Fbp (see Fig. S4 in the supplemental material). This genomic evidence and the Fbp-GlpX reciprocal complementation (i.e., the fact that only the double mutant shows a phenotype in vitro in the absence of glycerol) suggest that these are the main and possibly the only FBPases in B. abortus. An alternative to the third-FBPase hypothesis is the existence of an atypical gluconeogenesis less efficient than the classical one. Hypothetically, a fructose-6-P aldolase could take part in gluconeogenesis. This has been described for E. coli K-12, where ORF b0825 encodes an enzyme that catalyzes the reversible conversion of fructose-6-P to dihydroxyacetone and glyceraldehyde-3-P (58). This aldolase is different from that operating in the pentose-phosphate cycle, and its physiological role is uncertain. However, the only homolog in B. abortus (ORF BAB1_1813) is annotated as transaldolase, and in addition to the fact that it should represent the enzyme of the pentose-phosphate cycle, the identity (30%) is below the threshold considered to be significant (59). Obviously, a rigorous analysis of these two hypotheses requires enzymatic analysis of doubly deficient cells. Finally, the significance of the mucoid aggregates produced by the double FBPase mutant cannot be disregarded, since this phenotype suggests that the growth observed does not correspond to a natural condition.
Although it should be borne in mind that enzymatic analyses are necessary to reach definite conclusions, our results and those of previous works with B. abortus 2308 offer insight into some global models of the metabolism of these bacteria during intracellular life in the host.
A first model (the gluconeogenic model) can be proposed on the basis of the ability of B. abortus to grow in the defined medium of Gerhardt and Wilson and on proteomic studies in macrophages that suggest that B. abortus shifts to an amino acid-based metabolism in which the glutamate pool is increased (23). According to this model, molecules like glycerol, lactate, or pyruvate and amino acids channeled to oxaloacetate, keto-glutarate, or pyruvate are the main substrates, and molecules of 6 and 5 C are derived from the latter. In this regard, it is remarkable that dysfunction of two major FBPases did not bring about any perceptible attenuation either in cells or mice. Although the reduced growth of the BABΔfbpΔglpx mutant in vitro precludes a clear-cut conclusion, the contrasting in vivo and in vitro multiplication and the mucoid phenotype of the double mutant in the minimal medium are more consistent with models other than the gluconeogenic one. Moreover, two lines of evidence indicate that glucose (or closely related hexoses) is available in the host. First, a B. abortus 2308 (and B. suis 1330) GluP (glucose/galactose transporter) mutant has been identified as attenuated in signature-tagged experiments (15, 20, 20). Second, it was reported recently that the multiplication of B. abortus in alternatively activated macrophages increases when the intracellular glucose levels are artificially increased (60). Based on these observations, an almost opposite model proposes a main role for 6 C sugars in the replicative niche (and 5 C sugars if we assume that the evidence obtained in B. melitensis also applies to B. abortus; see the introduction). These sugars would provide triose-phosphate through the pentose phosphate cycle and also serve as precursors for biosynthesis of envelope polymers. This second model, however, does not account for the attenuation observed for the ppdK and mae mutants, which strongly suggests that molecules necessary for growth are derived from the TCA in vivo.
A third model proposes that there is a limited supply of 6 C (and 5 C) sugars that is compensated for by glutamate, alanine, and other amino acids. Those sugars would be used mostly or exclusively for biosynthesis of envelope polymers and for the pentose-phosphate cycle-dependent biosynthetic reactions. This model is consistent with the results of this and previous works. First, the different phenotype of the B. abortus double mutant BABΔfbpΔglpx in vivo and in vitro is more consistent with the hypothesis that classical gluconeogenesis is not extensively used in vivo. In addition, the B. abortus 2308 gluP mutant identified in signature-tagged mutagenesis studies is not clearly attenuated at 2 weeks postinfection and manifests its attenuation at a time (8 weeks) that corresponds to the chronic phase (20). This suggests that the bacteria do not depend totally on hexoses for intracellular biosynthetic processes and that this dependence is manifested at late times, perhaps as the result of changes in the replicative vacuole (see below). Indeed, the infection experiments performed in alternatively activated macrophages suggest that, although available, glucose is a limiting factor for B. abortus growth at least during the chronic phase (60). Proteomic studies carried out with B. abortus 2308 show that expression of two proteins of the dihydroxyacetone kinase complex (Dha) of the PEP-carbohydrate phosphotransferase system (PTS) is reduced throughout infection in macrophages (23). Although this has been interpreted to mean that reduced PTS expression may be the result of a short supply of sugars within the replicative niche, the Brucella PTS lacks the sugar permease unit and is likely to act as a regulatory system coordinating C and N metabolism (61). On the other hand, the signature-tagged mutagenesis and proteomic studies show attenuation of gltD (putatively encoding the small subunit of glutamate synthase) mutant (20) and an increment of enzymes involved in increasing the pool of glutamate (23) that is consistent with the model. Finally, this third model accounts for the attenuation of the ppdK and mae mutants and could also explain in part the differences between the mutants in these two genes that were observed in mice (Fig. 8). Since, according to the model, sugars are used mostly to construct envelope polymers and for pentose-phosphate cycle-derived precursors, additional molecules for biosynthesis must be derived from the TCA cycle. PpdK works to produce PEP, which is used to synthesize phenylalanine, tyrosine, tryptophan, glycerolipids, and other PEP-derived molecules. Mae supplies pyruvate for PpdK in vivo, but TCA would not be the only source of pyruvate. This is suggested by the fact that the mae mutant was both delayed in reaching the chronic phase of infection and present in lower numbers in the spleen during this phase, which contrasts with the inability of the ppdK mutant to generate chronic infections. Interestingly, it was recently shown that, when provided with multiple carbon sources, Mycobacterium tuberculosis differentially catabolizes each carbon source through the glycolytic, pentose phosphate, and/or TCA pathways to distinct metabolic fates, and it has been suggested that this ability reflects an adaptation to pathogenicity (62). Indeed, such ability could also be necessary for B. abortus to coordinately use the different substrates proposed for this model.
Although the last model fits the experimental evidence, it is obvious that it represents only a first approach to the situation in natural hosts, because we cannot assume that the niche is static during a chronic infection or uniform among different types of cells. In the above-mentioned signature-tagged mutagenesis studies, Hong et al. (20) presented evidence for the hypothesis that different sets of genes are required during the onset-acute phases and the chronic steady phase. This study identified three putative metabolic genes (gluP, glpD [see above], and gcvP) required during the chronic phase but not markedly during the acute phase of infection. Indeed, both the need for a functional ppdK for the infection to progress during the acute phase and the different phenotype of mae add further support to the hypothesis that different genes are required to different extents during the course of infection. Moreover, at least macrophages and trophoblastic cells have been clearly associated with B. abortus infection in cattle (63), and the physiological characteristics of these cells are different. Also, different spleen cells become colonized at different times after intraperitoneal inoculation of mice (64). Clearly, research is necessary to investigate these aspects of the relationship between metabolism and intracellular multiplication in B. abortus and in other species of the genus.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to E. Van Schaftingen for helpful discussions and suggestions.
This research was supported by grants from the Ministerio de Economía y Competitividad of Spain (AGL2011-30453–C04-00) and Fundación para la Investigación Médica Aplicada (FIMA), and, in part, by a grant from the Fond National de la Recherche Scientifique (FNRS) (convention FRFC 2452110, Belgium) and by the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office. A.Z.-R. was supported by a postdoctoral grant from FIMA, T.B. has a Ph.D. grant as Aspirant du FNRS, and L.P.-C. and R.C.-A. were funded in part by Departamento de Educación del Gobierno de Navarra (Programa ANABASID).
Footnotes
Published ahead of print 16 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01663-14.
REFERENCES
- 1.Barquero-Calvo E, Chaves-Olarte E, Weiss DS, Guzmán-Verri C, Chacón-Díaz C, Rucavado A, Moriyón I, Moreno E. 2007. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS One 2:e631. 10.1371/journal.pone.0000631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conde-Álvarez R, Arce-Gorvel V, Iriarte M, Mancek-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacón-Díaz C, Chaves-Olarte E, Martirosyan A, von Bargen K, Grilló MJ, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyón I, Gorvel JP. 2012. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog. 8:e1002675. 10.1371/journal.ppat.1002675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bolle X, Letesson JJ, Gorvel JP. 2012. Small GTPases and Brucella entry into the endoplasmic reticulum. Biochem. Soc. Trans. 40:1348–1352. 10.1042/BST20120156 [DOI] [PubMed] [Google Scholar]
- 4.Lacerda TL, Salcedo SP, Gorvel JP. 2013. Brucella T4SS: the VIP pass inside host cells. Curr. Opin. Microbiol. 16:45–51. 10.1016/j.mib.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 5.Pizarro-Cerdá J, Méresse S, Parton RG, Van der Goot G, Sola-Landa A, López-Goñi I, Moreno E, Gorvel JP. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. 2008. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic 9:678–694. 10.1111/j.1600-0854.2008.00718.x [DOI] [PubMed] [Google Scholar]
- 7.Barbier T, Nicolas C, Letesson JJ. 2011. Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett. 585:2929–2934. 10.1016/j.febslet.2011.08.011 [DOI] [PubMed] [Google Scholar]
- 8.Moreno E, Moriyón I. 2006. The genus Brucella, p 315–456 In Dworkin M, Falkow S, Rosenberg E, Scheleifer KH, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria, vol 5 Springer, New York, NY [Google Scholar]
- 9.Gerhardt P. 1958. The nutrition of brucellae. Bacteriol. Rev. 22:81–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson DC, McCullough WG. 1968. The glucose catabolism of the genus Brucella. I. Evaluation of pathways. Arch. Biochem. Biophys. 127:263–273. 10.1016/0003-9861(68)90225-7 [DOI] [PubMed] [Google Scholar]
- 11.Robertson DC, McCullough WG. 1968. The glucose catabolism of the genus Brucella. II. Cell-free studies with B. abortus (S-19). Arch. Biochem. Biophys. 127:445–456 [DOI] [PubMed] [Google Scholar]
- 12.Essenberg RC, Seshadri R, Nelson K, Paulsen I. 2002. Sugar metabolism by Brucellae. Vet. Microbiol. 90:249–261. 10.1016/S0378-1135(02)00212-2 [DOI] [PubMed] [Google Scholar]
- 13.Crasta OR, Folkerts O, Fei Z, Mane SP, Evans C, Martino-Catt S, Bricker B, Yu G, Du L, Sobral BW. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS One 3:e2193. 10.1371/journal.pone.0002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köhler S, Ouahrani-Bettache S, Layssac M, Teyssier J, Liautard JP. 1999. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect. Immun. 67:6695–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, Liautard JP. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U. S. A. 99:15711–15716. 10.1073/pnas.232454299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O'Callaghan D. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 68:1297–1303. 10.1128/IAI.68.3.1297-1303.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lestrate P, Delrue RM, Danese I, Didembourg C, Taminiau B, Mertens P, De Bolle X, Tibor A, Tang CM, Letesson JJ. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543–551. 10.1046/j.1365-2958.2000.02150.x [DOI] [PubMed] [Google Scholar]
- 18.Wu Q, Pei J, Turse C, Ficht TA. 2006. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 6:102. 10.1186/1471-2180-6-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essenberg RC, Candler C, Nida SK. 1997. Brucella abortus strain 2308 putative glucose and galactose transporter gene: cloning and characterization. Microbiology 143:1549–1555. 10.1099/00221287-143-5-1549 [DOI] [PubMed] [Google Scholar]
- 20.Hong PC, Tsolis RM, Ficht TA. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102–4107. 10.1128/IAI.68.7.4102-4107.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eskra L, Canavessi A, Carey M, Splitter G. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69:7736–7742. 10.1128/IAI.69.12.7736-7742.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Dahouk S, Jubier-Maurin V, Scholz HC, Tomaso H, Karges W, Neubauer H, Köhler S. 2008. Quantitative analysis of the intramacrophagic Brucella suis proteome reveals metabolic adaptation to late stage of cellular infection. Proteomics 8:3862–3870. 10.1002/pmic.200800026 [DOI] [PubMed] [Google Scholar]
- 23.Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, Michaud JF, Boucher L, Pedro I, Villeneuve A, Sitnikov D, Trudel K, Nassif N, Boudjelti D, Tomaki F, Chaves-Olarte E, Guzmán-Verri C, Brunet S, Cote-Martin A, Hunter J, Moreno E, Paramithiotis E. 2009. Intracellular adaptation of Brucella abortus. J. Proteome Res. 8:1594–1609. 10.1021/pr800978p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alton GG, Jones LM, Angus RD, Verger JM. 1988. Techniques for the brucellosis laboratory. INRA, Paris, France [Google Scholar]
- 25.Grilló MJ, Blasco JM, Gorvel JP, Moriyón I, Moreno E. 2012. What have we learned from brucellosis in the mouse model? Vet. Res. 43:29. 10.1186/1297-9716-43-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conde-Álvarez R, Grilló MJ, Salcedo SP, de Miguel MJ, Fugier E, Gorvel JP, Moriyón I, Iriarte M. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell Microbiol. 8:1322–1335. 10.1111/j.1462-5822.2006.00712.x [DOI] [PubMed] [Google Scholar]
- 27.Monreal D, Grilló MJ, González D, Marín CM, de Miguel MJ, López-Goñi I, Blasco JM, Cloeckaert A, Moriyón I. 2003. Characterization of Brucella abortus O-polysaccharide and core lipopolysaccharide mutants and demonstration that a complete core is required for rough vaccines to be efficient against Brucella abortus and Brucella ovis in the mouse model. Infect. Immun. 71:3261–3271. 10.1128/IAI.71.6.3261-3271.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palacios-Chaves L, Conde-Álvarez R, Gil-Ramírez Y, Zúñiga-Ripa A, Barquero-Calvo E, Chacón-Díaz C, Chaves-Olarte E, Vrce-Gorvel Gorvel JP, Moreno E, de Miguel MJ, Grilló MJ, Moriyón I, Iriarte M. 2011. Brucella abortus ornithine lipids are dispensable outer membrane components devoid of a marked pathogen-associated molecular pattern. PLoS One 6:e16030. 10.1371/journal.pone.0016030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerhardt P, Wilson JB. 1948. The nutrition of brucellae; growth in simple chemically defined media. J. Bacteriol. 56:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scupham AJ, Triplett EW. 1997. Isolation and characterization of the UDP-glucose 4′-epimerase-encoding gene, galE, from Brucella abortus 2308. Gene 202:53–59. 10.1016/S0378-1119(97)00453-8 [DOI] [PubMed] [Google Scholar]
- 31.Hallez R, Letesson JJ, Vandenhaute J, De Bolle X. 2007. Gateway-based destination vectors for functional analyses of bacterial ORFeomes: application to the Min system in Brucella abortus. Appl. Environ. Microbiol. 73:1375–1379. 10.1128/AEM.01873-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sieira R, Comerci DJ, Sanchez DO, Ugalde RA. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182:4849–4855. 10.1128/JB.182.17.4849-4855.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grilló MJ, Manterola L, de Miguel MJ, Muñoz PM, Blasco JM, Moriyón I, López-Goñi I. 2006. Increases of efficacy as vaccine against Brucella abortus infection in mice by simultaneous inoculation with avirulent smooth bvrS/bvrR and rough wbkA mutants. Vaccine 24:2910–2916. 10.1016/j.vaccine.2005.12.038 [DOI] [PubMed] [Google Scholar]
- 34.Donahue JL, Bownas JL, Niehaus WG, Larson TJ. 2000. Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli. J. Bacteriol. 182:5624–5627. 10.1128/JB.182.19.5624-5627.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu P, Wood D, Nester EW. 2005. Phosphoenolpyruvate carboxykinase is an acid-induced, chromosomally encoded virulence factor in Agrobacterium tumefaciens. J. Bacteriol. 187:6039–6045. 10.1128/JB.187.17.6039-6045.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osteras M, Driscoll BT, Finan TM. 1995. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J. Bacteriol. 177:1452–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marr AG, Olsen CB, Unger HS, Wilson JB. 1953. The oxidation of glutamic acid by Brucella abortus. J. Bacteriol. 66:606–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer E, Sauer U. 2003. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J. Biol. Chem. 278:46446–46451. 10.1074/jbc.M307968200 [DOI] [PubMed] [Google Scholar]
- 39.Ajl SL, Rust J, Jr, Wheat RW. 1956. Distribution of the tricarboxylic acid cycle enzymes in extracts of Escherichia coli. J. Cell Physiol. 47:317–339. 10.1002/jcp.1030470303 [DOI] [PubMed] [Google Scholar]
- 40.Smith RA, Gunsalus IC. 1954. Isocitratase: a nex tricarboxylic acid cleavage system. J. Am. Chem. Soc. 76:5002. 10.1021/ja01648a084 [DOI] [Google Scholar]
- 41.Kornberg HL, Krebs HA. 1957. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179:988–991. 10.1038/179988a0 [DOI] [PubMed] [Google Scholar]
- 42.Diehl P, McFadden BA. 1993. Site-directed mutagenesis of lysine 193 in Escherichia coli isocitrate lyase by use of unique restriction enzyme site elimination. J. Bacteriol. 175:2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diehl P, McFadden BA. 1994. The importance of four histidine residues in isocitrate lyase from Escherichia coli. J. Bacteriol. 176:927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehman A, McFadden BA. 1996. The consequences of replacing histidine 356 in isocitrate lyase from Escherichia coli. Arch. Biochem. Biophys. 336:309–315. 10.1006/abbi.1996.0562 [DOI] [PubMed] [Google Scholar]
- 45.Rehman A, McFadden BA. 1997. Serine319 and 321 are functional in isocitrate lyase from Escherichia coli. Curr. Microbiol. 34:205–211. 10.1007/s002849900169 [DOI] [PubMed] [Google Scholar]
- 46.Rehman A, McFadden BA. 1997. Lysine 194 is functional in isocitrate lyase from Escherichia coli. Curr. Microbiol. 35:14–17. 10.1007/s002849900203 [DOI] [PubMed] [Google Scholar]
- 47.Rehman A, McFadden BA. 1997. Cysteine 195 has a critical functional role in catalysis by isocitrate lyase from Escherichia coli. Curr. Microbiol. 35:267–269. 10.1007/s002849900251 [DOI] [PubMed] [Google Scholar]
- 48.Howard BR, Endrizzi JA, Remington SJ. 2000. Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 A resolution: mechanistic implications. Biochemistry 39:3156–3168. 10.1021/bi992519h [DOI] [PubMed] [Google Scholar]
- 49.Tugarinov V, Kay LE. 2005. Quantitative 13C and 2H NMR relaxation studies of the 723-residue enzyme malate synthase G reveal a dynamic binding interface. Biochemistry 44:15970–15977. 10.1021/bi0519809 [DOI] [PubMed] [Google Scholar]
- 50.Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12–50. 10.1128/MMBR.69.1.12-50.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno E, Gorvel JP. 2004. Invasion, intracellular trafficking and replication of Brucella organisms in professional and non-professional phagocytes, p 287 In López-Goñi I, Moriýon I. (ed), Brucella: molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom [Google Scholar]
- 52.Viadas C, Rodriguez MC, Sangari FJ, Gorvel JP, García-Lobo JM, López-Goñi I. 2010. Transcriptome analysis of the Brucella abortus BvrR/BvrS two-component regulatory system. PLoS One 5:e10216. 10.1371/journal.pone.0010216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang FC, Libby SJ, Castor ME, Fung AM. 2005. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 73:2547–2549. 10.1128/IAI.73.4.2547-2549.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKinney JD, Honer zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. 10.1038/35021074 [DOI] [PubMed] [Google Scholar]
- 55.Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, Garcia E. 2005. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect. Immun. 73:8353–8361. 10.1128/IAI.73.12.8353-8361.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown G, Singer A, Lunin VV, Proudfoot M, Skarina T, Flick R, Kochinyan S, Sanishvili R, Joachimiak A, Edwards AM, Savchenko A, Yakunin AF. 2009. Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli. J. Biol. Chem. 284:3784–3792. 10.1074/jbc.M808186200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujita Y, Yoshida K, Miwa Y, Yanai N, Nagakawa E, Kasahara Y. 1998. Identification and expression of the Bacillus subtilis fructose-1, 6-bisphosphatase gene (fbp). J. Bacteriol. 180:4309–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schurmann M, Sprenger GA. 2001. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 276:11055–11061. 10.1074/jbc.M008061200 [DOI] [PubMed] [Google Scholar]
- 59.Rost B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12:85–94. 10.1093/protein/12.2.85 [DOI] [PubMed] [Google Scholar]
- 60.Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TM, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Bäumler AJ, Santos RL, Tsolis RM. 2013. PPARγ-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 14:159–170. 10.1016/j.chom.2013.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dozot M, Poncet S, Nicolas C, Copin R, Bouraoui H, Maze A, Deutscher J, De Bolle X, Letesson JJ. 2010. Functional characterization of the incomplete phosphotransferase system (PTS) of the intracellular pathogen Brucella melitensis. PLoS One 5:e12679. 10.1371/journal.pone.0012679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. 2010. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17:1122–1131. 10.1016/j.chembiol.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 63.Enright FM. 1990. The pathogenesis and pathobiology of Brucella infection in domestic animals, p 301–320 In Nielsen KDJ. (ed), Animal brucellosis. CRC Press, Boca Raton, FL [Google Scholar]
- 64.Copin R, Vitry MA, Hanot Mambres D, Machelart A, DeTrez C, Vanderwinden JM, Magez S, Akira S, Ryffel B, Carlier Y, Letesson JJ, Muraille E. 2012. In situ microscopy analysis reveals local innate immune response developed around Brucella infected cells in resistant and susceptible mice. PLoS Pathog. 8:e1002575. 10.1371/journal.ppat.1002575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.