Abstract
Reiterative transcription is a reaction catalyzed by RNA polymerase, in which nucleotides are repetitively added to the 3′ end of a nascent transcript due to upstream slippage of the transcript without movement of the DNA template. In Escherichia coli, the expression of several operons is regulated through mechanisms in which high intracellular levels of UTP promote reiterative transcription that adds extra U residues to the 3′ end of a nascent transcript during transcription initiation. Immediately following the addition of one or more extra U residues, the nascent transcripts are released from the transcription initiation complex, thereby reducing the level of gene expression. Therefore, gene expression can be regulated by internal UTP levels, which reflect the availability of external pyrimidine sources. The magnitude of gene regulation by these mechanisms varies considerably, even when control mechanisms are analogous. These variations apparently are due to differences in promoter sequences. One of the operons regulated (in part) by UTP-sensitive reiterative transcription in E. coli is the carAB operon, which encodes the first enzyme in the pyrimidine nucleotide biosynthetic pathway. In this study, we used the carAB operon to examine the effects of nucleotide sequence at and near the transcription start site and spacing between the start site and −10 region of the promoter on reiterative transcription and gene regulation. Our results indicate that these variables are important determinants in establishing the extent of reiterative transcription, levels of productive transcription, and range of gene regulation.
INTRODUCTION
Usually during transcription, the nascent RNA transcript and the template strand of DNA move in tandem as an RNA-DNA hybrid. However, during transcription of a homopolymeric tract in the DNA template, the nascent transcript can slip (typically) one base upstream without movement of the DNA template within the active site of RNA polymerase (RNAP) (1, 2). This repositioning allows the same template base to specify an additional nucleotide in the transcript, and when transcript slippage occurs repetitively, the same template nucleotide can specify multiple extra residues. This reaction is called reiterative transcription (also known as RNAP stuttering, transcription slippage, and pseudotemplated transcription) and appears to be catalyzed by all RNAPs (2–5). Reiterative transcription can involve the repetitive addition of any of the four nucleoside triphosphate substrates and occurs during transcription initiation, elongation, and termination (2, 3, 6, 7). During initiation, when the length of the RNA-DNA hybrid can be shorter than that of the 8- to 9-bp hybrid that forms during elongation (8), a homopolymeric tract as short as three residues can enable reiterative transcription (9, 10). In contrast, longer homopolymeric tracts usually are required for reiterative transcription during elongation and termination (6, 7).
The physiological significance of reiterative transcription is that it plays a central role in regulating the expression of numerous prokaryotic, viral, and eukaryotic genes through an assortment of different mechanisms (3, 11). In Escherichia coli, expression of at least five operons is regulated by mechanisms involving the reiterative addition of U residues during transcription initiation (4, 12–15). Although these mechanisms can differ fundamentally (11), three examples include analogous mechanisms that regulate, in part, the expression of the pyrBI and carAB pyrimidine nucleotide biosynthetic operons and the galETKM (gal) galactose catabolic operon (12–14). The promoters of each of these operons (at least one when there are multiple promoters) include initially transcribed regions that contain a tract of three T·A base pairs located one or two bases downstream from the transcription start site (Fig. 1). These homopolymeric tracts, here referred to as (nontemplate strand) T tracts, enable a fraction of the nascent transcripts to enter the reiterative pathway. Entry into this pathway is enhanced by high intracellular concentrations of UTP (i.e., the reiterative substrate), resulting in a larger fraction of transcripts containing extra U residues. In each case, the addition of extra U residues causes the release of the nascent transcript from the transcription initiation complex, thereby repressing operon expression. This repression allows the cell to use UTP levels to control pyrBI, carAB, and gal expression and synthesize the encoded enzymes at optimal levels for growth. Although these control mechanisms are basically the same, the range of regulation afforded by them varies considerably, specifically, 2-fold, 3-fold, and 7-fold for the gal, carAB, and pyrBI operons, respectively (12–14). These different ranges presumably reflect important differences in promoter sequences, which could include different sequences at or near the transcription start site or the spacing between the start site and the −10 region (Fig. 1).
FIG 1.

Sequences of the pyrBI, carAB P1, and gal P2 promoter regions from the −10 region through the initially transcribed region. The −10 regions are labeled and underlined, and the transcription start sites are in boldface and marked with an asterisk.
In this study, we constructed mutant variants of the carAB P1 promoter, one of two carAB promoters and the only one that participates in reiterative transcription-mediated gene regulation (12). The mutations individually introduce the differing sequence elements of the gal and pyrBI promoters described above. We examined the effects of each mutation on reiterative transcription in vitro and reiterative transcription-mediated control of carAB expression in vivo. Our results show that both the sequence at or near the transcription start site and the spacing between the start site and −10 region have significant effects on the extent of reiterative and productive transcription and on the range of gene regulation. These effects appear, at least in part, to account for the different ranges of reiterative transcription-mediated regulation of carAB, pyrBI, and gal expression.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 strain CLT42 [F− car-94 Δ(argF-lac)U169 rpsL150 thiA1 relA1 deoC1 ptsF25 flbB5301 rbsR] (16) was used as the parent in the construction of bacteriophage lambda lysogens. Plasmid pDLC126 (12, 17) was used to construct carAp1::lacZ operon fusions. This plasmid contains a functional E. coli lacZ gene preceded by a strong ribosome binding site and a more-upstream unique BamHI cloning site. Plasmid pDLC126 does not contain a promoter from which the lacZ gene can be transcribed. Operon fusions were made by inserting a BamHI restriction fragment containing either the wild-type or a mutant carAB P1 promoter region into the unique BamHI cloning site of plasmid pDLC126. Fusion constructions were confirmed by DNA sequence analysis.
Restriction digests, ligations, and transformations.
Conditions for restriction digests, ligations, and transformations were as previously described (16).
DNA preparations and site-directed mutagenesis.
Plasmid DNA was isolated by using Qiagen plasmid kits. The carAB P1 promoter fragment, which contains nucleotides −58 to +40 of the promoter region (counting from the transcription start site) flanked by BamHI restriction sites, was prepared as previously described (12). Site-directed mutagenesis was used to introduce selected mutations at the transcription start site of the carAB P1 promoter fragment. Mutagenesis was performed by using a previously described PCR-based procedure (12). Wild-type and mutant promoter fragments were digested with BamHI prior to insertion into plasmid pDLC126.
In vitro transcription.
Purified RNA polymerase holoenzyme containing σ70 was prepared as previously described (18, 19). DNA templates were 207-bp or 208-bp fragments containing the carAB P1 promoter region (with either a wild-type or mutant promoter) and a downstream segment that included an efficient (∼98%) intrinsic transcription terminator, the pyrBI attenuator (20). The presence of the pyrBI attenuator facilitates RNA polymerase release from the DNA template in our multiple-round transcription assay, thereby increasing the synthesis of long (referred to as full-length) transcripts initiated at the carAB P1 promoter (12). All templates were prepared and their concentrations and purity determined as previously described (21). Transcription reaction mixtures (10 μl) contained 10 nM DNA template, 100 nM RNA polymerase, 20 mM Tris-acetate (pH 7.9), 10 mM magnesium acetate, 100 mM potassium glutamate, 0.2 mM Na2EDTA, 0.1 mM dithiothreitol, 400 μM (each) ATP, CTP, and GTP, and either 50 or 500 μM UTP. In the reaction mixture, one of the nucleoside triphosphates was 32P labeled (2 Ci/mmol). Reactions were initiated by addition of RNA polymerase, and the reaction mixtures were incubated at 37°C for 15 min. Heparin (1 μl of a 1 mg/ml solution) then was added to the mixture, and incubation was continued for an additional 10 min. Reactions were terminated by adding 10 μl of stop solution (7 M urea, 2 mM Na2EDTA, 0.25% [wt/vol] each of bromophenol blue and xylene cyanol) and placing the samples on ice. The samples were heated at 100°C for 3 min, and an equal volume of each sample was removed and run on a 25% (29:1 acrylamide-bisacrylamide)-50 mM Tris-borate (pH 8.3), 1 mM Na2EDTA sequencing gel containing 7 M urea (21). Transcripts were visualized by autoradiography and quantitated by scanning gels with a Molecular Dynamics PhosphorImager and by densitometry.
Transfer of carAp1::lacZ operon fusions from plasmids to the E. coli chromosome.
Wild-type and mutant carAp1::lacZ operon fusions carried on derivatives of plasmid pDLC126 were individually transferred to the chromosome of strain CLT42 by using phage lambda RZ5 (22). The presence of a single prophage at the lambda attachment site was confirmed by PCR analysis (23). In this procedure, the concentration of each primer was 500 nM.
Media and culture methods.
Cells used for enzyme assays and RNA isolations were grown at 37°C with shaking in N−C− medium (24) supplemented with 10 mM NH4Cl, 0.4% (wt/vol) glucose, 0.015 mM thiamine hydrochloride, 1 mM arginine, and either 1 mM uracil or 0.25 mM UMP. Cell growth was monitored as previously described, and samples were harvested during the exponential phase of growth at the same optical density (25).
Enzyme assays.
Cell extracts were prepared by sonic oscillation (22). β-Galactosidase activity was determined as previously described (12).
Isolation of cellular RNA and primer extension mapping.
Cellular RNA was isolated quantitatively as described by Wilson et al. (17). Primer extension mapping of the 5′ ends of carAp1::lacZ transcripts was performed as previously described (26), except that 45 μg of RNA from uracil-grown cells and 30 μg of RNA from UMP-grown cells were used for analysis. The different amounts of RNA, which were isolated from the same mass of cells, reflect the different levels of stable RNA in cells growing at different rates. The primer used in these experiments was 5′-CGCGGATCCAGCTGAATCAATGCAAATCTGC, which was labeled with 32P at the 5′ end. The 29 nucleotides at the 3′ end of this primer hybridize to nucleotides 24 to 52 in the carAp1::lacZ transcript. Each quantitative primer extension mapping experiment described in Results was performed independently two times with essentially the same results.
RESULTS
Effects of carAB P1 mutations that change the sequence of the transcription start site on reiterative transcription in vitro.
To assess the role of transcription start site sequence on reiterative transcription, we constructed two mutant versions of the carAB P1 promoter that changed the wild-type G start site to either A or AA (Fig. 2). These changes introduce the start site sequences found in the gal P2 and pyrBI promoters, respectively (Fig. 1). The wild-type and mutant promoter regions (from −58 to +40 counting from the wild-type transcription start site) were incorporated into 207/208-bp DNA templates that also included a strong (∼98% efficient) downstream intrinsic transcription terminator. These fragments do not include the carAB P2 promoter, which is located 68 bp downstream from promoter P1 (12). All detectable transcription initiation at the wild-type carAB P1 promoter occurs at the G residue (by convention, referring to the nontemplate strand sequence) located 7 bases downstream from the −10 region (Fig. 1) (12). Based on previous studies of preferred sites of transcription initiation in E. coli (26), it was expected that essentially all transcription initiation at both mutant promoters would also occur 7 bases downstream from the −10 region, at an A residue in this case (Fig. 2). This expectation was confirmed in experiments described below and by primer extension mapping of in vitro transcripts (data not shown). To examine effects on reiterative transcription, the DNA templates carrying either the wild-type or a mutant promoter were transcribed in vitro in reaction mixtures containing 400 μM each nucleoside triphosphate (NTP) except UTP, which was present at either 500 or 50 μM. The UTP concentrations were chosen to mimic levels found in cells grown under conditions of pyrimidine excess or limitation (27, 28). The reaction mixtures also included either [γ-32P]GTP or [γ-32P]ATP to label the 5′ ends of transcripts initiated at the wild-type and mutant promoters, respectively. Transcripts synthesized in each reaction were separated by gel electrophoresis, and radiolabeled transcripts were visualized by autoradiography (Fig. 3) and quantitated by phosphorimaging and densitometry.
FIG 2.

Sequences of carAB promoter P1 mutations used to assess the effects of transcription start site sequence and spacing between the start site and −10 region on reiterative transcription and gene regulation. The wild-type carAB promoter P1 sequence is shown and marked as described for Fig. 1, and arrows indicate the mutations introduced into this sequence. In two mutant promoters designed to examine start site sequence, the wild-type G start site was changed to either A or AA. In two other mutant promoters designed to examine the effects of spacing, either a C was inserted before the wild-type start site or the wild-type G start site was changed to CA.
FIG 3.

Comparison of reiterative transcription at the wild-type carAB P1 promoter and mutant promoters with changes in the sequence at the transcription start site. DNA templates containing the wild-type (wt) carAB P1 promoter (lanes 1 and 2) and the G-to-A (lanes 3 and 4) and G-to-AA (lanes 5 and 6) mutant carAB P1 promoters were transcribed in vitro in reaction mixtures containing 400 μM (each) ATP, CTP, GTP, and either 500 or 50 μM UTP, as indicated. Transcripts were radiolabeled at their 5′ ends with either [γ-32P]GTP or [γ-32P]ATP, as indicated, separated by polyacrylamide gel electrophoresis, and visualized by autoradiography. The lengths (in nucleotides) of transcripts initiated at the wild-type and mutant promoters are indicated at the left and right sides of the autoradiogram, respectively. Arrows indicate transcripts produced by simple abortive initiation: filled arrows indicate transcripts initiated at the wild-type promoter, and the open arrow indicates the AAUUUG transcript initiated at the G-to-AA promoter. Full-length transcripts are enclosed by a bracket and labeled; heterogeneity in the length of these transcripts is due to termination at multiple sites within the downstream intrinsic terminator (12).
The results with the wild-type promoter were essentially identical to our previously published characterization of in vitro transcription from the carAB P1 promoter (12). In the presence of 500 μM UTP, nearly all transcripts were included in a regularly spaced and progressively less intense ladder of transcripts with the general sequence GUUUn, where n = 1 to >30 (Fig. 3, lane 1). Ladder transcripts longer than 4 nucleotides were the products of reiterative transcription. Several minor transcripts were also detected, and these transcripts, containing 5, 6, 9, 10, and 11 nucleotides, were previously shown to be faithful copies of the DNA template generated by simple abortive initiation (i.e., normal release of short transcripts independent of reiterative transcription) (12). Furthermore, a low level of full-length transcripts, which were terminated at the downstream intrinsic terminator, was detected. These transcripts were previously shown to be faithful copies of the DNA template (12). In comparison, production of the GUUUn ladder was greatly diminished, while the levels of simple abortive transcripts (especially the 5-mer GUUUC) and full-length transcripts were significantly (≥1.5-fold) increased in the presence of 50 μM UTP (Fig. 3, lane 2). These results clearly demonstrate UTP-mediated control of entry into the nonproductive reiterative mode of transcription at the carAB P1 promoter. It also should be noted that the experiments described above illustrate that same-length transcripts with different nucleotide contents migrate at different rates in the gel. Each nucleotide in the transcript contributes differently but consistently to gel mobility (C>A = U>G), which allows the sequence of short (<15-nucleotide-long) transcripts to be determined by the spacing between adjacent bands in the gel as previously described (4). This technique is used extensively in the experiments described below.
Transcription from the G-to-A mutant promoter at 500 μM UTP also revealed a regularly spaced and progressively less intense ladder of transcripts with gel mobilities consistent with the general sequence AUUUn, where n ≥ 1 (Fig. 3, lane 3). This ladder also was shown to comigrate with a known AUUUn ladder and contain only A and U residues (data not shown). Therefore, ladder transcripts longer than 4 nucleotides were the products of reiterative transcription. This ladder was more intense and longer (apparently exceeding 100 nucleotides) than that observed with the wild-type promoter under the same reaction conditions. Very low levels of 5-mer and 6-mer transcripts (above the ladder transcripts AUUUU and AUUUUU in Fig. 3) also were detected and evidently were produced by simple abortive initiation. Full-length transcripts also were detected, but the level of these transcripts was much lower than that observed with the wild-type promoter. When the G-to-A promoter was transcribed in the presence of 50 μM UTP, the production of the AUUUn ladder was reduced roughly 3-fold but remained easily detectable. Additionally, the levels of the two minor aborted transcripts were increased slightly, and the level of full-length transcripts was increased nearly 4-fold (Fig. 3, lane 4). Taken together, these results indicate that reiterative transcription at the G-to-A promoter occurs much more extensively than it does at the wild-type promoter, resulting in a lower level of full-length transcript synthesis.
Transcription from the G-to-AA mutant promoter at 500 μM UTP again produced a regularly spaced ladder of transcripts, indicative of reiterative transcription and with gel mobilities consistent with the general sequence AAUUn, where n ≥ 1 (Fig. 3, lane 5). This ladder was shown to contain only A and U residues and to comigrate with the AAUUUn ladder produced in vitro by reiterative transcription at the pyrBI promoter (data not shown). Only ladder transcripts longer than 5 nucleotides were the products of reiterative transcription in this case. The intensity and length of the ladder produced at the G-to-AA promoter were intermediate between those produced at the wild-type and G-to-A promoters. The only other transcript detected with the G-to-AA promoter was a prominent 6-mer band that migrated slighter slower in the gel than the ladder transcript AAUUUU, indicating that this band was a simple aborted transcript with the sequence AAUUUG. Consistent with this assignment, this transcript was shown to comigrate with the AAUUUG aborted transcript produced at the pyrBI promoter (data not shown). When the UTP concentration was reduced to 50 μM, production of the AAUUUUn reiterative transcription ladder was greatly reduced, with only the 6-mer transcript being readily detectable and very low levels of the 7-mer and 8-mer transcripts observed (Fig. 3, lane 6). On the other hand, the levels of the AAUUUG aborted transcript and full-length transcripts were increased approximately 2-fold compared to the levels of these transcripts produced with 500 μM UTP. Overall, UTP-sensitive reiterative and nonreiterative transcription at the G-to-AA promoter was essentially identical to that at the pyrBI promoter (14), perhaps not surprisingly, because the sequences of the first 8 bp of the initially transcribed regions of these two promoters are identical (Fig. 1 and 2). Finally, comparing all three promoters, the extent of reiterative transcription at the G-to-AA promoter appeared to be similar to or slightly greater than that at the wild-type promoter and significantly less than that at the G-to-A promoter. These results suggest that the strength of base pairing between the DNA template and the 5′-end nucleotide(s) of the nascent transcript that precede the U-tract is inversely proportional to the extent of reiterative transcription.
Effects of carAB P1 mutations that change the sequence of the transcription start site on gene expression and regulation.
To measure the effects of the G-to-A and G-to-AA carAB P1 promoter mutations on gene expression and regulation, we constructed a set of carAp1::lacZ operon fusions that include either the wild-type or a mutant promoter region. Operon fusions were created by individually inserting a promoter region into plasmid pDLC126 and then transferring the fusion onto phage lambda RZ5 by recombination. The recombinant phage were used to infect strain CLT42 (car-94 ΔlacZYA), and lysogens carrying a single prophage at the lambda attachment site in the chromosome were isolated. These strains are pyrimidine (and arginine) auxotrophs, because the car-94 mutation inactivates the chromosomal carAB operon. Each lysogenic strain was grown in glucose-minimal salts (plus arginine) medium containing either uracil or UMP as the pyrimidine source, which provides a condition of pyrimidine excess or limitation, respectively (22). The level of lacZ-encoded ß-galactosidase in each culture was assayed as an indicator of expression from the cloned carAB P1 promoter (Table 1).
TABLE 1.
Effects of start site sequence on expression and regulation of carAp1::lacZ operon fusionsa
| Strain (carAp1 genotype) | ß-Galactosidase activity (nmol/min/mg) by pyrimidine levelb |
Regulation (fold) | |
|---|---|---|---|
| Excess | Limitation | ||
| CLT5174 (wild type) | 1,024 | 2,870 | 2.8 |
| CLT5189 (G to A) | 687 | 3,290 | 4.8 |
| CLT5190 (G to AA) | 1,100 | 2,780 | 2.5 |
Doubling times were 46 ± 2 min for cells grown on uracil and 68 ± 3 min for cells grown on UMP.
Values are the means from at least three independent determinations, with a variation of <8%.
In the wild-type promoter fusion strain (CLT5174), the level of ß-galactosidase in cells grown under conditions of pyrimidine limitation was 2.8-fold higher than that in cells grown with pyrimidine excess, as previously reported (12). In the G-to-A promoter fusion strain (CLT5189), the level of ß-galactosidase in cells grown under conditions of pyrimidine limitation was 4.8-fold higher than that in cells grown with pyrimidine excess, a range of regulation nearly twice that observed with the wild-type promoter fusion strain. This increase in range of regulation was due primarily to a lower level of expression of the mutant fusion operon under conditions of pyrimidine excess. In the case of the G-to-AA promoter fusion strain (CLT5190), the range of regulation (2.5-fold) and the levels of ß-galactosidase in cells grown under conditions of pyrimidine excess or limitation were approximately the same as the range of regulation and ß-galactosidase levels observed with the wild-type promoter fusion strain. A comparison of the results summarized above (Table 1) to the in vitro reiterative transcription analyses shown in Fig. 3 suggest that more extensive reiterative transcription, like that exhibited by the G-to-A promoter, permits greater repression of gene expression and a wider range of gene regulation.
Quantitative primer extension mapping of transcripts initiated at the mutant carAB P1 promoters with altered transcription start site sequence.
To further characterize expression and regulation of the wild-type and G-to-A and G-to-AA mutant carAp1::lacZ operon fusions, we used quantitative primer extension mapping to confirm the start sites and measure the levels of productive transcripts encoded by these operon fusions in lysogenic strains CLT5174 (wild type), CLT5189 (G to A), and CLT5190 (G to AA) grown under conditions of pyrimidine excess or limitation. Nonproductive reiterative transcripts synthesized in these strains cannot be detected by primer extension mapping. Furthermore, transcripts from the resident carAB operon of the parent strain CLT42 are eliminated by the car-94 mutation (12). As expected, our results show that nearly all transcripts initiated at the wild-type and mutant promoters are initiated at a G or A residue located 7 bases downstream from the −10 region (Fig. 4). The G-to-AA transcripts are one residue longer than wild-type (and G-to-A) transcripts due to the insertion of two mutant A residues and initiation at the first. In the case of the G-to-A promoter, we also observed a ladder of minor transcripts that was progressively longer than the dominant transcript. It is likely that these minor transcripts are produced by a small subset of transcripts containing extra U residues that, instead of being released from the initiation complex, are switched into the normal mode of transcript elongation.
FIG 4.

Levels of productive carAp1::lacZ transcripts initiated in vivo at the wild-type carAB P1 promoter and mutant promoters with changes in the sequence at the transcription start site. Cellular RNA was quantitatively isolated from exponentially growing cells of strains CLT5174 (wild type), CLT5189 (G to A), and CLT5190 (G to AA) grown in the presence of either uracil (R) or UMP (M) (i.e., conditions of pyrimidine excess or limitation, respectively), and transcript levels were measured by primer extension mapping as described in Materials and Methods. The figure shows an autoradiogram of a gel used to separate and analyze primer extension products. A dideoxy sequencing ladder (i.e., the four lanes marked G, A, T, and C) of the wild-type promoter region, which was used to identify both wild-type and mutant transcripts, was produced with the same DNA primer as that used for primer extension mapping. (Note that the sequence shown on the left is that of the template DNA strand, which is complementary to the sequence of the wild-type carAp1::lacZ transcript.) Primer extension product levels, which correspond to transcript levels, were measured with a PhosphorImager. Relative levels of transcripts and fold pyrimidine-mediated regulation are indicated.
With respect to transcript quantitation, our results showed that the relative levels of wild-type, G-to-A, and G-to-AA operon transcripts in cells grown with pyrimidine excess or limitation closely mirrored the operon-encoded ß-galactosidase levels included in Table 1 (Fig. 4). Accordingly, the ranges in pyrimidine-mediated regulation also were similar to those shown in Table 1. The ranges of regulation of transcript synthesis for the wild-type, G-to-A, and G-to-AA operons were 2.6-fold, 4.4-fold, and 2.3-fold, respectively. These results indicate that the observed pyrimidine-mediated regulation of wild-type and mutant carAp1::lacZ operon expression occurs essentially entirely at the level of transcription.
Effects of carAB P1 mutations that increase the spacing between the −10 region and transcription start site on reiterative transcription in vitro.
The carAB P1 and gal P2 promoters differ from the pyrBI promoter in that transcription initiation occurs either 7 or 8 bases downstream from the −10 region, respectively (Fig. 1). To assess the role of spacing between the −10 region and transcription start site on reiterative transcription, we constructed two additional mutant versions of the carAB P1 promoter in which either a C residue was inserted in front of the G start site of the wild-type promoter (+C mutant) or the G start site was changed to CA (G-to-CA mutant) (Fig. 2). Based on the fact that C is an extremely poor initiating nucleotide and other known preferences for transcription start site selection (26), we expected that essentially all transcription initiation at both mutant promoters would occur 8 bases downstream from the −10 region, at a G or A residue at the +C and G-to-CA promoters, respectively. Again, our expectation was confirmed in experiments described below and by primer extension mapping of in vitro transcripts (data not shown). Thus, the +C and G-to-CA promoters provided increased-spacing variants of the wild-type and previously described G-to-A promoters. The +C and G-to-CA promoters were analyzed by in vitro transcription with either 500 or 50 μM UTP as described above, and the results were compared to transcription from the corresponding shorter-spacing promoter (Fig. 5).
FIG 5.
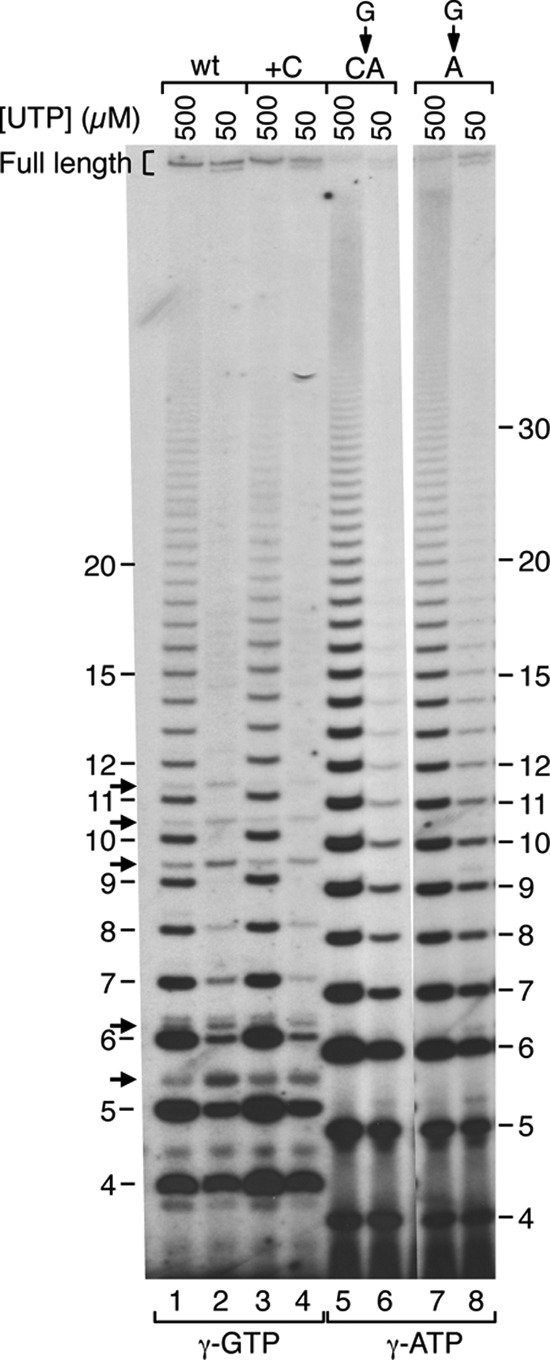
Comparison of reiterative transcription at wild-type and mutant carAB P1 promoters with different spacing between the −10 region and the transcription start site. DNA templates containing the wild-type (wt) carAB P1 promoter (lanes 1 and 2) and the +C (lanes 3 and 4), G-to-CA (lanes 5 and 6), and G-to-A (lanes 7 and 8) mutant carAB P1 promoters were transcribed in vitro, 32P labeled at their 5′ ends, and analyzed as described in the legend to Fig. 3. All lanes were from the same autoradiogram, but lanes 7 and 8 were repositioned to facilitate comparison to lanes 5 and 6. The lengths (in nucleotides) of GUUUn transcripts initiated at the wild-type and +C mutant promoters are indicated on the left, with arrows indicating transcripts produced by simple abortive initiation at the two promoters. The lengths of AUUUn transcripts initiated on the G-to-CA and G-to-A mutant promoters are indicated at the right. The [γ-32P]NTP used to label the 5′ ends of transcripts is indicated at the bottom, and full-length transcripts are enclosed by a bracket and labeled.
Transcription at the +C promoter produced a set of transcripts (Fig. 5, lanes 3 and 4) with gel mobilities identical to those of the transcripts synthesized with the wild-type promoter (Fig. 5, lanes 1 and 2). Furthermore, we showed that production of the regularly spaced ladder of transcripts initiated at the +C promoter (i.e., numbered transcripts in Fig. 5) required only GTP and UTP as substrates, consistent with the general transcript sequence of GUUUn (data not shown). These results indicate that the sequences of the transcripts initiated at the wild-type and +C promoters are identical; therefore, the start site at the +C promoters is located 8 nucleotides downstream from the −10 region. Quantitatively, the levels of transcript synthesis and UTP-mediated control of reiterative transcription with the wild-type and +C promoters were similar; however, there were two small but reproducible differences in transcript levels. First, the levels of 4-mer (GUUU) and 5-mer (GUUUU) transcripts initiated at the +C promoter were higher than the levels of these transcripts initiated at the wild-type promoter: 1.5-fold to 2-fold higher at 500 μM UTP and ≤1.5-fold higher at 50 μM UTP. The 4-mer transcripts apparently were produced by simple abortive initiation, while the 5-mer transcripts were the most abundant transcript produced by reiterative transcription. These results suggest that both simple abortive initiation and reiterative transcription were enhanced by a single base increase in the spacing between the −10 region and transcription start site of the wild-type promoter. Second, there was a modest (≤1.5-fold) reduction in the synthesis of full-length transcripts at the +C promoter which was typically slightly greater at 500 μM UTP. This reduction could be the consequence of the observed increases in abortive initiation and reiterative transcription.
Transcription from the G-to-CA promoter produced a set of transcripts (Fig. 5, lanes 5 and 6) that appeared to be identical to those initiated at the G-to-A promoter (Fig. 5, lanes 7 and 8) based on gel mobilities. Consistent with this assignment, we showed that the putative AUUUn ladder of transcripts initiated at the G-to-CA promoter contain only A and U residues (data not shown). These results demonstrate that the start site at the G-to-CA promoter is located 8 nucleotides downstream from the −10 region. As observed with the G-to-A promoter, UTP-mediated regulation of reiterative transcription at the G-to-CA promoter was readily apparent; however, the levels of transcripts initiated at the two promoters were different. At 500 μM UTP, the levels of reiterative transcripts initiated at the G-to-CA promoter were nearly 2-fold higher than those initiated at the G-to-A promoter. This higher level of reiterative transcript synthesis was accompanied by a nearly 2-fold decrease in the synthesis of full-length transcripts at the G-to-CA promoter. At 50 μM UTP, the production of reiterative transcripts at both promoters appeared similar; however, synthesis of full-length transcripts at the G-to-CA promoter was approximately 2-fold less than that at the G-to-A promoter. Therefore, our analysis of spacer-variant promoters, particularly the promoters with an A start site, suggest that the spacing between the −10 region and transcription start site influences the extent of reiterative transcription and production of full-length transcripts.
Effects of carAB P1 mutations that increase the spacing between the −10 region and the transcription start site on gene expression and regulation.
To measure the effects of spacing between the −10 region and transcription start site on gene expression and regulation, we constructed (as described above) two additional variants of strain CLT42 that carry a carAp1::lacZ operon fusion, including either the +C or G-to-CA carAB P1 promoter mutation. The resulting +C (CLT5237) and previously described wild-type promoter strains, as well as the resulting G-to-CA (CLT5238) and previously described G-to-A promoter strains, constitute pairs of strains that differ only in the spacing between the −10 region and transcription start site of the carAp1::lacZ operon. These four strains were grown in the minimal medium described above containing either uracil or UMP to provide conditions of pyrimidine excess and limitation, respectively. The level of lacZ-encoded ß-galactosidase in each culture was assayed as an indicator of expression from the carAB P1 promoter, and the ranges of pyrimidine-mediated regulation of carAp1::lacZ expression were calculated.
The results with each pair of comparable strains indicated that increasing the spacing between the −10 region and transcription start site from 7 to 8 bp resulted in an increase in the range of regulation (Table 2). For the pair of strains with a G start site, regulation occurred over a 2.8-fold range with the wild-type promoter strain compared to a 5.5-fold range with the +C promoter strain. The higher range of regulation with the +C promoter strain was due entirely to a >2-fold lower level of ß-galactosidase in cells grown under conditions of pyrimidine excess. Under conditions of pyrimidine limitation, ß-galactosidase levels were similar in both strains. For the pair of strains with an A start site, regulation occurred over a 4.8-fold range with the G-to-A promoter strain compared to a 7.1-fold range with the G-to-CA promoter strain. In this case, the ß-galactosidase levels in the G-to-CA promoter strain were significantly lower than those in the G-to-A promoter strain under both growth conditions. However, the fold decrease in the ß-galactosidase level was greater under conditions of pyrimidine excess, resulting in the wider range of regulation. It is also noteworthy that, in general, the levels of carAp1::lacZ expression shown in Table 2 are inversely proportional to the extent of reiterative transcription and directly proportional to the levels of full-length transcripts produced in vitro with DNA templates carrying the corresponding carAB P1 promoter (Fig. 5).
TABLE 2.
Effects of spacing between the −10 region and transcription start site on expression and regulation of carAp1::lacZ operon fusionsa
| Strain (carAp1 genotype) | ß-Galactosidase activity (nmol/min/mg) by pyrimidine levelb |
Regulation (fold) | |
|---|---|---|---|
| Excess | Limitation | ||
| CLT5174 (wild type) | 1,024 | 2,870 | 2.8 |
| CLT5237 (+C) | 460 | 2,520 | 5.5 |
| CLT5189 (G to A) | 687 | 3,290 | 4.8 |
| CLT5238 (G to CA) | 144 | 1,020 | 7.1 |
Doubling times were 46 ± 2 min for cells grown on uracil and 68 ± 3 min for cells grown on UMP.
Values are the means from at least three independent determinations, with a variation of <10%.
Quantitative primer extension mapping of transcripts initiated at the mutant carAB P1 promoters that increase the spacing between the −10 region and transcription start site.
To further characterize the productive transcripts initiated in vivo from the +C and G-to-CA promoters, we used quantitative primer extension mapping to analyze transcripts encoded by the carAp1::lacZ operon fusions in strains CLT5237 (+C) and CLT5238 (G to CA) (and, as a reference, wild-type promoter strain CLT5174) grown under conditions of pyrimidine excess or limitation (Fig. 6). With the exception of one sample, the data revealed that a single major transcript was initiated at both the +C and G-to-CA promoters and that these transcripts were the same length as the major transcript initiated at the wild-type promoter. This result confirmed that the major +C and G-to-CA transcripts were initiated 8 bases downstream from the −10 region (at G and A residues, respectively). In the sample from strain CLT5238 (G to CA) grown under conditions of pyrimidine excess, only low levels of two transcripts were detected, including the transcript described above, which initiates at the A residue located 8 bases from the −10 region. Low levels of this transcript were expected because of the high levels of nonproductive reiterative transcription that occur at the G-to-CA promoter in the presence of high levels of UTP (Fig. 5). The other low-level transcript appeared to be 1 base longer and also was detected in CLT5238 (G to CA) cells grown with pyrimidine limitation. This transcript most likely was initiated at a minor C start site located 7 bases downstream from the −10 region, the preferred start site location for E. coli RNA polymerase (26).
FIG 6.

Levels of productive carAp1::lacZ transcripts initiated in vivo at wild-type and mutant carAB P1 promoters with different spacing between the −10 region and transcription start site. Cellular RNA was quantitatively isolated from exponentially growing cells of strains CLT5174 (wild type), CLT5237 (+C), and CLT5238 (G to CA) grown in the presence of either uracil (R) or UMP (M), and transcript levels were analyzed by primer extension mapping as described above. The primer extension mapping data for strain CLT5189 (G to A) were taken from Fig. 4; the analysis of the data in Fig. 4 and 6 was identical. The dideoxy sequencing ladder (lanes G, A, T, and C) of the wild-type promoter region was generated as described in the legend to Fig. 4. Primer extension product levels, which correspond to transcript levels, were measured with a PhosphorImager, and the relative levels of transcripts and fold pyrimidine-mediated regulation are indicated.
To allow comparison of in vivo transcription from our two pairs of carAB P1 promoters that differ only in the spacing between the −10 region and transcription start site, equivalent primer extension mapping data obtained with strain CLT5189 (G to A) were included in Fig. 6. The data from the four strains showed that the relative levels of productive carAp1::lacZ-specific transcripts closely mirrored the relative levels of carAp1::lacZ-encoded ß-galactosidase shown in Table 2. Consequently, for each strain, the range of pyrimidine-mediated regulation determined from productive transcript levels was nearly the same as the range of regulation calculated from ß-galactosidase levels. Thus, the primer extension mapping data again show that a single-base increase in promoter spacing results in an increase in the range of pyrimidine-mediated regulation of carAp1::lacZ expression.
DISCUSSION
The goal of this study was to determine the effects of promoter sequence on mechanisms of gene regulation in E. coli that are based on reiterative transcription at T tracts during transcription initiation. This reaction, which is promoted by high intracellular levels of UTP, causes early transcript release and reduced gene expression. We focused on three analogous control mechanisms involved in regulating carAB, pyrBI, and gal expression through UTP-sensitive reiterative transcription at the carAB P1, pyrBI, and gal P2 promoters, respectively. The range of regulation for these mechanisms varies from 2-fold to 7-fold, and this variation presumably reflects differences in promoter sequences. We used the carAB P1 promoter (with an initially transcribed region sequence of 5′-GTTTG) as a platform to introduce sequence elements of the pyrBI and gal P2 promoters that changed the sequence at or near the transcription start site or the spacing between the promoter −10 region and the start site. Our results showed that both the sequence and spacing of the transcription start site significantly affect the extent of reiterative transcription, level of gene expression, and range of gene regulation.
To examine the effects of start site sequence (or, more accurately, the sequence of the initially transcribed region preceding the T tract), we changed the wild-type G start site to either A or AA, making the sequence resemble that of the gal P2 and pyrBI promoters, respectively. These changes are particularly relevant because ATP and GTP are the strongly preferred initiating NTPs in E. coli (26, 29–31). Our comparisons of the resulting promoters suggest that the importance of start site sequence is, at least in large part, its contribution to the base-pairing strength of the RNA-DNA hybrid formed by the first 4 or 5 residues at the 5′ end of the nascent transcript and the DNA template. Previous studies had, in fact, demonstrated that weak RNA-DNA base pairing facilitates reiterative transcription during transcription initiation and elongation (3, 11, 32, 33). Using different promoter contexts, we showed that an RNA-DNA hybrid containing the 5′-AUUU transcript allowed more extensive reiterative transcription, especially at high UTP concentrations, than that observed with stronger hybrids formed with 5′-GUUU and 5′-AAUUU transcripts. On the other hand, reiterative transcription was similar at the promoters specifying the 5′-GUUU and 5′-AAUUU transcripts, evidently reflecting similar calculated stabilities of the RNA-DNA hybrids formed with these two transcripts. Our results also demonstrated a direct correlation between nonproductive reiterative transcription and reduced gene expression in vivo and between the extent of reiterative transcription and range of gene regulation. For instance, changing the start site sequence from G to A increased the range of regulation nearly 2-fold. The wider range of regulation is achieved by increased nonproductive reiterative transcription at high UTP levels, resulting in decreased gene expression, coupled with a smaller effect on reiterative transcription and gene expression at low UTP levels.
The effects of spacing between the promoter −10 region and transcription start site were examined with promoters in which the spacing was either 7 or 8 bases, which corresponds to the spacing in the carAB P1 and gal P2 promoters or the pyrBI promoter, respectively. These two sites are the most preferred locations for transcription initiation in E. coli, with position 7 typically the better start site unless it is occupied by a poor initiating nucleotide (26, 29–31). The effect of spacing, observed with promoters with either GTTT or ATTT as the initially transcribed region, was significant. Increasing the spacing from 7 to 8 bases resulted in increased reiterative transcription in vitro (more obvious with A-initiated transcripts), reduced gene expression in vivo, and an increase in the range of gene regulation from 1.5-fold to 2-fold with promoters with ATTT and GTTT initially transcribed regions, respectively. Again, the wider range of regulation appeared to be due to a greater increase in the level of nonproductive reiterative transcription (and decreased gene expression) at high UTP concentrations than that observed at low UTP levels.
Taken together, our results show that the level of gene expression and the range of gene regulation established by conditional nonproductive reiterative transcription, like that occurring at the carAB P1, pyrBI, and gal P2 promoters, can be modulated by subtle changes in promoter sequence. We investigated only a limited number of changes in transcription start site sequence and spacing between the −10 region and the start site. There likely are other seemingly modest changes in promoter sequence and architecture that have significant effects on gene expression and regulation. A case in point is the greater than 2-fold different ranges of gene regulation observed with the G-to A mutant carAB P1 promoter and the gal P2 promoter (13) under identical conditions, even though these two promoters have the same initially transcribed region sequence (i.e., ATTT) and the same 7-base spacing between the −10 region and start site. Presumably, the different ranges of regulation are due to some other difference in the G-to-A and gal P2 promoters. The identification of this and other such modulatory promoter features obviously will require additional investigation.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants GM94466 and GM29466.
We thank Jeffery Vahrenkamp and Sylvia McPherson for assistance in analyzing data and valuable discussions.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Guo HC, Roberts JW. 1990. Heterogeneous initiation due to slippage at the bacteriophage 82 late gene promoter in vitro. Biochemistry 29:10702–10709. 10.1021/bi00499a019 [DOI] [PubMed] [Google Scholar]
- 2.Jacques JP, Kolakofsky D. 1991. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 5:707–713. 10.1101/gad.5.5.707 [DOI] [PubMed] [Google Scholar]
- 3.Anikin M, Molodtsov V, Temiakov D, McAllister WT. 2010. Transcript slippage and recoding, p 409–432 In Atkins JF, Gesteland RF. (ed), Recoding: expansion of decoding rules enriches gene expression. Nucleic acids and molecular biology, vol 24 Springer, New York, NY [Google Scholar]
- 4.Qi F, Turnbough CL., Jr 1995. Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J. Mol. Biol. 254:552–565. 10.1006/jmbi.1995.0638 [DOI] [PubMed] [Google Scholar]
- 5.Hausmann S, Garcin D, Delenda C, Kolakofsky D. 1999. The versatility of paramyxovirus RNA polymerase stuttering. J. Virol. 73:5568–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr JN, Wertz GW. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 75:6901–6913. 10.1128/JVI.75.15.6901-6913.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou YN, Lubkowska L, Hui M, Court C, Chen S, Court DL, Strathern J, Jin DJ, Kashlev M. 2013. Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J. Biol. Chem. 288:2700–2710. 10.1074/jbc.M112.429464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korzheva N, Mustaev A. 2001. Transcription elongation complex: structure and function. Curr. Opin. Microbiol. 4:119–125. 10.1016/S1369-5274(00)00176-4 [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y, Dylla SM, Turnbough CL., Jr 2001. A long T · A tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J. Bacteriol. 183:221–228. 10.1128/JB.183.1.221-228.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong XF, Reznikoff WS. 1993. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J. Mol. Biol. 231:569–580. 10.1006/jmbi.1993.1310 [DOI] [PubMed] [Google Scholar]
- 11.Turnbough CL., Jr 2011. Regulation of gene expression by reiterative transcription. Curr. Opin. Microbiol. 14:142–147. 10.1016/j.mib.2011.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X, Turnbough CL., Jr 1998. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J. Bacteriol. 180:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin DJ. 1994. Slippage synthesis at the galP2 promoter of Escherichia coli and its regulation by UTP concentration and cAMP·cAMP receptor protein. J. Biol. Chem. 269:17221–17227 [PubMed] [Google Scholar]
- 14.Liu C, Heath LS, Turnbough CL., Jr 1994. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 8:2904–2912. 10.1101/gad.8.23.2904 [DOI] [PubMed] [Google Scholar]
- 15.Tu AH, Turnbough CL., Jr 1997. Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription. J. Bacteriol. 179:6665–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roland KL, Powell FE, Turnbough CL., Jr 1985. Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J. Bacteriol. 163:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson HR, Archer CD, Liu J, Turnbough CL., Jr 1992. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J. Bacteriol. 174:514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgess RR, Jendrisak JJ. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634–4638. 10.1021/bi00692a011 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez N, Wiggs J, Chamberlin MJ. 1977. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch. Biochem. Biophys. 182:404–408. 10.1016/0003-9861(77)90521-5 [DOI] [PubMed] [Google Scholar]
- 20.Turnbough CL, Jr, Hicks KL, Donahue JP. 1983. Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 80:368–372. 10.1073/pnas.80.2.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi F, Liu C, Heath LS, Turnbough CL., Jr 1996. In vitro assay for reiterative transcription during transcriptional initiation by Escherichia coli RNA polymerase. Methods Enzymol. 273:71–85. 10.1016/S0076-6879(96)73007-0 [DOI] [PubMed] [Google Scholar]
- 22.Roland KL, Liu CG, Turnbough CL., Jr 1988. Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 85:7149–7153. 10.1073/pnas.85.19.7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765–5766. 10.1093/nar/22.25.5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alper MD, Ames BN. 1978. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J. Bacteriol. 133:149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Donahue JP, Heath LS, Turnbough CL., Jr 1993. Genetic evidence that promoter P2 is the physiologically significant promoter for the pyrBI operon of Escherichia coli K-12. J. Bacteriol. 175:2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Turnbough CL., Jr 1994. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J. Bacteriol. 176:2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen JT, Jensen KF, Poulsen P. 1991. Role of transcription pausing in the control of the pyrE attenuator in Escherichia coli. Mol. Microbiol. 5:327–333. 10.1111/j.1365-2958.1991.tb02113.x [DOI] [PubMed] [Google Scholar]
- 28.Neuhard J, Nygaard P. 1987. Purines and pyrimidines, p 445–473 In Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol 1 American Society for Microbiology, Washington, DC [Google Scholar]
- 29.Kim D, Hong JS, Qiu Y, Nagarajan H, Seo JH, Cho BK, Tsai SF, Palsson BØ. 2012. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 8:e1002867. 10.1371/journal.pgen.1002867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis DE, Adhya S. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol. Microbiol. 54:692–701. 10.1111/j.1365-2958.2004.04318.x [DOI] [PubMed] [Google Scholar]
- 31.Walker KA, Osuna R. 2002. Factors affecting start site selection at the Escherichia coli fis promoter. J. Bacteriol. 184:4783–4791. 10.1128/JB.184.17.4783-4791.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin DJ, Turnbough CL., Jr 1994. An Escherichia coli RNA polymerase defective in transcription due to its overproduction of abortive initiation products. J. Mol. Biol. 236:72–80. 10.1006/jmbi.1994.1119 [DOI] [PubMed] [Google Scholar]
- 33.Parks AR, Court C, Lubkowska L, Jin DJ, Kashlev M, Court DL. 2014. Bacteriophage λ N protein inhibits transcription slippage by E. coli RNA polymerase. Nucleic Acids Res. 42:5823–5829. 10.1093/nar/gku203 [DOI] [PMC free article] [PubMed] [Google Scholar]


