Abstract
Neisseria gonorrhoeae uses a type IV secretion system (T4SS) to secrete chromosomal DNA into the medium, and this DNA is effective in transforming other gonococci via natural transformation. In addition, the T4SS is important in the initial stages of biofilm development and mediates intracellular iron uptake in the absence of TonB. To better understand the mechanism of type IV secretion in N. gonorrhoeae, we examined the expression levels and localization of two predicted T4SS outer membrane proteins, TraK and TraB, in the wild-type strain as well as in overexpression strains and in a strain lacking all of the T4SS proteins. Despite very low sequence similarity to known homologues, TraB (VirB10 homolog) and TraK (VirB9 homolog) localized similarly to related proteins in other systems. Additionally, we found that TraV (a VirB7 homolog) interacts with TraK, as in other T4SSs. However, unlike in other systems, neither TraK nor TraB required the presence of other T4SS components for proper localization. Unlike other gonococcal T4SS proteins we have investigated, protein levels of the outer membrane proteins TraK and TraB were extremely low in wild-type cells and were undetectable by Western blotting unless overexpressed or tagged with a FLAG3 triple-epitope tag. Localization of TraK-FLAG3 in otherwise wild-type cells using immunogold electron microscopy of thin sections revealed a single gold particle on some cells. These results suggest that the gonococcal T4SS may be present in single copy per cell and that small amounts of T4SS proteins TraK and TraB are sufficient for DNA secretion.
INTRODUCTION
Neisseria gonorrhoeae is the causative agent of the sexually transmitted disease gonorrhea and is a strict pathogen of humans. Most strains of N. gonorrhoeae encode a type IV secretion system (T4SS) that secretes chromosomal DNA into the extracellular environment (1–3). N. gonorrhoeae is naturally transformable, and the DNA secreted by the T4SS can be taken up by other members of the population and incorporated into the chromosome by homologous recombination, thereby providing a mechanism of DNA donation independent of cell death and autolysis (4). The secreted DNA also acts in the initiation of biofilm development (5). The genes for the gonococcal T4SS are encoded on the gonococcal genetic island (GGI), a 57-kb genetic island that was likely horizontally acquired and is inserted in the chromosome near the replication terminus (1, 2, 6). T4SSs are a diverse family of transmembrane complexes that include both conjugation systems (such as F plasmid and the IncN plasmid pKM101) and effector translocator systems that deliver proteins or DNA-protein complexes directly into host cells (such as the well-studied VirB/D T4SS in Agrobacterium tumefaciens) (7–10). The fact that the gonococcal T4SS secretes DNA into the extracellular environment and not directly into a recipient or host cell distinguishes it from the other characterized T4SSs. The Ptl system in Bordetella pertussis is the only other example of contact-independent type IV secretion—in this case, of pertussis toxin (11).
Recent structural studies of the T4SS encoded by the IncN conjugative plasmid pKM101 and the IncW conjugative plasmid R388 have greatly increased our understanding of the structural biology of T4SSs (12–15). The 1.1-MDa core complex encoded by pKM101 is composed of 14 copies each of TraON, TraNN, and TraFN (the homologs of gonococcal TraK, TraV, and TraB, respectively), and a two-helix bundle in TraFN forms the outer membrane pore (12). The T4SS genes contained in the GGI exhibit limited sequence similarity to characterized homologs, although the gene organization is most similar to that of F plasmid (2, 16). Very little is known about the assembly, structure, expression, and localization of the T4SS in gonococci, and in this study, we focus on three structural proteins predicted to localize to the outer membrane of the gonococcal T4SS apparatus: TraK, TraV, and TraB. All three proteins are required for type IV secretion in gonococci (17). These proteins are of particular interest given that gonococcal DNA secretion does not require contact with a recipient cell, and it is possible that outer membrane proteins in the gonococcal T4SS may perform different functions or be subject to different selective pressures in gonococci compared with other characterized T4SSs. TraK, TraV, and TraB are also of interest given that they are highly conserved among sequenced gonococcal isolates (see Fig. S1 to S3 in the supplemental material). There are very few examples of conserved surface proteins in Neisseria, and many molecules in the gonococcal outer membrane undergo high-frequency phase and antigenic variation, including lipooligosaccharide, the type IV pilin subunit, and the opacity proteins (18, 19).
The present study addresses fundamental questions regarding the expression and localization of the gonococcal T4SS. We show that TraK localizes to the outer membrane in gonococci and interacts with the predicted lipoprotein TraV, as is the case in other T4SSs. An epitope-tagged variant of TraB (TraB-FLAG) also localizes to the outer membrane. Unexpectedly, TraK and TraB-FLAG were not detectable by Western blotting in wild-type (WT) cells during log-phase growth, despite the fact that gonococci secrete DNA throughout their growth cycle (20). A variant of TraK containing a triple C-terminal FLAG epitope, TraK-FLAG3, was detectable when expressed at WT levels, presumably due to signal amplification from the tandem epitope tag. In immunogold electron microscopy (EM) studies, overexpressed TraK-FLAG3 localized throughout the cell envelope on all cells, but expression from the native promoter resulted in low levels of TraK-FLAG3, with only some cells exhibiting a single gold particle in the outer membrane. We used flow cytometry to investigate whether TraK-FLAG3 might be differentially expressed at higher levels in a subset of the population, which could explain the apparently low expression levels in immunoblots. We found that a population of cells expressing traK-FLAG3 from the native locus was indistinguishable from the negative control, but increasing TraK-FLAG3 levels by 4-fold using an inducible promoter was sufficient to detect an increase in expression. Since we did not observe any subpopulations in the native expression strain producing TraK-FLAG3 at higher levels than in the negative control, it is unlikely that subpopulations exist with more than a 4-fold increase in TraK-FLAG3 expression. Collectively, these studies suggest that, despite low sequence similarity of the structural proteins to characterized homologs, the outer membrane pore of the gonococcal T4SS is similar to that in other systems. Since N. gonorrhoeae is only found within human hosts, the low-level expression of potentially surface-exposed components of the secretion apparatus is particularly interesting and may have important implications for the regulation and basic biology of this conserved transmembrane apparatus.
MATERIALS AND METHODS
Bacteria and growth conditions.
The bacterial strains used in this study are described in Table 1. Escherichia coli strains were grown on Luria-Bertani (LB) agar plates or in LB broth at 37°C (21). Gonococcal strains were grown on gonococcal base (GCB) agar plates (Difco) containing Kellogg's supplements (22) or in GCB liquid medium (GCBL) containing Kellogg's supplements and 0.042% NaHCO3 (cGCBL) (23). Erythromycin was used at 10 μg/ml for gonococci and 500 μg/ml for E. coli. Chloramphenicol was used at 10 μg/ml for gonococci and 25 μg/ml for E. coli. Ampicillin was used at 100 μg/ml for E. coli. Gene expression in gonococci was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). N. gonorrhoeae cultures were grown for 3 h (into the mid-late-logarithmic phase) before being harvested for analysis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Properties | Reference or source |
|---|---|---|
| Plasmids | ||
| pBT | Bacterial 2-hybrid bait plasmid for making C-terminal lambda phage cI fusions | Stratagene |
| pBT-traD | Bait plasmid encoding λcI-TraD (includes 450 bp downstream of traD); PCR of traD with BamHI-traD-F and XhoI-traD-R into pBT BamHI and XhoI sites | This work |
| pCK3 | Digestion of pMR12 with XmaI and ClaI, blunted and self-ligated (encodes MBP-TraKL44-I113, G231-G244) | This work |
| pCK4 | Digestion of pMR12 with SacI and SmaI, blunted and self-ligated (encodes MBP-TraKG231-G244) | This work |
| pCK5 | Digestion of pMR12 with SpeI and ClaI, blunted and self-ligated (encodes MBP-TraKD114-G244) | This work |
| pIDN3 | Cloning vector | 70 |
| pKH37 | lctP-aspC complementation construct | 71 |
| pKH86 | Bait plasmid encoding λcI-TraVS20-R193; PCR of traV with traV-EcoRI-F and traV-XhoI-R into pBT EcoRI and XhoI sites | This work |
| pKH87 | Target plasmid encoding RNAPα-TraKR53-G244; PCR of traK with traK-EcoRI-F and traK-XhoI-R into pTRG EcoRI and XhoI sites | This work |
| pKH103 | Bait plasmid encoding λcI-TraD (includes 86 bp downstream of traD); PCR of pBT-traD with traDdel-XhoI and traD-XhoI2, digested with XhoI and self-ligated | This work |
| pKH170 | Target plasmid encoding RNAPα-TraVS20-R193; traV on NotI-XhoI fragment of pKH86 into pKH87 digested with NotI and XhoI | This work |
| pKLD116 | Cloning vector for constructing His6-MBP fusions | 26 |
| pMR10 | PCR of MS11 chromosome with traK-RBS-EcoRI and traK-R-STOP into pIDN3 EcoRI and XhoI sites (contains full-length traK) | This work |
| pMR12 | PCR of MS11 chromosome with TraK-SspI-F and TraK-NheI-R into pKLD116 SspI and NheI sites (encodes MBP-TraKL44-G244 for protein purification) | This work |
| pMR21 | PCR of MS11 chromosome with delTraKF-EcoRV and TraKstartR-SalI into pIDN3 EcoRV and SalI sites (505 bp 5′ traK flanking DNA) | This work |
| pMR22 | PCR from MS11 chromosomal DNA with TraKendF-SalI and delTraKR-XhoI into pMR22 SalI-XhoI sites (690 bp 3′ traK flanking DNA, including last 15 bp of traK) | This work |
| pMR23 | PCR of MS11 chromosome with TraKendF-SalI and delTraKR-XhoI into pMR21 SalI and XhoI sites (traK deletion construct, which contains the last 15 bp of traK) | This work |
| pMR26 | PstI-XhoI fragment of pMR10 into PstI and XhoI sites of pKH37(traK overexpression construct) | This work |
| pMR73 | PCR from MS11 chromosome with TraBF-SalI and TraBR-XhoI into pIDN3 SalI and XhoI (contains full-length traB with 100 bp 3′ flanking sequence) | This work |
| pMR74 | PCR around pMR73 with TraB-FLAGCter-F and TraB-FLAGCter-R, digestion with BsaI, and self-ligation (contains traB-FLAG) | This work |
| pMR78 | XhoI-SpeI fragment of pMR74 into XhoI and SpeI sites of pKH37 (traB-FLAG overexpression construct) | This work |
| pMR81 | PCR of pCK5 with pCK5F-SpeI and pCK5R100 into pKLD116 SpeI and HindIII sites (encodes MBP-TraKD114-I146) | This work |
| pMR82 | PCR of pCK5 with pCK5F-SpeI and pCK5R200 into pKLD116 SpeI and HindIII sites (encodes MBP-TraKD114-D190) | This work |
| pMR83 | PCR of pCK5 with pCK5F-SpeI and pCK5R-end into pKLD116 SpeI-HindIII (encodes MBP-TraKD114-G244) | This work |
| pMR84 | PCR of pMR83 with pMR83F1-SpeI and pCK5R-end into pKLD116 SpeI and HindIII sites (encodes MBP-TraKS152-G244) | This work |
| pMR85 | PCR of pMR83 with pMR83F2-SpeI and pCK5R-end into pKLD116 SpeI and HindIII sites (encodes MBP-TraKS185-G244) | This work |
| pMR92 | Digestion of pMR84 with StuI and NheI, blunted, and self-ligated (encodes MBP-TraKS152-G193) | This work |
| pMR100 | PCR of linker-FLAG3 amplicon with FLAG3-EcoRI-F and FLAG3-HindIII-R into pIDN3 EcoRI and HindIII (plasmid for constructing C-terminal linker-FLAG3 fusions) | This work |
| pMR101 | PCR of traK from MS11 chromosomal DNA with traK-RBS-SacII and traK-EcoRI-R2 into the EcoRI and SacII sites of pMR100 (contains traK-FLAG3) | This work |
| pMR102 | SacII-XhoI fragment of pMR101 into the SacII and XhoI sites of pKH37 (IPTG-inducible traK-FLAG3 overexpression construct) | This work |
| pMR104 | PCR of traK from pMR101 with traK-RBS-SacII and traK-FLAG3-SalI-R into SacII and AccI sites of pMR22 (traK-FLAG3 with 3′ flanking DNA) | This work |
| pMR120 | traK-FLAG3 on SacII-SalI fragment of pMR101 into SacII and SalI sites of pMR68 (ATc-inducible traK-FLAG3 overexpression construct) | This work |
| pTRG | Bacterial 2-hybrid target plasmid for making C-terminal RNAPα subunit fusions | Stratagene |
| N. gonorrhoeae strains | ||
| MR535 | MS11 transformed with pMR23 (ΔtraK) | This work |
| MR537 | MS11 transformed with pMR26 (IPTG-inducible traK at lctP-aspC site) | This work |
| MR538 | ND500 transformed with pMR26 (IPTG-inducible traK at lctP-aspC site) | This work |
| MR565 | MS11 transformed with pMR78 (traB-FLAG at lctP-aspC site) | This work |
| MR600 | MS11 transformed with pMR102 (IPTG-inducible traK-FLAG3 at lctP-aspC site) | This work |
| MR602.1a | MS11 transformed with pMR104 (traK-FLAG3 at native locus) | This work |
| MR645 | ND500 transformed with pMR78 (ΔGGI, traB-FLAG at lctP-aspC site) | This work |
| MR646 | MS11 transformed with pMR74 (traB-FLAG at native locus) | This work |
| MR673 | MR602.1 transformed with pKH37 (traK-FLAG3 at native locus; Cmr) | This work |
| MR675 | MS11 transformed with pMR120 (ATc-inducible traK-FLAG3 at iga-trpB site) | This work |
| MS11 | WT N. gonorrhoeae | 72 |
| ND500 | MS11 ΔGGI | 2 |
For clarity, MR602.1 will be referred to as MR602 in this study.
DNA techniques.
The plasmids used in this study are described in Table 1, and the primer sequences are provided in Table 2. The QiaPrep miniprep kit (Qiagen) was used to isolate plasmid DNA. The QiaQuick gel extraction kit (Qiagen) was used to gel purify digested DNA. T4 DNA polymerase (New England BioLabs) was used to generate blunt DNA ends. Following ligation with T4 DNA ligase (New England BioLabs), DNA was transformed into chemically competent RapidTrans TAM1 E. coli cells (Active Motif). Transformants were screened for the expected plasmid construct by analysis of whole-cell lysates using the lysis solution of Kado and Liu (24). Additional screening was performed using restriction enzyme digestion and PCR.
TABLE 2.
PCR primers used in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| BamHI-traD-F | CGCGGGATCCATGAGTGCCCACTTCCCTGAAA |
| deltraKF-EcoRV | GGTGGTGATATCCGGCGGTCAATGACTGTATC |
| deltraK-XhoI-R | GGTGGTCTCGAGCGAAACTTGCCTTGCGAGAC |
| FLAG3-EcoRI-F | GGTGGTGAATTCGGTTCCGCTGGCTCCGCTGCTGG |
| FLAG3-HindIII-R | GGTGGTAAGCTTGTCCATGAAACCAACATAAGCAACTGG |
| pCK5F-SpeI | GGTGGTACTAGTGATGTGGCTGGAGCTAATCG |
| pCK5R100 | GGTGGTAAGCTTTCAGATGGCCCGTACATATTCTG |
| pCK5R200 | GGTGGTAAGCTTTCAATCAGCGGCGGTGTAACTTC |
| pCK5R-end | GGTGGTAAGCTTCCATGGCTAGCTCTCAAC |
| pMR83F1-SpeI | GGTGGTACTAGTATGATGCGTGGCACTACCG |
| pMR83F2-SpeI | GGTGGTACTAGTTACACCGCCGCTGATATGC |
| TraB-FLAGCter-F | GCCGGGTCTCCGACGACGACAAGTAGGATCCATCTATGAAAAAG |
| TraB-FLAGCter-R | GCCGGGTCTCCCGTCGTCCTTGTAGTCTTTGGTTTCGCCGGTATCAAT |
| TraB-SalI-F | GGTGGTGTCGACTATTATCAGCCGTCCGGGAG |
| TraB-XhoI-R | GGTGGTCTCGAGGTGCAGAGTGCTTCCAACAG |
| traDdel-XhoI | GCTCGAGGGCATTAATCGTGGGTATCTC |
| traD-XhoI2 | GCTCGAGGCACAATTAAGAATAAGGTATAAGCAGCT |
| traK-EcoRI-F | GCGAATTCTTGAGCAGAATCGCCATTGA |
| traK-EcoRI-R2 | GGTGGTGAATTCACCTCCCGGACGGCTGATAATAA |
| traK-endF-SalI | GGTGGTGTCGACCGTCCGGGAGGTTGAGATGAG |
| traK-FLAG3-SalI-R | AAGTAAGTTCTTGTCGACTTATTTATC |
| traK-NheI-R | GGTGGTGCTAGCTCTCAACCTCCCGGACGGCT |
| traK-R-STOP | GGTGGTCTCGAGTCAACCTCCCGGACGGCTGAT |
| traK-RBS-EcoRI | GGTGGTGAATTCCGAAGTTGAAACCGAGGAT |
| traK-RBS-SacII | GGTGGTCCGCGGCGAAGTTGAAACCGAGGAT |
| traK-SspI-F | GGTGGTAATATTAAGCAGAATCGCCATTGAAGG |
| traKstartR-SalI | GGTGGTGTCGACATCAATTACCTCGATTACCGCC |
| traK-XhoI-R | CAGCTCGAGAAATTGCACTGCCCACAATC |
| traV-EcoRI-F | CGGAATTCAACCTTAACCATGTCCGGTATC |
| traV-XhoI-R | GCTCGAGCTATCGAACGGTACCGGGAATAC |
| XhoI-traD-R | GCGCCTCGAGATTGCAAAGATTACTGAGATACCCACG |
Restriction sites are underlined.
Transformation of gonococci.
Gonococci were transformed with plasmid DNA using spot transformations (25). Plasmids smaller than 8,000 bp were linearized before transformation. Between 1 and 10 μg plasmid DNA in water was spotted onto a GCB plate and allowed to dry. Several piliated colonies of the appropriate gonococcal strain were streaked over the spot, and the plate was incubated overnight at 37°C with 5% CO2. A Dacron swab was used to transfer colonies from the spots onto plates containing 10 μg/ml chloramphenicol or erythromycin to select for transformants. Alternatively, in the absence of antibiotic selection, colonies were swabbed from the spots into GCBL, serially diluted, and plated on GCB agar. Individual transformants were then restreaked and screened by PCR.
Protein purification.
TraK was purified without its signal sequence (amino acids 44 to 244) as a C-terminal fusion to maltose binding protein (MBP) using pKLD116 (26). Plasmid pMR12 (encoding the malE-′traK fusion) was transformed into E. coli BL21 for expression and purification. Overnight cultures of E. coli BL21 containing pMR12 were used to inoculate 4 liters of LB broth containing 100 μg/ml ampicillin. Cultures were grown at 37°C with shaking to an optical density at 600 nm (OD600) of 0.5. The temperature was dropped to 21°C, 0.3 mM IPTG was added, and the cultures were grown overnight with shaking. The cells were harvested by centrifugation at 3,000 × g for 10 min and resuspended in 3 ml of amylose column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 10% glycerol) per 1 g (wet weight) of cells. Cells were lysed by three passages through a French press cell at 1,200 lb/in2, and the lysate was centrifuged at 20,200 × g for 30 min to remove unlysed cells. The supernatant was incubated with 1 ml amylose resin (New England BioLabs) at 4°C with mixing for 2 h and then loaded onto a gravity column. After the column had been washed with amylose column buffer, 1.5-ml fractions were eluted with 0.001, 0.01, 0.1, 1, and 10 mM maltose. The fractions were pooled and dialyzed overnight against nickel column buffer (20 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10% glycerol). Following incubation with 1 ml nickel resin (Sigma) at 4°C for 2 h with gentle shaking, the mixture was loaded onto a gravity column and washed with nickel column buffer containing 10 mM imidazole, and fractions were eluted with 30, 60, 100, and 150 mM imidazole. The purest fractions were pooled and dialyzed twice against storage buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% glycerol). The final preparation was 1.2 mg/ml and estimated to be 95% pure by densitometry analysis of a Coomassie-stained gel. MBP was purified in a similar manner from pKLD116 (26).
Monoclonal antibody production.
Purified MBP-TraK and MBP were sent to GenScript for monoclonal antibody production. Hybridoma lines were generated and screened by enzyme-linked immunosorbent assay (ELISA) to identify antibodies that reacted with MBP-TraK but not with the MBP tag purified from the empty vector. Three hybridoma lines were identified in this manner and named 2E5, 3H4, and 3B8. The hybridoma line 2E5 was selected for protein A/G affinity purification.
We sought to determine the epitopes for the TraK monoclonal antibodies by constructing a set of plasmids expressing different lengths of the traK gene fused to the 3′ end of the malE gene and assaying for antibody recognition by Western blotting of E. coli lysates. Coomassie staining of SDS-polyacrylamide gels confirmed that each MBP-TraK fusion was produced. The results of these experiments are summarized in Fig. S4 in the supplemental material. The epitope for the 3B8 antibody is located near the N terminus of TraK between amino acids Leu44 and Ile113 (pCK3 in Fig. S4). The epitopes for the 2E5 and 3H4 antibodies are located closer to the C terminus of TraK between amino acids Ser152 and Gly244 (pMR84 in Fig. S4). We expressed smaller portions of this region (in pMR85 or pMR92) and also tested a 30-amino-acid peptide spanning these regions (Leu179 to Ile208), but we did not observe recognition by the 2E5 or 3H4 antibodies in any of these cases (data not shown and Fig. S4).
These results suggest that the 2E5 and 3H4 antibodies recognize a similar epitope in a 92-amino-acid region near the C terminus of TraK. The epitope may be discontinuous given that neither antibody recognized a MBP-TraK fusion protein that contained smaller portions of this region. We were unable to use the 2E5 and 3H4 antibodies to detect TraK by immunofluorescence microscopy, suggesting that these antibodies may not recognize natively folded protein (data not shown). We were able to detect TraK in cell lysates of a traK overexpression strain using the 3B8 antibody by ELISA, suggesting that, in contrast to 2E5 and 3H4, the 3B8 antibody may recognize a native epitope (data not shown).
Western blots.
Neisseria strains were lysed by sonication (30 s of sonication at 35% amplitude with a 1-s on/off pulse using a Branson digital sonicator), SDS-loading dye was added, and the samples were boiled for approximately 5 min. If necessary, protein amounts were normalized using the Bradford assay prior to electrophoresis. Proteins were electrophoresed on either 10% or 12% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Bio-Rad) either for 1 h at 100 V or overnight at 30 V. Membranes were blocked with 5% milk in Tris-buffered saline containing 0.5% Tween 20 (TTBS) either for 1 h or overnight. Primary antibody in TTBS was added for 1 h, the membranes were washed with TTBS for 15 min, and then the secondary antibody in TTBS was added for an additional hour. Blots were then washed with TTBS for 30 min, developed using either the Immun-Star horseradish peroxidase (HRP) substrate kit (Bio-Rad) or the Immobilon Western chemiluminescent HRP substrate (Millipore), and exposed to film. For the primary antibodies, anti-chloramphenicol acetyltransferase (anti-CAT (Sigma) was used at 1:7000, anti-PilQ (H. S. Seifert) was used at 1:10,000, anti-SecY was used at 1:1000, anti-TraK was used between 1:500 and 1:1,000, and M2 anti-FLAG (Sigma) was used at 1:10,000. The anti-SecY antibody is a polyclonal antibody raised against the AKRQFNGRAGSQSTC peptide obtained from Genscript. SecY migrated with an apparent molecular mass of ∼30 kDa (27), and detection of SecY was used as an inner membrane fractionation control. The appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) were used at a 1:10,000 dilution.
Reverse transcriptase PCR.
Total RNA was isolated from log-phase gonococcal cultures that had been grown for 3 h. A 2-ml volume of vortexed culture was mixed with 2 ml −20°C methanol. The cells were harvested by centrifugation at 17,200 × g for 7 min at 4°C, and the pellet was frozen at −80°C for 20 min. The pellet was resuspended in 1 ml TRIzol, and 200 μl of chloroform was added. The tubes were mixed vigorously for 15 s, incubated for 3 min at room temperature, and centrifuged for 15 min at 12,000 × g at 4°C. The top aqueous phase was removed, 500 μl of isopropanol was added, and RNA was precipitated at room temperature for 10 min. To collect the RNA, the samples were centrifuged for 15 min at 12,000 × g at 4°C, washed once in 75% ethanol, and allowed to dry. After being resuspended in diethylpyrocarbonate (DEPC)-treated water, contaminating DNA was removed using the Turbo DNA-free kit (Applied Biosystems). The iScript cDNA synthesis kit (Bio-Rad) was used to reverse transcribe the RNA (350 ng RNA in a 40-μl reaction) in the presence or absence of reverse transcriptase (RT), and 1 μl of the resulting cDNA was used as a template for PCR. The traK gene was amplified using the primers traK-RT-F (GAAGCAGCAGTATTGGCTTCGCAA) and traK-RT-R (ATTGATGCCCATATCGCCGGTAGT). The rpoB gene was amplified using primers as previously described in reference 20.
Bacterial two-hybrid assay.
The BacterioMatchII two-hybrid system (Stratagene) was used following the manufacturer's instructions, with the following changes. BacterioMatchII E. coli cells were transformed with the appropriate target plasmid, and these cells (BacterioMatchII-pTarget) were made chemically competent. Briefly, E. coli cultures were inoculated from an overnight culture and grown to an OD600 of 0.5. The cells were centrifuged at 3,000 × g for 7 min, resuspended in 1/10 volume of cold TSS solution (10% polyethylene glycol 8 [PEG-8], 30 mM MgCl2, 5% dimethyl sulfoxide [DMSO] in LB), and frozen in aliquots at −80°C. For the two-hybrid assay, the BacterioMatchII-pTarget competent cells were thawed on ice, and 100-μl aliquots were added to prechilled round-bottom 14-ml tubes. The appropriate bait plasmid DNA (1.5 μg) was added to the cells, and the mixture was incubated on ice for 30 min. Following a 35-s heat shock at 42°C, the cells were incubated on ice for a further 2 min, and 900 μl warm LB was added. The tubes were incubated with shaking for 90 min at 37°C before being washed twice in M9+ His-dropout broth and allowed to grow in M9+ His-dropout broth for 2 h. A 200-μl volume was then plated on nonselective (minimal medium plates containing 25 μg/ml chloramphenicol and 12.5 μg/ml tetracycline) or selective plates (same as nonselective plates with the addition of 5 mM 3-amino-1,2,4-triazole [3-AT]) prepared according to the manufacturer's instructions. The plates were incubated overnight at 37°C and then for an additional 18 h at room temperature.
Subcellular fractionation.
To isolate the outer membrane fraction for TraK localization studies, gonococcal cells from 13 overnight GCB plates (containing 1 mM IPTG if appropriate) were swabbed into 4.5 ml cold phosphate-buffered saline (PBS), and the cells were harvested by centrifugation at 17,000 × g for 1 min. The cell pellet was resuspended in 3.75 ml cold lithium acetate buffer (0.2 M lithium chloride, 0.1 M lithium acetate, 10 mM EDTA [pH 6.0]) and rocked gently for 10 min. The cell suspension was then passed through a 22-gauge syringe approximately 16 times to induce membrane blebbing and centrifuged at 15,600 × g for 10 min at 4°C. The supernatant was removed and ultracentrifuged at 150,000 × g using a Beckman TLA110 rotor for 2 h at 4°C. To isolate the soluble and total membrane fractions, cells from one overnight GCB plate were used to inoculate 4-ml cultures in cGCBL at an OD540 of 0.25 and grown for 3 h into the log phase (with 1 mM IPTG added if necessary). The cells were harvested by centrifugation at 17,000 × g for 1 min, washed with cold PBS, and resuspended in 900 μl 0.01 M Tris-HCl (pH 7.0). The suspension was sonicated using a Branson digital sonicator at 40% amplitude for 20 s using a 1-s on/off pulse. Unlysed cells were removed by centrifugation at 12,000 × g for 10 min at 4°C, and the supernatant was removed for ultracentrifugation at 89,000 × g using a Beckman TLA120.2 rotor for 1.5 h. The supernatant was saved as the soluble fraction, and the pellet was washed and saved as the total membrane fraction.
The protocol described above was also used to fractionate TraB-FLAG strains, with the following changes. To isolate the outer membrane fraction, gonococcal cells were swabbed from 18 overnight GCB plates and harvested by centrifugation at 11,952 × g for 10 min at 4°C. Membrane blebbing was induced as above, and the cells were centrifuged again at 12,000 × g for 10 min at 4°C, filtered through a 0.45-μm-pore filter, and ultracentrifuged at 165,000 × g in a Beckman SW50.1 rotor for 3 h 45 min. To isolate total membrane and soluble fractions, harvested cells were resuspended in a 4-ml volume for sonication, as described above. Following the removal of unlysed cells, the supernatant was filtered through a 0.22-μm-pore filter and ultracentrifuged at 165,000 × g for 2 h 45 min. Following ultracentrifugation, the soluble fraction was concentrated approximately 2-fold using a centrifugal filter with a 10,000 molecular weight cutoff (MWCO) (Millipore).
This protocol was altered slightly to localize TraK-FLAG3. To isolate outer membranes, filtered supernatants were centrifuged at 150,000 × g in a TLA110 rotor for 2 h following treatment with lithium acetate buffer. To separate soluble and total membrane fractions, filtered lysates were centrifuged in a 1-ml volume in a TLA110 rotor for 1.5 h at 200,000 × g.
DNA secretion assay.
To monitor DNA secretion of N. gonorrhoeae, fluorescence-based secretion assays were performed essentially as described previously (28). Briefly, piliated colonies were selected and streaked out on GCB plates containing appropriate antibiotics. The next day, these cells were inoculated in prewarmed M199 medium (Life Technologies) containing Kellogg's supplement (22) and 0.042% NaHCO3 and grown for 2 h. After initial growth, cultures were diluted to an OD600 of ∼0.2. This growth/dilution cycle was repeated at least twice to reduce DNA background from lysed cells. Samples for zero hours after the end of log-phase growth (T0) were collected directly after the last dilution, and samples for 1 h after the end of log-phase growth (T1) samples were taken after another 2 h of growth. The amount of DNA was determined using PicoGreen (Molecular Probes/Invitrogen). PicoGreen was diluted 1:200 in 1× TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Directly before measurement, 100 μl of PicoGreen solution was mixed with 100 μl of supernatant from the cultures in a 96-well FluoroNunc plate (black, flat bottom). The excitation wavelength was set to 480 nm, and the emission was measured at 520 nm using the Tecan plate reader. Every measurement required a standard composed of random single-stranded DNA (ssDNA) at concentrations of 0, 0.025, 0.05, 0.1, 0.15, and 0.2 ng/μl. Strains ND500 (ΔGGI) and MS11 were included as negative and positive controls in all assays. Average secretion levels were obtained from at least three independent experiments performed, each with at least three independent cultures.
Immunofluorescence microscopy.
Cells were prepared for microscopy as previously described (29). A 1-ml volume of log-phase gonococcal cells was centrifuged at 17,000 × g for 1 min. The cells were washed once in PBS and resuspended in 4% paraformaldehyde in PBS for 15 min at room temperature followed by 15 min on ice. The cells were then washed three times with PBS before being resuspended in 0.5 ml GTE buffer (50 mM glucose, 1 mM EDTA, 20 mM Tris-HCl [pH 7.5]) at 4°C. A 100-μl volume of the cell suspension was added to the wells of an 8-well chamber slide (Lab-Tek), and the slides were incubated for 10 min at room temperature. Unattached cells were aspirated, and PBS-T (0.2% Triton X-100 in PBS) was added for 10 s. The PBS-T was removed, cold methanol was added to the samples, and the slide was incubated at −20°C for 10 min. The methanol was aspirated, and the slides were allowed to air dry and then incubated with PBS-T for 5 min. The slides were blocked with 5% skim milk powder in PBS-T for 1 h, followed by two washes with PBS for 10 s each. The M2 FLAG monoclonal antibody was diluted 1:150 in PBS-M (5% skim milk powder in PBS) and incubated on the slide for 1 h. The slides were washed twice with PBS for 10 s and once for 5 min. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse secondary antibody was diluted 1:100 in PBS-M containing 2 μg/ml DAPI (4′,6-diamidino-2-phenylindole) and incubated on the slides for 1 h. The slides were washed twice with PBS for 10 s and once for 5 min. The well supports were removed from the slides, and the slides were mounted and prepared for microscopy. A Zeiss Axio Imager 2 epifluorescence microscope was used to image the samples.
Preembedding immunogold EM.
Overnight plates of N. gonorrhoeae strains were used to inoculate two 3-ml cultures in cGCBL at an OD540 of 0.25 and allowed to grow for 3 h. IPTG (1 mM) was added if necessary. Cells were harvested by centrifugation at 2,900 × g for 5 min, washed once in cold PBS, and fixed in 4% formaldehyde in PBS for 15 min at room temperature followed by 15 min on ice. Cells were then washed three times with cold PBS, resuspended in cold GTE buffer for 15 min on ice, washed once with PBS-T, resuspended in cold methanol, and incubated at −20°C for 10 min. After two washes in PBS-T, the cells were incubated in PBS-T for 1 h with end-over-end mixing, washed twice in PBS, and resuspended in 60 μl PBS. All washes up until this point were performed by centrifuging the cells at 16,000 × g for 1 min, aspirating the supernatant, and resuspending the pellet in the next solution. The cell suspension was then embedded in agarose by mixing the suspension with an equal volume of 3% agarose in PBS at 55°C in an Eppendorf tube. The suspension was mixed several times and moved immediately to ice. The agarose plugs were removed from the tubes and saved overnight at 4°C submerged in PBS.
A microtome was used to cut 200-μm slices from the agarose plugs, and for all future steps, the agarose slices were incubated in a 500-μl volume of the appropriate reagent in a 24-well plate. Endogenous peroxidase activity was quenched by the addition of 3% H2O2 in PBS for 45 min at room temperature. After two washes in PBS, the agarose slices were blocked in PBS containing 0.05% Tween 20 and 5% normal goat serum (PBS-Tw-NGS) for 2 h at room temperature. The M2 anti-FLAG monoclonal antibody was diluted 1:400 in PBS-Tw-NGS and incubated with the agarose plugs overnight at 4°C. Cells were washed six times for 10 min with PBS containing 0.05% Tween 20 and three times for 10 min in PBS-T. The secondary antibody (biotinylated goat anti-mouse IgG) was diluted 1:200 in PBS-Tw-NGS and incubated with the cells overnight at 4°C. Additional washes were performed: six washes in PBS containing 0.05% Tween 20 and three washes in PBS.
Immunostaining was visualized using the Vectastain Elite ABC kit (Vector Laboratories) following the manufacturer's instructions. Diaminobenzidine (0.02% diaminobenzidine and 0.01% H2O2 in 0.05 M Tris-HCl buffer [pH 7.4]) was used as the final substrate, with an 8-min incubation time as described previously (30). Cells were washed extensively in 0.05 M Tris buffer and postfixed in 2.5% glutaraldehyde in PBS for 30 min. Cells were washed three times for 10 min with 0.2 M Tris-maleic acid buffer and rinsed twice for 5 min with water. Samples were resuspended in silver nitrate solution (185 mM hexamethylenetetramine, 12 mM silver nitrate, 5 mM sodium tetraborate in 0.1 M Tris-maleic acid buffer) for 10 min at 60°C in the dark. Cells were washed again in water, incubated in 0.05% gold chloride solution for 2 min at room temperature, and washed three times for 5 min in water. Following a 2-min incubation in 3% sodium thiosulfate, the cells were washed again with water and 0.01 M PBS. After silver enhancement, the agarose plugs were rinsed in 0.1 M phosphate buffer and postfixed in 1% osmium tetroxide in the same buffer for 1 h at room temperature. Following osmium postfixation, the samples were dehydrated in a graded ethanol series and then further dehydrated in propylene oxide and embedded in Epon epoxy resin. Ultrathin sections were cut with a Leica electron microscopy (EM) UC6 ultramicrotome and collected on 200-mesh copper grids. Ultrathin sections were observed with a Philips CM120 electron microscope, and images were captured with a MegaView III side-mounted digital camera.
Flow cytometry.
Cells were prepared for flow cytometry following essentially the same protocol used for immunofluorescence microscopy, with the following changes. Instead of 5% dry milk powder, 5% normal goat serum was used to block the cells. The M2 anti-FLAG antibody (Sigma) was used at a 1:750 dilution, and primary antibody staining was performed overnight at 4°C. An Alexa Fluor 647-conjugated secondary antibody (Invitrogen) was used at a 1:750 dilution. All steps were performed with the cells in solution, and cells were pelleted at 17,000 × g for 1 min between steps. Data were collected using a FACSCalibur flow cytometer.
RESULTS
TraK, TraV, and TraB are conserved between gonococcal strains but have limited similarity to homologs in other T4SSs.
Alignment of the TraK, TraV, and TraB amino acid sequences from eight sequenced gonococcal genomes containing the GGI demonstrated that these proteins are highly conserved between strains, with only a few variable amino acids across the collective sequences (see Fig. S1 to S3 in the supplemental material). However, sequence similarity between these proteins and their homologs in other T4SSs is very low. Gonococcal TraK is 23% identical to TraK from F plasmid (TraKF) and 22% identical to VirB9 from A. tumefaciens. Percent identity is similarly low for gonococcal TraV (23% identical to TraVF) and TraB (29% identical to TraBF and 18% identical to VirB10). The amino acid sequence of gonococcal TraV is almost four times longer than those of homologs in A. tumefaciens and pKM101, precluding similarity calculations (see Fig. S6 in the supplemental material). Alignment of the gonococcal T4SS proteins with homologs in A. tumefaciens, F plasmid, and pKM101 using AlignMe (for alignment of membrane proteins) revealed that despite the low sequence similarity, the hydrophobicity profiles of the amino acid sequences were often similar, especially between the gonococcal T4SS proteins and F plasmid homologs, suggesting that secondary structures may be conserved (see Fig. S5 to S7 in the supplemental material) (31).
TraK and TraB-FLAG are not detectable in WT cells, although traK is transcribed.
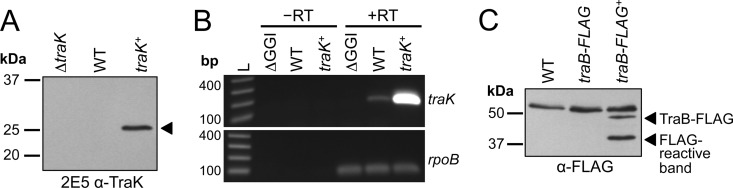
We sought to better characterize the predicted outer membrane proteins of the gonococcal T4SS and started by developing monoclonal antibodies against TraK. We were able to detect TraK (an ∼25-kDa band) in lysates of N. gonorrhoeae strain MR537, which inducibly overexpresses traK from a distant chromosomal locus (Fig. 1A). This band was not detected in the ΔtraK strain MR535 (Fig. 1A). Interestingly, we were unable to detect TraK in lysates from the WT strain MS11 (Fig. 1A). We were also unable to detect TraK in the WT strain when samples were taken over a time course at 2, 3, 4, or 6 h postinoculation, time points representing exponential-phase and early-stationary-phase growth (data not shown). We therefore investigated whether traK was being transcribed in WT cells by using two-step reverse transcription-PCR (RT-PCR). We were able to detect traK transcript in both the WT and traK+ strains but not in the GGI deletion strain ND500 (Fig. 1B). Amplification of the housekeeping gene rpoB was used to confirm that approximately equal amounts of RNA were used in each reaction, and reactions carried out in the absence of reverse transcriptase confirmed that there was no contaminating DNA (Fig. 1B). These results indicate that although traK is transcribed in WT cells, the TraK protein is present at low levels not detectable by Western blotting under standard laboratory growth conditions.
FIG 1.
Expression of TraK and TraB-FLAG in N. gonorrhoeae. (A) Western blot with the 2E5 TraK monoclonal antibody using lysates from gonococcal strains MR535 (ΔtraK), MS11 (WT), and MR537 (traK expressed from an IPTG-inducible promoter in the WT background [traK+]). This Western blot using the 2E5 anti-TraK antibody is representative of those obtained with the 3H4 and 3B8 anti-TraK monoclonal antibodies. The black triangle indicates TraK. Approximately 16.5 μg of protein from cell lysates was subjected to SDS-PAGE. (B) Detection of traK and rpoB mRNA by two-step RT-PCR. Total RNA was isolated from gonococcal strains ND500 (ΔGGI), MS11, and MR537. Reverse transcription reactions were carried out in the presence (+RT) or absence (−RT) of reverse transcriptase and amplified with primers to detect traK and rpoB. L, nucleotide ladder in base pairs. (C) Western blots using the anti-FLAG antibody (Sigma) to detect TraB-FLAG in gonococcal strains MS11 (WT), MR646 (traB-FLAG expressed from the native locus in the GGI [traB-FLAG]), and MR565 (traB-FLAG expressed from an IPTG-inducible promoter in the WT background, [traB-FLAG+]). Full-length TraB-FLAG and a smaller FLAG-reactive band are indicated. Approximately 15 μg of protein from cell lysates was subjected to SDS-PAGE.
We wondered whether other predicted T4SS outer membrane proteins were also present at low levels. TraB is encoded immediately downstream of traK and is also predicted to be an outer membrane component of the gonococcal T4SS. The addition of a C-terminal purification tag to the TraFN homolog in pKM101 did not interfere with core complex formation (13), so we investigated expression of TraB by adding a C-terminal FLAG epitope (TraB-FLAG). We expressed the epitope-tagged protein in Neisseria, either at the native locus in the GGI (traB-FLAG in strain MR646) or overexpressed from a distant locus (traB-FLAG+ in strain MR565). As was the case with TraK, we were unable to detect TraB-FLAG expressed at WT levels by Western blotting (Fig. 1C). We were able to detect TraB-FLAG in the overexpression strain MR565. We detected two FLAG-reactive bands in this strain, the larger of which (∼47 kDa) is the predicted size of the full-length TraB-FLAG protein (Fig. 1C). Multiple bands have also been observed in Western blots for the A. tumefaciens TraB homolog VirB10 (32–34). The significance and identity of the smaller ∼37-kDa band in gonococci are unknown, although we predict that it is a degradation product of TraB-FLAG. This smaller band was present even when cell lysates were prepared in the presence of protease inhibitors, suggesting that the TraB-FLAG processing or degradation event occurs during bacterial growth.
TraK localizes to the outer membrane in N. gonorrhoeae.
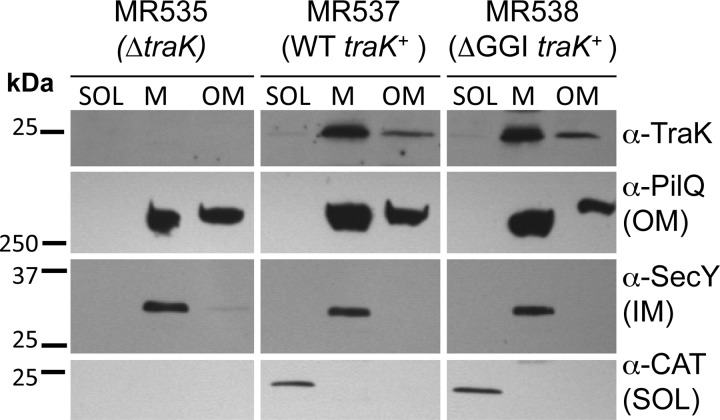
VirB9 localizes primarily to the outer membrane in A. tumefaciens, although it fails to accumulate to WT levels in the absence of the VirB7 lipoprotein (35, 36). TraKF also localizes to the outer membrane, and its localization is similarly perturbed in the absence of the lipoprotein TraVF (37). We investigated localization of gonococcal TraK in subcellular fractions of the traK overexpression strain MR537. We also investigated TraK localization in strain MR538, which ectopically expresses traK in a ΔGGI background, since we hypothesized that TraK might localize differently in the absence of other T4SS proteins. Strain MR535 contains a near-complete deletion of traK (with 15 bp at the 3′ end of the traK gene retained in order to not disrupt the ribosome binding site and start codon of the downstream gene traB) and was used as a negative control. Cells were separated into a soluble fraction containing cytoplasmic and periplasmic material, a total membrane fraction containing both the inner and outer membranes, and an outer membrane fraction prepared by inducing the formation of outer membrane vesicles (see Materials and Methods). In both MR537 and MR538, TraK localized to both the total membrane and outer membrane fractions, a fractionation pattern similar to that of the known outer membrane protein PilQ, indicating that TraK localizes to the outer membrane (Fig. 2). The fractionation pattern of TraK in MR538 was similar to that in MR537, suggesting that TraK localizes to the outer membrane independently of other T4SS proteins (Fig. 2). The cytoplasmic protein chloramphenicol acetyltransferase (CAT) was only detected in the soluble fraction, and the inner membrane protein SecY was only detected in the total membrane fraction, indicating that the outer membrane fraction was not contaminated with significant amounts of inner membrane or cytoplasmic proteins (Fig. 2).
FIG 2.
Subcellular localization of TraK in N. gonorrhoeae. Gonococcal cells were fractionated into soluble (SOL [both cytoplasm and periplasm]), total membrane (M [both inner and outer membranes]), and outer membrane (OM) fractions. An IPTG-inducible promoter was used to express traK from a distant chromosomal locus in either the WT N. gonorrhoeae strain MS11 (MR537) or an isogenic ΔGGI background (MR538). Strain MR535 contains a near-complete deletion of traK and does not encode chloramphenicol acetyltransferase (CAT). TraK was detected using the 2E5 anti-TraK monoclonal antibody. Antibodies against the outer membrane protein PilQ, the cytoplasmic protein CAT, and the inner membrane (IM) protein SecY were used as fractionation controls. Approximately 2.5 μg protein from each fraction was subjected to SDS-PAGE.
TraK interacts with TraV.
In A. tumefaciens, VirB9 is stabilized by interaction with VirB7 via a disulfide bond, and this outer membrane dimer has been proposed to serve as a nucleation point for T4SS formation (36, 38–41). Since TraK localized to the gonococcal outer membrane in the absence of other T4SS components, we investigated whether it interacted with the TraV lipoprotein. Gonococcal TraV is significantly larger than VirB7 (193 amino acids versus 55 amino acids), making it more similar in length to the F plasmid homolog TraVF (171 amino acids) (see Fig. S6 in the supplemental material) (42). Like TraVF, the mature gonococcal TraV protein contains three cysteines, one of which is found immediately after the predicted signal sequence cleavage site and is thus likely modified with a lipid anchor (42). If TraV and TraK do interact in gonococci, it is unlikely to be via a disulfide bridge, since the only cysteine in the TraK protein is located within the predicted Sec-dependent signal peptide (see Fig. S1 and S5 in the supplemental material).
We used a bacterial two-hybrid approach to investigate whether TraK and TraV interact. The putative coupling protein TraD was used as a negative control since it is not predicted to interact with TraK. We constructed bait and/or target plasmids encoding C-terminal fusions of the T4SS proteins to lambda phage cI protein or the RNA polymerase alpha (RNAPα) subunit, respectively. The predicted TraV lipobox and TraK signal sequences were excluded from these plasmids. Bait and target plasmids were cotransformed into the BacterioMatchII reporter E. coli strain, which is a hisB mutant. Interaction of the bait and target proteins enables transcriptional activation of the HIS3 gene, allowing growth in the presence of 3-AT, a competitive inhibitor of the HIS3 gene product. Growth was assayed on nonselective plates containing chloramphenicol and tetracycline to determine the number of cotransformants, as well as on selective plates containing the competitive inhibitor 3-AT in addition to the antibiotics. We observed growth on the nonselective plates for all cotransformations, indicating the successful cotransformation of the reporter strain with both bait and target plasmids (Table 3). However, we only detected colonies on the selective plates when cells were cotransformed with the TraV-containing bait plasmid (pKH86) and TraK-containing target plasmid (pKH87), indicating that TraK interacts with TraV. We did not observe colonies on the selective plates when cells were cotransformed with the TraD-containing bait plasmid (pKH103) and either TraK- or TraV-containing target plasmids (pKH87 or pKH170) (Table 3), confirming that TraD does not interact with either TraK or TraV. As an additional control, we confirmed that cotransformation of the reporter strain with the TraK- and TraV-containing plasmids along with the appropriate empty bait or target plasmids did not result in colonies on selective plates (Table 3).
TABLE 3.
Bacterial two-hybrid analysis of TraK interactions
| Bait plasmid | Target plasmid | CFU/ml ona: |
|
|---|---|---|---|
| Nonselective plates | Selective plates | ||
| pBT (empty vector) | pKH87 (TraKR53-G244) | 4,164 ± 879 | 0 |
| pKH86 (TraVS20-R193) | pTRG (empty vector) | 48 ± 11 | 0 |
| pKH86 (TraVS20-R193) | pKH87 (TraKR53-G244) | 953 ± 501 | 887 ± 391 |
| pKH103 (TraDM1-V680) | pKH87 (TraKR53-G244) | 966 ± 98 | 0 |
| pKH103 (TraDM1-V680) | pKH170 (TraVS20-R193) | 9,471 ± 2,316 | 0 |
The average ± standard deviation of four independent experiments is reported. Nonselective plates contained minimal medium plus 25 μg/ml chloramphenicol (Cam) and 12.5 μg/ml tetracycline (Tet). Selective plates contained minimal medium plus 25 μg/ml Cam, 12.5 μg/ml Tet, and 5 mM 3-AT.
Full-length TraB-FLAG localizes to the outer membrane in N. gonorrhoeae.
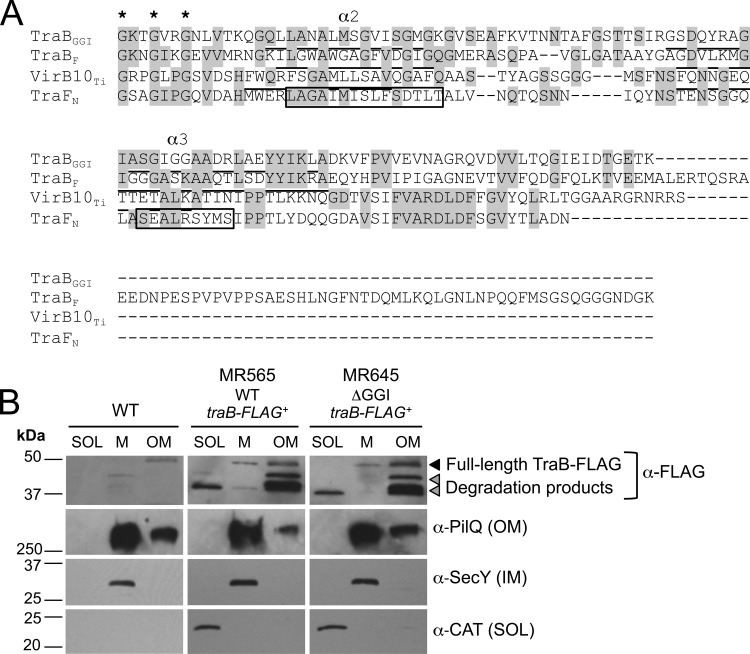
In pKM101 and F plasmid, TraK and TraV homologs associate with a third protein, TraB, to form a core T4SS complex (13, 37). In pKM101, two α helices (α2 and α3) near the C terminus of the TraB homolog (TraFN) form a two-helix bundle that crosses the outer membrane (indicated by boxes in Fig. 3A), and this antenna projection domain is conserved in the A. tumefaciens homolog VirB10 (12, 43, 44). When we aligned the C termini of several TraB homologs and used Phyre2 to predict secondary structures in gonococcal TraB (45), we observed that similar secondary structures were conserved in gonococcal TraB despite low sequence similarity (Fig. 3A; see Fig. S7 in the supplemental material), suggesting that gonococcal TraB may have a similar membrane topology to its TraFN homolog. Given that secondary structures appear to be conserved between gonococcal TraB and TraFN, we hypothesized that TraB would also localize to both membranes. Gonococcal cells were fractionated into a soluble fraction (cytoplasm and periplasm), a total membrane fraction (inner and outer membranes), and an outer membrane fraction (generated by inducing the formation of outer membrane vesicles). TraB-FLAG was localized in three strains: MR565, which expresses traB-FLAG from an IPTG-inducible promoter in the WT background; MR645, which expresses traB-FLAG from an IPTG-inducible promoter in the ΔGGI background; and the WT strain, which served as a negative control.
FIG 3.
TraB-FLAG. (A) Alignment of the C termini of TraB homologs. The highly conserved GxxGxxG motif characterized by Banta et al. (44) that is involved in channel gating in A. tumefaciens is indicated with asterisks. The alpha helices (α2 and α 3) identified by Chandran et al. (12) in the TraFN crystal structure are indicated by boxes. The predicted alpha helices in VirB10, TraBGGI, and TraBF are underlined. (B) Subcellular localization of TraB-FLAG in N. gonorrhoeae. Gonococcal cells were fractionated into soluble (SOL [both cytoplasm and periplasm]), total membrane (M [both inner and outer membranes]), and outer membrane (OM) fractions. An IPTG-inducible promoter was used to express traB-FLAG in either the WT N. gonorrhoeae strain MS11 (MR565) or an isogenic ΔGGI strain (MR645). The WT strain MS11 served as the negative control and does not encode chloramphenicol acetyltransferase (CAT). TraB-FLAG was detected using the commercially available anti-FLAG monoclonal antibody. Full-length TraB-FLAG is indicated (black triangle) along with two smaller FLAG-reactive bands (gray triangles). Antibodies against the outer membrane protein PilQ, the cytoplasmic protein chloramphenicol acetyltransferase (CAT), and the inner membrane (IM) protein SecY were used as fractionation controls. Approximately 3.5 μg of protein from each fraction was subjected to SDS-PAGE.
Interestingly, the two FLAG-reactive bands fractionated differently. In both MR565 and MR645, full-length TraB-FLAG (the larger ∼47-kDa band [black triangle in Fig. 3B]) fractionated to both the total membrane and outer membrane fractions, a pattern similar to that of the outer membrane protein PilQ, suggesting that it primarily associates with the outer membrane (Fig. 3B). SecY and CAT served as additional fractionation controls and localized to the total membrane and cytoplasmic fractions, respectively (Fig. 3B). Because the FLAG tag was added to the C terminus of TraB, it is likely that the smaller (∼37-kDa) TraB-FLAG product does not contain the N-terminal predicted inner membrane helix (α1 in Fig. S7 in the supplemental material) and therefore may not associate with the inner membrane. We observed that the smaller FLAG-reactive band (∼37 kDa) localized to the soluble and outer membrane fractions in both MR565 and MR645, but not the total membrane fraction (lower gray triangle in Fig. 3B). We also observed an additional FLAG-reactive band (∼45 kDa) that appeared only in the outer membrane fraction (upper gray triangle in Fig. 3B). A possible explanation for the fractionation pattern of these smaller TraB-FLAG products could be that they localize to the periplasm and that some periplasmic material is incorporated into the outer membrane vesicles produced by this fractionation method (46).
TraK-FLAG3.
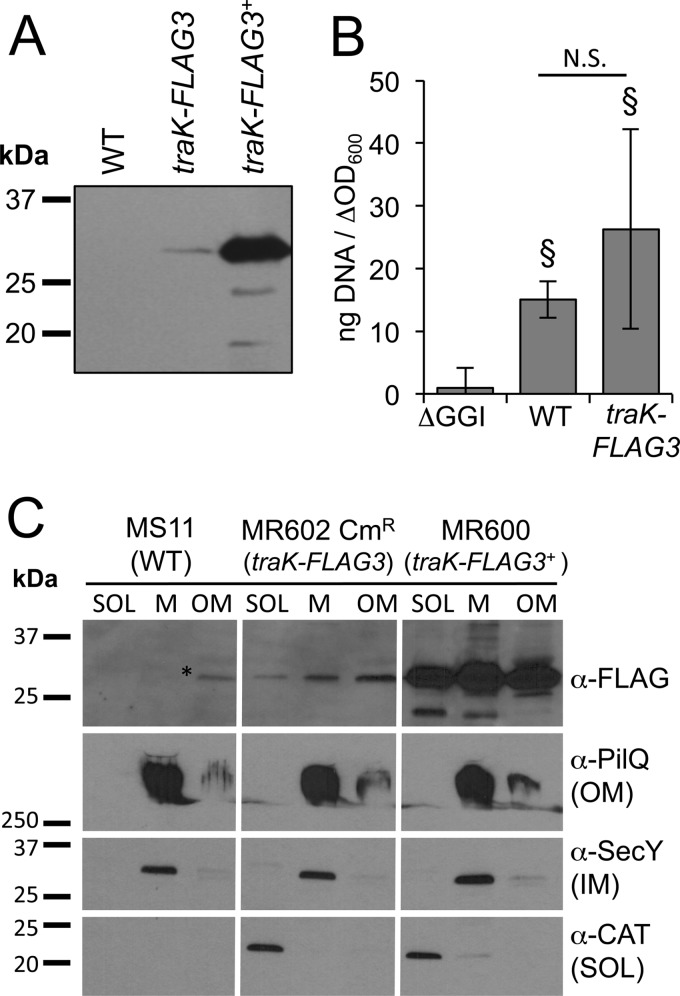
Because we were unable to detect TraK and TraB-FLAG in WT cells using the monoclonal antibodies we had developed, we sought to develop a more sensitive detection method that would allow us to detect one of these proteins expressed from the native locus. We added a linker sequence and three tandem copies of the FLAG epitope to the C terminus of TraK (TraK-FLAG3) (29). The FLAG3 epitope has been used successfully to amplify detection of FtsZ in Streptococcus pneumoniae (29), and we hypothesized that the triple epitope would also allow us to detect TraK-FLAG3 in WT gonococcal cells. We constructed a strain in which traK-FLAG3 was incorporated into the GGI and expressed from the native promoter (MR602) as well as a strain in which traK-FLAG3 was overexpressed from an IPTG-inducible promoter at a distant chromosomal locus (MR600). We were able to detect TraK-FLAG3 by Western blotting using the FLAG antibody in both strains (Fig. 4A). DNA secretion assays demonstrated that MR602 secretes DNA at comparable levels to the WT strain, and significantly higher levels of DNA were present in the culture medium compared to the amounts in the medium from GGI deletion strain ND500. These results suggest that TraK-FLAG3 is functional as part of the secretion apparatus (Fig. 4B).
FIG 4.
Addition of a FLAG3 epitope to the C terminus of TraK (TraK-FLAG3). (A) Western blot for TraK-FLAG3 using the FLAG antibody in MS11 (WT), MR602 (traK-FLAG3 at the native locus), and MR600 (traK-FLAG3 expressed from an IPTG-inducible promoter). Approximately 16.5 μg of protein from cell lysates was subjected to SDS-PAGE. (B) Fluorometric detection of secreted DNA in supernatants of gonococcal strains ND500 (ΔGGI), MS11, and MR602. §, Student's two-tailed t test P < 0.05 compared to ND500, N.S., not significantly different by the Student two-tailed t test. (C) Subcellular localization of TraK-FLAG3 in gonococcal strains MS11 (WT [negative control]), MR673 (traK-FLAG3 at the native locus, MR602 transformed with pKH37 encoding chloramphenicol acetyltransferase [CAT]), and MR600 (traK-FLAG3+). Cells were fractionated into a soluble fraction (SOL [cytoplasm and periplasm]), a total membrane fraction (M [inner and outer membranes]), and an outer membrane (OM) fraction. Antibodies against the outer membrane protein PilQ, the cytoplasmic protein chloramphenicol acetyltransferase (CAT), and the inner membrane (IM) protein SecY were used as fractionation controls. Approximately 2 μg of protein from each fraction was subjected to SDS-PAGE. CmR, chloramphenicol resistant.
Subcellular fractionation of TraK-FLAG3.
We performed subcellular fractionation studies to localize TraK-FLAG3 in the cell. In the traK-FLAG3 overexpression strain (MR600), TraK-FLAG3 localized to the total membrane and outer membrane fractions, as was the case with TraK (Fig. 2), but it also localized to the soluble fraction (Fig. 4C), suggesting that the C-terminal epitope tag has some effect on protein localization. When traK-FLAG3 was expressed from the native locus, we observed a similar fractionation pattern to that in the overexpression strain, with TraK-FLAG3 localizing to the total membrane and outer membrane fractions, as well as to the soluble fraction (Fig. 4C). Antibodies against the outer membrane protein PilQ, the inner membrane protein SecY, and the cytoplasmic protein CAT were used as fractionation controls. These results, in combination with those in Fig. 2, indicate that TraK localizes to the outer membrane, regardless of whether it is overexpressed from a distant locus or epitope tagged and expressed from the native locus. The addition of the FLAG3 tag did subtly affect protein localization since TraK-FLAG3 was also present in the soluble fraction (Fig. 4C).
Localization of TraK-FLAG3 in gonococcal cells.
We asked whether the T4SS localized to a specific region of the gonococcal cell. We were unable to detect TraK-FLAG3 expressed from the WT locus in MR602 using standard immunofluorescence microscopy techniques (data not shown), although TraK-FLAG3 localized in punctate spots outside the DAPI-stained region in the overexpression strain MR600 (see Fig. S8 in the supplemental material). These results suggested that TraK-FLAG3 localizes outside the cytoplasm and are consistent with the subcellular fractionation data for TraK and TraK-FLAG3 (Fig. 2 and 4C).
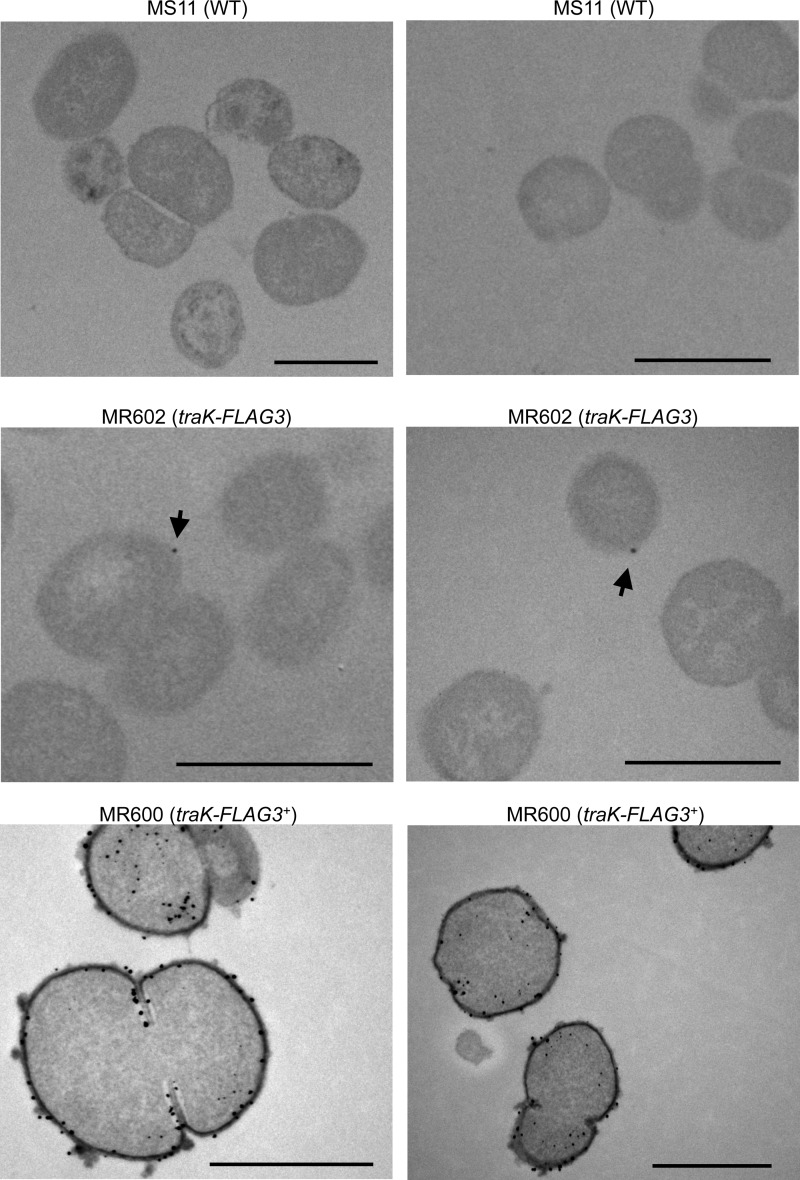
To increase sensitivity and resolution, we turned to thin-section immunogold electron microscopy to investigate localization of TraK-FLAG3. We had limited success using standard immunogold electron microscopy protocols, so we optimized a preembedding technique for use with Neisseria. In this protocol, antibody staining is performed prior to sample embedding and dehydration, which helps to maintain antigenicity. In strain MR600, the majority of cells we visualized had multiple gold particles that localized around the periphery of the cell and seemed to associate with both the inner and outer membranes (Fig. 5). Occasionally, we observed cells with gold particles in the cytoplasm (Fig. 5). The appearance of some membrane blebbing in this sample may suggest that high-level ectopic expression of TraK-FLAG3 generates a periplasmic stress in gonococci (Fig. 5). We saw very little staining in the MS11 negative control, and the gold particles that were present were localized throughout the cell (Fig. 5). In strain MR602, we observed that some, but not all of the cells contained a single gold particle, and the gold particle was associated with the outside the cell (Fig. 5). We never observed more than one gold particle per cell. These results are consistent with the low expression levels of TraK-FLAG3 (Fig. 5) and suggest that each gonococcus may produce one or zero T4SS apparatuses.
FIG 5.
Preembedding immunogold electron microscopy for TraK-FLAG3 in MS11 (WT), MR602 (traK-FLAG3 at native locus), and MR600 (traK-FLAG3 expressed from an IPTG-inducible promoter) using the FLAG antibody. Two representative images are shown for each strain. Arrows indicate gold particles in MR602. Scale bars indicate 1 μm.
Expression of TraK-FLAG3 within gonococcal populations.
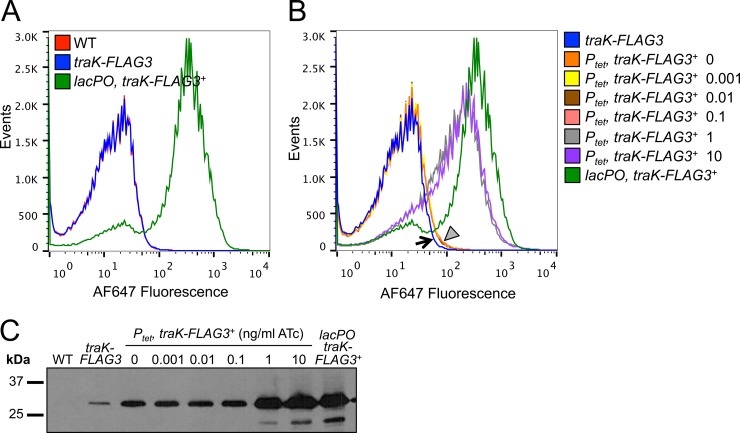
One possible explanation for our inability to detect TraK and TraB-FLAG in WT cells by Western blotting could be differential expression within the population. If only a small percentage of cells in the population express the T4SS, but express it at high levels, proteins such as TraK would become diluted in pooled lysates. We used flow cytometry to investigate the TraK-FLAG3 expression profile in MS11 (no FLAG tag), MR602 (traK-FLAG3 at the native locus), and MR600 (traK-FLAG3 overexpression strain) using the FLAG antibody and an Alexa Fluor 647-conjugated secondary antibody (Fig. 6A). The majority of the MR600 population exhibited high levels of fluorescence, as expected given that traK-FLAG3 expression is induced with IPTG in this strain (Fig. 6A). We were unable to resolve MR602 (expressing traK-FLAG3 at native levels) from the negative control (the WT strain MS11), but we did not observe any subpopulations in the MR602 sample that exhibited high levels of fluorescence (Fig. 6A).
FIG 6.
TraK-FLAG3 expression in gonococcal populations. (A) Expression of TraK-FLAG3 in populations of gonococcal strains MS11 (WT), MR602 (traK-FLAG3), and MR600 (traK-FLAG3 expressed from the IPTG-inducible lacPO promoter). Fixed bacteria were permeabilized and then incubated with the FLAG antibody and an Alexa Fluor 647 (AF647)-conjugated secondary antibody, and 150,000 events were analyzed by flow cytometry. (B) Determination of the TraK-FLAG3 limit of detection by flow cytometry by expression of traK-FLAG3 from the anhydrotetracycline (ATc)-inducible promoter Ptet. Cells were prepared as in panel A, and TraK-FLAG3 expression was analyzed in strains MR602 (traK-FLAG3), MR675 (traK-FLAG3 expressed from Ptet), and MR600 (traK-FLAG3 expressed from the IPTG-inducible promoter lacPO). MR675 was grown in the presence of different concentrations of ATc (ng/ml), and MR600 was grown in the presence of 1 mM IPTG. The black arrow indicates MR602, while the gray triangle indicates 4 samples that overlap one another (MR675 grown in the presence of 0, 0.001, 0.01, or 0.1 ng/ml ATc). (C) Western blot for TraK-FLAG3 in the same strains analyzed by flow cytometry in panel B using the FLAG monoclonal antibody. Approximately 12 μg of protein from cell lysates was subjected to SDS-PAGE, representing protein from approximately 1.6 × 107 cells.
We sought to determine the limit of detection in the flow cytometry assay, since it was possible that TraK-FLAG3 levels could still vary within the population but remain below the limit of detection for the assay. Previously, we showed that the anhydrotetracycline (ATc)-inducible Ptet promoter exhibited a broad dose-response curve in response to inducer in gonococci (47). We expressed traK-FLAG3 under the control of Ptet at the iga-trpB chromosomal site and grew the resulting strain (MR675) in the presence of increasing amounts of ATc (Fig. 6B and C). We observed similar amounts of TraK-FLAG3 in MR675 when the cells were uninduced or grown in the presence of 0.001, 0.01, or 0.1 ng/ml ATc (Fig. 6C). The amount of TraK-FLAG3 in these samples was approximately 4-fold higher than the amount of TraK-FLAG3 in the MR602 native expression strain (Fig. 6C [densitometry analysis performed using ImageJ]) (48). We were able to distinguish the MR675 populations induced with low levels of ATc from MR602 by using flow cytometry, suggesting that our limit of detection is approximately 4-fold higher than the amount of TraK-FLAG3 produced from the native locus (Fig. 6B). TraK-FLAG3 levels were significantly higher when strain MR675 was grown in the presence of 1 or 10 ng/ml ATc or 1 mM IPTG (in the case of lacPO), and these populations were easily distinguishable from MR602 by flow cytometry (Fig. 6B and C). These results are consistent with a model in which all of the cells in the population express TraK-FLAG3 at low levels. Although we are unable to rule out the possibility that some variation may occur below the limit of detection, the fact that we are able to detect a 4-fold increase in TraK-FLAG3 levels compared to WT suggests that such variation is limited (Fig. 6B).
DISCUSSION
This study represents the first in-depth analysis of the outer membrane proteins in the gonococcal T4SS. Although the content and organization of the genes in the GGI are most similar to that of E. coli F plasmid, sequence similarity is very low (2). This, in combination with the fact that the gonococcal T4SS is different from other characterized T4SSs and secretes DNA directly into the environment, prompted us to investigate gonococcal TraK, TraV, and TraB. All three proteins are required for type IV secretion in gonococci (17), and homologs of these proteins have been shown to interact with one another to form a core complex in a diverse set of T4SSs (13, 37, 49–52). We found that TraK and TraB localize to the outer membrane in gonococci and that their localization is not perturbed in the absence of other T4SS proteins (Fig. 2 and 3). Furthermore, TraK, TraV, and TraB are highly conserved between gonococcal strains (see Fig. S1 to S3 in the supplemental material), suggesting that they may be rare examples of nonvariable outer membrane proteins in gonococci.
The two-helix bundle identified in the pKM101 homolog of TraB is conserved in gonococcal TraB despite low sequence similarity (12) (Fig. 3A), so it is possible that a portion of gonococcal TraB may be surface exposed. Western blots to detect TraB-FLAG in a gonococcal overexpression strain revealed two specific bands (Fig. 1), and we predict that the smaller FLAG-reactive band (which localized to the soluble and outer membrane fractions) represents a periplasmic C-terminal TraB-FLAG cleavage product. In A. tumefaciens, VirB10 undergoes a conformational change in response to ATP utilization by the inner membrane T4SS ATPases VirB11 and VirD4 that is thought to be crucial for substrate secretion and can be monitored based on the susceptibility of VirB10 to an exogenously added protease (53). It is possible that the C-terminal TraB cleavage product could be a result of a similar conformational change in gonococci. However, the smaller C-terminal cleavage product is present in gonococci regardless of whether TraB-FLAG is expressed in the WT or the ΔGGI background (i.e., regardless of whether the T4SS ATPases are present) (Fig. 3), which could suggest that the smaller FLAG-reactive band is just a degradation product resulting from overexpression and not indicative of a conformational change.
A surprising result of this study was the observation that neither TraK nor TraB-FLAG was detectable by Western blotting in WT cells during log-phase growth, although traK is transcribed (Fig. 1). Signal amplification from a triple FLAG epitope added to the C terminus of TraK allowed us to detect TraK-FLAG3 in WT cells (29), a result consistent with the hypothesis that TraK is present in WT cells but produced at levels not detectable by standard Western blotting methods (Fig. 4). We used the TraK-FLAG3 allele to investigate localization of the T4SS in the cell and its expression in the population. Previous microscopy studies of T4SS proteins in other bacteria have reported both polar (54–57) and helical (58) localization patterns. N. gonorrhoeae is a coccus, making it difficult to identify cell poles, except in diplococci. However, proteins such as the NalP autotransporter localize asymmetrically in Neisseria and are capable of polar localization when expressed in E. coli, and it has been proposed that specific regions of the Neisseria cell membrane could be equivalent to the poles of rod-shaped bacteria as a result of division in alternate perpendicular plates (59, 60). When TraK-FLAG3 was overexpressed (in MR600), it localized throughout the cell envelope (Fig. 5). When TraK-FLAG3 was produced from the native locus (in MR602), a single gold particle was observed in some cells and localized to the outer membrane (Fig. 5). The low expression levels of TraK-FLAG3 make it difficult to make conclusions about localization patterns of the T4SS, but it seems clear from these results that the T4SS apparatus is present at a very low copy number per cell.
We also investigated whether the apparent low expression levels of the T4SS were due to differential expression in a subset of the population by using flow cytometry on strains expressing TraK-FLAG3. We hypothesized that the elaboration of a T4SS apparatus and secretion of chromosomal DNA would constitute a large metabolic cost and that expression of the T4SS by a small subpopulation would minimize this cost while still providing a mechanism for horizontal gene transfer. The fluorescence profile of the MR602 population (expressing traK-FLAG3 at the native locus) was indistinguishable from that of the negative control, suggesting that TraK-FLAG3 levels in this strain are below the limit of detection by flow cytometry, although we did not observe any MR602 subpopulations exhibiting high levels of fluorescence (Fig. 6). A population of cells expressing approximately 4-fold-higher levels of TraK-FLAG3 than MR602 was clearly distinguishable by flow cytometry, suggesting that variation below the limit of detection does not exceed 4-fold. These results are consistent with a model in which all gonococci in the population express low levels of the structural T4SS proteins.
Gonococci secrete DNA throughout their growth cycle, so it is puzzling that TraK and TraB are not readily detectable (20). The traK and traB genes are contained on a long transcript that includes 17 other genes, most of which are predicted to be structural components of the secretion apparatus (2). Previously characterized gonococcal T4SS proteins, such as TraI (the relaxase) and TraG (an inner membrane mating pair formation homolog), are each encoded on a different transcript and are both detectable in WT cells by Western blotting (61, 62). We identified a possible σ70 promoter upstream of ltgX, the first gene in the long transcript containing traK and traB. The −35 and −10 hexamers (TTCATA and TTTAAT, respectively) are close to consensus sequences and are separated by 16 bp. We were also able to identify putative ribosome binding sites upstream of traK and traB (GAGGT and GGAGG, respectively) as well as upstream of other genes in the long transcript, many of which have start and stop codons that overlap or are only separated by a few base-pairs, suggesting that they are cotranscribed and may be cotranslated (2). All of the predicted surface-exposed T4SS proteins (such as TraB, TraN, and TraK) are encoded together on this long transcript. Given that N. gonorrhoeae is a strict human pathogen and any surface-exposed protein would be exposed to the host immune system, it is possible that expression of this particular T4SS transcript is regulated to prevent high-level expression of conserved proteins throughout infection.
Little is known about the regulation of the T4SS in gonococci. Gonococci expressing type IV pili secrete more DNA than nonpiliated cells, but the precise mechanism by which this occurs is unknown (20). Optimal expression of T4SS genes in other bacteria often requires an environmental stimulus, such as the phenolic plant hormone acetosyringone in the case of A. tumefaciens or acidic conditions that mimic intracellular infection of macrophages in the case of Brucella suis (63–65). Expression of F plasmid genes is dependent on growth phase and is subject to regulation by global regulators such as Hfq and H-NS (66–68). Gonococci are highly adapted for survival within their sole human host, and it is possible that high-level expression of the T4SS may be induced by conditions that mimic a certain stage or type of infection. There is evidence for expression of the gonococcal T4SS during intracellular infection since strains encoding the T4SS were found to survive intracellular infections of ME180 cervical cells in the absence of TonB-dependent iron transport, which is required for intracellular growth of gonococcal strains lacking the T4SS (69).
Altogether, these results indicate that TraK and TraB are conserved outer membrane proteins in the gonococcal T4SS and suggest that, despite its low similarity to other characterized systems and its distinct mechanism of DNA secretion, the gonococcal T4SS apparatus is likely structurally similar to characterized homologs. The low levels of TraK and TraB in the cell suggest that very few copies of these structural components are necessary for DNA secretion. We predict that high-level expression of these proteins may be regulated in gonococci. Identification of the conditions or regulators that induce increased expression of the T4SS will provide important insight regarding the context and biological significance of type IV secretion in gonococcal pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health (NIH) grant AI047958 to J.P.D. M.E.R. was supported in part by NIH training grant T32 GM07215.
We thank H. S. Seifert for the PilQ antibody and M. E. Winkler for the FLAG3 construct. Immunogold electron microscopy was performed at the University of Wisconsin Medical School Electron Microscope Facility with the assistance of Ben August. We also thank Elizabeth Heath-Heckman, Eva Ziegelhoffer, and Terry Glaab for assistance and advice regarding immunofluorescence microscopy.
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01825-14.
REFERENCES
- 1.Dillard JP, Seifert HS. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263–278. 10.1046/j.1365-2958.2001.02520.x [DOI] [PubMed] [Google Scholar]
- 2.Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704–1721. 10.1111/j.1365-2958.2005.04521.x [DOI] [PubMed] [Google Scholar]
- 3.Ramsey ME, Woodhams KL, Dillard JP. 2011. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria. Front. Microbiol. 2:61. 10.3389/fmicb.2011.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton HL, Dillard JP. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376–385. 10.1111/j.1365-2958.2005.04964.x [DOI] [PubMed] [Google Scholar]
- 5.Zweig MA, Schork S, Koerdt A, Siewering K, Sternberg C, Thormann K, Albers SV, Molin S, van der Does C. 2014. Secreted single-stranded DNA is involved in the initial phase of biofilm formation by Neisseria gonorrhoeae. Environ. Microbiol. 16:1040–1052. 10.1111/1462-2920.12291 [DOI] [PubMed] [Google Scholar]
- 6.Dominguez NM, Hackett KT, Dillard JP. 2011. XerCD-mediated site-specific recombination leads to loss of the 57-kilobase gonococcal genetic island. J. Bacteriol. 193:377–388. 10.1128/JB.00948-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808. 10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149. 10.1038/nrmicro753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1–15. 10.1016/S0378-1097(03)00430-0 [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJJ, Farrand SK, Winans SC. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885–3895. 10.1128/JB.182.14.3885-3895.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrivastava R, Miller JF. 2009. Virulence factor secretion by Bordetella species. Curr. Opin. Microbiol. 12:88–93. 10.1016/j.mib.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. 10.1038/nature08588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. 10.1126/science.1166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera-Calzada A, Fronzes R, Savva CG, Chandran V, Lian PW, Laeremans T, Pardon E, Steyaert J, Remaut H, Waksman G, Orlova EV. 2013. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 32:1195–1204. 10.1038/emboj.2013.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. 10.1038/nature13081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pachulec E. 2010. The type IV secretion systems of Neisseria gonorrhoeae. Ph.D. thesis University of Groningen, Groningen, Netherlands [Google Scholar]
- 18.Cornelissen CN, Hollander A. 2011. TonB-dependent transporters expressed by Neisseria gonorrhoeae. Front. Microbiol. 2:117. 10.3389/fmicb.2011.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer TF. 1991. Evasion mechanisms of pathogenic Neisseriae. Behring Inst. Mitt. 88:194–199 [PubMed] [Google Scholar]
- 20.Salgado-Pabón W, Du Y, Hackett KT, Lyons KM, Arvidson CG, Dillard JP. 2010. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae. J. Bacteriol. 192:1912–1920. 10.1128/JB.01357-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Kellogg DS, Jr, Peacock WL, Jr, Deacon WE, Brown L, Pirkle CL. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morse SA, Bartenstein L. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc. Soc. Exp. Biol. Med. 145:1418–1421. 10.3181/00379727-145-38025 [DOI] [PubMed] [Google Scholar]
- 24.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillard JP. 2011. Genetic manipulation of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. Chapter 4:Unit 4A2. 10.1002/9780471729259.mc04a02s00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocco CJ, Dennsion KL, Klenchin VA, Rayment I, Escalante-Semerena JC. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231–237. 10.1016/j.plasmid.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bessonneau P, Besson V, Collinson I, Duong F. 2002. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 21:995–1003. 10.1093/emboj/21.5.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain S, Zweig M, Peeters E, Siewering K, Hackett KT, Dillard JP, van der Does C. 2012. Characterization of the single stranded DNA binding protein SsbB encoded in the gonoccocal genetic island. PLoS One 7:e35285. 10.1371/journal.pone.0035285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wayne KJ, Sham LT, Tsui HC, Gutu AD, Barendt SM, Keen SK, Winkler ME. 2010. Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae. J. Bacteriol. 192:4388–4394. 10.1128/JB.00578-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deardorff AS, Romer SH, Deng Z, Bullinger KL, Nardelli P, Cope TC, Fyffe RE. 2013. Expression of postsynaptic Ca2+-activated K+ (SK) channels at C-bouton synapses in mammalian lumbar-motoneurons. J. Physiol. 591:875–897. 10.1113/jphysiol.2012.240879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamm M, Staritzbichler R, Khafizov K, Forrest LR. 2013. Alignment of helical membrane protein sequences using AlignMe. PLoS One 8:e57731. 10.1371/journal.pone.0057731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward JE, Jr, Dale EM, Nester EW, Binns AN. 1990. Identification of a VirB10 protein aggregate in the inner membrane of Agrobacterium tumefaciens. J. Bacteriol. 172:5200–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar RB, Xie YH, Das A. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36:608–617. 10.1046/j.1365-2958.2000.01876.x [DOI] [PubMed] [Google Scholar]
- 34.Thorstenson YR, Kuldau GA, Zambryski PC. 1993. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J. Bacteriol. 175:5233–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirasu K, Kado CI. 1993. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett. 111:287–294. 10.1111/j.1574-6968.1993.tb06400.x [DOI] [PubMed] [Google Scholar]
- 36.Fernandez D, Spudich GM, Zhou XR, Christie PJ. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178:3168–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris RL, Hombs V, Silverman PM. 2001. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42:757–766. 10.1046/j.1365-2958.2001.02667.x [DOI] [PubMed] [Google Scholar]
- 38.Spudich GM, Fernandez D, Zhou XR, Christie PJ. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. U. S. A. 93:7512–7517. 10.1073/pnas.93.15.7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baron C, Thorstenson YC, Zambryski PC. 1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson LB, Hertzel AV, Das A. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. U. S. A. 93:8889–8894. 10.1073/pnas.93.17.8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das A, Anderson LB, Xie YH. 1997. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J. Bacteriol. 179:3404–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris RL, Silverman PM. 2002. Roles of internal cysteines in the function, localization, and reactivity of the TraV outer membrane lipoprotein encoded by the F plasmid. J. Bacteriol. 184:3126–3129. 10.1128/JB.184.11.3126-3129.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. 2009. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 71:779–794. 10.1111/j.1365-2958.2008.06565.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banta LM, Kerr JE, Cascales E, Giuliano ME, Bailey ME, McKay C, Chandran V, Waksman G, Christie PJ. 2011. An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect. J. Bacteriol. 193:2566–2574. 10.1128/JB.00038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 46.Schwechheimer C, Sullivan CJ, Kuehn MJ. 2013. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 52:3031–3040. 10.1021/bi400164t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsey ME, Hackett KT, Kotha C, Dillard JP. 2012. New complementation constructs for inducible and constitutive gene expression in Neisseria gonorrhoeae and Neisseria meningitidis. Appl. Environ. Microbiol. 78:3068–3078. 10.1128/AEM.07871-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophoton. Int. 11:36–42 [Google Scholar]
- 49.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. 2008. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J. Bacteriol. 190:2161–2171. 10.1128/JB.01341-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das A, Xie YH. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758–763. 10.1128/JB.182.3.758-763.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99:11405–11410. 10.1073/pnas.172390699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, Vogel JP. 2006. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62:1278–1291. 10.1111/j.1365-2958.2006.05446.x [DOI] [PubMed] [Google Scholar]
- 53.Cascales E, Christie PJ. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. U. S. A. 101:17228–17233. 10.1073/pnas.0405843101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar RB, Das A. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523–1532. 10.1046/j.1365-2958.2002.02829.x [DOI] [PubMed] [Google Scholar]
- 55.Judd PK, Kumar RB, Das A. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. U. S. A. 102:11498–11503. 10.1073/pnas.0505290102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 26:2540–2551. 10.1038/sj.emboj.7601696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeong KC, Vincent CD, Buford E, Vogel JP. 2006. Subcellular localization of the Dot/Icm type IV secretion proteins, p 192–194 In Cianciotto NP, Kwaik YA, Edelstein PH, Fields BS, Geary DF, Harrison TG, Joseph CB, Ratcliff RM, Stout JE. (ed), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC [Google Scholar]
- 58.Aguilar J, Zupan J, Cameron TA, Zambryski PC. 2010. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. U. S. A. 107:3758–3763. 10.1073/pnas.0914940107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westling-Häggström B, Elmros T, Normark S, Winblad B. 1935. Growth pattern and cell division in Neisseria gonorrhoeae. J. Bacteriol. 29:333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain S, van Ulsen P, Benz I, Schmidt MA, Fernandez R, Tommassen J, Goldberg MB. 2006. Polar localization of the autotransporter family of large bacterial virulence proteins. J. Bacteriol. 188:4841–4850. 10.1128/JB.00326-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salgado-Pabón W, Jain S, Turner N, van der Does C, Dillard JP. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol. Microbiol. 66:930–947. 10.1111/j.1365-2958.2007.05966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohler PL, Chan YA, Hackett KT, Turner N, Hamilton HL, Cloud-Hansen KA, Dillard JP. 2013. Mating pair formation homologue TraG is a variable membrane protein essential for contact-independent type IV secretion of chromosomal DNA by Neisseria gonorrhoeae. J. Bacteriol. 195:1666–1679. 10.1128/JB.02098-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stachel SE, Nester EW, Zambryski PC. 1986. A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc. Natl. Acad. Sci. U. S. A. 83:379–383. 10.1073/pnas.83.2.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stachel SE, Zambryski PC. 1986. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325–333. 10.1016/0092-8674(86)90653-7 [DOI] [PubMed] [Google Scholar]
- 65.Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O'Callaghan D. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U. S. A. 99:1544–1549. 10.1073/pnas.032514299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Will WR, Frost LS. 2006. Hfq is a regulator of F-plasmid TraJ and TraM synthesis in Escherichia coli. J. Bacteriol. 188:124–131. 10.1128/JB.188.1.124-131.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Will WR, Frost LS. 2006. Characterization of the opposing roles of H-NS and TraJ in transcriptional regulation of the F-plasmid tra operon. J. Bacteriol. 188:507–514. 10.1128/JB.188.2.507-514.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Will WR, Lu J, Frost LS. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54:769–782. 10.1111/j.1365-2958.2004.04303.x [DOI] [PubMed] [Google Scholar]
- 69.Zola TA, Strange HA, Domínguez NM, Dillard JP, Cornelissen CN. 2010. Type IV secretion machinery promotes ton-independent intracellular survival of Neisseria gonorrhoeae within cervical epithelial cells. Infect. Immun. 78:2429–2437. 10.1128/IAI.00228-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamilton HL, Schwartz KJ, Dillard JP. 2001. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 183:4718–4726. 10.1128/JB.183.16.4718-4726.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. 2007. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J. Bacteriol. 189:5421–5428. 10.1128/JB.00531-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swanson J. 1972. Studies on gonococcus infection II. Freeze-fracture, freeze-etch studies on gonococci. J. Exp. Med. 136:1258–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.